Abstract
Two-phase sampling design, where biomarkers are subsampled from a phase-one cohort sample representative of the target population, has become the gold standard in biomarker evaluation. Many two-phase case–control studies involve biased sampling of cases and/or controls in the second phase. For example, controls are often frequency-matched to cases with respect to other covariates. Ignoring biased sampling of cases and/or controls can lead to biased inference regarding biomarkers' classification accuracy. Considering the problems of estimating and comparing the area under the receiver operating characteristics curve (AUC) for a binary disease outcome, the impact of biased sampling of cases and/or controls on inference and the strategy to efficiently account for the sampling scheme have not been well studied. In this project, we investigate the inverse-probability-weighted method to adjust for biased sampling in estimating and comparing AUC. Asymptotic properties of the estimator and its inference procedure are developed for both Bernoulli sampling and finite-population stratified sampling. In simulation studies, the weighted estimators provide valid inference for estimation and hypothesis testing, while the standard empirical estimators can generate invalid inference. We demonstrate the use of the analytical variance formula for optimizing sampling schemes in biomarker study design and the application of the proposed AUC estimators to examples in HIV vaccine research and prostate cancer research.
Keywords: AUC, Biased sampling, Frequency match, Inverse probability weighting, ROC curve, Two-phase studies
1. Introduction
Recent advances in lab techniques have provided researchers with a rich resource of biomarkers potentially useful for disease diagnosis and risk prediction. It is essential to use proper statistical methods to rigorously evaluate these biomarkers. The receiver operating characteristics curve (ROC) is a standard graphic tool to characterize a biomarker's classification accuracy. The area under the ROC curve (AUC) has been commonly used to gauge and compare biomarker's performance. In this paper, we consider the evaluation and comparison of biomarkers with respect to AUC for a binary disease outcome using data from two-phase sampling designs. In the first phase, a cohort sample representative of the target population relevant to clinical application is drawn, from which participants' disease status and easy to measure covariates are obtained; in the second phase, a subsample is drawn randomly, without replacement from the phase-one cohort sample for biomarker measurement, where sampling probability of each individual can depend on other covariates available. In particular, we consider studies where cases and controls in the second phase are separately sampled from the phase-one cohort. This is different from, for example, a case–cohort design for failure time, where cases and a random cohort are separately sampled. These types of designs prospectively collect bio-specimens before outcome ascertainment to minimize systematic difference in specimen collection, and retrospectively sample cases and controls for measuring biomarkers from the stored specimens to save costs. They have been proposed as gold standards for biomarker evaluation (Pepe and others, 2008).
In the second phase of a two-phase study, oftentimes cases and/or controls are not simple random samples from their respective distributions. For example, controls are often frequency-matched to cases with respect to other covariates, such as an individual's demographic characteristics (i.e., gender, age group, etc.); or cases and controls can be randomly sampled within some covariate strata. The effect of biased sampling on biomarker evaluation varies with the parameter of interest. Janes and Pepe (2009) showed that frequency matching does not invalidate inference on a biomarker's classification accuracy within matching covariate stratum; however, when the parameter of interest is the biomarker's classification accuracy in the general population, ignoring biased sampling can lead to invalid inference (Pepe and others, 2012). Inverse probability weighting provides a natural solution to account for biased sampling of cases and/or controls when evaluating marker classification performance in the population, e.g., in Pepe and others (2012) for binary disease outcome, and in Cai and Zheng (2011) for failure time outcome. Similar strategies of weighting to estimate classification accuracy under biased sampling have been adopted by other authors in a different problem setting: for correcting verification bias when true disease status (instead of the biomarker) is ascertained only from a subset of subjects (He and others, 2009).
However, asymptotic theories for estimators of AUC or difference in AUC for binary disease outcome in two-phase studies where expensive biomarkers are only measured in a subset of the study cohort, are lacking, despite the commonality of AUC in characterizing and comparing diagnostic tests in biomarker research. Our research in this paper aims to fill in this gap. In particular, we will develop inverse-probability-weighted (IPW) AUC estimator in two-phase study designs and develop inference procedures for comparing two diagnostic biomarkers. The closed-form expressions of asymptotic variances we develop for the proposed estimators will be valuable for understanding the implication of frequency matching on efficiency of biomarker evaluation.
This paper will consider two types of two-phase sampling designs: the finite-population stratified sampling (Neyman, 1938) and the Bernoulli sampling (Manski and Lerman, 1977). The former design is commonly used in biomarker research and is the major focus of this paper. The latter design has the advantage of simplicity and will be introduced as a pathway for studying the results in finite-population stratified sampling design. The two designs differ in how individuals are sampled for biomarker measurement in phase two. Finite-population stratified sampling requires pre-specification of a finite number of covariate strata: fixed number of cases and/or controls are then sampled from each stratum. It has the advantage that the number of cases and controls sampled from each stratum in phase two can be fixed at the outset. In Bernoulli sampling, each individual is selected with a known sampling probability that can depend on one's disease status and covariate value, independently of other individuals. It works in more general settings without the need to pre-specify a finite number of strata, e.g., when outcome and/or auxiliary covariates observed in phase one are continuous. The number of cases and/or controls sampled in phase two are random in Bernoulli sampling design. Despite the differences between the two designs, theoretical results of their estimators are closely connected.
In Section 2, we will start with the problem setting in Bernoulli sampling design and propose IPW estimators of AUC and difference in AUC using estimated sampling weights. We will then investigate estimation under finite-population stratified sampling design and show the connection in theoretical results of AUC estimators between the two designs. In Section 3, we conduct simulation studies to demonstrate performance of our estimators and subsequent inference procedures, compared with the empirical AUC estimator and hypothesis test ignoring biased sampling. In Section 4, we demonstrate, using a numerical example, the use of the analytical variance formula to guide the optimization of biomarker sampling scheme. The application of our proposed AUC estimators will be demonstrated by data examples in HIV vaccine research and prostate cancer research in Section 5. Concluding remarks are made in Section 6.
2. Methods
Let 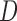 be a binary outcome of interest to differentiate. In this paper, we consider
be a binary outcome of interest to differentiate. In this paper, we consider  to be a binary disease outcome, with value 0 and 1 indicating non-diseased and diseased, respectively. We use subscript
to be a binary disease outcome, with value 0 and 1 indicating non-diseased and diseased, respectively. We use subscript  and
and  to indicate case and control, respectively. Let
to indicate case and control, respectively. Let 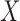 be a continuous biomarker that is expensive to measure such as a lab assay, assuming that increase in
be a continuous biomarker that is expensive to measure such as a lab assay, assuming that increase in 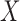 is associated with increased likelihood of disease. Suppose we have data collected from a two-phase study to evaluate the marker's classification accuracy. In the first phase, subjects' disease status and covariates that are easy to measure such as demographics are collected from a random sample of size
is associated with increased likelihood of disease. Suppose we have data collected from a two-phase study to evaluate the marker's classification accuracy. In the first phase, subjects' disease status and covariates that are easy to measure such as demographics are collected from a random sample of size 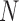 from the target population, with
from the target population, with  and
and  the number of cases and controls, respectively. In the second phase, the phase-one cohort is further subsampled to measure the biomarker value. Our first objective is to estimate AUC for marker
the number of cases and controls, respectively. In the second phase, the phase-one cohort is further subsampled to measure the biomarker value. Our first objective is to estimate AUC for marker 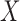 :
:  . In addition, suppose there is another continuous marker
. In addition, suppose there is another continuous marker  measured together with marker
measured together with marker  . Let
. Let 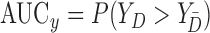 . Our second objective is to make inference about the difference in AUC between markers
. Our second objective is to make inference about the difference in AUC between markers 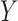 and
and  , i.e.,
, i.e.,  .
.
2.1. Bernoulli sampling
2.1.1. Evaluation of a single marker.
First, consider the standard Bernoulli sampling where in phase two individuals are selected independently of others with pre-specified probabilities. For case 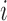 in the phase-one cohort, let
in the phase-one cohort, let 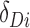 be the indicator that one's biomarker value is collected in the second phase, with
be the indicator that one's biomarker value is collected in the second phase, with 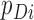 the corresponding sampling probability. Similarly, for control
the corresponding sampling probability. Similarly, for control 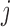 in the phase-one cohort, let
in the phase-one cohort, let 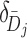 and
and 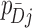 indicate whether he/she is sampled in the second phase and the corresponding sampling probability. Note that
indicate whether he/she is sampled in the second phase and the corresponding sampling probability. Note that 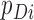 and
and 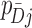 are individual-specific, whose values can depend on covariate value for case
are individual-specific, whose values can depend on covariate value for case 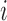 and control
and control  . For example, suppose phase-two sampling probabilities of cases and controls depend on discrete covariate strata: among phase-one samples, cases are allocated into
. For example, suppose phase-two sampling probabilities of cases and controls depend on discrete covariate strata: among phase-one samples, cases are allocated into 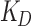 strata with
strata with 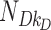 cases in stratum
cases in stratum 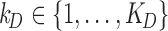 , and controls are allocated into
, and controls are allocated into  strata with
strata with 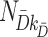 controls in stratum
controls in stratum 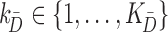 ; in the second phase, cases in stratum
; in the second phase, cases in stratum 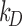 are independently sampled with probability
are independently sampled with probability 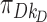 and controls in stratum
and controls in stratum 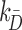 are independently sampled with probability
are independently sampled with probability 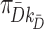 . Let
. Let 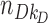 and
and 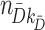 denote the number of phase-two cases and controls sampled from strata
denote the number of phase-two cases and controls sampled from strata 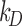 and
and 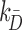 . They are random numbers with expected values
. They are random numbers with expected values  and
and 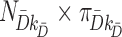 , respectively. We have
, respectively. We have 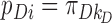 for case
for case 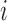 belonging to stratum
belonging to stratum 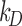 and
and 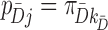 for control
for control 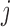 belonging to stratum
belonging to stratum 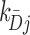 .
.
For estimation of population AUC, when phase-two case and control samples are representative of their respective populations, standard empirical AUC estimator (Bamber, 1975) 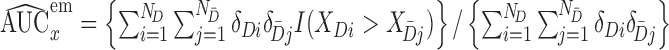 using phase-two biomarker data provides a valid estimate. However, when phase-two case and control samples are not representative of their respective populations,
using phase-two biomarker data provides a valid estimate. However, when phase-two case and control samples are not representative of their respective populations, 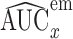 can be severely biased. For example, it is common in biomarker study designs that simple random samples of cases are drawn in the second phase, while controls are matched to cases by covariate strata such that control biomarkers are not representative of their population. To take care of the biased sampling, we propose the use of a weighted estimator of AUC based on the idea of inverse-probability weighting (Horvitz and Thompson, 1952). In particular, we construct IPW versions for the numerator and denominator of the empirical AUC estimator, where the contribution of each participant to a case–control pair is weighted by inverse of the estimated sampling probability of the participant in phase two:
can be severely biased. For example, it is common in biomarker study designs that simple random samples of cases are drawn in the second phase, while controls are matched to cases by covariate strata such that control biomarkers are not representative of their population. To take care of the biased sampling, we propose the use of a weighted estimator of AUC based on the idea of inverse-probability weighting (Horvitz and Thompson, 1952). In particular, we construct IPW versions for the numerator and denominator of the empirical AUC estimator, where the contribution of each participant to a case–control pair is weighted by inverse of the estimated sampling probability of the participant in phase two:
 |
(2.1) |
where  and
and  indicate estimated phase-two sampling probabilities for case
indicate estimated phase-two sampling probabilities for case  and control
and control  . For continuous covariate,
. For continuous covariate,  and
and 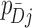 can be estimated by parametric models such as the logistic regression model. For discrete covariate strata, their empirical estimates can be derived. That is, for a case
can be estimated by parametric models such as the logistic regression model. For discrete covariate strata, their empirical estimates can be derived. That is, for a case 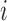 belonging to stratum
belonging to stratum 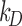 , his/her sampling probability
, his/her sampling probability 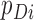 is estimated with
is estimated with  , the proportion of phase-one cases sampled in phase two from stratum
, the proportion of phase-one cases sampled in phase two from stratum 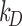 ; similarly one can estimate
; similarly one can estimate 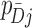 for control
for control 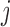 in stratum
in stratum 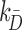 with
with 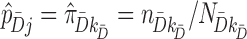 .
.
