Abstract
TBX15 is a T-box transcription factor essential for development, also proposed as a marker in prostate cancer; and, recently, its antiapoptotic function indicates a role in carcinogenesis. Regulation of TBX15 is uncovered. In this study, we investigated the regulation of TBX15 expression in human cancer cells, by analyzing the regulatory function of a 5’-distal conserved region of TBX15. Bisulfite sequencing showed high methylation of the CpG island contained in this region that was not correlated with TBX15 mRNA levels, in the cancer cell lines analyzed; however, after 5-aza-dC treatment of TPC-1 cells an increase of TBX15 expression was observed. We also found a significant response of TBX15 to TNF-α activation of the NF-κB pathway using five cancer cell lines, and similar results were obtained when NF-κB was activated with PMA/ionomycin. Next, by luciferase reporter assays, we identified the TBX15 regulatory region containing two functional NF-κB binding sites with response to NF-κBp65, mapping on the -3302 and -3059 positions of the TBX15 gene. Moreover, a direct interaction of NF-κBp65 with one of the two NF-κB binding sites was indicated by ChIP assays. In summary, we provide novel data showing that NF-κB signaling up-regulates TBX15 expression in cancer cells. Furthermore, the link between TBX15 and NF-κB found in this study may be important to understand cancer and development processes.
Introduction
TBX15 is a member of the conserved T-box gene family that is essential for many developmental processes [1]. TBX15 encodes a transcription factor involved in the development of the skeleton [2]. In humans, the Cousin syndrome is characterized by craniofacial and skeletal defects with scapular and pelvic hipoplasia and is caused by TBX15 mutations [3, 4]. The Tbx15-deficient mouse, droopy ear, and engineered Tbx15-null mutants presented similar phenotype than the Cousin syndrome in addition to pigment pattern alterations [5, 6, 7]. More recently, it has been reported that TBX15 is involved in adipocyte differentiation and mitochondrial respiration [8, 9]. Also, accumulating evidences indicate a role of T-box genes in differentiation, proliferation and apoptosis, which are relevant processes in carcinogenesis [10, 11], in addition to the altered expression of diverse T-box genes associated with a variety of cancers [12].
The information about TBX15 expression is mainly based on developmental studies, TBX15 being expressed in the mesenchyme during limb development, in mesenchymal precursor cells, chondrocytes and skeletal musculature [2]. Also, differentiation of the distinct adipocyte lineages showed differential expression of TBX15 [9]. To date, the regulatory mechanisms of TBX15 remain poorly understood, but considering the dynamic and temporal expression of TBX15 regulatory systems that are tissue and stage specific can be anticipated. There are also some hints suggesting that both genetic and epigenetic factors would regulate TBX15 transcription. One study carried out in human placentas, indicated that PDX1 regulates TBX15 in a methylation-dependent manner [13]; and methylation of TBX15 has also been proposed as a prognostic marker for prostate cancer [14].
Recently we found that TBX15 has an antiapoptotic role and its expression was altered in cancer cells [15]. These results are correlated with our previous observation that a region in chromosome 1p12 containing the TBX15 gene was associated with thyroid cancer susceptibility in humans [16]. Thus, to understand the implication of TBX15 on cell proliferation and survival related to carcinogenesis is important, and it requires uncovering the regulation of TBX15 expression. In this study, we investigate the regulatory function of a highly conserved region in the distal promoter of TBX15. This region extends the CG rich regulatory sequence of TBX15 found in human placentas by Chelbi et al. [13]. We identified a NF-κB responsive region with a positive effect in the transcription of TBX15 in human cancer cells.
Material and Methods
Cell culture
The human cell lines used for this study were from papillary thyroid cancer: TPC-1 and BCPAP; from follicular thyroid cancer: WRO and CGTH; from anaplastic thyroid cancer: FRO, 8305, BHT-101 and BHT-101 IκBαSR; from cervical cancer: HeLa; and the human embryonic kidney cell line HEK293. The TPC-1 and BHT-101 cell lines were provided by Dr. R. Melillo and Dr. M. Santoro (Università Degli Studi Di Napoli, Naples, Italy), and the WRO and FRO cells by Dr. R. Ciampi (University of Pisa, Pisa, Italy). The HeLa cell line was obtained from the Centro Nacional de Investigaciones Oncológicas (CNIO, Madrid, Spain) and the HEK293 was provided by Dr. F. Rosselli (Gustave Roussy, Paris, France). BHT-101 IκBαSR (constitutively express an IκBα super-repressor) cell line has been generated by infecting BHT-101 WT cells with a lentivirus encoding a super-repressor form of IkBα. All laboratories guaranteed the identity of these cell lines, providing the cell lines with a low number of passages. The human thyroid cancer cell lines BCPAP, CGTH and 8305C were purchased from the Leibniz-Institut DSMZ—Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH cell bank (DSMZ). The DSMZ assured cell lines identity by regular analysis using the authentication method based in short tandem repeat DNA fingerprint. The TPC-1, HeLa and HEK293 cell lines were grown in DMEM medium (Sigma-Aldrich) containing 10% heat-inactivated FCS (Gibco®) and 2.5μg/mL of Plasmocin™ (InvivoGen). BHT-101 and BHT-101 IκBαSR cell lines were grown in DMEM medium (Sigma-Aldrich) containing 20% heat-inactivated FCS (Gibco®), glutamine and 2.5μg/mL of Plasmocin™ (InvivoGen). WRO and FRO cells were grown in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% heat-inactivated FCS (Gibco®), 1% no essential amino acids (Sigma-Aldrich), 1% sodium pyruvate (Sigma-Aldrich) and 2.5μg/mL of Plasmocin™ (InvivoGen). BCPAP, CGTH and 8305C cells were grown in RPMI 1640 medium (Sigma-Aldrich) containing 10% heat-inactivated FCS (Gibco®) and 2.5μg/mL of Plasmocin™ (InvivoGen).
The two mouse embryonic fibroblast cell lines, p65+/+MEF and p65-/-MEF, were kindly provided by Dr. G. Franzoso (The Gwen Knapp Center for Lupus and Immunology research, The University of Chicago, USA). These cells were grown in DMEM medium (Sigma-Aldrich) containing 10% heat-inactivated FCS (Gibco®) and 2.5 μg/mL of Plasmocin™ (InvivoGen).
