Abstract
Objective
The Foundational Model of Anatomy (FMA) is an ontology that represents canonical anatomy at levels ranging from the entire body to biological macromolecules, and is rapidly become the primary reference ontology for human anatomy, and a template for model organisms. Prior to this work, the FMA was developed in a knowledge modeling language known as Protégé Frames. Frames is an intuitive representational language, but is no longer the industry standard. Recognizing the need for an official version of the FMA in the more modern semantic web language OWL2 (hereafter referred to as OWL), the objective of this work was to create a generalizable Frames-to-OWL conversion tool, to use the tool to convert the FMA to OWL, to “clean up” the converted FMA so that it classifies under an EL reasoner, and then to do all further development in OWL.
Methods
The conversion tool is a Java application that uses the Protégé knowledge representation API for interacting with the initial Frames ontology, and uses the OWL-API for producing new statements (axioms, etc.) in OWL. The converter is relation centric. The conversion is configurable, on a property-by-property basis, via user-specifiable XML configuration files. The best conversion, for each property, was determined in conjunction with the FMA knowledge author. The convertor is potentially generalizable, which we partially demonstrate by using it to convert our Ontology of Craniofacial Development and Malformation as well as the FMA. Post-conversion cleanup involved using the Explain feature of Protégé to trace classification errors under the ELK reasoner in Protégé, fixing the errors, then re-running the reasoner.
Results
We are currently doing all our development in the converted and cleaned-up version of the FMA. The FMA (updated every 3 months) is available via our FMA web page http://si.washington.edu/projects/fma, which also provides access to mailing lists, an issue tracker, a SPARQL endpoint (updated every week), and an online browser. The converted OCDM is available at http://www.si.washington.edu/projects/ocdm. The conversion code is open source, and available at http://purl.org/sig/software/frames2owl. Prior to the post-conversion cleanup 73% of the more than 100,000 classes were unsatisfiable. After correction of six types of errors no classes remained unsatisfiable.
Conclusion
Because our FMA conversion captures all or most of the information in the Frames version, is the only complete OWL version that classifies under an EL reasoner, and is maintained by the FMA authors themselves, we propose that this version should be the only official release version of the FMA in OWL, supplanting all other versions. Although several issues remain to be resolved post-conversion, release of a single, standardized version of the FMA in OWL will greatly facilitate its use in informatics research and in the development of a global knowledge base within the semantic web. Because of the fundamental nature of anatomy in both understanding and organizing biomedical information, and because of the importance of the FMA in particular in representing human anatomy, the FMA in OWL should greatly accelerate the development of an anatomically based structural information framework for organizing and linking a large amount of biomedical information.
Keywords: OWL, Frames, ontology, Foundational Model of Anatomy, Ontology of Craniofacial Development and Malformation
1 Introduction
The Foundational Model of Anatomy (FMA) (1,2) is an ontology that represents canonical anatomy at levels ranging from the entire body to biological macromolecules. With over a hundred thousand concepts and over a million relations between concepts the FMA is one of the largest and most complex biomedical ontologies in existence. Because of its extensive coverage of anatomy and because of the fundamental importance of anatomy in both understanding and organizing biomedical information the FMA is rapidly becoming the reference standard for representing human anatomy, and a template for model organisms, with most of the existing ontologies and terminologies incorporating or aligning to the FMA for their own anatomy axes.
Prior to the work described in this report the FMA was represented in a modeling formalism referred to as Protégé Frames (3) which derives from the Open Knowledge Base Connectivity (OKBC) specification (4). Although Frames was the representation of choice when we began the FMA project, the current preferred representation is OWL (Web Ontology Language) (5). OWL has gained a wide following in recent years, in part because it is the representation of choice for the semantic web (6) and as such facilitates the creation of a worldwide interconnected web of knowledge. There has therefore has been an increasing demand, both by us and by other FMA users, to convert the FMA to OWL so that it may more easily contribute to the global knowledge base.
The conversion of the FMA into OWL has been attempted several times in the past (7-11). Our original intension was to extend the implementations from one or more of these earlier efforts. However, these efforts were either incomplete, did not classify under a reasoner, or were simply unavailable to us. We thus decided to do our own conversion, with the added advantage that since we are also the authors of the FMA we not only have domain-specific insight into the ontological formalisms we are trying to represent, but also are able to modify the output of the conversion to be consistent under reasoning.
No other authors of conversion tools had this kind of control and domain expertise, with the result that our converted and post-processed FMA is the only conversion that captures all or most of the information in the original Frames model while also classifying under a reasoner. For these reasons we propose that our OWL version of the FMA should replace any existing OWL conversions, and should become the official release version of the FMA.