In Bernoulli sampling, the true sampling probability for each individual is known, but using “estimated” weight can improve efficiency (Web Appendix A&B, see supplementary material). This has also been recommended in other problem settings such as the weighted likelihood estimators (Robins and others, 1994; Breslow and Wellner, 2007). Intuitively it holds because using known sampling weights only involves data for subjects sampled at phase two but estimation of the weights allows incorporation of phase-one data available for all subjects, e.g., the number of phase-one cases/controls in each strata in scenarios where sampling probability in phase two varies across discrete strata.
Suppose we model sampling probabilities of the biomarker among cases and controls separately with finite-dimensional parameters 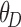 and
and  . Let
. Let  and
and  be maximum likelihood estimators. Asymptotic distribution of the IPW AUC estimator based on corresponding estimates of
be maximum likelihood estimators. Asymptotic distribution of the IPW AUC estimator based on corresponding estimates of 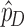 and
and 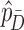 is stated below in Theorem 1 (proof in Web Appendix B, see supplementary material).
is stated below in Theorem 1 (proof in Web Appendix B, see supplementary material).
Theorem 1. —
Suppose
as
and
for each case and control; then
converges asymptotically to a normal random variable with mean 0 and variance
where
are information matrices for estimating
and
,
,
,
, and
.
2.1.2. Comparison of two markers.
For marker  measured together with
measured together with 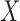 , we can similarly estimate its AUC as
, we can similarly estimate its AUC as 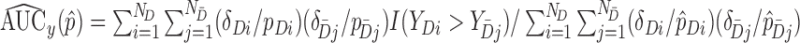 , and estimate the difference in AUC between the two markers with
, and estimate the difference in AUC between the two markers with 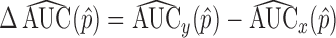 . Asymptotic distribution of
. Asymptotic distribution of 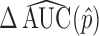 is shown in Theorem 2 (proof in Web Appendix C, see supplementary material).
is shown in Theorem 2 (proof in Web Appendix C, see supplementary material).
Theorem 2. —
Suppose
as
and
for every case and control; then
converges asymptotically to a normal random variable with mean 0 and variance
Previously, many authors have studied inference for comparing AUC between paired markers (Hanley and McNeil, 1983; DeLong and others, 1988; Wieand and others, 1989; Obuchowski and McClish, 1997). These tests were developed, however, for scenarios where cases and controls are randomly sampled from their respective distributions, and thus are not applicable for settings when there is biased sampling associated with cases and/or controls. In contrast, IPW estimator of 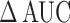 and its analytical variance as presented in Theorem 2 can be used to construct Wald tests for equal AUC between markers, as will be shown later in simulation studies.
and its analytical variance as presented in Theorem 2 can be used to construct Wald tests for equal AUC between markers, as will be shown later in simulation studies.
2.2. Finite-population stratified sampling
Now we consider the finite-population stratified sampling design, the design commonly used in biomarker studies. Again suppose cases and controls among phase-one samples are allocated into 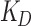 and
and 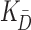 strata, respectively, with number
strata, respectively, with number 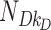 and
and  in each stratum. Fixed numbers of cases
in each stratum. Fixed numbers of cases 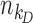 and controls
and controls  are then sampled in phase two from these covariate strata to measure the biomarker
are then sampled in phase two from these covariate strata to measure the biomarker  . Sampling fractions
. Sampling fractions 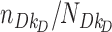 and
and 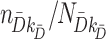 can be random.
can be random.
Let 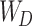 be the stratum indicator among cases taking unique values
be the stratum indicator among cases taking unique values  and
and 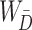 be the stratum indicator among controls taking unique values
be the stratum indicator among controls taking unique values 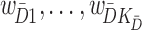 . Compute
. Compute 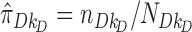 and
and 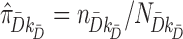 . In finite-population stratified sampling, sampling probability of a case or control is constant within their corresponding stratum, i.e.,
. In finite-population stratified sampling, sampling probability of a case or control is constant within their corresponding stratum, i.e., 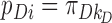 for case
for case 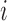 in stratum
in stratum 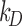 and
and  for control
for control  in stratum
in stratum  . The IPW estimator of AUC for marker
. The IPW estimator of AUC for marker 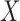 (2.1) can be equivalently represented as
(2.1) can be equivalently represented as
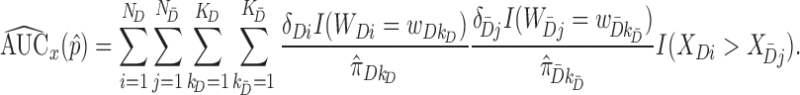 |
(2.2) |
Suppose, as  , sampling fractions for cases among stratum
, sampling fractions for cases among stratum 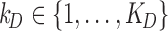 converge with
converge with  , and sampling fractions for controls among stratum
, and sampling fractions for controls among stratum  converge with
converge with 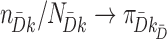 . Then the asymptotic variance of the IPW AUC estimator (2.2) in finite-population stratified sampling is identical to the asymptotic variance of
. Then the asymptotic variance of the IPW AUC estimator (2.2) in finite-population stratified sampling is identical to the asymptotic variance of  in Bernoulli sampling, if, in phase two of the latter design, cases in stratum
in Bernoulli sampling, if, in phase two of the latter design, cases in stratum  are sampled independently with probability
are sampled independently with probability 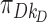 and controls in stratum
and controls in stratum 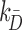 are sampled independently with probability
are sampled independently with probability 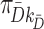 , with sampling probabilities estimated empirically. A proof is given in Web Appendix C (see supplementary material). The equality in asymptotic variance of
, with sampling probabilities estimated empirically. A proof is given in Web Appendix C (see supplementary material). The equality in asymptotic variance of 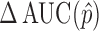 between the two designs can be similarly derived. The same argument on efficiency of weighted likelihood estimators in Cox regression comparing the two designs was made earlier (Breslow and Wellner, 2007).
between the two designs can be similarly derived. The same argument on efficiency of weighted likelihood estimators in Cox regression comparing the two designs was made earlier (Breslow and Wellner, 2007).
3. Simulation studies
In this section, we conduct simulation studies to investigate performance of the proposed IPW estimators of AUC and  . We consider a binary disease outcome
. We consider a binary disease outcome 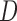 with prevalence
with prevalence 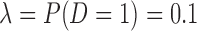 in the population. Let
in the population. Let 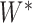 be a continuous covariate that follows the standard normal distribution among controls
be a continuous covariate that follows the standard normal distribution among controls  and
and 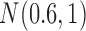 among cases
among cases 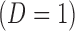 . Let
. Let 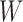 be a discrete covariate stratum derived from
be a discrete covariate stratum derived from 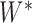 with three levels:
with three levels: 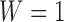 if
if 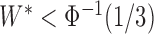 ,
,  if
if 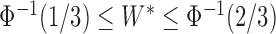 , and
, and 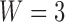 if
if 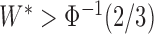 , where
, where 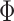 is the CDF of the standard normal distribution. We consider two biomarkers
is the CDF of the standard normal distribution. We consider two biomarkers  and
and  , where
, where  are jointly normally distributed conditional on
are jointly normally distributed conditional on 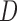 , with
, with 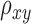 ,
, 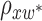 , and
, and 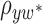 the correlations between
the correlations between 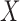 and
and 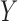 , between
, between  and
and  , and between
, and between 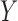 and
and  , respectively, conditional on
, respectively, conditional on  . Among controls,
. Among controls, 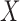 and
and 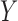 each follows the standard normal distribution. Among cases,
each follows the standard normal distribution. Among cases,  follows
follows  and
and 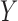 follows
follows 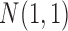 . The ROC curve based on
. The ROC curve based on 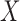 and
and 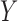 individually is thus
individually is thus  with
with 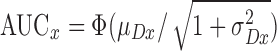 and
and 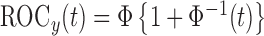 with
with  .
.
We generate data from two-phase studies. In the first phase,  subjects are randomly sampled from the population, whose
subjects are randomly sampled from the population, whose 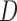 and
and 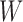 values are measured. In the second phase, we considered both Bernoulli sampling and finite-population stratified sampling of cases and controls for measuring markers
values are measured. In the second phase, we considered both Bernoulli sampling and finite-population stratified sampling of cases and controls for measuring markers 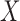 and
and 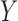 , assuming that they are measured on the same set of subjects. In Bernoulli sampling, cases are sampled independently with a constant probability
, assuming that they are measured on the same set of subjects. In Bernoulli sampling, cases are sampled independently with a constant probability 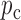 , and controls are sampled independently with a probability that depends on the stratum
, and controls are sampled independently with a probability that depends on the stratum 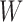 . In particular, the sampling probability for a control in stratum
. In particular, the sampling probability for a control in stratum  equals
equals 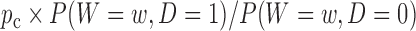 . This ensures that, on average, biomarkers are measured on equal numbers of cases and controls within each stratum. In finite-population stratified sampling,
. This ensures that, on average, biomarkers are measured on equal numbers of cases and controls within each stratum. In finite-population stratified sampling, 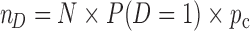 cases are sampled without replacement, and then within each
cases are sampled without replacement, and then within each  stratum, the same number of controls as cases in that stratum are drawn without replacement. This type of sampling design where simple random samples of cases are drawn in the second phase while controls are matched to cases by covariate strata is common in biomarker research. We also investigate other scenarios where both case and control sampling probabilities in phase two vary across strata. The comparative performance of various estimators is similar to the results we will present below (results omitted).
stratum, the same number of controls as cases in that stratum are drawn without replacement. This type of sampling design where simple random samples of cases are drawn in the second phase while controls are matched to cases by covariate strata is common in biomarker research. We also investigate other scenarios where both case and control sampling probabilities in phase two vary across strata. The comparative performance of various estimators is similar to the results we will present below (results omitted).
Based on 5000 Monte Carlo simulations in each setting, we evaluate performance of AUC estimators for individual markers. We compute 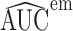 , and
, and 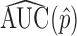 with sampling probabilities estimated empirically among cases and among controls conditional on sampling strata. These estimators are compared with respect to bias, efficiency, coverage of 95% Wald confidence intervals (CIs) based on analytical variance estimates, and the power to test
with sampling probabilities estimated empirically among cases and among controls conditional on sampling strata. These estimators are compared with respect to bias, efficiency, coverage of 95% Wald confidence intervals (CIs) based on analytical variance estimates, and the power to test 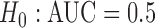 . We also evaluate performance of corresponding estimators for
. We also evaluate performance of corresponding estimators for 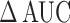 . We compare Wald tests based on
. We compare Wald tests based on  and the common Delong–Delong test (DeLong and others, 1988) with respect to type-I error rate and power for testing
and the common Delong–Delong test (DeLong and others, 1988) with respect to type-I error rate and power for testing 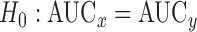 . The latter is implemented in the R package pROC (Robin and others, 2011).
. The latter is implemented in the R package pROC (Robin and others, 2011).
3.1. Evaluate performance of a single marker
Table 1 gives performance of 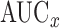 estimators for
estimators for 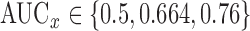 , with
, with 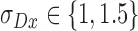 ,
,  , and the constant case sampling rate
, and the constant case sampling rate 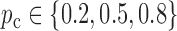 , for both Bernoulli sampling and finite-population stratified sampling. For both designs, the empirical AUC estimator is biased (with 5–15% relative bias), while
, for both Bernoulli sampling and finite-population stratified sampling. For both designs, the empirical AUC estimator is biased (with 5–15% relative bias), while 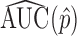 has minimum bias. While negative biases for empirical AUC estimator were observed for the particular simulation settings presented here, in general this estimator can have both positive and negative biases depending on the setting. Coverage of 95% Wald CIs for
has minimum bias. While negative biases for empirical AUC estimator were observed for the particular simulation settings presented here, in general this estimator can have both positive and negative biases depending on the setting. Coverage of 95% Wald CIs for 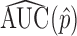 is close to the nominal level, while the CIs based on the empirical AUC has an undercoverage problem. The Wald test for
is close to the nominal level, while the CIs based on the empirical AUC has an undercoverage problem. The Wald test for 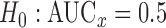 based on
based on 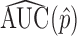 has type-I error close to the nominal level, while the test based on empirical AUC has inflated type-I error, the inflation getting worse with the increase in sample size. The
has type-I error close to the nominal level, while the test based on empirical AUC has inflated type-I error, the inflation getting worse with the increase in sample size. The  estimators in the two different sampling designs have similar variances.
estimators in the two different sampling designs have similar variances.
Table 1.
Performance of different 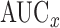 estimators for the bi-normal marker model described in Section 3. Disease prevalence is
estimators for the bi-normal marker model described in Section 3. Disease prevalence is 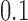 . Biomarker
. Biomarker  is standard normal among controls. By
is standard normal among controls. By 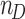 and
and 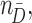 we indicate the expected number of cases and controls sampled in phase two for Bernoulli sampling and exact number of cases and controls sampled for finite-population stratified sampling. Results are based on 5000 Monte Carlo simulations
we indicate the expected number of cases and controls sampled in phase two for Bernoulli sampling and exact number of cases and controls sampled for finite-population stratified sampling. Results are based on 5000 Monte Carlo simulations
| Bernoulli sampling |
FPS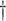 sampling sampling |
||||||
|---|---|---|---|---|---|---|---|
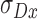
|
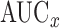
|
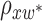
|

