Abstract
Here, we present a novel scaffolding architecture of an inducible regulatory device. This dual control system is completely silent in the off stage and is coupled to the regulation of gene expression at both the transcriptional and translational levels. This system also functions as an AND gate. We demonstrated the effectiveness of the cumate-riboswitch dual control system for the control of pamamycin production in Streptomyces albus. Placing the cre recombinase gene under the control of this system permitted the construction of synthetic devices with non-volatile memory that sense the signal and respond by altering DNA at the chromosomal level, thereby producing changes that are heritable. In addition, we present a library of synthetic inducible promoters based on the previously described cumate switch. With only one inducer and different promoters, we demonstrate that simultaneous modulation of the expression of several genes to different levels in various operons is possible. Because all modules of the AND gates are functional in bacteria other than Streptomyces, we anticipate that these regulatory devices can be used to control gene expression in other Actinobacteria. The features described in this study make these systems promising tools for metabolic engineering and biotechnology in Actinobacteria.
Keywords: Inducible promoter, Dual control, AND gate, Memory switch, Non-volatile memory
Highlights
-
•
A novel scaffolding architecture of an inducible regulatory device for Actinobacteria is presented.
-
•
Dual control system, for the first time, couple the regulation of gene expression at both transcriptional and translational levels in Actinobacteria.
-
•
Designed devices are completely silent in the absence of both inputs and are highly inducible.
-
•
Effectiveness of the dual control system for the control of pamamycin production in Streptomyces albus was demonstrated.
1. Introduction
Synthetic biology is a relatively young discipline that aims to create new and/or reprogram existing cellular functions with useful properties using principles of mechanical engineering (Boyle and Silver, 2012, Davidson et al., 2012, Keasling, 2012, Singh, 2014). These goals can be achieved with the use of well-defined, orthogonal, and interchangeable controlling elements (promoters, ribosomal binding sites, operators, and terminators) that can be joined into regulatory devices (Egbert and Klavins, 2012, Lucks et al., 2011, Salis et al., 2009, Herai et al., 2004, Zhang et al., 2015, Yansura and Henner, 1984, Weinmann et al., 1994, Wieland and Hartig, 2008). These devices can then be used for the construction of predictable and quantitatively controllable programmable gene circuits with balanced expression of genes from the “ground up.” Regulatory devices and logic gates function as digital systems with Boolean logic that can sense and process information and respond accordingly. Therefore, using genetic circuits, we can program the cell of interest to respond in a particular way by sensing different extra- and intracellular inputs. However, any imbalance or leakiness within the regulatory circuits often leads to undesirable behavior in the designed biological system.
To date, dozens of synthetic regulatory devices and logic gates have been described (Elowitz and Leibier, 2000, Blount et al., 2012, Gardner et al., 2000, Stricker et al., 2008, Danino et al., 2010, Siuti et al., 2013, Nielsen et al., 2013, Brophy and Voigt, 2014, Hoynes-O’Connor and Moon, 2015). Most of these devices are based on transcriptional regulators characterized by combinatorial logic and are functional only in E. coli or other model chassis strains that they were developed for. Thus, these devices are not insulated from the host. Therefore, there is a scarcity of the aforementioned tools in less studied but industrially important bacteria, such as Actinobacteria, that are known producers of bioactive natural products (Katz and Baltz, 2016). Genes, products of which are involved in the biosynthesis of these compounds, are in most cases organized into clusters that may contain several operons in which expression is governed by different promoters to different levels. Therefore, to express the operons heterologously in various workhorse strains, there is an urgent need for inducible regulatory devices that are characterized by different induction strengths. Thus, regulatory devices that are used for the construction of multiple gene circuits or for the control of the expression of genes in gene clusters should be tight enough and should not induce cross-regulation, as this will lead to misregulation and an undesirable response. Furthermore, these devices should be capable of being modified so that the expression of genes in complex operons can be altered to different levels simultaneously. In addition, any leakage of an inducible system can be harmful for the cell. To date, there are no regulatory devices that fit all of the aforementioned criteria. Thus, there is a permanent need to design and invent new well-defined regulatory BioBricks, devices and logic gates with improved characteristics that are independent of their environment and may be reused and combined in plug-and-play fashion for the construction of more complicated gene circuits and systems. Modifications of the expression of genes in gene clusters responsible for the production of bioactive natural compounds is also a desirable property.
During the previous years, new types of regulatory devices have been developed. These devices include riboswitches (Winkler and Breaker, 2005, Wieland and Hartig, 2008, Topp et al., 2010, Rudolph et al., 2013, Serganov and Nudler, 2013), ribozymes (Wieland and Hartig, 2008, Doherty and Doudna, 2001, Ogawa and Maeda, 2008, Yen et al., 2006, Endo et al., 2014) and other small regulatory non-coding RNAs (Kang et al., 2014, Qi and Arkin, 2014, Papenfort and Vanderpool, 2015) that control the expression of genes at the transcriptional or translational level and overcome the primary drawback of transcriptional regulators because they do not require additional proteins for regulation. Several logic gates based on these types of regulators have been described (Sharma et al., 2008, Hemphill and Deiters, 2013, Chappell et al., 2015, Shen et al., 2015). But, synthetic regulatory RNAs have never been used before for the simultaneous regulation of the expression of a single gene in combination with transcriptional regulators. However, during the preparation of this manuscript, an article describing the simultaneous regulation of gene expression at the transcriptional and translational level has been published (Morra et al., 2015). No other examples are known. Such an approach may extend the list of available controlling elements for the precise regulation of gene expression and the construction of modular regulatory devices and multiple input circuits. In addition, regulation of the expression of a single gene at both the transcriptional and translational level can severely improve the tightness of the regulatory device. All inducible systems known to date, including the recently characterized dual transcriptional–translational cascade (Morra et al., 2015), are characterized by a certain level of basal expression in the off stage. This basal expression can be a problem for the extremely active meganucleases and site-specific recombinases, such as CRISPR-Cas9 and Cre (Wu et al., 2014, Kirchner and Schneider, 2015, Siegl et al., 2010, Herrmann et al., 2012). To our knowledge, there are no inducible switches that can efficiently control the expression of these genes. Therefore, there is an urgent need for new well-defined, non-leaky inducible systems.
Here, we present the construction of several modular dual control systems that, for the first time, couple the regulation of gene expression at both the transcriptional and translational levels in Actinobacteria. These systems are based on previously described cumate or resorcinol switches that are responsible for the regulation of gene expression at the transcriptional level (Horbal et al., 2014) and the theophylline riboswitch (Rudolph et al., 2013) or the hammerhead ribozyme (Horbal et al., 2014) that govern translation initiation. The newly designed regulatory devices that function in the form of an AND gate and are implemented as a Boolean function are completely silent in the absence of both inputs and, at the same time, are highly inducible. Furthermore, the AND gates are modular because a combination of diverse regulated promoters with the operators and riboswitches provide functionality to the systems. These gates are an example of volatile cellular memory that requires the permanent presence of inputs to sustain the output. To endow our logic gates with non-volatile synthetic memory, we placed the cre recombinase gene under the control of the dual control system. As a result, a memory switch was created that could sense signals and convert them into changes at the chromosomal level. These changes are recorded permanently (like on a “USB stick”) and cannot be lost. In addition to dual control systems, we constructed a library of synthetic cumate-inducible promoters that are characterized by different strengths and permit the simultaneous modulation of expression of multiple genes in different operons to various levels. Based on the described features, we envision that these AND logic gates and memory switches will enable the rational engineering of complex cellular processes in Streptomyces and other Actinobacteria.
2. Materials and methods
2.1. Bacterial strains and culture conditions
The bacterial strains used in this study are listed in Table 1. E. coli strains were grown in Luria–Bertani (LB) broth medium. When required, antibiotics (Roth, Germany; Sigma, USA) were added to cultures at the following concentrations: 75 μg ml−1 ampicillin, 50 μg ml−1 kanamycin, 50 or 120 μg ml−1 hygromycin, and 50 μg ml−1 apramycin.
Table 1.