The purpose of this paper is to describe our conversion and post-conversion methods. Since so many people use the FMA, either for content development or for informatics research, and since the conversion is a major change from the Frames version, we feel it is important to describe our methods so others can understand the ontological formalisms we chose and the various tradeoffs we made.
In the remainder of this paper we first describe our conversion methods and their potential generalization to other Frame-based ontologies, which we have partially verified by using it not only to convert the FMA, but also to convert our Ontology of Craniofacial Development and Malformation (OCDM) (12). We then describe our post-conversion work that resulted in an FMA that classifies under an EL reasoner. Next, we describe changes likely to be needed by applications to use the converted FMA, illustrated with changes we needed to make to our own applications. Finally, we speculate about the potential impact that the converted FMA can have on the emerging life sciences semantic web (13) for representing and organizing biomedical knowledge and data. At the end of the paper we provide links for obtaining the converter program, the converted FMA and OCDM, and an online browser for FMA viewing.
2 Conversion Procedure
Our overall conversion approach is similar to earlier efforts. In particular it is based on that of Golbreich, et al. (7-9), but was also informed by those of Noy, et al. (10) and Dameron, et al. (11) as well as unpublished investigations by Alan Ruttenberg and by Robert Hoehndorf.
The purpose of the conversion was first and foremost a translation in syntax into a model that is readable, operable, editable by current OWL ontology tools, and amenable to post-processing as we describe in section 3. We endeavored to capture everything that was said in the Frames model, albeit often with subtle differences. We attempted to do this in a very generic way that would also work across several other ontologies that we are presently working with, in particular the OCDM.
Our approach is configurable, but in broad brush-strokes. We do not support frame-by-frame configuration. Rather, our implementation is relation centric. It allows a user to dictate how relationship types are transformed into OWL constructs, by associating specialized converters. This is a far more manageable way to configure the conversion of the FMA, as there are over 150 types of relationship but around 200,000 frames.
We attempted to capture everything, even if doing so might result in a computationally intractable model (e.g. a model upon which a classifier might not complete in a reasonable amount of time). We attempted to fix the problems that were easily identifiable in Frames prior to conversion (facet violations). We did not try to correct errors that were not flagged by Protégé, and we did not try to detect logical issues, such as unsatisfiable classes, in the OWL model being constructed. Many of these issues were addressed post-conversion (section 3).
Very little was added during the translation. As was the case with cleaning the model, much was left as to-do items once we had the FMA in OWL. An important example of this pertains to sufficient conditions. As sufficient conditions are not present in the Frames model, they were not generated in our OWL translation (e.g. only necessary conditions were generated, complete logical class definitions have yet to be constructed).
Our converter is written in Java and configured via an associated XML file. It uses the Protégé Frames Application Programming Interface (API) to interrogate the original FMA in Frames. The OWL API is used to generate the OWL classes, individuals, properties, axioms, etc., and to write the resulting model out to a file. Use of the APIs is an example of one of the changes that we wanted to make to the prior conversion tool most similar to our own, but for which we were unable to obtain the source code (7). That conversion required that the FMA, originally stored in a Protégé Frames database backend, first be converted into the Protégé flat file format CLIPS. The CLIPS file was then used as input. The conversion of the FMA to CLIPS is time and resource consuming, sometimes introduces errors, and in our view is not necessary. So our converter runs directly over the native FMA database via the Protégé API.
2.1 Frames and OWL: apples and oranges
Before we talk further about our specific methods, we mention a few terminological distinctions between Frames and OWL, drawing some rough analogies between the two modeling formalisms. Anatomical concepts are represented as classes, and the notion of a “class” exists in both Frames and in OWL. In Frames a class is a collection of “instances”, whereas in OWL it is a potential collection of “individuals”. In Frames, relationships are referred to as “slots” whereas in OWL they are called “properties”. “Facets” are used in Frames to constrain the allowed values for a slot. In OWL there are similar constructs, referred to as facets or “restrictions”. They should not, however, be viewed as constraints, but rather as rules of inference.
As we proceed we will suggest where the above modeling constructs are different, but for now we briefly mention a fundamental difference in the assumptions made by Frames vs. OWL. Most notably we refer to the differences between the closed-world assumption made in a Frames model, and the open-world assumption made in OWL. From a closed-world point of view, only those facts explicitly stated in a model are true. From an open-world perspective, at least those facts stated in a model (explicitly or implicitly) are true. The latter leaves open the possibility that there are additional facts about those entities discussed in the model, that are true but not yet stated (perhaps not yet known).