|
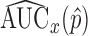
|
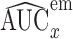
|

|
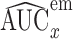
|
|
|||||||
| 1.00 | 0.50 | 0.30 | 100 |
 0.06 0.06 |
 3.89 3.89 |
 0.05 0.05 |
 3.90 3.90 |
| 250 | 0.00 |
 3.85 3.85 |
 0.04 0.04 |
 3.87 3.87 |
|||
| 400 | 0.01 |
 3.84 3.84 |
 0.04 0.04 |
 3.89 3.89 |
|||
| 0.50 | 100 |
 0.01 0.01 |
 6.51 6.51 |
 0.14 0.14 |
 6.63 6.63 |
||
| 250 |
 0.03 0.03 |
 6.54 6.54 |
 0.09 0.09 |
 6.58 6.58 |
|||
| 400 |
 0.04 0.04 |
 6.56 6.56 |
 0.04 0.04 |
 6.55 6.55 |
|||
| 1.50 | 0.50 | 0.30 | 100 | 0.01 |
 2.99 2.99 |
 0.06 0.06 |
 3.08 3.08 |
| 250 |
 0.01 0.01 |
 3.03 3.03 |
 0.07 0.07 |
 3.08 3.08 |
|||
| 400 |
 0.04 0.04 |
 3.05 3.05 |
 0.05 0.05 |
 3.05 3.05 |
|||
| 0.50 | 100 | 0.02 |
 5.05 5.05 |
 0.08 0.08 |
 5.17 5.17 |
||
| 250 |
 0.03 0.03 |
 5.10 5.10 |
 0.08 0.08 |
 5.14 5.14 |
|||
| 400 |
 0.05 0.05 |
 5.12 5.12 |
 0.03 0.03 |
 5.12 5.12 |
|||
| 1.00 | 0.664 | 0.30 | 100 |
 0.05 0.05 |
 3.57 3.57 |
 0.05 0.05 |
 3.58 3.58 |
| 250 | 0.00 |
 3.54 3.54 |
 0.04 0.04 |
 3.57 3.57 |
|||
| 400 | 0.01 |
 3.53 3.53 |
 0.05 0.05 |
 3.59 3.59 |
|||
| 0.50 | 100 |
 0.02 0.02 |
 6.01 6.01 |
 0.11 0.11 |
 6.10 6.10 |
||
| 250 |
 0.02 0.02 |
 6.04 6.04 |
 0.08 0.08 |
 6.08 6.08 |
|||
| 400 |
 0.04 0.04 |
 6.05 6.05 |
 0.05 0.05 |
 6.05 6.05 |
|||
| 1.50 | 0.664 | 0.30 | 100 | 0.03 |
 2.73 2.73 |
 0.06 0.06 |
 2.83 2.83 |
| 250 | 0.00 |
 2.77 2.77 |
 0.07 0.07 |
 2.82 2.82 |
|||
| 400 |
 0.03 0.03 |
 2.80 2.80 |
 0.04 0.04 |
 2.80 2.80 |
|||
| 0.50 | 100 | 0.03 |
 4.63 4.63 |
 0.08 0.08 |
 4.76 4.76 |
||
| 250 |
 0.01 0.01 |
 4.68 4.68 |
 0.07 0.07 |
 4.74 4.74 |
|||
| 400 |
 0.05 0.05 |
 4.72 4.72 |
 0.02 0.02 |
 4.71 4.71 |
|||
| 1.00 | 0.76 | 0.30 | 100 |
 0.04 0.04 |
 3.05 3.05 |
 0.05 0.05 |
 3.07 3.07 |
| 250 | 0.00 |
 3.03 3.03 |
 0.04 0.04 |
 3.05 3.05 |
|||
| 400 | 0.01 |
 3.02 3.02 |
 0.06 0.06 |
 3.07 3.07 |
|||
| 0.50 | 100 |
 0.02 0.02 |
 5.14 5.14 |
 0.09 0.09 |
 5.20 5.20 |
||
| 250 |
 0.02 0.02 |
 5.16 5.16 |
 0.07 0.07 |
 5.20 5.20 |
|||
| 400 |
 0.04 0.04 |
 5.17 5.17 |
 0.05 0.05 |
 5.18 5.18 |
|||
| 1.50 | 0.76 | 0.30 | 100 | 0.04 |
 2.31 2.31 |
 0.06 0.06 |
 2.42 2.42 |
| 250 | 0.01 |
 2.36 2.36 |
 0.06 0.06 |
 2.41 2.41 |
|||
| 400 |
 0.03 0.03 |
 2.40 2.40 |
 0.04 0.04 |
 2.39 2.39 |
|||
| 0.50 | 100 | 0.03 |
 3.95 3.95 |
 0.07 0.07 |
 4.06 4.06 |
||
| 250 |
 0.01 0.01 |
 4.00 4.00 |
 0.06 0.06 |
 4.05 4.05 |
|||
| 400 |
 0.04 0.04 |
 4.04 4.04 |
 0.02 0.02 |
 4.02 4.02 |
|||
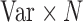
|
|||||||
| 1.00 | 0.50 | 0.30 | 100 | 9.68 | 8.26 | 9.50 | 7.70 |
| 250 | 3.88 | 3.42 | 3.74 | 3.15 | |||
| 400 | 2.43 | 2.16 | 2.38 | 2.02 | |||
| 0.50 | 100 | 9.00 | 8.07 | 8.62 | 6.83 | ||
| 250 | 3.55 | 3.31 | 3.41 | 2.65 | |||
| 400 | 2.17 | 2.03 | 2.16 | 1.66 | |||
| 1.50 | 0.50 | 0.30 | 100 | 9.72 | 8.85 | 9.56 | 8.16 |
| 250 | 3.88 | 3.65 | 3.79 | 3.29 | |||
| 400 | 2.36 | 2.18 | 2.38 | 2.10 | |||
| 0.50 | 100 | 9.15 | 8.60 | 8.72 | 7.28 | ||
| 250 | 3.72 | 3.58 | 3.52 | 2.88 | |||
| 400 | 2.24 | 2.16 | 2.25 | 1.82 | |||
| 1.00 | 0.664 | 0.30 | 100 | 8.03 | 7.62 | 8.00 | 7.21 |
| 250 | 3.23 | 3.16 | 3.14 | 2.94 | |||
| 400 | 2.04 | 2.01 | 2.00 | 1.88 | |||
| 0.50 | 100 | 7.27 | 7.80 | 7.05 | 6.62 | ||
| 250 | 2.91 | 3.21 | 2.79 | 2.56 | |||
| 400 | 1.79 | 1.97 | 1.78 | 1.62 | |||
| 1.50 | 0.664 | 0.30 | 100 | 8.15 | 8.01 | 8.09 | 7.39 |
| 250 | 3.30 | 3.32 | 3.23 | 3.01 | |||
| 400 | 2.03 | 2.01 | 2.03 | 1.92 | |||
| 0.50 | 100 | 7.64 | 8.08 | 7.35 | 6.84 | ||
| 250 | 3.13 | 3.36 | 2.96 | 2.70 | |||
| 400 | 1.92 | 2.05 | 1.89 | 1.71 | |||
| 1.00 | 0.76 | 0.30 | 100 | 6.04 | 6.13 | 6.06 | 5.85 |
| 250 | 2.44 | 2.54 | 2.38 | 2.39 | |||
| 400 | 1.55 | 1.63 | 1.52 | 1.53 | |||
| 0.50 | 100 | 5.38 | 6.43 | 5.32 | 5.58 | ||
| 250 | 2.17 | 2.66 | 2.08 | 2.14 | |||
| 400 | 1.34 | 1.64 | 1.33 | 1.35 | |||
| 1.50 | 0.76 | 0.30 | 100 | 6.17 | 6.38 | 6.24 | 5.97 |
| 250 | 2.52 | 2.65 | 2.47 | 2.42 | |||
| 400 | 1.57 | 1.62 | 1.55 | 1.54 | |||
| 0.50 | 100 | 5.73 | 6.52 | 5.69 | 5.70 | ||
| 250 | 2.38 | 2.74 | 2.25 | 2.22 | |||
| 400 | 1.48 | 1.69 | 1.45 | 1.41 | |||
| Coverage of 95% CI | |||||||
| 1.00 | 0.50 | 0.30 | 100 | 94.3 | 83.4 | 94.1 | 84.2 |
| 250 | 94.2 | 67.9 | 94.7 | 67.5 | |||
| 400 | 94.5 | 53.3 | 94.4 | 51.6 | |||
| 0.50 | 100 | 94.2 | 63.9 | 94.4 | 63.2 | ||
| 250 | 94.6 | 28.2 | 95.0 | 24.4 | |||
| 400 | 94.7 | 10.2 | 95.0 | 8.00 | |||
| 1.50 | 0.50 | 0.30 | 100 | 93.8 | 88.0 | 94.1 | 88.5 |
| 250 | 93.9 | 78.0 | 94.5 | 78.7 | |||
| 400 | 94.5 | 69.3 | 94.4 | 68.7 | |||
| 0.50 | 100 | 94.3 | 76.3 | 94.8 | 77.4 | ||
| 250 | 94.5 | 50.3 | 95.1 | 49.7 | |||
| 400 | 94.5 | 31.0 | 94.9 | 28.8 | |||
| 1.00 | 0.664 | 0.30 | 100 | 93.9 | 86.2 | 94.0 | 86.6 |
| 250 | 94.1 | 70.6 | 94.7 | 70.6 | |||
| 400 | 94.2 | 56.7 | 94.5 | 55.3 | |||
| 0.50 | 100 | 94.2 | 68.6 | 94.7 | 68.6 | ||
| 250 | 94.8 | 33.4 | 95.1 | 30.0 | |||
| 400 | 94.8 | 13.5 | 95.1 | 10.5 | |||
| 1.50 | 0.664 | 0.30 | 100 | 94.1 | 90.1 | 94.2 | 90.2 |
| 250 | 94.2 | 79.9 | 94.3 | 81.2 | |||
| 400 | 94.5 | 71.5 | 94.4 | 71.1 | |||
| 0.50 | 100 | 94.1 | 79.8 | 94.8 | 81.3 | ||
| 250 | 94.3 | 55.4 | 94.9 | 54.8 | |||
| 400 | 94.6 | 35.0 | 94.8 | 33.5 | |||
| 1.00 | 0.76 | 0.30 | 100 | 93.8 | 88.1 | 93.7 | 88.6 |
| 250 | 94.1 | 73.8 | 94.9 | 73.3 | |||
| 400 | 94.3 | 60.6 | 94.7 | 58.8 | |||
| 0.50 | 100 | 94.1 | 73.3 | 94.3 | 73.3 | ||
| 250 | 94.9 | 38.4 | 95.0 | 36.2 | |||
| 400 | 94.9 | 18.0 | 95.0 | 14.4 | |||
| 1.50 | 0.76 | 0.30 | 100 | 94.2 | 91.5 | 94.1 | 91.5 |
| 250 | 94.6 | 81.7 | 94.4 | 83.1 | |||
| 400 | 94.2 | 74.4 | 94.4 | 74.5 | |||
| 0.50 | 100 | 94.5 | 82.9 | 94.3 | 84.0 | ||
| 250 | 94.1 | 59.8 | 95.0 | 60.0 | |||
| 400 | 94.4 | 39.8 | 94.7 | 39.3 | |||
Power for testing 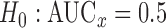
|
|||||||
| 1.00 | 0.50 | 0.30 | 100 | 5.7 | 16.6 | 5.9 | 15.8 |
| 250 | 5.8 | 32.1 | 5.3 | 32.5 | |||
| 400 | 5.5 | 46.7 | 5.6 | 48.4 | |||