Cells were placed in culture flasks and incubated in a humidified atmosphere (5% CO2) at 37°C. All experiments were performed at passages <20–25.
Prediction of transcription factor binding sites
A DNA sequence of approximately 1kb in the 5-flanking region of the TBX15 gene was obtained from Ensembl GRCh38 human genome database (http://www.ensembl.org/). Prediction of transcription factor binding sites was assessed using four online prediction softwares with a score cutoff > 85%, MatInspector (www.genomatix.de), rVista 2.0 (http://rvista.dcode.org), TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) and CONSITE (http://consite.genereg.net/).
DNA isolation and bisulfite sequencing
Phenol-chloroform total DNA extraction was performed from TPC-1, BCPAP, WRO, CGTH, FRO, 8305 and HeLa cell lines. Genomic DNA was modified with sodium bisulfite using the EZ DNA Methylation-Startup™ Kit (Zymo Research, USA) and following the manufacturer’s instructions. Using bisulfite-modified DNA, the genomic region covering the CpG island of the TBX15 distal promoter (-3852/-3381 from the TBX15 transcription start site) was amplified in two overlapping fragments (C1, 250bp and C2, 288bp) followed by sequence analysis. The primers sequences were designed using the DNA Methylation Analysis PCR Primer Database [17]. Each PCR reaction was performed in a total volume of 25μL containing 50–100 ng of bisulfite-modified DNA of each cell line, 1 x ZymoTaq™ PreMix (Zymo Research, USA) and 1mM of each primer. PCR reaction containing Universal Methilated Human DNA Standard (Zymo Research, USA) was carried out as control reaction. The amplification conditions were: 95°C for 10 min; 40 cycles of 95°C for 35s, annealing temperature (Ta) for 30s, and 72°C for 1min; and a final extension step for 7 min at 72°C. The primers and their annealing temperature (Ta) are listed in S1 Table. The amplified PCR products were separated by agarose gel electrophoresis, purified and sequenced. Methylation levels were estimated using the semiquantitative method described by Jiang et al., [18], applying the following formula: % Methylation = 100 X numberC/(numberC + numberT). Percentages were rounded off to the ranges: 0%, 25%, 50%, 75% and 100%.
Genomic DNA demethylation
TPC-1 cells were seeded in 6-well plates at 105 cells/well (roughly 20% of confluence). After 24h, the DNA demethylating reagent 5-aza-2′-deoxycytidine (5-aza-dC, Sigma-Aldrich) was added to the final concentrations of 2 μM or 5μM and the cells were growing for 96h. Fresh 5-aza-dC was replaced every 24 hours. On day 5, 5-aza-dC treated cells were harvested for RNA extraction and RT-qPCR analysis.
TNFα and phorbol 12-myristate 13-acetate (PMA)/ionomycin (I) stimulation
TNFα (Peprotech) stock solution aliquots in culture medium were 106 U/mL TNFα. FRO, CGTH, TPC-1, HeLa and MEF cells were seeded in 6-well plates (105 cells/well). When cells were attached, culture medium was replaced with fresh medium containing 2000 U/mL TNFα as a final concentration, or fresh medium for control cultures. After 24h of TNFα treatment, cells were harvested for RNA extraction and RT-qPCR analysis.
BHT-101 and BHT-101 IκBαSR cells were seeded in 6-well plates (4x105 cells/well). When cells were attached, culture medium was replaced with fresh medium containing 2000 U/mL TNFα, or 2 μM PMA (Sigma-Aldrich) plus 400 ng/mL I (Sigma-Aldrich) as a final concentration, or fresh medium for control cultures. After 3h of treatment, cells were harvested for RNA extraction and RT-qPCR analysis.
RNA isolation and RT-qPCR analysis
Isolation of total RNA from cell samples was performed using TRIzol® Reagent (Ambion®, Life Technologies™), following manufacturer’s protocol.
cDNA synthesis was carried out with 1 μg of total RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche). The quantitative expression of the TBX15, CXCL1 and RPL27 genes was analyzed by Real Time PCR using LightCycler® 480 SYBRGreen I Master according to the manufacturer's protocol. Gene specific primers are listed in S1 Table. Each real-time PCR reaction was performed for 5 min at 95°C, 45 cycles at 95°C for 10s, 58°C for 15s, 72°C for 25s, and 78°C for 5s. The specificity of the reaction was verified by dissociation curve analysis using a cycle of 95°C for 5s followed by constant increase of temperature between 65°C and 95°C. Reactions were carried out in a LightCycler® 480 System. Relative expression levels of the TBX15 and CXCL1 genes were normalized to the housekeeping gene RPL27 (internal control) and were calculated using the 2nd Derivative Max Method. Each real-time PCR reaction was performed in triplicate.
Construction of the TBX15 promoter reporter plasmids
Five different fragments of the 5-flanking region of the human TBX15 gene were generated by PCR using the specific primers listed in S1 Table that included the restriction sites for BglII and KpnI (New England Biolabs) to direct cloning into the firefly luciferase vector pGL4.26 (Promega). PCR conditions were 95°C for 5 min followed by 35 cycles at 95°C for 45s, 65°C for 45s and 72°C for 1min 45s. The reporter constructs were: P1 (-3478/+206), P2 (-3478/-2865), P2rev (-2865/-3478), P3 (-3427/-2865) and P4 (-3587/-3283). The constructs were confirmed by restriction mapping and sequencing.
Site-directed mutants of the reporter construct P2 were generated using the QuickChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies), following the manufacturer’s indications. The three putative NF-κB binding sites of the TBX15 promoter contained in the construct P2 were mutated using primers with EcoRI, KpnI or kBS2 restriction sequences to substitute part of the kBS1, kBS2 or kBS3 sequences, respectively. The mutated P2 constructs comprised three NF-κB single mutated plasmids (P2mutS1, P2mutS2 and P2mutS3) and a NF-κB triple mutated plasmid (P2mutS1+S2+S3). Mutated reporter constructs were verified by sequencing.