Strains and plasmids used in this study.
| Bacterial strains and plasmids | Description | Source or reference |
|---|---|---|
| S. albus J100 | Isoleucine and valine auxotrophic derivative of S. albus G (DSM 40313) lacking SalI-restriction activity | Salas J., Oviedo, Spain |
| S. lividans TK24 | Derivative of S. lividans TK21 that contains mutation in the rpsL gene and is resistant to spectinomycin | (Kieser et al., 2000) |
| E. coli DH5α | routine cloning | MBI Fermentas |
| E. coli ET12567 (pUZ8002) | conjugative transfer of DNA | (Kieser et al., 2000) |
| pUC57 | Apr, general cloning vector | MBI Fermentas |
| pUC19 | Apr, general cloning vector | MBI Fermentas |
| pTOS | Amr; VWB-based integrative vector | (Herrmann et al., 2012) |
| pTOShyg | pTOS with aac(3)IV substituted with hyg | This work |
| pSET152 | Amr; ϕC31-based integrative vector | |
| pHET152 | pSET1529 with aac(3)IV substituted with hyg | This work |
| pKC1139 | Amr; E. coli–Streptomyces shuttle vector | (Kieser et al., 2000) |
| pOJ10700 | HygR; vector containing the hyg gene and oriT flanked by frt sites | (Gust et al., 2004) |
| pGUS | Promoter probe vector containing promoterless gusA | (Myronovskyi et al., 2011) |
| pGUSPA3Ribozyme | Derivative of pGUSbezRBS containing gusA gene fused with HHR and PA3 synthetic promoter | This work |
| pGUSP21Oper | Derivative of pGUS containing the P21 promoter fused to the cmt operator | (Horbal et al., 2014) |
| pGCymRP21 | Derivative of pGUSP21Oper containing the cymR gene | This work |
| pGCymRPA2 | Derivative of pGUS containing PA2 inducible promoter fused with gusA | This work |
| pGCymRPA3 | Derivative of pGUS containing PA3 inducible promoter fused with gusA | This work |
| pGCymRPA4 | Derivative of pGUS containing PA4 inducible promoter fused with gusA | This work |
| pGCymRPA8 | Derivative of pGUS containing PA8 inducible promoter fused with gusA | This work |
| pJETEgfpMcherry | Derivative of pJET1.2, containing egfp under the control of PA3 promoter and the mCherry gene under the control of PA8 | This work |
| pGCymPA3egfpA8RFP | Derivative of pGCymRp21, containing egfp under the control of PA3 promoter and mCherry gene under the control of PA8 | This work |
| pJETA3mcherry | Derivative of pJETEgfpMcherry, containing mCherry gene under the control of PA3 promoter | This work |
| pJETA3cherryA8gfp | Derivative of pJETEgfpMcherry, containing the mCherry gene under the control of PA3 promoter and egfp under – PA8 | This work |
| pGCymRPA2A3RFPA8gfp | Derivative of pGCymRp21, containing egfp under the control of PA8 promoter and the mCherry gene under the control of PA3 | This work |
| pGUST_ermE_E* | Derivative of pGUS containing gusA gene fused with theophylline riboswitch | |
| pGCymRibos | Derivative of pGCymRP21, containing the gusA gene under the control of P21-cmt promoter and theophylline riboswitch | This work |
| pGUSRolRibos | Derivative of pGUSRolRPA3, containing the gusA gene under the control of PA3-rolO promoter and theophylline riboswitch | This work |
| pGUSPA3Ribozyme | Derivative of pGUSbezRBS containing gusA gene fused with HHR and PA3 synthetic promoter | (Horbal et al., 2014) |
| pGCymRibozyme | Derivative of pGUSRolRPA3, containing the gusA gene under the control of P21-cmt promoter and theophylline HHR | This work |
| pALCre | pAL1 derivative with cre(a) gene under tipA promoter | (Fedoryshyn et al., 2008) |
| pKCHygCymRibos | Derivative of pKH1139, containing the gusA gene under the control of P21-cmt promoter and theophylline riboswitch | This work |
| pH1139 | pKC1139 with aac(3)IV substituted with hyg | Myronovskyi et al., 2014 |
| pKHygCymRibosCre | Derivative of pKH1139, containing the cre gene under the control of P21-cmt promoter and theophylline riboswitch | This work |
| pCreInt | Derivative of pSET152, containing aac(3)IV surrounded with direct loxP sites | (Fedoryshyn et al., 2008) |
| pGCymRibosCre | Derivative of pGCymRibos, containing the cre gene under the control of P21-cmt promoter and theophylline riboswitch | This work |
| pGRolRibosCre | Derivative of pGUSRolRibos, containing the cre gene under the control of PA3-rolO promoter and theophylline riboswitch | This work |
| pGusP21Riboswitch | Derivative of pGCymRibos, containing the gusA gene under the control of P21 promoter and theophylline riboswitch | This work |
| pKCHygP21Riboswitch | Derivative of pKHygCymRibos, containing the gusA gene under the control of P21 promoter and theophylline riboswitch | This work |
| pKCHygCymRibosPamJ | Derivative of pKHygCymRibos, containing the pamJ gene under the control of P21-cmt promoter, CymR and theophylline riboswitch | This work |
| pTOShyg | Derivative of pTOS with aac(3)IV substituted with hyg | This work |
| pTOShygCymRibosCre | Derivative of pTOShyg, containing the cre gene under the control of P21-cmt promoter and theophylline riboswitch | This work |
| pTOShygRolRibosCre | Derivative of pTOShyg, containing the cre gene under the control of PA3-rolO promoter and theophylline | This work |
For conjugation, Streptomyces albus and Streptomyces lividans strains were grown on oatmeal or mannitol soy (MS) agar (Kieser et al., 2000) for sporulation. For glucuronidase activity measurements, strains were grown in liquid tryptic soy broth (TSB).
2.2. Recombinant DNA techniques
Chromosomal DNA from Streptomyces strains and plasmid DNA from E. coli were isolated using standard protocols (Kieser et al., 2000, Sambrook and Russell, 2001). Restriction enzymes and molecular biology reagents were used per the manufacturer's protocol (NEB, England; Thermo Scientific, Germany).
Construction of the plasmids is described in Supplementary material section.
2.3. Assessment of the promoter strength and activity of regulatory devices (GUS assay)
For direct detection of glucuronidase activity, 1-to 5-day plates were flooded with 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc) solution and incubated at 28 °C for 1–4 h. The 1 M X-Gluc stock solution was prepared in dimethyl sulfoxide. The final concentration of the X-Gluc dilution used for flooding plates was 20 or 200 mM.
For the quantitative measurement of GusA activity, 1 ml of 24-h seed cultures of the S. lividans TK24 or S. albus J100 recombinant strains was inoculated into 25 ml of TSB. The cells were grown for 1 or 2 days. A 5-ml aliquot of the culture was harvested by centrifugation (6000g for 10 min) and used for the measurement of glucuronidase activity as described in Horbal et al. (2014). All measurements were normalized for weight, and the results reported were from three independent experiments. Microsoft Excel was used for statistical analyses.
2.4. Analysis of enhanced green fluorescence protein (EGFP) and mCherry reporter fluorescence
S. lividans TK24 and S. albus recombinants were grown and treated as described in Horbal et al. (2014). A 200-µl sample of the supernatant was transferred in black 96-well plates, and EGFP fluorescence was measured at 520 nm after excitation at 485 nm using a plate reader (POLARstar Omega, BMG Labtech). mCherry fluorescence was measured at 610 nm after excitation at 587 nm using the same plate reader.
2.5. Analysis of pamamycin production
For pamamycin production, 2 ml of a 2-day-old pre-culture was inoculated into 50 ml of TSB media and grown for 5 days at 30 °C with agitation at 200 rpm. The biomass was separated from the supernatant by centrifugation at 10,000 rpm for 10 min. Metabolites were extracted with ethyl acetate from cultural liquid and with an acetone:methanol (1:1) mixture from biomass. Samples were evaporated, dissolved in 450 μl of methanol:DMSO (1:1) and subjected to LC–MS analysis (Rebets et al., 2015). Chemically synthesized pamamycin 607 was used as a standard. Metabolites were separated on Waters BEH C18 column (100 mmx2.1 mm, 1.7 μm, column temperature 45 °C) using an UPLC-ESI-MS instrument (Dionex Ultimate 3000, Thermo Fisher Scientific GmbH and amaZon Speed ETD, Bruker). Samples were eluted with solvent A: ammonium formate buffer 90 mM and solvent B: 80:20 acetonitrile/100 mM ammonium formate buffer in a multistep gradient. 0.2 min 20% B, 20% B to 97%B in 2.8 min, 97%B to 100% B in 6 min, 2 min 100% B, 100% B to 20% B in 1 min, 3 min equilibration at 20% B. Flow 0.55 mL/min. Detection for quantitative analysis in srm-mode on Amazon MS-spectrometer using the ms2 fragment m/z=396 from parent ion of pamamycin m/z=608 [M+H]+. Pamamycin 607 reference sample concentrations were c=10, 5, 2.5 and 1 μg/ ml.
3. Results
3.1. Library of cumate-inducible promoters with a wide range of strength
In most cases, secondary metabolite gene clusters and other metabolic pathways carry a number of operons in which expression is governed from different promoters to various levels (Trefzer et al., 2002, Sosio et al., 2004, Bihlmaier et al., 2006). To modulate gene expression in such complicated systems and to reconstitute their expression successfully in heterologous hosts, a library of constitutive and inducible promoters is needed. Therefore, we constructed several inducible promoters that are characterized by different strengths based on the cumate switch and our library of constitutive promoters (Siegl et al., 2013).
We chose four variable in strength constitutive promoters from the library: PA2 (5.4 units/g), PA3 (30.0 units/g), PA4 (17.5 units/g) and PA8 (9.7 units/g) (Siegl et al., 2013). These promoters were fused to the cmt operator and the gusA gene that codes for the highly sensitive beta-d-glucuronidase reporter protein and cloned into the plasmid containing the CymR regulator. As a result, the plasmids pGCymRPA2, pGCymRPA3, pGCymRPA4, pGCymRPA8 (Table 1, for construction details see Supplementary materials) were constructed and transformed into S. albus. Newly constructed promoters were indeed inducible and were characterized by different dynamic ranges: PA2 has the highest and PA8 has the lowest (Fig. S1). In addition, based on these results, we postulate that there is no direct or predictable correlation between the strength of the constitutive promoter and its inducible derivative. The constructed PA2 inducible promoter is a prominent instance, since it was constructed on the basis of the weakest PA2 constitutive promoter and after fusion with the cmt operator appeared to be medium one (Fig. S1). This indicates that nucleotides between the promoter and start codon of each gene of interest can influence the strength of the promoter and/or the initiation of transcription. A similar observation was described for a set of modified PesaR promoters that contained a second EsaR binding site located in different positions. The strength of the promoters was strongly dependent on the location of the second operator binding site (Shong and Collins, 2013).