| 0.50 | 100 | 5.8 | 36.1 | 5.6 | 36.8 | ||
| 250 | 5.4 | 71.8 | 5.0 | 75.6 | |||
| 400 | 5.3 | 89.8 | 5.0 | 92.0 | |||
| 1.50 | 0.50 | 0.30 | 100 | 6.2 | 12.0 | 5.9 | 11.5 |
| 250 | 6.1 | 22.0 | 5.5 | 21.3 | |||
| 400 | 5.5 | 30.7 | 5.6 | 31.3 | |||
| 0.50 | 100 | 5.7 | 23.7 | 5.2 | 22.6 | ||
| 250 | 5.5 | 49.7 | 4.9 | 50.3 | |||
| 400 | 5.5 | 69.0 | 5.1 | 71.2 | |||
| 1.00 | 0.664 | 0.30 | 100 | 96.7 | 89.2 | 97.1 | 90.3 |
| 250 | 100 | 99.9 | 100 | 99.9 | |||
| 400 | 100 | 100 | 100 | 100 | |||
| 0.50 | 100 | 98.3 | 74.1 | 98.5 | 74.5 | ||
| 250 | 100 | 98.2 | 100 | 99.1 | |||
| 400 | 100 | 100 | 100 | 100 | |||
| 1.50 | 0.664 | 0.30 | 100 | 97.4 | 92.0 | 97.70 | 92.4 |
| 250 | 100 | 100 | 100 | 100 | |||
| 400 | 100 | 100 | 100 | 100 | |||
| 0.50 | 100 | 98.0 | 82.3 | 98.4 | 83.8 | ||
| 250 | 100 | 99.3 | 100 | 99.7 | |||
| 400 | 100 | 100 | 100 | 100 | |||
| 1.00 | 0.76 | 0.30 | 100 | 100 | 100 | 100 | 100 |
| 250 | 100 | 100 | 100 | 100 | |||
| 0.50 | 100 | 100 | 100 | 100 | 100 | ||
| 250 | 100 | 100 | 100 | 100 | |||
| 1.50 | 0.76 | 0.30 | 100 | 100 | 100 | 100 | 100 |
| 250 | 100 | 100 | 100 | 100 | |||
| 0.50 | 100 | 100 | 100 | 100 | 100 | ||
| 250 | 100 | 100 | 100 | 100 | |||
 Finite-population stratified sampling.
Finite-population stratified sampling.
3.2. Compare performance between markers
Table 2 shows performance of  estimators for both types of sampling designs for settings where
estimators for both types of sampling designs for settings where 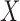 and
and  have the same variance and same correlation with
have the same variance and same correlation with 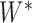 conditional on
conditional on 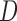 :
:  and
and 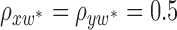 , where we have
, where we have 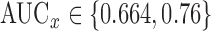 ,
, 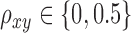 , and
, and  . The IPW
. The IPW 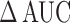 estimator has good performance: minimum bias, coverage of 95% CI and type-I error for testing the equivalence in AUC close to nominal level. When markers
estimator has good performance: minimum bias, coverage of 95% CI and type-I error for testing the equivalence in AUC close to nominal level. When markers 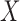 and
and 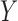 have exactly the same distribution (consequently same ROC curve and AUC), the empirical estimator of
have exactly the same distribution (consequently same ROC curve and AUC), the empirical estimator of 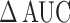 is also unbiased: the biases in
is also unbiased: the biases in 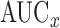 and
and 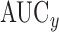 are equal and thus cancel out, due to the equality in the distribution of the two markers and in their correlation with the matching stratum. The coverage of its 95% CI is close to the nominal level. Type-I error for testing the equivalence in AUC using the Delong–Delong test is also close to the nominal level. When
are equal and thus cancel out, due to the equality in the distribution of the two markers and in their correlation with the matching stratum. The coverage of its 95% CI is close to the nominal level. Type-I error for testing the equivalence in AUC using the Delong–Delong test is also close to the nominal level. When 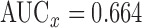 and
and 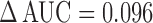 , the empirical
, the empirical 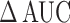 estimator has small bias, with a magnitude much smaller compared with that of the
estimator has small bias, with a magnitude much smaller compared with that of the 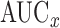 estimator; its 95% CI has good coverage when sample size is small but slight undercoverage when sample size gets large
estimator; its 95% CI has good coverage when sample size is small but slight undercoverage when sample size gets large  . Despite the bias in empirical
. Despite the bias in empirical 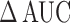 , the Delong–Delong test for equivalence in AUC can have advantage in power compared with the IPW estimator in this particular setting, due to the positive bias in AUC difference.
, the Delong–Delong test for equivalence in AUC can have advantage in power compared with the IPW estimator in this particular setting, due to the positive bias in AUC difference.
Table 2.
Performance of various estimators of  for scenarios where the two markers have same variability and same correlation with covariate
for scenarios where the two markers have same variability and same correlation with covariate 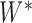 conditional on
conditional on 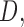 for the bi-normal model described in Section 3. Disease prevalence is
for the bi-normal model described in Section 3. Disease prevalence is  . Marker
. Marker  and
and  is each standard normal among controls. Here we have
is each standard normal among controls. Here we have 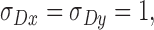
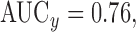
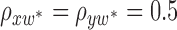 . By
. By 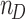 and
and  we indicate, respectively, the expected number of cases and controls sampled in phase two for Bernoulli sampling and exact number of cases and controls sampled for finite-population stratified sampling. Results are based on 5000 Monte Carlo simulations
we indicate, respectively, the expected number of cases and controls sampled in phase two for Bernoulli sampling and exact number of cases and controls sampled for finite-population stratified sampling. Results are based on 5000 Monte Carlo simulations
| Bernoulli sampling |
FPS sampling sampling |
||||||
|---|---|---|---|---|---|---|---|
|
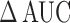
|