Luciferase reporter assays
HEK293 cells were seeded in 6-well plates (105 cells/well) and transfected using Lipofectamine® 2000 Transfection Reagent (Life Technologies), following the standard manufacturer’s protocol. Cells were transfected with 0.5 μg/well of the appropriate TBX15 promoter reporter construct and 0.25μg/well of renilla luciferase reporter (Promega) as internal control for transfection efficiency. After 24h of TNFα stimulation, cells were lysated in PLB buffer, and subsequently assayed for luciferase activity. When the expression of p65 was required, cells were cotransfected with promoter reporter construct and the pCDNA3.p65 expression plasmid (kindly provided by Dr. Leonardi, Naples, Italy) by using 0.15μg/well of luciferase reporter construct, 0.05μg/well of renilla reporter and 0.8μg/well of pCDNA3.p65 (or empty pcDNA3.1 as negative control). After 24h, cells were lysated and assayed for luciferase activity. Luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (Promega). All transfections were performed in duplicate, and experiments were repeated at least three times. The firefly luciferase activity was normalized according to the renilla luciferase activity.
Chromatin immunoprecipitation (ChIP)
The ChIP assay was as described by Nowak et al. [19] with modifications, using TPC and CGTH and solutions specified in S2 Table. Cells were seeded in plates at 5x106 cells/plate with growth medium supplemented with 0.5% bovine serum albumin. After 24 h, cells were stimulated with 2000 U/mL TNFα for 30 min, washed at room temperature with PBS and after with PBS/Mg. TNFα stimulated cells were cross-linked with 2 mM disuccinimidyl glutarate (DSG, Fisher Scientific) for 45 min, washed with PBS, and fixed with 1% freshly prepared solution of formaldehyde in PBS/Mg (v/v) for 15 min at room temperature. Then fixed cells were washed with PBS, scraped, transferred into eppendorf tubes and lysed in 900 μL of L1 buffer for 15 min on ice. Nuclei were precipitated by centrifugation and suspended in 500 μL of SDS lysis buffer at room temperature. The samples were sonicated on a Digital Branson Sonifier® (Branson Ultrasonics Danbury, USA) on ice during 7 min in cycles of 30s on/30s off. Low ionic strength dilution buffer was added to the soluble chromatin to a final volume of 900 μL with 20 μL of magnetic protein G beads (Protein G Mag Sepharose Xtra, GE Healthcare Life Sciences). p65 antibody-magnetic beads complexes were prepared by adding 4 μg of p65 antibody (NFKB p65 sc-372-G of Santa Cruz Biotechnology) to a final volume of 500 μL of low ionic strength dilution buffer with 20μL of magnetic beads, and overnight incubation at 4°C with rotation. Then washed with low ionic strength dilution buffer and added to sheared chromatin suspension and incubated with rotation at 4°C overnight. 10 μL of chromatin suspension were removed before immunoprecipitation to determine the imput quantity of DNA. Immunocomplexes were captured using a magnetic stand (MagRack 6, GE Healthcare Life Sciences), and washed twice with 500μL of low ionic strength dilution buffer, high-salt wash buffer, LiCl wash buffer, and twice with TE. Precipitated complexes were eluted with 400 μL of elution buffer for 30 min at room temperature with rotation. Samples were de-cross-linked in 200 mM NaCl, 50 mM Tris pH 6.8, 10mM EDTA adding 200 μg/mL proteinase K and incubated at 65°C for 2 h 30 min. DNA was extracted by phenol-chloroform, precipitated using ammonium acetate/ethanol and used for qPCR analysis. PCR primers and conditions for quantitative PCR are shown in S1 Table.
Statistical analysis
The data were expressed as means ± standard deviations (SD) and analyzed by Student's t-test. p-value <0.05 was considered to indicate statistical significance.
Results
The information about TBX15 regulation is very scarce. To our knowledge there is only a study in the literature describing a CG rich sequence approximately 3 kb upstream of the TBX15 transcription start point (TSS) with a possible role in TBX15 expression. The study was carried out in human placentas showing that methylation of such CG rich sequence was altered in pathological placentas, and its methylation status was related to TBX15 expression [13]. Databases pointed out that this region contains a CpG island and is highly conserved in vertebrates (UCSC Genome Browser), suggesting its functional importance. In Fig 1A we represented the conservative region in the 5’-distal position of TBX15. Hence, we hypothesized that TBX15 regulation signals take place in that sequence, and then we explored such regulatory activity in human cancer cells.
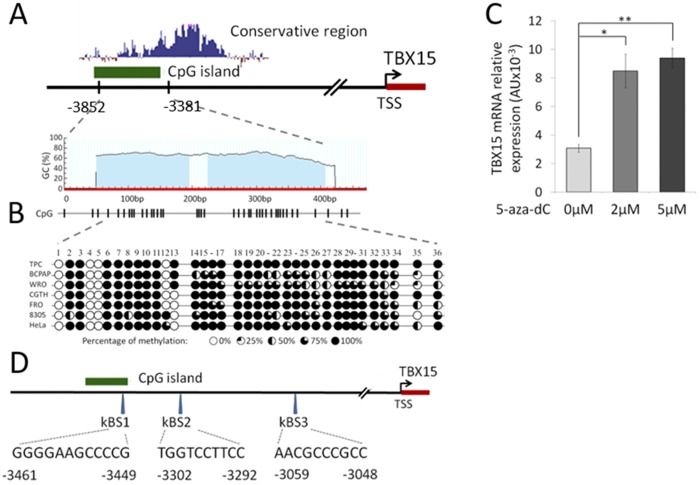
Fig 1. Methylation of the TBX15 promoter in cancer cell lines.
(A) Schematic diagram of TBX15 promoter indicating the CpG island and the conservative area in the 5’flanking region. TSS: transcription start site. (B) Methylation of the studied CpG island in HeLa and six thyroid cancer cell lines. The bisulfite conversion and sequencing analysis cover 36 CpG sites in the studied CpG island. Each row represents the results for each cell line where the circles indicate the methylation status of each CpG site. (C) Treatment of TPC-1 cells with 5-aza-dC increased TBX15mRNA. qRT-PCR analysis and mRNA levels normalized to RPL27, showing the mean±SD of mRNA levels of two independent experiments in triplicates. * indicates p-value < 0.05 and ** p-value < 0.01. (D) Location of the in silico predicted NF-κB binding sites (kBS1, kBS2 and kBS3) in the studied region of the TBX15 promoter.