3.2. Controllable synthetic gene circuits with several operons
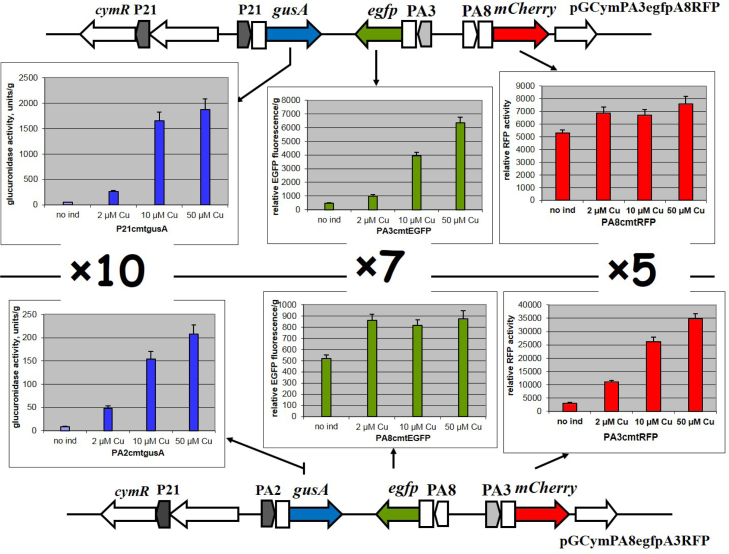
To investigate whether using different cumate-inducible promoters from our library and only one inducer simultaneously to modulate expression of diverse genes in different operons to various levels is possible, we constructed two plasmids, pGCymPA3egfpA8RFP and pGCymRPA2A3RFPA8gfp (Table 1 and Fig. 1). These plasmids contained two synthetic gene circuits that are comprised of three operons. Three reporter genes, gusA (Myronovskyi et al., 2011), egfp (Sun et al., 1999) and mCherry (Persson et al., 2013), were placed under the control of different cumate-inducible promoters. Both plasmids contain the cymR regulatory gene under the control of the P21-constitutive promoter. In the pGCymPA3egfpA8RFP plasmid, the gusA reporter is under the control of the P21-inducible promoter, whereas the expression of two other reporter genes, egfp and mCherry, is governed from the PA3 and PA8 promoters, respectively (Fig. 1). In another synthetic gene circuit that is located on the pGCymRPA2A3RFPA8gfp plasmid, the same reporter genes are under the control of other inducible promoters. Namely, the expression of gusA is controlled by the PA2 promoter, egfp – from PA8 and mCherry – from PA3 (Fig. 1). In the presence of the same concentration of cumate, the expression level of the same reporter genes from different promoters differs significantly. For instance, in the presence of 50 µM cumate, glucuronidase activity reaches approximately 1900 units/g when the gusA gene is under the control of the P21 inducible promoter, whereas the activity is only 200 units/g when the expression of the same reporter is under the control of the weaker PA2 promoter (Fig. 1). Thus, the P21 promoter promotes expression that is nearly 10 times higher the expression level promoted by the PA2 promoter. The same is true for the two other reporter genes, egfp and mCherry, where the difference in gene expression due to the promoter strength reaches 5-7 times (Fig. 1).
Fig. 1.
Schematic depiction of two synthetic gene circuits and results of measurements of GusA activity, EGFP and mCherry fluorescence. The following abbreviations are used: cymR – gene coding for the repressor; gusA – glucuronidase reporter gene; egfp – enhanced green fluorescence gene; mCherry – red fluorescence; P21, PA3, PA8, PA2 – synthetic promoters. The cmt operator sites are denoted by rectangles. Error bars indicate the standard deviations of triplicate experiments.
3.3. Hyper-inducible variant of the theophylline riboswitch
Inspired by the fact that RNA does not require a protein cofactor for the control of gene expression, synthetic biologists have created new types of regulatory devices that are known as riboswitches. There are several synthetic RNA switches that have been developed for E. coli (Topp et al., 2010, Ausländer and Fussenegger, 2014, Pedrolli et al., 2015), and only one, theophylline riboswitch, is known for Streptomycetes (Rudolph et al., 2013). The latter functions at the translational level and is characterized by low basal expression (Rudolph et al., 2013). Most riboswitches are involved in the regulation of the expression of metabolic enzymes; therefore, they do not confer high expression levels in the on stage (Winkler and Breaker, 2005, Serganov and Nudler, 2013). Due to this drawback, their applications are limited.
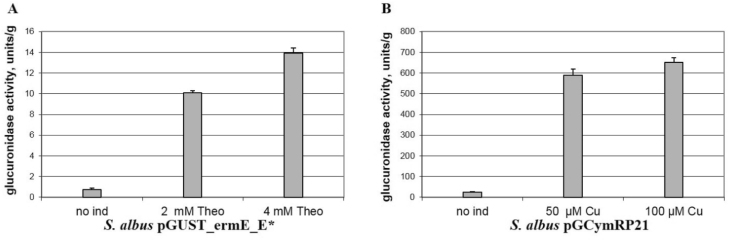
Because theophylline riboswitch is tight and hyper-inducible (Rudolph et al., 2013), we decided to compare the regulation efficiency of this system with the cumate toggle. To address this aim, plasmids that contained the gusA reporter gene under the control of the riboswitch (pGUST_ermE_E*) or only the cumate toggle (pGCymRP21) (Table 1) were transferred into S. albus. In the absence of an inducer (theophylline or cumate), the basal expression of the gusA gene for the strain that contains only the riboswitch was on average 25 times lower than that observed for the strain carrying the cumate switch (Fig. 2). This finding suggests that the riboswitch system provides more tight control over gene expression. In contrast, the activation ratio for the cumate switch was much higher compared to the riboswitch. The difference in expression was approximately 40–45 times (Fig. 2). Thus, the level of gene expression that can be achieved using the theophylline riboswitch in the on stage is quite low. Therefore, we decided to attempt to improve the former.
Fig. 2.
Glucuronidase activity in cell lysates of recombinant Streptomyces albus strains. (A) Activity in S. albus containing gusA under the control of the ErmE* promoter fused to the theophylline riboswitch. (B) Activity in S. albus containing gusA under the control of the P21 promoter fused to the cmt operator in the presence of CymR. The strains were grown in TSB medium for 2 days. Error bars indicate the standard deviations of triplicate experiments.
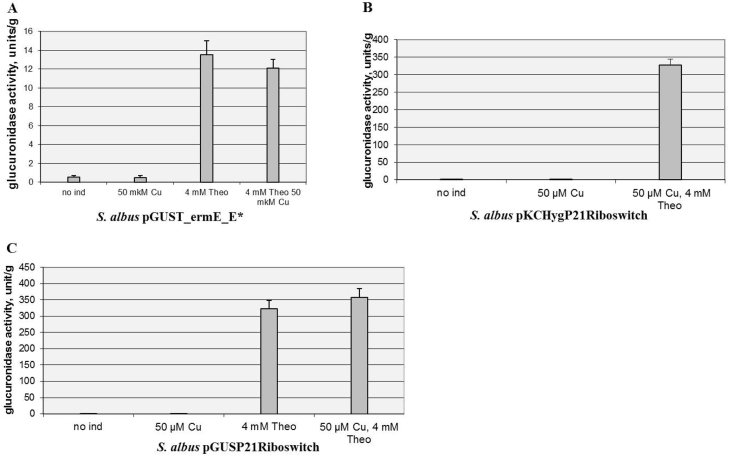
To address this aim, the ermE* promoter in the theophylline riboswitch was substituted with the 2 times stronger P21 promoter from our library of synthetic promoters that was also fused to the cmt operator. As a result, integrative pGUSP21Riboswitch and replicative pKHygP21Riboswitch (Table 1) plasmids were constructed. In these plasmids, gusA expression is controlled by the P21-cmt promoter at the transcriptional level and by the riboswitch at the translational level. As a negative control, the plasmid pGUST_ermE_E* contained only a riboswitch fused to the ermE* promoter was used. These constructs were transferred into S. albus. Both inducible systems provided approximately the same basal level of gusA expression in the absence of theophylline. However, the toggle-containing riboswitch fused with the P21-cmt promoter was 20–25 times more highly inducible than the control version of the riboswitch (Fig. 3). This disparity in expression is related not only to the difference in the promoter strength between P21 and ermE* but also for some other unknown reasons. Taking into account that direct fusion of the riboswitch sequence, without the 5′ UTR region, to the P21-cmt promoter led to a variant that was completely tight and not inducible at all (data not shown), we supposed that the distance, which is now increased because of the cmt operator placed in between the promoter and the 5′ UTR region of the riboswitch, or the sequence of these nucleotides itself, played a key role in creating hyper-inducible variants of the riboswitch. In any case, combining the theophylline riboswitch with the P21-cmt promoter and the 5′ UTR region yielded a tight expression system with high levels of expression of the gene of interest in the on stage that has not been described before for this type of riboswitch.