|

|

|

|
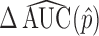
|

|
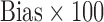
|
|||||||
| 0.76 | 0.00 | 0.00 | 100 | 0.12 | 0.11 | 0.12 | 0.13 |
| 250 | 0.03 | 0.02 | 0.07 | 0.09 | |||
| 400 |
 0.00 0.00 |
 0.00 0.00 |
0.05 | 0.07 | |||
| 0.50 | 100 | 0.08 | 0.06 |
 0.01 0.01 |
0.01 | ||
| 250 | 0.03 | 0.03 |
 0.02 0.02 |
 0.02 0.02 |
|||
| 400 |
 0.01 0.01 |
0.01 |
 0.03 0.03 |
 0.03 0.03 |
|||
| 0.664 | 0.096 | 0.00 | 100 | 0.12 | 0.98 | 0.14 | 1.03 |
| 250 | 0.03 | 0.90 | 0.08 | 0.97 | |||
| 400 | 0.01 | 0.88 | 0.05 | 0.94 | |||
| 0.50 | 100 | 0.11 | 0.96 | 0.00 | 0.90 | ||
| 250 | 0.04 | 0.92 |
 0.01 0.01 |
0.86 | |||
| 400 |
 0.01 0.01 |
0.89 |
 0.03 0.03 |
0.85 | |||
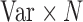
|
|||||||
| 0.76 | 0.00 | 0.00 | 100 | 11.66 | 13.31 | 11.94 | 13.63 |
| 250 | 4.78 | 5.48 | 4.72 | 5.34 | |||
| 400 | 2.94 | 3.42 | 2.92 | 3.36 | |||
| 0.50 | 100 | 6.44 | 7.26 | 6.42 | 7.05 | ||
| 250 | 2.51 | 2.88 | 2.62 | 2.91 | |||
| 400 | 1.59 | 1.84 | 1.62 | 1.82 | |||
| 0.664 | 0.096 | 0.00 | 100 | 13.79 | 14.68 | 13.92 | 14.97 |
| 250 | 5.59 | 6.02 | 5.50 | 5.86 | |||
| 400 | 3.42 | 3.75 | 3.42 | 3.69 | |||
| 0.50 | 100 | 7.55 | 7.96 | 7.57 | 7.73 | ||
| 250 | 2.94 | 3.16 | 3.08 | 3.21 | |||
| 400 | 1.85 | 2.00 | 1.91 | 2.01 | |||
| Coverage of 95% CI | |||||||
| 0.76 | 0.00 | 0.00 | 100 | 94.6 | 94.8 | 94.8 | 95.1 |
| 250 | 94.7 | 94.8 | 94.7 | 95.0 | |||
| 400 | 95.0 | 94.7 | 94.7 | 95.1 | |||
| 0.50 | 100 | 94.8 | 94.8 | 94.7 | 95.0 | ||
| 250 | 95.3 | 94.9 | 94.6 | 94.8 | |||
| 400 | 95.2 | 95.0 | 95.0 | 95.0 | |||
| 0.664 | 0.096 | 0.00 | 100 | 94.8 | 94.4 | 94.8 | 94.6 |
| 250 | 94.3 | 94.0 | 94.7 | 93.8 | |||
| 400 | 94.9 | 93.3 | 94.8 | 93.1 | |||
| 0.50 | 100 | 94.6 | 94.2 | 94.5 | 94.5 | ||
| 250 | 95.2 | 93.5 | 94.4 | 93.1 | |||
| 400 | 95.1 | 92.4 | 94.6 | 92.8 | |||
Power for testing 
|
|||||||
| 0.76 | 0.00 | 0.00 | 100 | 5.4 | 5.1 | 5.2 | 4.8 |
| 250 | 5.3 | 5.2 | 5.3 | 5.0 | |||
| 400 | 5.0 | 5.2 | 5.3 | 4.9 | |||
| 0.50 | 100 | 5.2 | 5.1 | 5.3 | 4.9 | ||
| 250 | 4.7 | 5.1 | 5.4 | 5.1 | |||
| 400 | 4.8 | 5.0 | 5.0 | 5.0 | |||
| 0.664 | 0.096 | 0.00 | 100 | 46.7 | 48.9 | 46.7 | 49.4 |
| 250 | 82.5 | 85.6 | 83.3 | 86.5 | |||
| 400 | 95.4 | 96.6 | 95.9 | 96.9 | |||
| 0.50 | 100 | 71.4 | 75.1 | 70.7 | 75.5 | ||
| 250 | 97.6 | 98.8 | 97.6 | 98.6 | |||
| 400 | 100 | 100 | 99.9 | 100 | |||
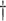 Finite-population stratified sampling.
Finite-population stratified sampling.
Table 3 shows results of  estimators for settings again with equal variance between
estimators for settings again with equal variance between 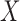 and
and 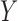 conditional on
conditional on 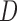 , i.e.,
, i.e.,  . However, unlike Table 2 where both markers have the same correlation with the covariate stratum, here we fix
. However, unlike Table 2 where both markers have the same correlation with the covariate stratum, here we fix 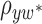 to be 0.5 but vary
to be 0.5 but vary 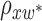 from 0.4 to 0.1. When markers
from 0.4 to 0.1. When markers 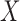 and
and 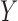 have the same distribution and AUC, the empirical estimator of
have the same distribution and AUC, the empirical estimator of  is biased because the magnitude of bias is different between
is biased because the magnitude of bias is different between 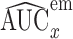 and
and  due to the difference in correlation between each marker and the covariate stratum. Bias is also observed when
due to the difference in correlation between each marker and the covariate stratum. Bias is also observed when  . Corresponding 95% CI based on
. Corresponding 95% CI based on  has an undercoverage problem. The Delong–Delong test also has an inflated type-I error rate. The inflation gets more severe as the difference between
has an undercoverage problem. The Delong–Delong test also has an inflated type-I error rate. The inflation gets more severe as the difference between 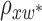 and
and  increases: type-I error can become similar to power for some settings in Table 3 or become even larger than power in some other constructed settings (details omitted). This is due to the bias in the empirical
increases: type-I error can become similar to power for some settings in Table 3 or become even larger than power in some other constructed settings (details omitted). This is due to the bias in the empirical  estimator such that the observed difference between two markers equivalent in AUC can appear similar or even larger compared with the observed difference between two markers that differ in AUC. In practice, when difference in correlations between marker and stratification variable exists, its magnitude is likely on the small to medium side, and thus we expect some inflation of type-I error when applying Delong–Delong's test but not extreme. In contrast, the IPW estimator of
estimator such that the observed difference between two markers equivalent in AUC can appear similar or even larger compared with the observed difference between two markers that differ in AUC. In practice, when difference in correlations between marker and stratification variable exists, its magnitude is likely on the small to medium side, and thus we expect some inflation of type-I error when applying Delong–Delong's test but not extreme. In contrast, the IPW estimator of 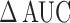 are approximately unbiased with coverage of 95% CIs close to the nominal level; corresponding Wald tests for equivalence in AUC between markers have well-controlled type-I error rates.
are approximately unbiased with coverage of 95% CIs close to the nominal level; corresponding Wald tests for equivalence in AUC between markers have well-controlled type-I error rates.
Table 3.
Performance of different estimators of  for scenarios where the two markers have same variability but different correlation with covariate
for scenarios where the two markers have same variability but different correlation with covariate 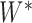 conditional on
conditional on 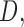 for the bi-normal marker model described in Section 3. Disease prevalence is
for the bi-normal marker model described in Section 3. Disease prevalence is 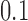 . Marker
. Marker 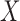 and
and 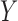 is each standard normal among controls. Here we have
is each standard normal among controls. Here we have 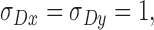

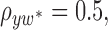
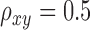 . By
. By  and
and  we indicate, respectively, the expected number of cases and controls sampled in phase two for Bernoulli sampling and exact number of cases and controls sampled for finite-population stratified sampling. Results are based on 5000 Monte Carlo simulations
we indicate, respectively, the expected number of cases and controls sampled in phase two for Bernoulli sampling and exact number of cases and controls sampled for finite-population stratified sampling. Results are based on 5000 Monte Carlo simulations
| Bernoulli sampling |
FPS sampling sampling |
||||||
|---|---|---|---|---|---|---|---|

|
|
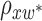
|

|
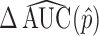
|

|

|

|

|
|||||||
| 0.76 | 0.00 | 0.40 | 100 | 0.07 |
 0.99 0.99 |
0.01 |
 1.02 1.02 |
| 250 |
 0.01 0.01 |
 1.07 1.07 |
0.03 |
 1.04 1.04 |
|||
| 400 |
 0.04 0.04 |
 1.09 1.09 |
0.01 |
 1.05 1.05 |
|||
| 0.30 | 100 | 0.03 |
 2.04 2.04 |
 0.03 0.03 |
 2.13 2.13 |
||
| 250 |
 0.01 0.01 |
 2.10 2.10 |
0.01 |
 2.10 2.10 |
|||
| 400 |
 0.01 0.01 |
 2.10 2.10 |
0.02 |
 2.09 2.09 |
|||
| 0.20 | 100 | 0.11 |
 2.98 2.98 |
0.03 |
 3.09 3.09 |
||
| 250 | 0.04 |
 3.08 3.08 |
0.02 |
 3.09 3.09 |
|||
| 400 | 0.03 |
 3.09 3.09 |
0.01 |
 3.09 3.09 |
|||
| 0.10 | 100 | 0.11 |
 4.03 4.03 |
0.02 |
 4.10 4.10 |
||
| 250 | 0.02 |
 4.10 4.10 |
0.04 |
 4.09 4.09 |
|||
| 400 | 0.03 |
 4.11 4.11 |
0.01 |
 4.12 4.12 |
|||
| 0.664 | 0.096 | 0.40 | 100 | 0.09 |
 0.29 0.29 |
0.03 |
 0.30 0.30 |
| 250 | 0.01 |
 0.36 0.36 |
0.04 |
 0.33 0.33 |
|||
| 400 |
 0.02 0.02 |
 0.39 0.39 |
0.02 |
 0.35 0.35 |
|||
| 0.30 | 100 | 0.03 |
 1.52 1.52 |
 0.03 0.03 |
 1.62 1.62 |
||
| 250 |
 0.02 0.02 |
 1.59 1.59 |
0.01 |
 1.58 1.58 |
|||
| 400 |
 0.02 0.02 |
 1.59 1.59 |
0.01 |
 1.57 1.57 |
|||
| 0.20 | 100 | 0.12 |
 2.62 2.62 |
0.03 |
 2.76 2.76 |
||
| 250 | 0.04 |
 2.74 2.74 |
0.01 |
 2.75 2.75 |
|||
| 400 | 0.02 |
 2.76 2.76 |
0.01 |
 2.75 2.75 |
|||
| 0.10 | 100 | 0.11 |
 3.85 3.85 |
0.03 |
 3.92 3.92 |
||
| 250 | 0.01 |
 3.93 3.93 |
0.04 |
 3.92 3.92 |
|||
| 400 | 0.02 |
 3.94 3.94 |
0.01 |
 3.95 3.95 |
|||