A 5’-distal conserved region of TBX15 is highly methylated in cancer cells
First we investigated whether the level of methylation at the CpG island located 3 kb upstream of the TBX15 TSS was related to TBX15 transcription in cancer cells. Bisulphite sequencing of this CpG island, position -3852 bp to -3381bp of the TBX15 gene, was performed in HeLa cells and six thyroid cancer cell lines (TPC-1, BCPAP, WRO, CGTH, FRO and 8305). As shown in Fig 1B, high levels of methylation along all CG sites analyzed were found in the seven cell lines (range 69%-89%); however, the levels of TBX15 mRNA were much more different between these cell lines (S3 Table), being unfeasible to establish a correlation between methylation at the analyzed CpG island and transcription of TBX15 in the studied cell lines. For example, in the cell line CGTH the TBX15 mRNA expression was 22-fold higher than in TPC-1 (65x10-3 AU and 3.04x10-3 AU, respectively; S3 Table), but both cell lines presented similar methylation levels in the CpG island evaluated (Fig 1B). On the other hand, treatment of TPC-1 cells with the demethylating agent 5-aza-dC increased 2.6-fold the level of TBX15 mRNA compared with untreated cells (Fig 1C), which would indicate a modulation of TBX15 expression by epigenetic factors; most likely, other regulatory processes may also control the TBX15 expression, as was indicated in human placentas [13].
Next, we searched for cis-regulatory elements in the upstream region of TBX15 using four independent programs (see Materials and Methods). Three putative NF-κB binding sites were identified in the area containing the CpG island analyzed above, and are referred as kBS1, kBS2 and kBS3 in this study (Fig 1D). Other potential transcription factors binding sites were also predicted, but only by one or two of these programs. Based on these data and taking into account that NF-κB is an important factor in cancer, we considered NF-κB as a potentially relevant factor to regulate TBX15 transcription.
Activation of NF-κB by TNF-α increases TBX15 mRNA expression in cancer cells
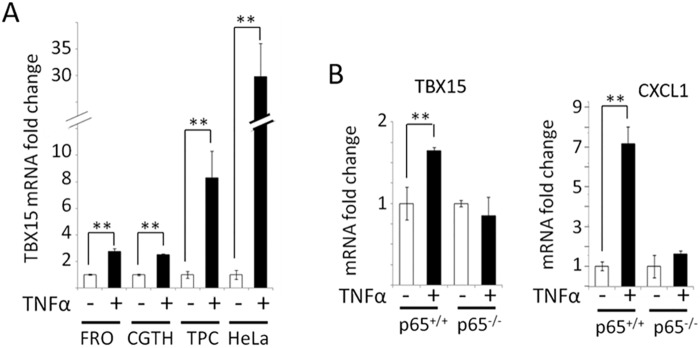
To investigate if NF-κB was involved in the regulation of TBX15 expression we stimulated HeLa cells and three thyroid cancer cell lines with TNF-α to activate NF-κB, and subsequently we measured the TBX15 mRNA levels. As shown in Fig 2A, treatment of cells with TNF-α significantly increased the levels of TBX15 mRNA, compared to untreated cells (increase ranging from 31.8- to 2.5-fold, depending of the cell line). The induction of TBX15 expression by TNF-α was comparable to the induction of CXCL1 expression measured in the same experiments, which was used as positive control for TNF-α activation of the NF-κB pathway (S1 Fig). To further confirm that TBX15 expression was controlled by NF-κB activation, we analyzed the amount of TBX15 mRNA in TNF-α stimulated p65+/+ and p65-/- MEF cells, and we found that TNF-α increased the level of TBX15 mRNA in p65+/+ but not in p65-/- MEF cells (Fig 2B). In these experiments, activation of NF-κBp65 by TNF-α was verified by the induction of CXCL1 expression in p65+/+ MEF cells after TNF-α stimulation. Thus, these data demonstrate that NF-κBp65 up-regulates TBX15 transcription in MEF cells, and indicate that activated NF-κBp65 has a positive effect in TBX15 regulation in human cancer cells.
Fig 2. Expression of TBX15mRNA in different cell lines after stimulation with TNF-α.
(A) qRT-PCR of TBX15mRNA analysis in FRO, CGTH, TPC-1 and HeLa cells after 2h of TNF-α treatment. (B) qRT-PCR of TBX15mRNA (left) and CXCL1mRNA (right) analysis in p65+/+ and p65-/- MEF cells after TNF-α treatment. Data were normalized to RPL27 and expressed as fold change referred to untreated cells whose TBX15mRNA or CXCL1mRNA relative expression was defined as 1. Data are mean±SD of mRNA levels of two independent experiments in triplicates. ** indicates p-value < 0.01.
Effect of NF-κB inhibition by IκBα super-repressor in TBX15 mRNA expression
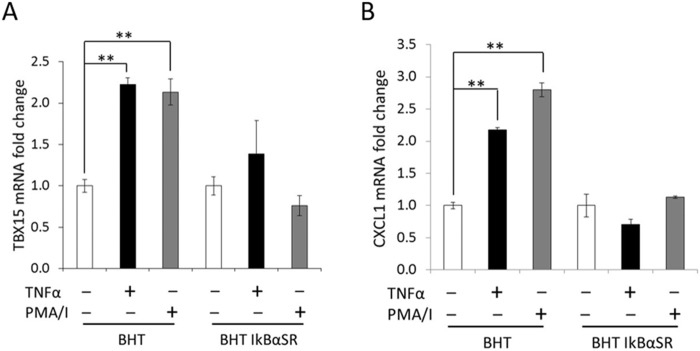
An additional evidence of the implication of NF-κB in TBX15 regulation was provided using an IκBα super-repressor (IκBαSR) to inhibit NF-κB activation. Then, we analyzed the TBX15 expression in BHT-101 and BHT-101 IκBαSR cells after TNF-α or PMA/I stimulation. BHT-101 IκBαSR cells constitutively express an IκBαRS sequestering NF-κB so is unable to response to activation stimuli. As shown in Fig 3A, after treatment with TNF-α or PMA/I, the levels of TBX15 mRNA in BHT-101 IκBαSR cells were similar than in untreated cells. However, as expected, stimulation of BHT cells showed an increase of TBX15 mRNA. In these experiments the expression of CXCL1 was also evaluated and used as a positive control for activation of the NF-κB pathway, obtaining equivalent results than in the analysis of TBX15 expression (Fig 3B). These results support the role of NF-κB canonical pathway in TBX15 regulation in cancer cells.
Fig 3. Expression of TBX15mRNA and CXCL1mRNA in BHT cell lines after stimulation with TNF-α and PMA/I.