Fig. 3.
Glucuronidase activity in cell lysates of recombinant S. albus strains. (A) Activity in S. albus containing gusA under the control of the ErmE* promoter fused to the theophylline riboswitch. (B) Activity in S. albus containing gusA under the control of the P21 synthetic promoter fused to the cmt operator and theophylline riboswitch in the absence of CymR. All of the components of the system are located on the replicative pKC1139 vector. (C) Activity in S. albus containing gusA under the control of the P21 synthetic promoter fused to the cmt operator and theophylline riboswitch in the absence of CymR. All of the components of the system are located on the integrative pSET152 vector. The strains were grown in TSB medium for 24 hours. Error bars indicate the standard deviations of triplicate experiments.
3.4. Dual control system based on the cumate toggle and theophylline riboswitch
To sum up the aforementioned data, we conclude that both riboswitch variants and the cumate toggle are leaky (Fig. 3). Thus far, there is no inducible system that is completely tight and prevents expression in the off stage.
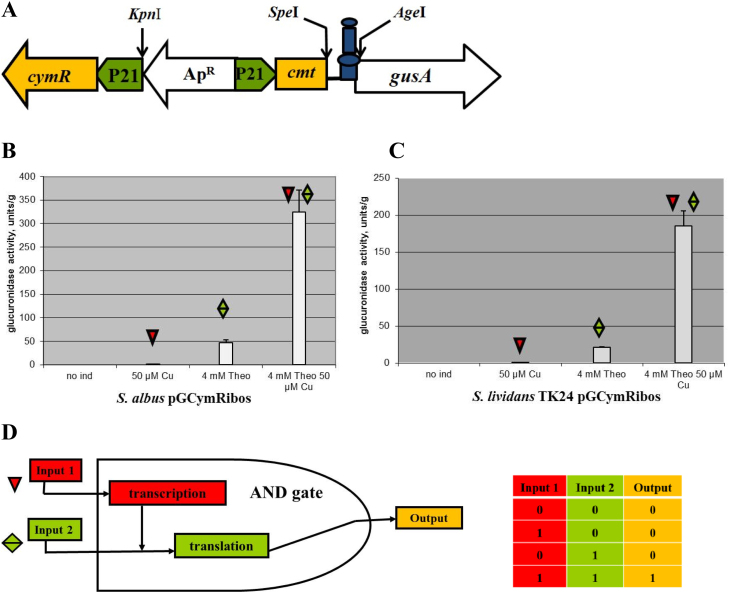
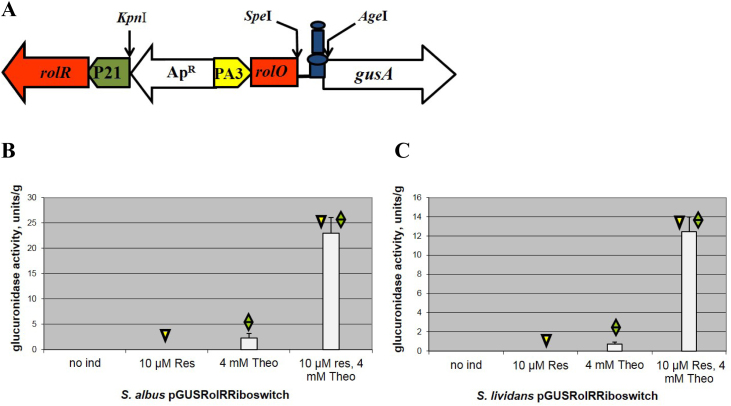
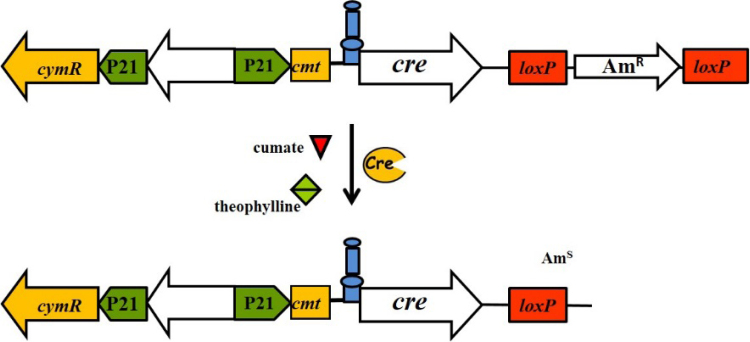
Therefore, we decided to examine whether a dual control system that will control gene expression at two levels, the transcriptional and translational, will not show signs of leakage. To address this aim, cumate toggle and the hyper-inducible riboswitch were used, as both systems are quite tight. As a result, the pGCymRibos (Table 1, Fig. 4A and S2) plasmid that is a derivative of the pGCymRP21 plasmid was constructed. In this plasmid, the transcription of the reporter gusA gene is under the control of the hybrid promoter, P21-cmt, consequently, the CymR regulator, which is also present on the plasmid, and translation is governed by the theophylline riboswitch that is fused with the start codon of the reporter (Figs. 4A and S2). As a control, the pGUST_ermE_E* plasmid that contained only a riboswitch that was fused with the ermE* promoter and the gusA reporter was used. Both plasmids were transferred into S. albus and S. lividans TK24. The cumate-riboswitch expression system was completely tight in the absence of both inducers because no GusA activity was detected in S. albus and S. lividans – even after 24 hours of incubation with substrate (Figs. 4B and C, S3). At the same time, the system is highly inducible; however, this system does not reach the expression level of the cumate switch alone in the induced stage (Fig. 2B) or even the level of the non-repressed P21-cmt promoter. This system has a much higher induction level (on average 23–25 times higher) compared to the riboswitch alone. In fact, the level of induction is approximately the same as in the case of the hyper-inducible variant of the riboswitch (Fig. 3, Fig. 4). In the S. albus strain, the activity of glucuronidase was, on average, 2 times higher than that in S. lividans (Fig. 4B and C). This difference in the numbers of GusA activity in these strains is related to the presence of two attB-like sites for the integration of vectors using the ϕC31 system in the genome of S. albus. Only one attB-like site is present in S. lividans (Bilyk and Luzhetskyy, 2014). Finally, the created dual control system, that is not known for any other type of bacteria except for Streptomycetes and E. coli, has the digital logic and function of an AND gate (Fig. 4D) because the highest output is achieved in the presence of both inducers.
Fig. 4.
Cumate-riboswitch dual control system. (A) Schematic representation of the cumate-riboswitch dual control system: cymR – gene coding for the TetR repressor, cmt – operator sequence of the cumate degradation operon, P21 – synthetic promoter, theophylline riboswitch, gusA – reporter gene, ApR – ampicillin resistance genes, KpnI, SpeI and AgeI – sites for endonucleases of restriction. (B) Glucuronidase activity in cell lysates of recombinant S. albus strains containing gusA under the control of the P21 promoter fused to the cmt operator and theophylline riboswitch in the presence of CymR. (C) Glucuronidase activity in cell lysates of recombinant S. lividans strains containing gusA under the control of the P21 promoter fused to the cmt operator and theophylline riboswitch in the presence of CymR. (D) Schematic representation of an AND gate and the outputs in the presence of different combinations of input signals. The strains were grown in TSB medium for 24 h. Error bars indicate the standard deviations of triplicate experiments.
3.5. Effect of an inducer on the cumate expression system
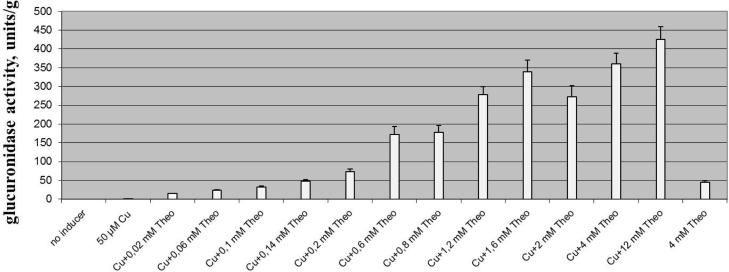
In the newly created dual control system, there is a theoretical possibility to tune the expression of the gene of interest using either cumate or theophylline, or both inducers simultaneously. To verify this, the S. albus strain containing the cumate-riboswitch dual control system (pGCymRibos) was grown in TSB medium containing different concentrations of theophylline, ranging from 20 μM to 12 mM. The concentration of cumate was stable (50 μM). Glucuronidase activity increases as theophylline concentration increases, suggesting that the system is dose-dependent. The inducibility of the system is approximately linear from 0.1 to 1.6 mM (Fig. 5).
Fig. 5.
Modulation of gusA gene expression in S. albus using the cumate-riboswitch dual control system. The strain was grown in liquid TSB medium for 20 h. The concentration of cumate was constant – 50 µM. Error bars indicate the standard deviations of the three independent experiments.