|
|||||||
| 0.76 | 0.00 | 0.40 | 100 | 6.63 | 7.18 | 6.47 | 6.98 |
| 250 | 2.51 | 2.78 | 2.53 | 2.74 | |||
| 400 | 1.61 | 1.77 | 1.63 | 1.78 | |||
| 0.30 | 100 | 6.62 | 6.98 | 6.63 | 6.81 | ||
| 250 | 2.57 | 2.75 | 2.66 | 2.69 | |||
| 400 | 1.62 | 1.75 | 1.62 | 1.67 | |||
| 0.20 | 100 | 6.64 | 6.90 | 6.44 | 6.34 | ||
| 250 | 2.56 | 2.65 | 2.64 | 2.57 | |||
| 400 | 1.66 | 1.72 | 1.62 | 1.59 | |||
| 0.10 | 100 | 6.75 | 7.00 | 6.40 | 5.88 | ||
| 250 | 2.56 | 2.71 | 2.51 | 2.33 | |||
| 400 | 1.57 | 1.66 | 1.61 | 1.46 | |||
| 0.664 | 0.096 | 0.40 | 100 | 7.85 | 7.92 | 7.61 | 7.71 |
| 250 | 2.97 | 3.08 | 2.97 | 3.04 | |||
| 400 | 1.88 | 1.95 | 1.92 | 1.97 | |||
| 0.30 | 100 | 7.92 | 7.78 | 7.81 | 7.66 | ||
| 250 | 3.05 | 3.06 | 3.14 | 3.03 | |||
| 400 | 1.93 | 1.96 | 1.91 | 1.88 | |||
| 0.20 | 100 | 7.82 | 7.67 | 7.70 | 7.23 | ||
| 250 | 3.06 | 2.98 | 3.11 | 2.90 | |||
| 400 | 1.96 | 1.92 | 1.91 | 1.80 | |||
| 0.10 | 100 | 7.97 | 7.81 | 7.57 | 6.68 | ||
| 250 | 3.02 | 3.02 | 2.95 | 2.63 | |||
| 400 | 1.86 | 1.85 | 1.90 | 1.67 | |||
| Coverage of 95% CI | |||||||
| 0.76 | 0.00 | 0.40 | 100 | 94.6 | 94.1 | 95.0 | 94.2 |
| 250 | 95.0 | 92.0 | 95.0 | 93.2 | |||
| 400 | 95.1 | 90.5 | 94.4 | 91.2 | |||
| 0.30 | 100 | 94.5 | 91.4 | 94.7 | 90.7 | ||
| 250 | 94.7 | 85.7 | 94.8 | 85.6 | |||
| 400 | 94.7 | 79.1 | 94.8 | 79.1 | |||
| 0.20 | 100 | 94.2 | 86.7 | 94.7 | 86.9 | ||
| 250 | 95.0 | 73.6 | 94.8 | 74.1 | |||
| 400 | 94.2 | 60.2 | 94.9 | 60.7 | |||
| 0.10 | 100 | 94.3 | 79.4 | 94.3 | 81.1 | ||
| 250 | 95.1 | 57.7 | 94.8 | 57.4 | |||
| 400 | 95.0 | 38.1 | 95.1 | 37.2 | |||
| 0.664 | 0.096 | 0.40 | 100 | 94.5 | 95.2 | 94.7 | 95.0 |
| 250 | 95.2 | 94.8 | 95.0 | 94.9 | |||
| 400 | 95.3 | 94.5 | 94.5 | 94.4 | |||
| 0.30 | 100 | 94.5 | 92.6 | 94.7 | 92.6 | ||
| 250 | 95.0 | 90.1 | 94.5 | 89.8 | |||
| 400 | 94.7 | 86.7 | 94.6 | 87.4 | |||
| 0.20 | 100 | 94.1 | 89.1 | 94.6 | 88.9 | ||
| 250 | 95.1 | 80.0 | 94.5 | 79.9 | |||
| 400 | 94.2 | 70.3 | 94.7 | 70.6 | |||
| 0.10 | 100 | 94.5 | 82.2 | 94.2 | 83.5 | ||
| 250 | 94.9 | 63.3 | 94.8 | 64.2 | |||
| 400 | 95.0 | 46.1 | 94.8 | 45.7 | |||
Power for testing 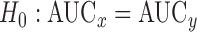
|
|||||||
| 0.76 | 0.00 | 0.40 | 100 | 5.4 | 5.6 | 5.0 | 7.9 |
| 250 | 5.0 | 7.9 | 5.0 | 6.7 | |||
| 400 | 4.9 | 9.5 | 5.6 | 8.8 | |||
| 0.30 | 100 | 5.5 | 8.5 | 5.3 | 14.2 | ||
| 250 | 5.3 | 14.2 | 5.2 | 14.3 | |||
| 400 | 5.3 | 20.9 | 5.2 | 20.7 | |||
| 0.20 | 100 | 5.8 | 13.1 | 5.0 | 26.3 | ||
| 250 | 5.0 | 26.3 | 5.2 | 25.7 | |||
| 400 | 5.8 | 39.6 | 5.1 | 39.2 | |||
| 0.10 | 100 | 5.7 | 20.3 | 4.9 | 42.0 | ||
| 250 | 4.9 | 42.0 | 5.2 | 42.5 | |||
| 400 | 5.0 | 61.7 | 4.9 | 62.6 | |||
| 0.664 | 0.096 | 0.40 | 100 | 70.2 | 64.7 | 97.8 | 95.8 |
| 250 | 97.8 | 95.8 | 97.9 | 96.3 | |||
| 400 | 99.8 | 99.6 | 99.9 | 99.6 | |||
| 0.30 | 100 | 69.4 | 54.2 | 97.5 | 90.1 | ||
| 250 | 97.5 | 90.1 | 97.5 | 90.1 | |||
| 400 | 99.8 | 98.3 | 99.9 | 98.6 | |||
| 0.20 | 100 | 70.0 | 43.1 | 97.7 | 79.9 | ||
| 250 | 97.7 | 79.9 | 97.7 | 80.3 | |||
| 400 | 99.9 | 94.2 | 99.8 | 94.8 | |||
| 0.10 | 100 | 70.6 | 31.7 | 97.8 | 64.4 | ||
| 250 | 97.8 | 64.4 | 98.1 | 65.8 | |||
| 400 | 99.9 | 83.8 | 99.9 | 84.8 | |||
 Finite-population stratified sampling.
Finite-population stratified sampling.
Table 4 presents results for settings with 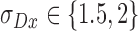 . That is, the variability of marker
. That is, the variability of marker 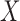 is larger than that of
is larger than that of 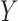 among cases. Note that when
among cases. Note that when 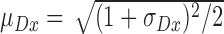 , we have
, we have  , although the two markers can have different ROC curves when
, although the two markers can have different ROC curves when 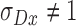 . When this happens, the Delong–Delong test has an inflated type-I error, even when correlation with the stratification variable is the same for both markers; whereas the Wald test based on the IPW estimator of
. When this happens, the Delong–Delong test has an inflated type-I error, even when correlation with the stratification variable is the same for both markers; whereas the Wald test based on the IPW estimator of  has a well-controlled type-I error rate. For both the scenarios with
has a well-controlled type-I error rate. For both the scenarios with 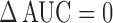 and
and 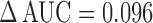 , the empirical
, the empirical 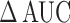 estimator is biased; corresponding 95% CI undercovers the true parameter value. In contrast, the IPW estimators have minimum bias and good coverage of 95% CIs.
estimator is biased; corresponding 95% CI undercovers the true parameter value. In contrast, the IPW estimators have minimum bias and good coverage of 95% CIs.
Table 4.
Performance of different estimators of 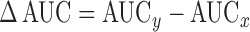 for scenarios where the two markers have different variability among cases, for the bi-normal model described in Section 3. Disease prevalence is
for scenarios where the two markers have different variability among cases, for the bi-normal model described in Section 3. Disease prevalence is 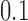 . Marker
. Marker  and
and 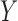 are each standard normal among controls. Here we have
are each standard normal among controls. Here we have 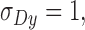
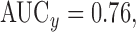
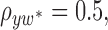
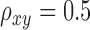 . By
. By 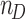 and
and 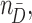 we indicate, respectively, the expected number of cases and controls sampled in phase two for Bernoulli sampling and exact number of cases and controls sampled for finite-population stratified sampling. Results are based on 5000 Monte Carlo simulations
we indicate, respectively, the expected number of cases and controls sampled in phase two for Bernoulli sampling and exact number of cases and controls sampled for finite-population stratified sampling. Results are based on 5000 Monte Carlo simulations
| Bernoulli sampling |
FPS sampling sampling |
|||||||
|---|---|---|---|---|---|---|---|---|
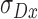
|
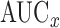
|
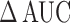
|
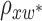
|