(A) qRT-PCR of TBX15mRNA analysis in BHT and BHT IkBαSR cells after 3h of TNF-α or PMA/I treatment. (B) qRT-PCR of CXCL1mRNA analysis in BHT and BHT IkBαSR cells after 3h of TNF-α or PMA/I treatment. Data were normalized to RPL27 and expressed as fold change referred to untreated cells whose TBX15mRNA or CXCL1mRNA relative expression was defined as 1. Data are mean ± SD of mRNA levels in triplicates. ** indicates p-value < 0.01.
Identification of a TBX15 regulatory region containing NF-κB binding sites
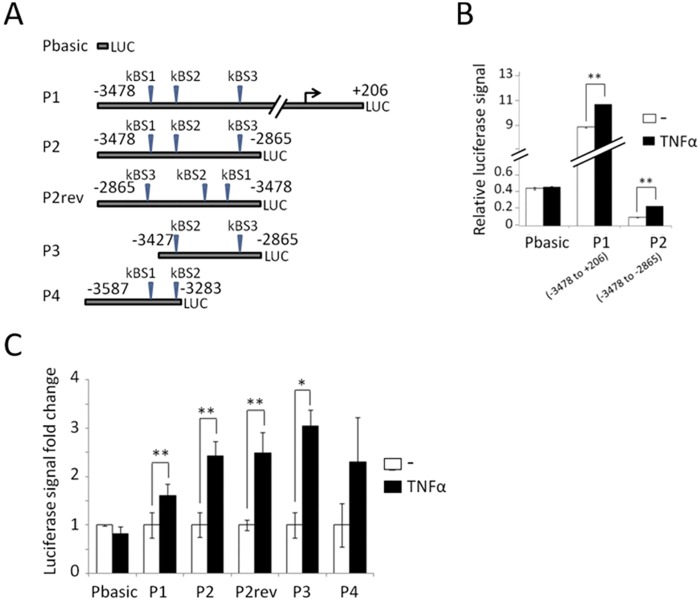
Luciferase reporter experiments were performed to study the TBX15 promoter activity and to identify the regulatory region that responds to NF-κB. Five 5’-fragments of the human TBX15 gene were cloned into the pGL4 luciferase reporter plasmid: a 3.68 kb fragment containing the three predicted NF-κB binding sites and the TSS (P1: -3478/+206) and four fragments from the 5’-flanking region of TBX15 (P2: -3478/-2865, P2rev: -2865/-3478, P3: -3427/-2865 and P4: -3587/-3283) (Fig 4A). Following transfection of each construct in HEK293 cells, luciferase activity was evaluated in TNF-α -stimulated and non-stimulated cells and transfected cells with pGL4-basic were used as negative control. Analysis of luciferase activity in non-stimulated cells served to assess the promoter activity. As shown in Fig 4B, promoter activity was observed in the 3.68 kb P1 fragment of the TBX15 promoter, but not in its 5’-flanking region represented by the P2 fragment. On the other hand, P1 and P2 fragments showed similar response to the endogenous activation of NF-κB by TNF-α (Fig 4B), and the response to the activated NF-κB was also observed in the rest of constructs, P2rev, P3 and P4 (Fig 4C). These results are in agreement with our indication before of activation of constitutive NF-κB regulating TBX15 in different cell lines, and locate the regulatory region in the 5’-flanking position of the TBX15 gene from -3478 to -2865. Since the P2 fragment contains the three predicted NF-κB binding sites in the 5’-distal region of TBX15, and because both P2 and P2rev showed similar response to NF-κB activation, we propose that the -3478/-2865 region of TBX15 would integrate NF-κB signals in an orientation-independent manner to modulate positively the expression of TBX15.
Fig 4. Functional analysis of the 5’-flanking region of the human TBX15 gene.
(A) Representation of the different luciferase reporter constructs used in the analysis, with predicted NF-κB binding sites (kBS1, kBS2 and kBS3). (B) The TBX15 promoter and its 5’-flanking region activity measured by luciferase in TNF-α treated and untreated cells. Vector expressing the Renilla luciferase was co-transfected as an internal reference to correct the transfection efficiency. (C) Luciferase analysis of the different reporter constructions. Data was expressed as fold change referred to untreated cells as 1. At least three independent experiments were performed with duplicates. Bars represent the mean±SD. * indicates p-value < 0.05 and ** p-value < 0.01.
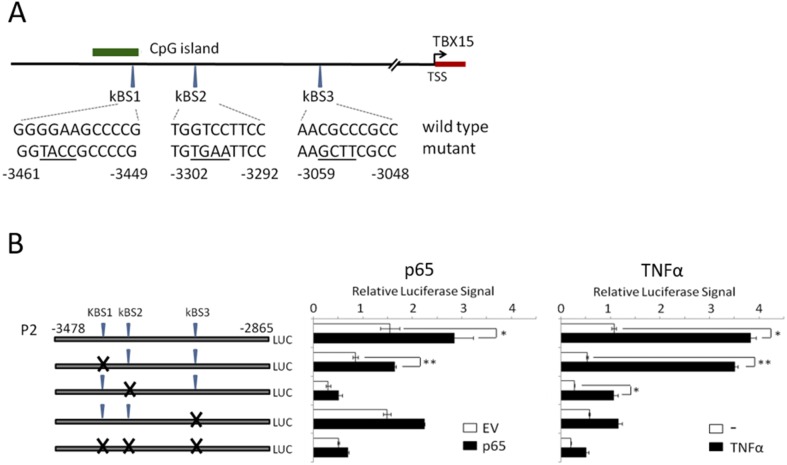
The NF-κB responsive region of the 5’-flanking region of TBX15 was further analyzed by introducing mutations in the P2 reporter construct to alter each of the predicted NF-κB binding site followed by luciferase assays. The mutated plasmids that were evaluated and their altered sequences are indicated in Fig 5A. The reporter assays performed in HEK293 cells along with a NF-κBp65 expression plasmid or a control empty vector, allowed identifying the NF-κBp65-responsive sites in the 5’-distal region of TBX15. The results are illustrated in Fig 5B. In agreement with our finding of NF-κBp65 up-regulating TBX15 in MEF cells, cotransfection of the NF-κBp65 expression vector increased 2-fold the activity of the P2 wild type construct compared to the empty vector; however, no increase of activity was detected in the P2 triple mutant construct. The kBS1 mutant plasmid showed a similar increase of activity than the P2 wild type plasmid, after transfection with the NF-κBp65 expression vector. In contrast, the single kBS2 and kBS3 mutant plasmids were not activated by NF-κBp65, indicating that kBS2 and kBS3 sites are NF-κBp65 responsive elements. Furthermore, we observed that in the reporter assays with the control empty vector, the kBS2 mutant plasmid presented a reduced activity of >80% (Fig 5B), indicating the contribution of the kBS2 site to the basal expression of TBX15. Additional evidence of kBS2 and kBS3 as the NF-κB responsive elements in the 5’-flanking region of TBX15 was provided by the luciferase reporter assays with the same P2 mutant constructs under TNF-α -stimulated conditions. Similar to ectopic NF-κBp65, activation of endogenous NF-κB with TNF-α increased the activity of the P2 wild type and the kBS1 mutant constructs, whereas much less response was observed with the kBS2 mutant construct and no response was observed with the kBS3 mutant construct (Fig 5B). It is interesting to point out that the activity of the P2 wild type plasmid after TNF-α stimulation was higher than with the ectopic expression of NF-κBp65 (4-fold and 2-fold, respectively, compared to control). Altogether, these results indicate that kBS2 and kBS3 are functional NF-κB motifs that respond to NF-κB increasing TBX15 expression.