3.6. Dual control system based on the resorcinol toggle and theophylline riboswitch
Another dual control system based on resorcinol and riboswitch was constructed (Fig. 6A and S5). In this system, the PA3 promoter, which is approximately 2–3 times weaker than P21, is fused with the rolO operator, the theophylline riboswitch and the gusA reporter. Thus, in the pGUSRolRibos plasmid (Table 1 and Fig. S5), gusA transcription is under the control of the RolR regulator, and translation is governed by the riboswitch (Fig. 6A). This plasmid was transferred into S. albus and S. lividans TK24. The resorcinol based dual control system, similar to the cumate-based system, is completely silent in the off stage because no glucuronidase activity was detected in the absence of both inducers or even in the presence of only resorcinol (Fig. 6B and C). The inducibility of this system is much weaker compared to the cumate-based dual control system. However, this system still has a higher (for S. albus) inducibility than the system with the riboswitch alone (Fig. 2A and 6B). Finally, this regulatory switch also functions as an AND gate: the switch senses input signals, processes them and provides certain outputs.
Fig. 6.
Resorcinol-riboswitch dual control system. (A) Schematic representation of the resorcinol-riboswitch dual control system: rolR – gene coding for the TetR repressor; rolO – operator sequence of the resorcinol degradation operon; P21, PA3 – synthetic promoters; theophylline riboswitch; gusA – reporter gene, ApR – ampicillin resistance genes, KpnI, SpeI and AgeI – sites for endonucleases. (B) Glucuronidase activity in cell lysates of recombinant S. albus strains containing gusA under the control of the PA3 promoter fused to the rolO operator and theophylline riboswitch in the presence of RolR. (C) Glucuronidase activity in cell lysates of recombinant S. lividans strains containing gusA under the control of the PA3 promoter fused to the rolO operator and theophylline riboswitch in the presence of RolR. The strains were grown in TSB medium for 24 hours. Error bars indicate the standard deviations of triplicate experiments.
3.7. Dual control systems based on the cumate switch and hammerhead ribozyme
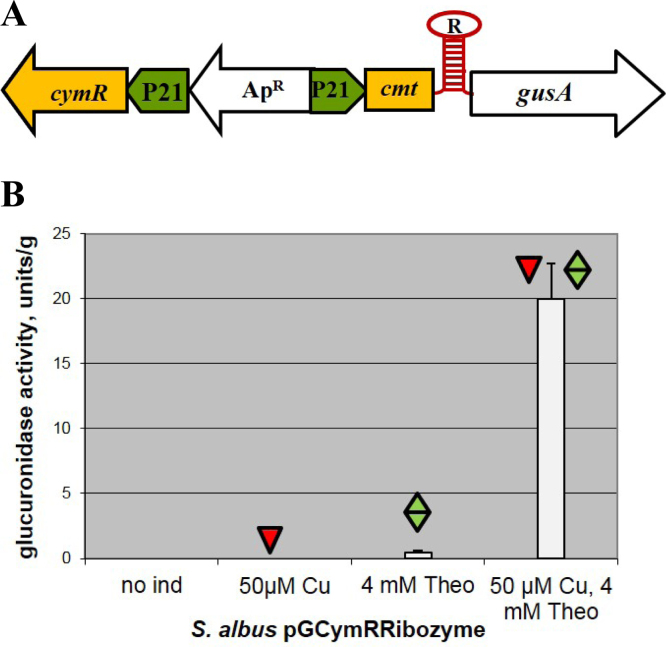
In our previous article on synthetic regulatory devices (Horbal et al., 2014), we succeeded in the construction of a hammerhead ribozyme (HHR)-based expression system that was characterized by non-leakage in the off stage. However, the activation of this system was extremely poor. We suppose that the possible reason for this poor activation may be the location of the ribozyme sequence in the transcriptional start site (directly downstream of the promoter). Therefore, we decided to integrate the constitutive 5′-untranslated region (ATACGACTCACTATA) in between the promoter and the ribozyme. This 5′-untranslated region is widely used for the construction of riboswitch-based inducible systems (Topp et al., 2010). Afterwards, the modified ribozyme was used for the construction of a dual control system based on the cumate switch. As a result, the plasmid pGCymRibozyme (Table 1 and Fig. 7A) was obtained. In this plasmid, expression of the gusA gene is under the control of the hybrid P21-cmt promoter, the CymR regulator, and the theophylline HHR, which is fused with the start codon of the reporter (Fig. 7A). As a control, the pGUSPA3Ribozyme plasmid, which contains the gusA gene fused only to the ribozyme and the PA3 promoter (Horbal et al., 2014), was used. All of the plasmids were transferred into the S. albus strain. We succeeded in the construction of this dual control system. The system is not leaky in the absence of both theophylline and cumate. In contrast, the presence of both inducers activates glucuronidase expression (Fig. 7B). Thus, the cloning of the constitutive 5′-UTR region upstream of the ribozyme improved the inducibility of the system by 1000 times and provided a system that functions as an AND gate with digital logic. This system is not leaky and may be used for the precise control of gene expression.
Fig. 7.
Cumate-ribozyme dual control system. (A) Schematic representation of the cumate-ribozyme dual control system: cymR – gene coding for the TetR repressor, cmt – operator sequence of the cumate degradation operon, P21 – synthetic promoter, R – theophylline ribozyme, gusA – reporter gene, ApR – ampicillin resistance genes. (B) Glucuronidase activity in cell lysates of recombinant S. albus strains containing gusA under the control of the P21 promoter fused to the cmt operator and theophylline ribozyme in the presence of CymR. The strains were grown in TSB medium for 24 hours. Error bars indicate the standard deviations of triplicate experiments.
3.8. Dual control system for the regulation of pamamycin biosynthesis
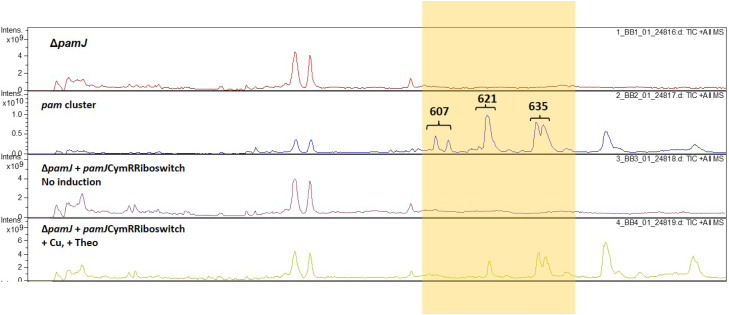
We used one of the created dual control systems to regulate the production of pamamycin. Pamamycin is produced by S. alboniger and is a macrodiolide antibiotic of polyketide origin that is active against gram-positive bacteria and some fungi (McCann and Pogell, 1979). Because the compound is extremely toxic to streptomycetes, we succeeded to perform its heterologous expression only in S. albus (Rebets et al., 2015). We predict that regulatory system that will allow to perform precise control over pamamycin production, namely turn it on and off at a certain time point, will give an opportunity to express this cluster in any other Streptomyces strains as well. To investigate the efficiency of our dual control system, we placed the pamJ gene, which codes for the ketoacyl-acyl carrier protein synthase II involved in the closure of hydroxy acids S and L into a macrodiolide ring (Rebets et al., 2015), under the control of the CymR regulator and the theophylline switch. The plasmid pKCHygCymRibosPamJ (Table 1), which is based on the replicative vector pH1139 (Table 1), was constructed. This plasmid was used to complement the S. albus R2 ΔPamJ mutant, which does not produce pamamycin (Rebets et al., 2015). The S. albus R2 ΔPamJ strain was used as a negative control. Pure pamamycin served as a positive control. HPLC-MS analysis of antibiotic production in the recombinant strains revealed that the S. albus R2 ΔPamJ mutant containing intact copies of the pamJ gene under the control of CymR and the riboswitch failed to produce any pamamycin in the absence of both inducers. However, cultivation of the strain in the medium with both cumate and theophylline present restored the pamamycin biosynthesis (Fig. 8).
Fig. 8.
Metabolite profile of the S. albus recombinant strains. Cu – denotes cumate and Theo – theophylline.