|

|

|
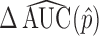
|

|
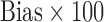
|
||||||||
| 1.50 | 0.76 | 0.00 | 0.50 | 100 | 0.04 |
 1.11 1.11 |
0.04 |
 1.08 1.08 |
| 250 | 0.01 |
 1.13 1.13 |
 0.01 0.01 |
 1.15 1.15 |
||||
| 400 | 0.01 |
 1.12 1.12 |
 0.02 0.02 |
 1.16 1.16 |
||||
| 0.30 | 100 | 0.04 |
 2.69 2.69 |
 0.03 0.03 |
 2.79 2.79 |
|||
| 250 | 0.01 |
 2.75 2.75 |
 0.01 0.01 |
 2.77 2.77 |
||||
| 400 |
 0.01 0.01 |
 2.76 2.76 |
 0.00 0.00 |
 2.76 2.76 |
||||
| 2.00 | 0.76 | 0.00 | 0.50 | 100 |
 0.07 0.07 |
 2.02 2.02 |
0.10 |
 1.83 1.83 |
| 250 |
 0.02 0.02 |
 1.95 1.95 |
0.02 |
 1.91 1.91 |
||||
| 400 | 0.02 |
 1.90 1.90 |
 0.00 0.00 |
 1.93 1.93 |
||||
| 0.30 | 100 | 0.01 |
 3.19 3.19 |
 0.04 0.04 |
 3.27 3.27 |
|||
| 250 |
 0.00 0.00 |
 3.22 3.22 |
 0.01 0.01 |
 3.23 3.23 |
||||
| 400 |
 0.00 0.00 |
 3.21 3.21 |
 0.02 0.02 |
 3.24 3.24 |
||||
| 1.50 | 0.664 | 0.096 | 0.50 | 100 | 0.05 |
 0.43 0.43 |
0.05 |
 0.39 0.39 |
| 250 | 0.01 |
 0.45 0.45 |
0.00 |
 0.46 0.46 |
||||
| 400 | 0.01 |
 0.44 0.44 |
 0.01 0.01 |
 0.47 0.47 |
||||
| 0.30 | 100 | 0.04 |
 2.29 2.29 |
 0.02 0.02 |
 2.38 2.38 |
|||
| 250 |
 0.00 0.00 |
 2.35 2.35 |
0.00 |
 2.35 2.35 |
||||
| 400 |
 0.01 0.01 |
 2.35 2.35 |
0.01 |
 2.35 2.35 |
||||
| 2.00 | 0.664 | 0.096 | 0.50 | 100 |
 0.04 0.04 |
 1.46 1.46 |
0.11 |
 1.27 1.27 |
| 250 |
 0.02 0.02 |
 1.40 1.40 |
0.03 |
 1.36 1.36 |
||||
| 400 | 0.03 |
 1.35 1.35 |
0.00 |
 1.37 1.37 |
||||
| 0.30 | 100 | 0.01 |
 2.86 2.86 |
 0.03 0.03 |
 2.93 2.93 |
|||
| 250 |
 0.01 0.01 |
 2.90 2.90 |
 0.00 0.00 |
 2.90 2.90 |
||||
| 400 |
 0.00 0.00 |
 2.89 2.89 |
 0.01 0.01 |
 2.90 2.90 |
||||
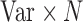
|
||||||||
| 1.50 | 0.76 | 0.00 | 0.50 | 100 | 6.84 | 7.62 | 6.57 | 7.29 |
| 250 | 2.69 | 3.05 | 2.71 | 3.01 | ||||
| 400 | 1.67 | 1.90 | 1.70 | 1.92 | ||||
| 0.30 | 100 | 6.92 | 7.55 | 6.85 | 7.26 | |||
| 250 | 2.67 | 2.94 | 2.68 | 2.81 | ||||
| 400 | 1.67 | 1.84 | 1.67 | 1.77 | ||||
| 2.00 | 0.76 | 0.00 | 0.50 | 100 | 7.27 | 7.98 | 6.91 | 7.82 |
| 250 | 2.89 | 3.21 | 2.82 | 3.17 | ||||
| 400 | 1.81 | 2.00 | 1.77 | 2.01 | ||||
| 0.30 | 100 | 7.32 | 8.07 | 7.20 | 7.73 | |||
| 250 | 2.87 | 3.17 | 2.85 | 3.03 | ||||
| 400 | 1.79 | 1.98 | 1.79 | 1.92 | ||||
| 1.50 | 0.664 | 0.096 | 0.50 | 100 | 7.93 | 8.39 | 7.68 | 8.05 |
| 250 | 3.14 | 3.37 | 3.17 | 3.35 | ||||
| 400 | 1.91 | 2.07 | 1.98 | 2.12 | ||||
| 0.30 | 100 | 8.07 | 8.36 | 7.98 | 8.13 | |||
| 250 | 3.14 | 3.27 | 3.12 | 3.15 | ||||
| 400 | 1.95 | 2.04 | 1.95 | 1.98 | ||||
| 2.00 | 0.664 | 0.096 | 0.50 | 100 | 8.47 | 8.92 | 8.03 | 8.71 |
| 250 | 3.34 | 3.57 | 3.28 | 3.56 | ||||
| 400 | 2.05 | 2.18 | 2.06 | 2.24 | ||||
| 0.30 | 100 | 8.44 | 8.96 | 8.26 | 8.63 | |||
| 250 | 3.33 | 3.52 | 3.28 | 3.39 | ||||
| 400 | 2.07 | 2.18 | 2.06 | 2.14 | ||||
| Coverage of 95% CI | ||||||||
| 1.50 | 0.76 | 0.00 | 0.50 | 100 | 94.8 | 93.6 | 95.0 | 94.1 |
| 250 | 94.6 | 91.9 | 94.6 | 92.2 | ||||
| 400 | 94.6 | 90.5 | 94.7 | 90.3 | ||||
| 0.30 | 100 | 94.2 | 88.0 | 94.3 | 88.1 | |||
| 250 | 94.8 | 78.8 | 94.8 | 78.5 | ||||
| 400 | 94.6 | 68.3 | 94.9 | 68.4 | ||||
| 2.00 | 0.76 | 0.00 | 0.50 | 100 | 94.7 | 91.4 | 95.3 | 92.3 |
| 250 | 94.8 | 87.8 | 94.6 | 86.9 | ||||
| 400 | 94.7 | 83.5 | 94.4 | 83.5 | ||||
| 0.30 | 100 | 94.3 | 86.4 | 94.6 | 85.6 | |||
| 250 | 94.4 | 73.6 | 94.6 | 73.7 | ||||
| 400 | 94.5 | 61.8 | 94.6 | 60.9 | ||||
| 1.50 | 0.664 | 0.096 | 0.50 | 100 | 94.7 | 94.7 | 94.7 | 95.0 |
| 250 | 94.8 | 94.0 | 94.3 | 94.2 | ||||
| 400 | 94.9 | 93.9 | 94.4 | 93.6 | ||||
| 0.30 | 100 | 94.3 | 90.3 | 94.4 | 89.7 | |||
| 250 | 94.6 | 84.2 | 94.9 | 84.1 | ||||
| 400 | 94.5 | 77.4 | 94.8 | 77.0 | ||||
| 2.00 | 0.664 | 0.096 | 0.50 | 100 | 94.3 | 93.0 | 95.1 | 93.7 |
| 250 | 94.5 | 91.2 | 94.8 | 91.0 | ||||
| 400 | 94.7 | 89.5 | 94.5 | 89.2 | ||||
| 0.30 | 100 | 94.6 | 88.7 | 94.6 | 87.9 | |||
| 250 | 94.3 | 79.3 | 94.5 | 79.3 | ||||
| 400 | 94.7 | 70.2 | 94.6 | 70.0 | ||||
Power for testing 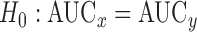
|
||||||||
| 1.50 | 0.76 | 0.00 | 0.50 | 100 | 5.2 | 6.3 | 5.0 | 5.8 |
| 250 | 5.4 | 8.0 | 5.4 | 7.7 | ||||
| 400 | 5.4 | 9.5 | 5.3 | 9.6 | ||||
| 0.30 | 100 | 5.8 | 11.8 | 5.7 | 11.8 | |||
| 250 | 5.2 | 21.1 | 5.2 | 21.3 | ||||
| 400 | 5.4 | 31.6 | 5.1 | 31.5 | ||||
| 2.00 | 0.76 | 0.00 | 0.50 | 100 | 5.3 | 8.5 | 4.70 | 7.5 |
| 250 | 5.2 | 12.1 | 5.4 | 13.0 | ||||
| 400 | 5.3 | 16.4 | 5.6 | 16.5 | ||||
| 0.30 | 100 | 5.7 | 13.4 | 5.4 | 14.2 | |||
| 250 | 5.6 | 26.3 | 5.4 | 26.2 | ||||
| 400 | 5.5 | 38.1 | 5.4 | 39.0 | ||||
| 1.50 | 0.664 | 0.096 | 0.50 | 100 | 68.9 | 61.1 | 69.9 | 62.3 |
| 250 | 97.0 | 94.8 | 97.1 | 94.6 | ||||
| 400 | 99.9 | 99.6 | 99.8 | 99.3 | ||||
| 0.30 | 100 | 68.6 | 44.4 | 69.6 | 44.4 | |||
| 250 | 97.1 | 81.9 | 97.6 | 82.0 | ||||
| 400 | 99.9 | 95.2 | 99.9 | 95.4 | ||||
| 2.00 | 0.664 | 0.096 | 0.50 | 100 | 66.1 | 49.1 | 67.9 | 50.9 |
| 250 | 96.2 | 87.5 | 96.6 | 87.6 | ||||
| 400 | 99.8 | 97.6 | 99.6 | 97.6 | ||||
| 0.30 | 100 | 66.4 | 37.4 | 66.9 | 37.2 | |||
| 250 | 96.3 | 72.2 | 96.9 | 73.0 | ||||
| 400 | 99.8 | 89.9 | 99.9 | 89.9 | ||||
 Finite-population stratified sampling.
Finite-population stratified sampling.
In Web Appendix H (see supplementary material), we also present additional simulation results when biomarkers follow gamma distributions conditional on disease status. The conclusion regarding the performance of the IPW and the empirical AUC estimators is similar to the bi-normal marker model.
4. Implication on efficiency of sampling schemes
The analytical variance formula we developed in Section 2 will be valuable to biomarker researchers for studying the impact of the sampling scheme on the efficiency of biomarker performance estimators. We demonstrate that using an example comparing two designs with the same number of participants measuring disease outcome, covariate, and biomarker. The setting is similar to that in Section 3, with disease prevalence  . Covariate
. Covariate 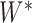 and biomarker
and biomarker 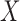 are bivariate normal with correlation
are bivariate normal with correlation 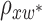 conditional on
conditional on 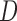 . Among controls
. Among controls 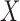 and
and  are each standard normal; among cases
are each standard normal; among cases 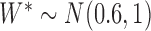 and
and  . Let
. Let  be a binary covariate stratum derived from
be a binary covariate stratum derived from  , with
, with 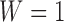 if
if 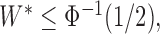 and
and  otherwise. We compare two sampling designs. Both are two-phase studies with a random cohort sample of size
otherwise. We compare two sampling designs. Both are two-phase studies with a random cohort sample of size 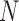 drawn in the first phase. In the second phase, both designs include all cases from a phase-one sample, i.e.,
drawn in the first phase. In the second phase, both designs include all cases from a phase-one sample, i.e., 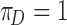 ; a simple random sample of controls of size
; a simple random sample of controls of size  are drawn without replacement in Design 1, simple random samples of controls with the same number as cases are drawn without replacement from each
are drawn without replacement in Design 1, simple random samples of controls with the same number as cases are drawn without replacement from each  stratum in Design 2. In Design 1, phase-two case and control samples are representative of their respective distributions; thus the empirical estimator of AUC is valid and is considered for this design. In contrast, phase-two case and control samples in Design 2 are not representative of their respective distributions; thus we use the proposed
stratum in Design 2. In Design 1, phase-two case and control samples are representative of their respective distributions; thus the empirical estimator of AUC is valid and is considered for this design. In contrast, phase-two case and control samples in Design 2 are not representative of their respective distributions; thus we use the proposed 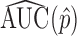 with empirically estimated sampling weights conditional on sampling strata for Design 2.
with empirically estimated sampling weights conditional on sampling strata for Design 2.
Following Theorem 1 and Section 2.4, asymptotic variance of 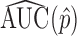 in Design 2 equals
in Design 2 equals
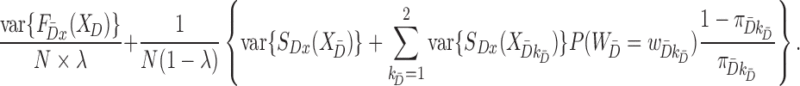 |
(4.1) |
Since  in Design 1 can be thought of as an
in Design 1 can be thought of as an 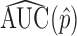 estimator where there is only one sampling stratum for cases and for controls, its asymptotic variance can be similarly derived as
estimator where there is only one sampling stratum for cases and for controls, its asymptotic variance can be similarly derived as
 |
(4.2) |
for  . The result also follows DeLong and others (1988).
. The result also follows DeLong and others (1988).
Figure 1 shows the relative asymptotic efficiency of  in Design 2 versus
in Design 2 versus  in Design 1 for two different
in Design 1 for two different  values, as
values, as 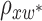 changes from
changes from  0.9 to 0.9. A common U-shape is observed, where Design 2 is more efficient as the magnitude of the correlation increases, whereas Design 1 can be more efficient when the correlation is small. Comparing variance formulae of the two estimators, their difference arises from two components: (i) the difference between
0.9 to 0.9. A common U-shape is observed, where Design 2 is more efficient as the magnitude of the correlation increases, whereas Design 1 can be more efficient when the correlation is small. Comparing variance formulae of the two estimators, their difference arises from two components: (i) the difference between  and
and  , and (ii) the difference between
, and (ii) the difference between 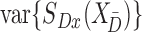 and
and 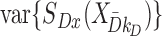 . Note that when
. Note that when 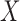 and
and  are not correlated among controls, the difference in variance between the two estimators is solely due to component (i): simple random sampling without replacement from controls is more efficient since the weighted average of
are not correlated among controls, the difference in variance between the two estimators is solely due to component (i): simple random sampling without replacement from controls is more efficient since the weighted average of  for
for 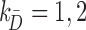 is larger than
is larger than  in our example. As
in our example. As 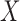 and
and  become more correlated, variance of
become more correlated, variance of 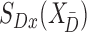 conditional on the covariate stratum becomes smaller compared with its variance among all controls, and thus stratified sampling is more efficient. This is consistent with the use of stratified sampling in survey sampling as a possible way to increase efficiency in estimating parameters such as the population total when a heterogeneous population can be divided into strata with homogeneous units (Cochran, 2007). The same pattern can be observed when biomarkers conditional on disease status follow gamma distributions (Web Supplementary Figure S3). In practice, researchers can evaluate efficiency of different sampling schemes based on prior knowledge in the relationship between marker, covariate, and disease.
conditional on the covariate stratum becomes smaller compared with its variance among all controls, and thus stratified sampling is more efficient. This is consistent with the use of stratified sampling in survey sampling as a possible way to increase efficiency in estimating parameters such as the population total when a heterogeneous population can be divided into strata with homogeneous units (Cochran, 2007). The same pattern can be observed when biomarkers conditional on disease status follow gamma distributions (Web Supplementary Figure S3). In practice, researchers can evaluate efficiency of different sampling schemes based on prior knowledge in the relationship between marker, covariate, and disease.
Fig. 1.