Fig 5. Identification of functional NF-κB responsive elements in the 5’-flanking region of the human TBX15 gene.
(A) Location and sequence of the predicted NF-κB binding sites (kBS1, kBS2 and kBS3) in the studied region of the TBX15 promoter, with indication of altered sequence in mutated reporter constructs (underlined). Site directed mutagenesis was performed using P2 reporter construct to obtain the mutated reporter constructs. (B) Luciferase analysis of each mutated reporter constructs. Left panel, co-tansfected with the pCDNA3-p65 expression vector (p65) or the pCDNA3 empty vector (EV). Right panel, in nonstimulated or stimulated TNF-α cells. At least three independent experiments were performed with duplicates. Bars represent the mean±SD. * indicates p-value < 0.05 and ** p-value < 0.01.
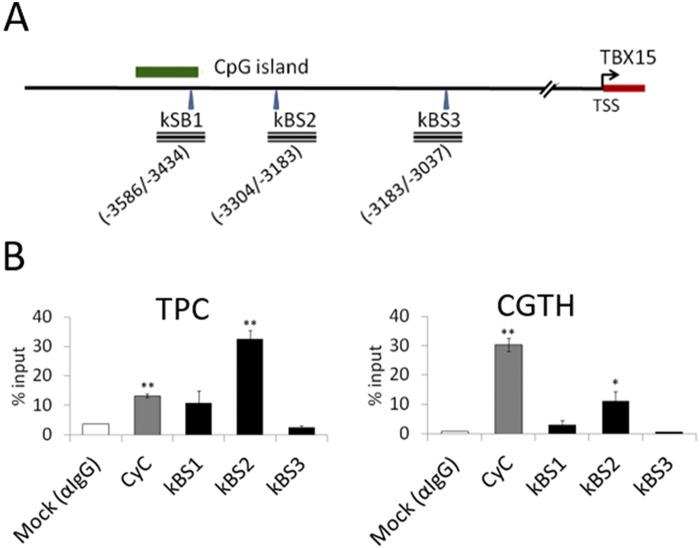
Next, in vivo association of NF-κBp65 to the 5’-flanking region of TBX15 was assessed by ChIP analysis in TPC and CGTH cells after NF-κB activation by TNF-α treatment (Fig 6). Using a NF-κBp65 antibody, the amplification with specific primer of the kBS1, kBS2 and kBS3 regions showed that NF-κBp65 binds to the kBS2 region, but not to the kBS1 or kBS3 (Fig 6B). These results, together with the analysis before of the lack of induction of TBX15 expression in p65-/- MEF cells, indicate that NF-κBp65 acts as a positive regulator of TBX15 by direct interaction with a 5’-distal regulatory region of this gene.
Fig 6. Chromatin inmunoprecipitation (ChIP) assays.
(A) The PCR amplified regions comprising the predicted NFκB binding sites (kBS1, kBS2 and kBS3) are indicated as rectangles, with location of these specific DNA segments indicated by numbers. (B) TPC and CGTH cells were treated with TNF-α and ChIP assays performed using IgG (mock) and NFκBp65 antibodies. Imnmunoprecipitated DNA was analyzed by qPCR with primers targeted to the predicted NFκB binding sites (kBS1, kBS2 and kBS3). The quantitative data reflect occupancy calculated as % input and represented as mean±SD of two independent experiments. *: P<0.05; **: P<0.01.
Discussion
TBX15 is a T-box gene that encodes a transcription factor being crucial in development processes [1]. More recently, we have shown an antiapoptotic role of TBX15, together with its altered expression in thyroid cancer cells [15]. Others had also reported that TBX15 controls the number of mesenchymal precursor cells and chondrocytes during development of the skeleton of the limbs [2]. These data indicate a role of TBX15 in cell proliferation, and support the assumption of its implication in carcinogenesis, as it was previously indicated for some members of the T-box gene family [10, 11]. To understand the function of TBX15 is crucial to disclose the mechanisms that regulate TBX15. In this study, we provide novel data showing that NF-κB signaling pathway upregulates TBX15 expression in cancer cells.