3.9. Precise control of cre recombinase gene expression – memory switch
Cre belongs to the family of site-specific recombinases that catalyze the recombination between its two recognition sites: loxP (Herrmann et al., 2012, Nagy, 2000, Fedoryshyn et al., 2008). These enzymes are extremely active, and to our knowledge to date, there is no inducible system that efficiently controls cre expression in Actinobacteria. However, precise control of cre expression is important because Cre is widely used for the construction of conditional mutants, reporter strains, and memory switches (Nagy, 2000, Mao et al., 2001, Maranhao and Ellington, 2013). Considering that our dual control system does not appear to be leaky, as evidenced by the lack of glucuronidase activity in the off stage, we decided to use it for the regulation of cre expression. The plasmids pKHygCymRibosCre, pTOShygCymRibosCre and pTOShygRolRibosCre (Table 1) were constructed. The pKHygCymRibosCre plasmid is a derivative of the replicative pH1139 vector and thus contains a thermosensitive ori pSG5 replicon that allows the plasmid to be eliminated under non-permissive conditions. Two other plasmids are derivatives of the integrative pTOS vector (Table 1). Overall, the expression of the cre gene is governed by dual control systems that are based on riboswitches and the CymR (Fig. 9) or RolR regulator. These plasmids were transferred into the S. albus strain that contained the pCreInt construct (Table 1). In the pCreInt plasmid, the apramycin resistance gene is surrounded with directly oriented loxP sites. Thus, apramycin resistance serves as reporters of Cre recombinase activity. This indicates that when cre expression is not induced, clones should be apramycin resistant. However, when expression of the gene is activated, the ratio between apramycin-resistant and apramycin-sensitive clones will be changed by up to 100%. To measure Cre activity, recombinant strains were grown on solid MS medium in the absence or presence of different concentrations of one of the inducers or a combination of several inducers. In the absence of both inducers, no apramycin-sensitive clones were detected after two passages of non-selective conditions for all of the recombinant strains. In the presence of only cumate or resorcinol, the ratio between apramycin-resistant and apramycin-sensitive colonies changed. Specifically, in the S. albus strain where the cre gene is under the control of the cumate-based dual control system located on the replicative plasmid, the percentage of apramycin-sensitive clones was 54% Table 2. (Table 3). For the strain that has cre under the control of the cumate dual control system but is based on an integrative vector, the percentage of apramycin-sensitive clones was 75.7% (Table 4). For strains with cre under the control of the resorcinol-based dual control system, the percentage of apramycin-sensitive clones was 0% (Table 5). There is an absence of apramycin-sensitive clones in the latter case because in this system, the cre gene is under the control of the PA3 promoter, which is 2–3 times weaker than the P21 promoter. Induction with only theophylline led to higher levels of cre expression. As a result, the percentage of apramycin-sensitive clones changed from 98.8% to 12.4% when induction was controlled by only theophylline (Table 3, Table 4, Table 5). Addition of both inducers for the cumate-riboswitch-based dual control systems converted all clones from apramycin-resistant to apramycin-sensitive (Table 3, Table 4). In contrast, even in the presence of both inducers, some apramycin-resistant colonies (7%) were still observed when cre gene expression was under the control of the resorcinol-riboswitch-based system (Table 5). This finding confirms that expression from the PA3 promoter is weaker than that from the P21 promoter.
Fig. 9.
Schematic representation of cre gene expression under the control of a cumate-riboswitch-based dual control system.
Table 2.
Primers used in this work.
| Primers | Sequence 5′-3′ | Purpose |
|---|---|---|
| CymRForw | AAAAAGCTAGCTCTAGTGGAAGAGCGCCCAATAC | fusion |
| CymRPA2operRev | AAAAAACTAGTATAATACAAACAGACCAGATTGTCTGTTTGTTGGTTGCATCCTAATTTCCACCATTGTGAGGTAGCCTCCACTGACGAAAGGGCCTCGTGATACGCC | semisynthetic promoters with the cmt operator |
| CymRPA3operRev | AAAAAACTAGTATAATACAAACAGACCAGATTGTCTGTTTGTTCATCTGATCCTACATCAGGCGTTAGTTTTGGAGCCCTGCTAGACGAAAGGGCCTCGTGATACGCC | -/-/- |
| CymRPA4operRev | AAAAAACTAGTATAATACAAACAGACCAGATTGTCTGTTTGTTTCTTGTATCCTACTGGAAGCTGTAGCGATATAGCCGCTTTTGACGAAAGGGCCTCGTGATACGCC | -/-/- |
| CymRPA8operRev | AAAAAACTAGTATAATACAAACAGACCAGATTGTCTGTTTGTTTTGAAGATCCTAGCCCTCACATTGATCTGACAGCCTCTATAGACGAAAGGGCCTCGTGATACGCC | -/-/- |
| RiboswitchForward | AAAAAACTAGTAATACGACTCACTATAGGTTC | Riboswitch-gusA amplification |
| RiboswitchRev | AAAAAGATATCTCACTGCTTCCCGCCCTGCTG | |
| CreCymRiboswForw | AAAAAACTAGTAATACGACTCACTATAGGTTCCGGTGATACCAGCATCGTCTTGATGCCCTTGGCAGCACCCTGCTAAGGAGGCAACAAGATGTCCAACCTGCTGACCGTC | Riboswitch-cre |
| CreCymRiboswitRev | AAAAAGATATCTCATCAGTCGCCGTCTTCCAG | amplification |
| RibozymelinkerForw | AAAAAACTAGTAATACGACTCACTATAGGTTCCTTCTCCTTCGGTACATCCAGCTG | Ribozyme |
| RibozymelinkerRev | AAAAAGATATCTTATCACTGCTTCCCGCCCTGCTGCGG | |
| HygRVSAmForw | GTGCAATACGAATGGCGAAAAGCCGAGCTCATCGGTCAGCCCGTAGAGATTGGCGATCCC | Substitution of apramycin resistance gene with hygromycin |
| HygRVSAmRev | TCATGAGCTCAGCCAATCGACTGGCGAGCGGCATCGCATCAGGCGCCGGGGGCGGTGTC |
Table 3.
Apramycin marker excision using Cre recombinase.
| S. albuspKChygRCymRRibosCre | Am resistant colonies (%) | Am sensitive colonies (%) |
|---|---|---|
| No induction | 100 | 0 |
| +Cumate | 54 | 46 |
| +Theophylline | 23 | 77 |
| +Cumate,+Theophylline | 0 | 100 |
Table 4.
Apramycin marker excision using Cre recombinase.
| S. albuspTOShygRCymRRibosCre | Am resistant colonies (%) | Am sensitive colonies (%) |
|---|---|---|
| +Cumate 50 mkM | 75.7 | 24.3 |
| +Theophylline | 1.1 | 98.9 |
| +Cumate,+Theophylline | 0 | 100 |
Table 5.
Apramycin marker excision using Cre recombinase.
| S. albuspTOShygRolRRibosCre | Am resistant colonies (%) | Am sensitive colonies (%) |
|---|---|---|
| +Resorcinol 10 mkM | 100 | 0 |
| +Theophylline | 34 | 66 |
| +Resorcinol+Theophylline | 6.8 | 93.4 |
4. Discussion
Precise and balanced control of gene expression in synthetic circuits constructed from the “ground up” or in heterologously expressed metabolic pathways is one of the primary prerequisites of successful engineering of biological systems in synthetic biology or metabolic engineering. In this article, we have presented a novel scaffolding architecture of an inducible controlling device, named the dual control system, which is not known for other bacteria, except for Actinobacteria and E. coli. The architecture is composed of a TetR type regulator and a theophylline riboswitch or ribozyme and regulates gene expression at both the transcriptional and translational levels. In addition, we constructed a library of cumate-inducible promoters with a wide range of strengths that allow the expression of different genes to be modulated to various levels using only one inducer in circuits that have complex genetic organization.
In this work, we expanded the repertoire of cumate-inducible promoters. We clearly showed that, by using diverse concentrations of the same inducer and different promoters from our library, fine-tuning the expression of three reporter genes in different operons to various levels in two synthetic gene circuits is possible. Furthermore, this system can modulate gene expression in two ways. The first way is based on promoter strength. The second way is based on the concentration of the inducer. Such features of cumate-inducible promoters increase the chances of obtaining expected expression levels for the gene of interest and in this way overcome bottlenecks and the production of toxins, non-detrimental proteins, and natural products.
In addition, we succeeded in improving the induction ratio of the theophylline riboswitch and the HHR 20–25 and 1000 times, respectively. In the case of ribozymes, the induction ratio was improved by simple addition of the constitutive 5′-UTR and using a stronger promoter. The reason for the improved riboswitch induction is still unknown and requires additional investigations. However, we propose that the improved induction may be due to the increasing distance between the promoter and the riboswitch. The HHR-based system had a lower induction level in the on stage than the riboswitch. However, the one advantage of the HHR-based system compared to the riboswitch-based system is that after the binding of theophylline to ribozyme, the latter catalyzes self-cleavage of its 5′-end. Therefore, the ribozyme does not influence the efficiency of translation of the transcript.
For the first time, we demonstrated that the dual control system that orchestrates gene expression at the transcriptional and translational levels may be used for accurate and efficient regulation of gene expression in Streptomycetes and in other Actinobacteria. This system provides a new type of regulatory device. The combination of gene regulation at the transcriptional and translational levels allows us to construct several different regulatory devices that produce no detectable output in the absence of both input signals. To our knowledge, this is the first example of completely tight systems described for Actinobacteria among those known to date. During the writing of this manuscript, another article on dual control systems called RiboTite and developed for E. coli was published (Morra et al., 2015). However, this system is still characterized by a certain level of basal expression. This is in contrast to our regulatory devices that are completely tight because no detectable output can be detected by the gusA reporter. The dual control systems that we constructed have digital logic and function as AND gates, that were not known before for Actinobacteria. This finding indicates that the highest output is obtained only in the simultaneous presence of both inducers.
Several variants of dual control systems that will expand the portfolio of available controlling devices were constructed. The first system is based on the RolR regulator and the riboswitch. Only few of the aforementioned elements and the PA3-rolO promoter are required to construct the resorcinol-riboswitch system. This system is not leaky, is easy to use, is inexpensive and is highly inducible.