Efficiency of 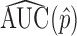 in finite-population stratified sampling (FPS) of controls relative to the empirical AUC estimator in simple random sampling (SRS) without replacement of controls. Efficiency = asymptotic variance of empirical AUC estimator in SRS (equation (4.2))/asymptotic variance of
in finite-population stratified sampling (FPS) of controls relative to the empirical AUC estimator in simple random sampling (SRS) without replacement of controls. Efficiency = asymptotic variance of empirical AUC estimator in SRS (equation (4.2))/asymptotic variance of 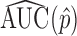 in FPS (equation (4.1)).
in FPS (equation (4.1)).
5. Example
5.1. RV144 example
We illustrate the proposed methodology for estimating and comparing AUC with a real example of biomarker study from the RV144 Thailand HIV vaccine trial. The trial included 16 402 participants aged 18–30 who were 1:1 randomized into a vaccine and a placebo arm. Among vaccine recipients in the RV144 trial who were not yet infected at week 26, an immune response study was conducted to assess vaccine-induced immune response based on peak immunogenicity at week 26 following a finite-population stratified sampling design (Haynes and others, 2012). Around 1.8% vaccinees were censored before the end of the study and were treated as non-infected for subsequent sampling. The study includes all 41 vaccinees infected after week 26 visits. The control vaccinees were selected from a stratified random sample of vaccinees free of HIV-1 infection at 42 months, within strata constructed by gender, number of vaccinations received, and per-protocol status, with five times the number of cases within each stratum.
Two of the primary assays studied, the binding of IgG antibodies to variable regions 1 and 2 (V1V2) of the gp120 Env, and the binding of plasma IgA antibodies to Env, were found to correlate significantly with infection risk (Haynes and others, 2012). Here we evaluate and compare AUC of the two markers. In this application, 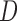 will be HIV infection at 42 months, and
will be HIV infection at 42 months, and 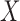 and
and  are V1V2 and IgA measures at week 26, respectively.
are V1V2 and IgA measures at week 26, respectively.
First, the naive empirical AUC estimate is 0.573 (95% 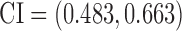 ) for V1V2, and 0.596 (95%
) for V1V2, and 0.596 (95%  ) for IgA. There is around 4% relative increase in AUC for IgA compared with V1V2, although the difference is not statistically significant (
) for IgA. There is around 4% relative increase in AUC for IgA compared with V1V2, although the difference is not statistically significant (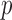 -value
-value  0.774 based on the Delong–Delong test). With empirically estimated sampling weights conditional on the matching stratum, the
0.774 based on the Delong–Delong test). With empirically estimated sampling weights conditional on the matching stratum, the 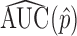 equals 0.588 (95%
equals 0.588 (95% 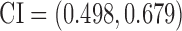 ) for V1V2 and 0.589 (95%
) for V1V2 and 0.589 (95% 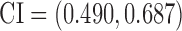 ) for IgA. The difference in AUC between the two markers becomes even smaller after accounting for the sampling scheme with a
) for IgA. The difference in AUC between the two markers becomes even smaller after accounting for the sampling scheme with a  -value of 0.997 based on the Wald test. In this example, adjusting for the sampling scheme makes a small difference in point estimates due to the relatively small variability in sampling weights of controls across strata; the observation that the IPW estimate of the AUC difference between IgA and V1V2 is smaller than the empirical estimate is consistent with observations made in simulation studies, where bias in
-value of 0.997 based on the Wald test. In this example, adjusting for the sampling scheme makes a small difference in point estimates due to the relatively small variability in sampling weights of controls across strata; the observation that the IPW estimate of the AUC difference between IgA and V1V2 is smaller than the empirical estimate is consistent with observations made in simulation studies, where bias in  can make two markers look more different when they have equal AUC.
can make two markers look more different when they have equal AUC.
5.2. Prostate cancer study example
In the second example, we consider a prospective study conducted by the Early Detection Research Network aimed to assess a urine biomarker for prostate cancer, the Prostate Cancer Antigen 3 (PCA3) (Deras and others, 2008). This study involved 570 men enrolled at four North American sites scheduled for prostate biopsy, with a prostate cancer  prevalence of 36.6%. Urinary PCA3
prevalence of 36.6%. Urinary PCA3  and serum PSA (prostate-specific agent,
and serum PSA (prostate-specific agent,  ) are obtained from every participant using specimens collected before biopsy. Each biomarker is log-transformed and standardized to have mean zero and variance 1 among subjects without prostate cancer. Among those with prostate cancer, PCA3 and PSA have mean 0.64 and 0.42, and variance 1.0 and 0.83, respectively. Pearson correlations with age are 0.40 for PCA3 and 0.15 for PSA among those without prostate cancer, and 0.24 for PCA3 and 0.28 for PSA among those with prostate cancer. Increase in age also appears to be associated with increased risk of prostate cancer.
) are obtained from every participant using specimens collected before biopsy. Each biomarker is log-transformed and standardized to have mean zero and variance 1 among subjects without prostate cancer. Among those with prostate cancer, PCA3 and PSA have mean 0.64 and 0.42, and variance 1.0 and 0.83, respectively. Pearson correlations with age are 0.40 for PCA3 and 0.15 for PSA among those without prostate cancer, and 0.24 for PCA3 and 0.28 for PSA among those with prostate cancer. Increase in age also appears to be associated with increased risk of prostate cancer.
To illustrate application of our methodology, we perform a finite-population stratified sampling based on age strata generated by the first and second tertiles of age distribution among controls, i.e., age  60, 60–67,
60, 60–67,  67 years. We randomly sampled 120 cases and then sampled the same number of controls as cases within each age stratum. Based on the case–control sample, the empirical AUC estimate is 0.553 (95%
67 years. We randomly sampled 120 cases and then sampled the same number of controls as cases within each age stratum. Based on the case–control sample, the empirical AUC estimate is 0.553 (95%  ) for PSA and 0.645 (95%
) for PSA and 0.645 (95% 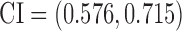 ) for PCA3, with a difference not statistically significant (
) for PCA3, with a difference not statistically significant ( with 95% CI (
with 95% CI ( 0.004, 0.188), and
0.004, 0.188), and  -value
-value  0.061 based on the Delong–Delong test). In contrast,
0.061 based on the Delong–Delong test). In contrast, 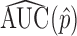 with empirically estimated sampling weights conditional on the stratum equals 0.553 (95%
with empirically estimated sampling weights conditional on the stratum equals 0.553 (95% 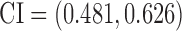 ) for PSA and 0.671 (95%
) for PSA and 0.671 (95% 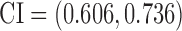 ) for PCA3, with a significant difference detected (
) for PCA3, with a significant difference detected (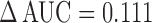 with 95% CI (0.017, 0.206),
with 95% CI (0.017, 0.206),  -value
-value  0.021 based on the Wald test). Note that there is a significant difference between the two markers if we estimate the empirical AUC based on the full cohort sample (
0.021 based on the Wald test). Note that there is a significant difference between the two markers if we estimate the empirical AUC based on the full cohort sample ( , 95%
, 95% 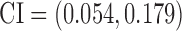 ,
,  -value
-value  0.0003 based on Delong–Delong's test). Thus, with the case–control sample only, we would have identified the significant difference with the weighted estimator, but have missed it using the empirical estimator.
0.0003 based on Delong–Delong's test). Thus, with the case–control sample only, we would have identified the significant difference with the weighted estimator, but have missed it using the empirical estimator.
6. Concluding remarks
In this paper, we developed methods for assessing a biomarker's classification accuracy for a binary disease as characterized by the area under the ROC curve in two-phase sampling designs. Finite-population stratified sampling of biomarkers from a phase-one cohort representative of the target population has become increasingly common in recent years with the availability of large clinical trials and cohort studies. But statistical methods to properly handle the design components when assessing biomarkers are not yet well developed. Empirical area under the ROC curve estimated from the phase-two biomarker samples was often reported in applied literatures even in the presence of biased sampling of cases and/or controls. We showed in this paper that empirical estimators of AUC ignoring the biased sampling scheme can lead to severely biased estimates of classification accuracy and invalid inference for comparing biomarkers. We investigated an inverse-sampling-probability-weighted estimator that achieves unbiased estimation of AUC and developed asymptotic variance formulae applicable to inference in finite-population stratified sampling, through its connection with the IPW estimator in the standard Bernoulli sampling design. The analytical variance formulae we developed will provide valuable guidance to biomarker researchers on optimizing the sampling scheme in designing future biomarker studies, in order to achieve better efficiency in evaluating biomarkers for classification accuracy. For Bernoulli sampling, we observed analytically and numerically that using estimated sampling weights is more efficient even when the true sampling weights are known by design (Web Supplementary Appendix A–C, F–H, see supplementary material). In particular, using estimated weights can lead to much improved efficiency for estimating performance of a single marker. In contrast, the improvement due to weights estimation is relatively minor for comparing performance of paired markers; when sample size is small, the CI of the  estimator based on known sampling weights can have slightly better coverage than that based on estimated weights.
estimator based on known sampling weights can have slightly better coverage than that based on estimated weights.
In this paper, we considered simple design-based weights, the estimation of which does not require more information beyond the sampling strata. In etiology studies, it has been shown that when auxiliary variables are available from a phase-one cohort, they can be used to further adjust the weights for potential efficiency gains in estimating the disease odds ratio or hazard ratio (Breslow and others, 2009). It is interesting in future work to investigate the impact of weight adjustments using auxiliary variables on biomarker performance measure such as the AUC. Finally, the inverse-probability-weighting methods can be naturally applied to other performance measures such as the points on the ROC curve and the partial area under the ROC curve.
Supplementary material
Supplementary Material is available here.
Funding
This work was supported by the U.S. National Institutes of Health grants R01 GM106177-01 and R01 GM54438. Funding to pay the Open Access publication charges for this article was provided by NIH grant R01 106177.
Supplementary Material
Acknowledgments
Conflict of Interest: None declared.
References
- Bamber D. (1975). The area above the ordinal dominance graph and the area below the receiver operating characteristic graph. Journal of Mathematical Psychology 124, 387–415. [Google Scholar]
- Breslow N. E., Lumley T., Ballantyne C. M., Chambless L. E., Kulich M. (2009). Improved Horvitz–Thompson estimation of model parameters from two-phase stratified samples: applications in epidemiology. Statistics in Biosciences 11, 32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow N. E., Wellner J. A. (2007). Weighted likelihood for semiparametric models and two-phase stratified samples, with application to cox regression. Scandinavian Journal of Statistics 341, 86–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T., Zheng Y. (2011). Evaluating prognostic accuracy of biomarkers in nested case–control studies. Biostatistics 131, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran W. G. (2007) Sampling Techniques. John Wiley & Sons, New York. [Google Scholar]
- DeLong E. R., DeLong D. M., Clarke-Pearson D. L. (1988). Comparing the areas under two or more correlated roc curves: a nonparametric approach. Biometrics 44, 837–845. [PubMed] [Google Scholar]
- Deras I. L., Aubin S. M. J., Blase A., Day J. R., Koo S., Partin A. W., Ellis W. J., Marks L. S., Fradet Y., Rittenhouse H.. and others (2008). Pca3: a molecular urine assay for predicting prostate biopsy outcome. The Journal of Urology 1794, 1587–1592. [DOI] [PubMed] [Google Scholar]
- Hanley J. A., McNeil B. J. (1983). A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 1483, 839–843. [DOI] [PubMed] [Google Scholar]
- Haynes B. F., Gilbert P. B., McElrath M. J., Zolla-Pazner S., Tomaras G. D., Alam S. M., Evans D. T., Montefiori D. C., Karnasuta C., Sutthent R.. and others (2012). Immune-correlates analysis of an HIV-1 vaccine efficacy trial. New England J. of Medicine 36614, 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Lyness J. M., McDermott M. P. (2009). Direct estimation of the area under the roc curve in the presence of verification bias. Statistics in Medicine 283, 361–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz D. G., Thompson D. J. (1952). A generalization of sampling without replacement from a finite universe. Journal of the American Statistical Association 47260, 663–685. [Google Scholar]
- Janes H., Pepe M. S. (2009). Adjusting for covariate effects on classification accuracy using the covariate-adjusted roc curve. Biometrika 962, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manski C. F., Lerman S. R. (1977). The estimation of choice probabilities from choice based samples. Econometrica: Journal of the Econometric Society, 458, 1977–1988. [Google Scholar]
- Neyman J. (1938). Contribution to the theory of sampling human populations. Journal of the Acoustical Society of America 33201, 101–116. [Google Scholar]
- Obuchowski N. A., McClish D. K. (1997). Sample size determination for diagnostic accuracy studies involving binormal roc curve indices. Statistics in Medicine 1613, 1529–1542. [DOI] [PubMed] [Google Scholar]
- Pepe M. S., Fan J., Seymour C. W., Li C., Huang Y., Feng Z. (2012). Biases introduced by choosing controls to match risk factors of cases in biomarker research. Clinical Chemistry 588, 1242–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe M. S., Feng Z., Janes H., Bossuyt P. M., Potter J. D. (2008). Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. Journal of the National Cancer Institute 10020, 1432–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J., Müller M. (2011). pROC: an open-source package for r and s+ to analyze and compare roc curves. BMC Bioinformatics 121, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins J. M., Rotnitzky A., Zhao L. P. (1994). Estimation of regression coefficients when some regressors are not always observed. Journal of the Acoustical Society of America 89427, 846–866. [Google Scholar]
- Wieand S., Gail M. H., James B. R., James K. L. (1989). A family of nonparametric statistics for comparing diagnostic markers with paired or unpaired data. Biometrika 763, 585–592. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