To study TBX15 regulation, we center our attention in a 5’-conserved region of TBX15 that showed regulatory functions in placentas [13]. This region contains three putative NF-κB responsive elements as revealed by our in silico sequence analysis (Fig 1D). Of importance, we found that TBX15 mRNA expression was increased in response to TNF-α in HeLa and three thyroid cancer cell lines; moreover, the analysis of the TBX15 expression in p65-/- MEF cells stimulated with TNF-α indicated that the effect was mediated by NF-κB. Further evidences of a role of NF-κB in TBX15 regulation were given by using BHT-101 IκBαSR cells that stably expressed an IκBα super-repressor that inhibits NF-κB. These cells when stimulated with TNF-α or PMA/I presented basal levels of TBX15 mRNA, which was in contrast with the induction of TBX15 mRNA found in stimulated BHT-101, indicating that NF-κB signaling was required to induce TBX15 expression. We found that a fragment of 600 bp in the 5’-flanking region of TBX15 that containing the three predicted NF-κB binding sites had regulatory activity, responding to TNF-α or to ectopic expression of NF-κBp65. Functional analysis of this region using reporter constructs with mutations in the three putative NF-κB binding sites showed that two of the three sites, located on -3302 and -3059 of the TBX15 gene (kBS2 and kBS3, respectively; Fig 5A), were functional. We demonstrated that the common NF-κB subunit p65 is important for the positive response of these motifs to TNF-α. In addition, ChIP assays indicated a direct interaction of NF-κBp65 with the kBS2 binding site in TPC1 and CGTH cells. Here, we propose that a cooperative action of the two functional NF-κB motifs takes place to upregulate TBX15. The fact that reporter assays showing higher activity after stimulation with TNF-α than with NF-κBp65, also suggest that, besides p65, other factors activated by TNF-α including different NF-κB subunits would recognize these functional motifs. In this context it is feasible to assume that the two functional NF-κB motifs identified in this study integrate the NF-κB signals required to positively modulate the transcription of TBX15. Most likely, additional factors would be needed to control TBX15 transcription acting in a cell type-dependent manner. Thus, in pathological placentas, TBX15 is downregulated by PDX1 in a methylation-dependent manner [13]; and differential TBX15 expression was found in adipocytes [9]. In the present study we show that demethylation with 5-aza-dC resulted in an increase of the TBX15 mRNA expression in TPC-1 thyroid cancer cells; however, no correlation was found between methylation levels in the CpG island of TBX15 analyzed and the TBX15 mRNA expression, in nine thyroid cancer cell lines and HeLa cells. We cannot discard a possible association between the methylation status of other CG sites along the TBX15 promoter and the expression of TBX15. Of interest, methylation status of the TBX15 promoter related to clinicopathological features of prostate cancer was reported, although the study did not mention TBX15 expression [14]. Therefore, at present, the significance of methylation in the regulation of TBX15 needs to be investigated. Nevertheless, it seems that TBX15 undergoes up- or down-regulation depending on the cell type and conditions, with expected effects on its target genes. More studies are needed to determine the genetic and epigenetic mechanisms of TBX15 regulation.
A relevant issue in the present study is the link between TBX15 and NF-κB, and this novel discovery would be important in development and cancer processes. NF-κB plays a central role in many cellular processes via the regulation of its target genes [20, 21]. Also, the activation of NF-κB in many types of cancer has been extensively reported, including thyroid cancer [22, 23]; and, the effects of NF-κB in cancer are related to the activation of cell proliferation and inhibition of apoptosis [24, 25]. Interestingly, we have also reported an antiapoptotic function of TBX15 [15] that together with the present study indicates that, by activating TBX15, NF-κB may promote the antiapoptotic activity of TBX15, which points out TBX15 as a possible downstream gene in the antiapoptotic activity of NF-κB. The expression of TBX15 in cancer is unknown, but because NF-κB modulated the expression of TBX15 we speculate that alterations of TBX15 expression would be expected in different types of cancer with consequences in cancer development. To this regard, TBX15 expression is altered in thyroid cancer cell lines ([15], and S3 Table) and the activation of NF-κB in cancer cells and tumor of the thyroid is well known [26, 27].
In development processes, there are some hints that also invite us to think of TBX15 as a possible downstream target of NF-κB. During embryonic limb formation, NF-κB acts to transmit growth factor signals between the ectoderm and the mesenchyme [28], and TBX15 controls the number of mesenchymal precursor cells [2]; therefore, TBX15 could be a downstream factor of the ectoderm and the mesenchyme communication mediated by NF-κB.
In conclusion, here we have demonstrated the NF-κB mediated upregulation of TBX15 in cancer cells, identifying two NF-κB functional motifs in the 5’-flanking region of TBX15. This novel finding together with our previous discovery of an antiapoptotic function of TBX15, support the implication of TBX15 in carcinogenesis. Here we also propose a link between TBX15 and NF-κB. So, further investigation is warranted to understand the role of TBX15 in cancer and development processes, and in relation to NF-κB.
Supporting Information
Data were normalized to RPL27 and expressed as fold change referred to untreated cells whose CXCL1mRNA relative expression was defined as 1. Data are mean ± SD of mRNA levels of two independent experiments in triplicates. ** indicates p-value < 0.01.
(DOCX)
(DOCX)
For luciferase constructions, restriction enzymes sites are in bold. For point mutation construction primers, changed nucleotides are marked in bold letter. BS-PCR: bisulfite polymerase chain reaction. Ta: annealing temperature.
(DOCX)
Data are normalized to the reference gene RPL27 and expressed as mean ± SD.
(DOCX)
Acknowledgments
We thank Dr. Domenico Somma for his helpful scientific and technical contributions to this work, and Cristian Valiente for his technical support. We also thank the guidance of Dr. Anna Portela in ChIP protocol.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
JA was supported by a predoctoral fellowship (PIF) from the Universitat Autònoma de Barcelona. This research was supported by grant support: Spanish Ministry of Education and Science (project SAF2007-6338) and the Generalitat de Catalunya (CIRIT; 2009SGR-725).