Other versatile, portable, completely silent and highly inducible dual control systems were created based on the CymR repressor and the riboswitch or ribozyme. Again, these two inducible systems use several simple controlling elements. In the cumate-riboswitch system these elements are the following: the P21-cmt promoter, the CymR regulator and riboswitch. In the cumate-ribozyme system, the HHR is used. All of the parts of the aforementioned systems are located in the cis position on one integrative vector; therefore, these systems do not require any additional elements or cofactors to control gene expression and are genetically stable – even without selective pressure. The aforementioned systems are also characterized by different activation ratios, thereby inducing various expression levels for the gene of interest. Thus, the applications of these types of systems are manifold. Since no detectable expression of the reporter gusA gene was recorded for these devices in the off stage, we postulate that the systems are tight and not leaky. This feature makes these devices ideal for the regulation of genes encoding site-specific recombinases, transposases, and meganucleases, which are extremely active – even at low concentrations. This assumption is confirmed by the successful application of cumate- or resorcinol-riboswitch systems for the control of cre gene expression in S. albus. We did not obtain any apramycin-sensitive clones after two passages of recombinant strains in the absence of both inducers. At the same time, cre and gusA expression was inducible to different levels depending on the presence of various concentrations of one or both inducers. This finding suggests that the systems are dose-dependent. This feature of the systems allows gene expression to be modulated using the same promoter and different concentrations of inducers, namely cumate (or resorcinol) or theophylline alone, or a combination of the two. Thus, our dual control systems allow for accurate control of gene expression – even of highly active proteins. This is a unique characteristic of these regulatory devices that is not known for any other Streptomycetes-inducible systems constructed thus far. Furthermore, placing cre gene expression under the control of these regulatory devices led to the construction of memory switches that sense input signals and respond to these signals by inducing changes at the chromosomal level. As a result, these changes are “recorded on the chromosome” like on a USB stick. Therefore, they are permanent and heritable. Furthermore, these changes will never be lost; thus, they can be checked at any time. This is an example of non-volatile memory that does not require the permanent presence of an input signal to maintain the output. Such a memory switch provides an excellent model for creating different types of biosensors that are able to detect – pollutants, such as heavy metals and drugs. For this reason, only simple substitution of the cymR regulator and the P21-cmt promoter with any other TetR repressor, which may sense any other ligand and respective promoter, is needed. In addition, to make the output of the system more easily detectable, substitution of the apramycin resistance gene with gusA is required. Depending on the concentration of pollutants, the ratio of blue and white colonies will differ. The advantage of such memory switch-based sensors is that they will be able to detect even pollutants that are present not permanently but transiently in the environment, thereby “saving the data” on the chromosome. Thus, the investigator will be able to check the results at any time.
Furthermore, we demonstrated the efficiency of the cumate-riboswitch-based dual control system for the regulation of the biosynthesis of a highly toxic natural product – pamamycin. No pamamycin production was detected in the absence of both inducers. Thus, our dual control system should find it's application in metabolic engineering and heterologous expression of toxic metabolites, in the fine tuning of the expression of genes that constitute key steps or bottlenecks of the biosynthetic pathways and construction of metabolic pathways from the “ground-up”, when the precise control over gene expression is needed. In addition, a full tightness of the system in the off stage gives an opportunity to turn on the production of a natural product of interest at the desired time point, for example in case of toxic products it might be after the biomass accumulation. Such a programmed behavior of the dual control system allows accurately to control production of toxic products and in this way to overcome their toxicity for the producing cells.
We also postulate that the aforementioned regulatory devices are modular based on the fact that we created several different functional dual control systems using various combinations of synthetic particles, regulatory elements and devices that we had constructed earlier (Horbal et al., 2014, Siegl et al., 2013). Thus, simple substitution of one inducible promoter with the other one that differs in its strength will alter the induction level of the device. Furthermore, by taking into the account the functionality of the separate elements of these systems in different bacterial strains (Wieland and Hartig, 2008, Topp et al., 2010, Horbal et al., 2014, Siegl et al., 2013), we believe that these regulatory instruments are highly versatile. In addition, compounds that are used as inducers in these systems do not undergo metabolic conversion in streptomycetes. Altogether, these features make these systems ideal for overcoming existing intracellular regulatory networks and the overexpression or modulation of the transcription of genes that constitute bottlenecks in biosynthetic pathways. In addition, these systems may be used for the construction of conditional knockouts and the study of the functions of vital genes. Therefore, we assume that the regulatory devices described here will be among the most indispensable instruments of molecular tool kits used for the metabolic engineering and synthetic biology of Actinobacteria.
Acknowledgment
This work was supported by the European Commission under the 7th Framework Program through the “Collaborative Project” action “STREPSYNTH” Grant no. 613877 and through the ERC starting Grant EXPLOGEN no. 281623 to AL.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ymben.2016.03.008.
Appendix A. Supplementary material
Supplementary material
References
- Ausländer S., Fussenegger M. Synthetic biology: Toehold gene switches make big footprints. Nature. 2014;516(7531):333–334. doi: 10.1038/516333a. [DOI] [PubMed] [Google Scholar]
- Bihlmaier C., Welle E., Hofmann C., Welzel K., Vente A., Breitling E., Müller M., Glaser S., Bechthold A. Biosynthetic gene cluster for the polyenoyltetramic acid alpha-lipomycin. Antimicrob. Agents Chemother. 2006;50(6):2113–2121. doi: 10.1128/AAC.00007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilyk B., Luzhetskyy A. Unusual site-specific DNA integration into the highly active pseudo-attB of the Streptomyces albus J1074 genome. Appl. Microbiol. Biotechnol. 2014;98(11):5095–5104. doi: 10.1007/s00253-014-5605-y. [DOI] [PubMed] [Google Scholar]
- Blount B.A., Weenink T., Ellis T. Construction of synthetic regulatory networks in yeast. FEBS Lett. 2012;586(15):2112–2121. doi: 10.1016/j.febslet.2012.01.053. [DOI] [PubMed] [Google Scholar]
- Boyle P.M., Silver P.A. Parts plus pipes: synthetic biology approaches to metabolic engineering. Metab. Eng. 2012;14(3):223–232. doi: 10.1016/j.ymben.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy J.A., Voigt C.A. Principles of genetic circuit design. Nat. Methods. 2014;11(5):508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J., Takahashi M.K., Lucks J.B. Creating small transcription activating RNAs. Nat. Chem. Biol. 2015;11(3):214–220. doi: 10.1038/nchembio.1737. [DOI] [PubMed] [Google Scholar]
- Danino T., Mondragón-Palomino O., Tsimring L., Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463(7279):326–330. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E.A., Windram O.P., Bayer T.S. Building synthetic systems to learn nature's design principles. Adv. Exp. Med. Biol. 2012;751:411–429. doi: 10.1007/978-1-4614-3567-9_19. [DOI] [PubMed] [Google Scholar]
- Doherty E., Doudna J.A. Ribozyme structures and mechanisms. Annu. Rev. Biophys. Biomol. Struct. 2001;30:457–475. doi: 10.1146/annurev.biophys.30.1.457. [DOI] [PubMed] [Google Scholar]
- Egbert R.G., Klavins E. Fine-tuning gene networks using simple sequence repeats. Proc. Natl. Acad. Sci. USA. 2012;109(42):16817–16822. doi: 10.1073/pnas.1205693109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M.B., Leibier S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403(6767):335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Endo K., Parr C., Saito H. Expanding the synthetic ribonucleoprotein world in cells. Nat. Methods. 2014;11:1105–1106. doi: 10.1038/nmeth.3148. [DOI] [PubMed] [Google Scholar]
- Fedoryshyn M., Welle E., Bechthold A., Luzhetskyy A. Functional expression of the Cre recombinase in actinomycetes. Appl. Microbiol. Biotechnol. 2008;78(6):1065–1070. doi: 10.1007/s00253-008-1382-9. [DOI] [PubMed] [Google Scholar]
- Gardner T.S., Cantor C.R., Collins J.J. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403(6767):339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Gust B., Chandra G., Jakimowicz D., Yuqing T., Bruton C.J., Chater K.F. Lambda red-mediated genetic manipulation of antibiotic producing Streptomyces. Adv. Appl. Microbiol. 2004;54:107–128. doi: 10.1016/S0065-2164(04)54004-2. [DOI] [PubMed] [Google Scholar]
- Hemphill J., Deiters A. DNA computation in mammalian cells: microRNA logic operations. J. Am. Chem. Soc. 2013;135(28):10512–10518. doi: 10.1021/ja404350s. [DOI] [PubMed] [Google Scholar]