References
- 1.Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet 2005;39:219–239. [DOI] [PubMed] [Google Scholar]
- 2.Singh MK, Petry M, Haenig B, Lescher B, Leitges M, Kispert A. The T-box transcription factor Tbx15 is required for skeletal development. Mech Develop 2005;122:131–144. [DOI] [PubMed] [Google Scholar]
- 3.Dikoglu E, Simsek-Kiper PO, Utine GE, Campos-Xavier B, Boduroglu K, Bonafé L et al. Homozygosity for a novel truncating mutation confirms TBX15 deficiency as the cause of Cousin Syndrome. Am J Med Genet 2013;161A:3161–3165. 10.1002/ajmg.a.36173 [DOI] [PubMed] [Google Scholar]
- 4.Lausch E, Hermanns P, Farin HF, Alanay Y, Unger S, Nikkel S et al. TBX15 mutations cause craniofacial dysmorphism, hypoplasia of scapula and pelvis, and short stature in Cousin syndrome. Am J Hum Genet 2008;83:649–655. 10.1016/j.ajhg.2008.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Candille SI, Van Raamsdonk CD, Chen C, Kuijper S, Chen-Tsai Y, Russ A et al. Dorsoventral patterning of the mouse coat by Tbx15. PLoS Biology 2004; 2:E3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuijper S, Beverdam A, Kroon C, Brouwer A, Candille S, Barsh G et al. Genetics of shoulder girdle formation: roles of Tbx15 and aristaless-like genes. Development 2005;132:1601–1610. [DOI] [PubMed] [Google Scholar]
- 7.Singh MK, Petry M, Haenig B, Lescher B, Leitges M, Kispert A. The T-box transcription factor Tbx15 is required for skeletal development. Mech Develop 2005;122:131–144. [DOI] [PubMed] [Google Scholar]
- 8.Gesta S, Bezy O, Mori MA, Macotela Y, Lee KY, Kahn CR. Mesodermal developmental gene Tbx15 impairs adipocyte differentiation and mitochondrial respiration. Proc Natl Acad Sci USA 2011;108:2771–2776. 10.1073/pnas.1019704108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gburcik V, Cawthorn WP, Nedergaard J, Timmons JA, Cannon B. An essential role for Tbx15 in the differentiation of brown and "brite" but not white adipocytes. Am J Physiol-Endoc M 2012;303:E1053–1060. [DOI] [PubMed] [Google Scholar]
- 10.Peres J, Davis E, Mowla S, Bennett DC, Li JA, Wansleben S et al. The Highly Homologous T-box transcription factors, TBX2 and TBX3, have distinct roles in the oncogenic process. Genes Cancer 2010;1:272–282. 10.1177/1947601910365160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, Ma X, Cheung KF, Li X, Tian L, Wang S et al. Epigenetic inactivation of T-box transcription factor 5, a novel tumor suppressor gene, is associated with colon cancer. Oncogene 2010;29:6464–6474. 10.1038/onc.2010.370 [DOI] [PubMed] [Google Scholar]
- 12.Papaioannou VE. The T-box gene family: emerging roles in development, stem cells and cancer. Development 2014; 141:3819–3833. 10.1242/dev.104471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chelbi ST, Doridot L, Mondon F, Dussour C, Rebourcet R, Busato F et al. Combination of promoter hypomethylation and PDX1 overexpression leads to TBX15 decrease in vascular IUGR placentas. Epigenetics 2011; 6:247–255. [DOI] [PubMed] [Google Scholar]
- 14.Kron K, Liu L, Trudel D, Pethe V, Trachtenberg J, Fleshner N et al. Correlation of ERG Expression and DNA Methylation Biomarkers with Adverse Clinicopathologic Features of Prostate Cancer. Clin Cancer Res 2012;18:2896–2904. 10.1158/1078-0432.CCR-11-2901 [DOI] [PubMed] [Google Scholar]
- 15.Arribas J, Giménez E, Marcos R, Velázquez A. Novel antiapoptotic effect of TBX15: overexpression of TBX15 reduces apoptosis in cancer cells. Apoptosis 2015; 10:1338–46. [DOI] [PubMed] [Google Scholar]
- 16.Baida A, Akdi M, González-Flores E, Galofré P, Marcos R, Velázquez A. Strong association of chromosome 1p12 loci with thyroid cancer susceptibility. Cancer Epidemiol Biomarkers Prev 2008;17:1499–1504. 10.1158/1055-9965.EPI-07-0235 [DOI] [PubMed] [Google Scholar]
- 17.Li LC and Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002; 18:1427–1431. [DOI] [PubMed] [Google Scholar]
- 18.Jiang M, Zhang Y, Fei J, Chang X, Fan W, Qian X et al. Rapid quantification of DNA methylation by measuring relative peak heights in direct bisulfite-PCR sequencing traces. Laboratory investigation; a journal of technical methods and pathology 2010; 90: 282–290. 10.1038/labinvest.2009.132 [DOI] [PubMed] [Google Scholar]
- 19.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques 2005; 39:715–725. [DOI] [PubMed] [Google Scholar]
- 20.Joyce D, Albanese C, Steer J, Fu M, Bouzahzah B, Pestell RG. NF-kB and cell-cycle regulation: the cyclin connection. Cytokine and Growth Factor Reviews 2001; 12:73–90. [DOI] [PubMed] [Google Scholar]
- 21.Hayden MS, Ghosh S. Shared Principles in NF-κB Signaling. Cell 2008; 132:344–362. 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- 22.Escárcega RO, Fuentes-Alexandro S, García-Carrasco M, Gatica A, Zamora A. The transcription factor nuclear factor-kappa B and cancer. Clin Oncol (R Coll Radiol) 2007; 19:154–61. [DOI] [PubMed] [Google Scholar]
- 23.Pacifico F, Leonardi A. NF-kappaB in solid tumors. Biochem Pharmacol 2006; 72:1142–1452. [DOI] [PubMed] [Google Scholar]
- 24.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nature Immunology 2002; 3:221–227. [DOI] [PubMed] [Google Scholar]
- 25.Pacifico F, Mauro C, Barone C, Crescenzi E, Mellone S, Monaco M et al. Oncogenic and anti-apoptotic activity of NF-kappa B in human thyroid carcinomas. J Biol Chem 2004; 279:54610–54619. [DOI] [PubMed] [Google Scholar]
- 26.Pacifico F, Leonardi A. Role of NF-kappaB in thyroid cancer. Mol Cell Endocrinol 2010; 321:29–35. 10.1016/j.mce.2009.10.010 [DOI] [PubMed] [Google Scholar]
- 27.Xing M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat Rev Cancer 2013; 13:184–199. 10.1038/nrc3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bushdid PB, Brantley DM, Yull FE, Blaeuer GL, Hoffman LH, Niswander L et al. Inhibition of NF-kappaB activity results in disruption of the apical ectodermal ridge and aberrant limb morphogenesis. Nature 1988; 392:615–618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data were normalized to RPL27 and expressed as fold change referred to untreated cells whose CXCL1mRNA relative expression was defined as 1. Data are mean ± SD of mRNA levels of two independent experiments in triplicates. ** indicates p-value < 0.01.
(DOCX)
(DOCX)
For luciferase constructions, restriction enzymes sites are in bold. For point mutation construction primers, changed nucleotides are marked in bold letter. BS-PCR: bisulfite polymerase chain reaction. Ta: annealing temperature.
(DOCX)
Data are normalized to the reference gene RPL27 and expressed as mean ± SD.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.