- Herai S., Hashimoto Y., Higashibata H., Maseda H., Ikeda H., Omura S., Kobayashi M. Hyper-inducible expression system for streptomycetes. Proc. Natl. Acad. Sci. USA. 2004;101(39):14031–14035. doi: 10.1073/pnas.0406058101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann S., Siegl Th, Luzhetska M., Petzke L., Jilg C., Welle E., Erb A., Leadlay P.F., Bechthold A., Luzhetskyy A. Site-specific recombination strategies for engineering actinomycete genomes. Appl. Environ. Microbiol. 2012;78:1804–1812. doi: 10.1128/AEM.06054-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbal L., Fedorenko V., Luzhetskyy A. Novel and tightly regulated resorcinol and cumate-inducible expression systems for Streptomyces and other actinobacteria. Appl. Microbiol. Biotechnol. 2014;98(20):8641–8655. doi: 10.1007/s00253-014-5918-x. [DOI] [PubMed] [Google Scholar]
- Hoynes-O’Connor A., Moon T.S. Programmable genetic circuits for pathway engineering. Curr. Opin. Biotechnol. 2015;36:115–121. doi: 10.1016/j.copbio.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Kang Z., Zhang C., Zhang J., Jin P., Zhang J., Du G., Chen J. Small RNA regulators in bacteria: powerful tools for metabolic engineering and synthetic biology. Appl. Microbiol. Biotechnol. 2014;98(8):3413–3424. doi: 10.1007/s00253-014-5569-y. [DOI] [PubMed] [Google Scholar]
- Katz L., Baltz R.H. Natural product discovery: past, present, and future. J. Ind. Microbiol. Biotechnol. 2016 doi: 10.1007/s10295-015-1723-5. (ahead of publishing) [DOI] [PubMed] [Google Scholar]
- Keasling J.D. Synthetic biology and the development of tools for metabolic engineering. Metab. Eng. 2012;14(3):189–195. doi: 10.1016/j.ymben.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Kieser T., Bibb M.J., Buttner M.J., Chater K.F., Hopwood D.A. 2nd edn. John Innes Foundation; Norwich: 2000. Practical Streptomyces Genetics. [Google Scholar]
- Kirchner M., Schneider S. CRISPR-Cas: from the bacterial adaptive immune system to a versatile tool for genome engineering. Angew. Chem. Int. Ed. Engl. 2015;54(46):13508–13514. doi: 10.1002/anie.201504741. [DOI] [PubMed] [Google Scholar]
- Lucks J.B., Qi L., Mutalik V.K., Wang D., Arkin A.P. Versatile RNA sensing transcriptional regulators for engineering genetic networks. Proc. Natl. Acad. Sci. USA. 2011;108(21):8617–8622. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Fujiwara Y., Chapdelaine A., Yang H., Orkin S.H. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97(1):324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- Maranhao A.C., Ellington A.D. Endowing cells with logic and memory. Nat. Biotechnol. 2013;31(5):413–415. doi: 10.1038/nbt.2573. [DOI] [PubMed] [Google Scholar]
- McCann P.A., Pogell B.M. Pamamycin: a new antibiotic and stimulator of aerial mycelia formation. J. Antibiot. 1979;32(7):673–678. doi: 10.7164/antibiotics.32.673. [DOI] [PubMed] [Google Scholar]
- Morra R., Shankar J., Robinson C.J., Halliwell S., Butler L., Upton M., Hay S., Micklefield J., Dixon N. Dual transcriptional-translational cascade permits cellular level tunable expression control. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv912. (ahead of printing) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myronovskyi M., Welle E., Fedorenko V., Luzhetskyy A. Beta-glucuronidase as a sensitive and versatile reporter in actinomycetes. Appl. Environ. Microbiol. 2011;77(15):5370–5383. doi: 10.1128/AEM.00434-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26(2):99–109. [PubMed] [Google Scholar]
- Nielsen A.A., Segall-Shapiro T.H., Voigt C.A. Advances in genetic circuit design: novel biochemistries, deep part mining, and precision gene expression. Curr. Opin. Chem. Biol. 2013;17(6):878–892. doi: 10.1016/j.cbpa.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Ogawa A., Maeda M. An artificial aptazyme based riboswitch and its cascading system in E. coli. ChemBioChem. 2008;9(2):206–209. doi: 10.1002/cbic.200700478. [DOI] [PubMed] [Google Scholar]
- Papenfort K., Vanderpool C.K. Target activation by regulatory RNAs in bacteria. FEMS Microbiol. Rev. 2015;39(3):362–378. doi: 10.1093/femsre/fuv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrolli D., Langer S., Hobl B., Schwarz J., Hashimoto M., Mack M. The ribB FMN riboswitch from Escherichia coli operates at the transcriptional and translational level and regulates riboflavin biosynthesis. FEBS J. 2015;282(16):3230–3242. doi: 10.1111/febs.13226. [DOI] [PubMed] [Google Scholar]
- Persson J., Chater K.F., Flärdh K. Molecular and cytological analysis of the expression of Streptomyces sporulation regulatory gene whiH. FEMS Microbiol. Lett. 2013;341(2):96–105. doi: 10.1111/1574-6968.12099. [DOI] [PubMed] [Google Scholar]
- Qi L.S., Arkin A.P. A versatile framework for microbial engineering using synthetic non-coding RNAs. Nat. Rev. Microbiol. 2014;12(5):341–354. doi: 10.1038/nrmicro3244. [DOI] [PubMed] [Google Scholar]
- Rebets Y., Brötz E., Manderscheid N., Tokovenko B., Myronovskyi M., Metz P., Petzke L., Luzhetskyy A. Insights into the pamamycin biosynthesis. Angew. Chem. Int. Ed. Engl. 2015;54(7):2280–2284. doi: 10.1002/anie.201408901. [DOI] [PubMed] [Google Scholar]
- Rudolph M.M., Vockenhuber M.P., Suess B. Synthetic riboswitches for the conditional control of gene expression in Streptomyces coelicolor. Microbiology. 2013;159(Pt 7):1416–1422. doi: 10.1099/mic.0.067322-0. [DOI] [PubMed] [Google Scholar]
- Salis H.M., Mirsky E.A., Voigt A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27(10):946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; NY: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Serganov A., Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V., Nomura Y., Yokobayashi Y. Engineering complex riboswitch regulation by dual genetic selection. J. Am. Chem. Soc. 2008;130(48):16310–16315. doi: 10.1021/ja805203w. [DOI] [PubMed] [Google Scholar]
- Shen S., Rodrigo G., Prakash S., Majer E., Landrain T.E., Kirov B., Daròs J.A., Jaramillo A. Dynamic signal processing by ribozyme-mediated RNA circuits to control gene Expression. Nucleic Acids Res. 2015;43(10):5158–5170. doi: 10.1093/nar/gkv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shong J., Collins C.H. Engineering the esaR promoter for tunable quorum sensing-dependent gene expression. ACS Synth. Biol. 2013;2(10):568–575. doi: 10.1021/sb4000433. [DOI] [PubMed] [Google Scholar]
- Siegl T., Petzke L., Welle E., Luzhetskyy A. I-SceI endonuclease: a new tool for DNA repair studies and genetic manipulations in streptomyces. Appl. Microbiol. Biotechnol. 2010;87(4):1525–1532. doi: 10.1007/s00253-010-2643-y. [DOI] [PubMed] [Google Scholar]
- Siegl T., Tokovenko B., Myronovkyi M., Luzhetskyy A. Design, construction and characterization of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab. Eng. 2013;19:98–106. doi: 10.1016/j.ymben.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Singh V. Recent advancements in synthetic biology: current status and challenges. Gene. 2014;535(1):1–11. doi: 10.1016/j.gene.2013.11.025. [DOI] [PubMed] [Google Scholar]
- Siuti P., Yazbek J., Lu T.K. Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol. 2013;31(5):448–452. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- Sosio M., Kloosterman H., Bianchi A., de Vreugd P., Dijkhuizen L., Donadio S. Organization of the teicoplanin gene cluster in Actinoplanes teichomyceticus. Microbiology. 2004;150(1):95–102. doi: 10.1099/mic.0.26507-0. [DOI] [PubMed] [Google Scholar]
- Stricker J., Cookson S., Bennett M.R., Mather W.H., Tsimring L.S., Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456(7221):516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Kelemen G.H., Fernández-Abalos J.M., Bibb M.J. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2) Microbiology. 1999;145(Pt9):2221–2227. doi: 10.1099/00221287-145-9-2221. [DOI] [PubMed] [Google Scholar]
- Topp S., Reynoso C.M., Seeliger J.C., Goldlust I.S., Desai S.K., Murat D., Shen A., Puri A.W., Komeili A., Bertozzi C.R., Scott J.R., Gallivan J.P. Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl. Environ. Microbiol. 2010;76(23):7881–7884. doi: 10.1128/AEM.01537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trefzer A., Pelzer S., Schimana J., Stockert S., Bihlmaier C., Fiedler H.P., Welzel K., Vente A., Bechthold A. Biosynthetic gene cluster of simocyclinone, a natural multihybrid antibiotic. Antimicrob. Agents Chemother. 2002;46(5):1174–1182. doi: 10.1128/AAC.46.5.1174-1182.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann P., Gossen M., Hillen W., Bujard H., Gatz C. A chimeric transactivator allows tetracycline-responsive gene expression in whole plants. Plant J. 1994;5:559–569. doi: 10.1046/j.1365-313x.1994.5040559.x. [DOI] [PubMed] [Google Scholar]
- Wieland M., Hartig J.S. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew. Chem. Int. Ed. Engl. 2008;47(14):2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- Winkler W.C., Breaker R.R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Wu X., Kriz A.J., Sharp A. Target specificity of the CRISPR-Cas9 system. Angew. Chem. Int. Ed. Engl. 2014;2(2):59–70. doi: 10.1007/s40484-014-0030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yansura D.G., Henner D.J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen L., Magnier M., Weissleder R., Stockwell B.R., Mulligan R.C. Identification of inhibitors of ribozyme self-cleavage in mammalian cells via high throughput screening of chemical libraries. RNA. 2006;12:797–806. doi: 10.1261/rna.2300406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liu Y.J., Cui G.Z., Cui Q. A novel arabinose-inducible genetic operation system developed for Clostridium cellulolyticum. Biotechnol. Biofuels. 2015;8:36. doi: 10.1186/s13068-015-0214-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material