So, what are some of the consequences of the aforementioned assumptions? A thorough study of the differences in interpretation, between Frames and OWL models, is beyond the scope of this paper. But we give a brief example here to highlight the sorts of model mismatches that can occur. Suppose we say, in Frames, that a heart has, as parts, a right atrium, a left atrium, a right ventricle, and a left ventricle. If that is all we say about a heart, then it follows from the model that a right arm cannot be a part of a heart. That is because facts not asserted, in a closed world model, are deemed to be untrue. A consequence of this assumption is that a Frames model is considered to be complete. If the same statements (and only those statements) were made about a heart in OWL, it would not follow that a heart could not have a right arm. It must have the four chambers, but the open world model allows that there may be additional unknown or unstated facts.
In the following sections we describe the specific methods we used in the conversion, with discussion in those cases where differences between Frames and OWL required special attention.
2.2 Creating the classes
The converter starts from the non-systems (i.e. not built into Protégé) classes and performs a depth first search of the class hierarchy. For each class in Frames, a corresponding OWL class is created, and an rdfs:subClassOf link is asserted between each child and parent class. IRIs for created classes are chosen based on a strategy assigned within the configuration file. For the FMA, IRIs are generated based on the value of the slot ‘FMAID’.
While generating classes in OWL, rdfs:labels are also assigned. Presently these are based on the unique identifier in Frames (:NAME). This strategy might not be appropriate for ontologies other than the FMA, and should eventually be moved to the configuration file (indicating which slot the label should be derived from). We are also considering the pros and cons of having multiple rdfs:labels per class, some based on the preferred name, some on the synonyms (much like is done in the SKOS vocabulary (14)).
Also while generating new OWL classes, if the configuration flag disjoint-siblings = true, we create disjointness axioms between sibling classes (using OWL 2 syntax, not pairwise disjointness). Though sibling disjointness is not explicitly stated in the FMA, it is an assumption of the authors.
2.3 Copying the slot values
As each class is created, the converter visits its slots in Frames, and makes sure all values are converted into property values in OWL. Note that these properties have not yet been created (e.g. property declarations have not yet been added to the OWL model), but we do so in a subsequent step.
Decisions were made on how to convert slot values into property values on a slot-by-slot basis (as opposed to value-by-value) and were then done across the board for that slot (as opposed to converting slot values via one approach in some part of the ontology, and another approach elsewhere). Earlier when we referred to our conversion as using “broad brush-strokes”, this is what we meant. Each slot was considered, in consultation with the original content author (Mejino), to determine how it should best be converted.
In Frames, slots connecting classes to other classes have a different interpretation or semantics than they do in OWL. For example, if in Frames the class Heart has_part Right_atrium this is not interpreted as “a part of the class of all hearts is the class of all right atriums”. Rather, it is understood to be a statement about the potential instances of a class, i.e. “every heart has a part that is a right atrium” (the FMA represents only canonical anatomy, not malformations or variations). In OWL this is most closely converted to existential restrictions (e.g. ‘some’).
The following are the default strategies of the converter (alternative strategies are discussed in the next section):
• For slots with value type Cls (classes in Frames), we convert these to OWL object property existential restrictions as noted above.
For example, the assertion that a tooth (fma12516) must have a constitutional part that is an enamel of tooth (fma55629) looks like this:
fma:fma12516 rdfs:subClassOf (fma:constitutional_part some fma:fma55629)
In RDF/XML:
• For slots with values that are not objects (i.e. not class or instance type) we convert these to DataHasValue expressions. This strategy applies to slots whose value types are string, integer, float, Boolean, etc. as well as those that are of the Protégé type ‘symbol’. The latter is basically a string value type with an enumerated list of allowed strings (value set).
Here is an example of how we converted the Frames symbol type slot “dimension” to OWL, by creating a new datatype (in Manchester OWL syntax):
DataProperty: dimension
Annotations:
FMAID “85827”^^xsd:string
Characteristics:
Functional
Range:
{”0-dimension”^^xsd:string , “1-dimension”^^xsd:string , “2-dimension”^^xsd:string, “3-dimension”^^xsd:string}
• Slots with value type Instance in the FMA are all reified relationships in Frames. Reified relationships are a means of adding attributes to relationships, or of constructing n-ary relationships, in a language supporting only unattributed, binary, relations. Our default conversion of these to OWL follows a suggestion from the W3C 2006 working group - pattern 1 (15) and is the same solution used by Golbreich et. al. (8). This approach is akin to RDF reification and is not always ideal (since the connection between the property and its ‘primary’ value is indirect). But in some cases it was the only solution.
An example of this approach is illustrated below, again for the tooth (fma:fma12516) and its related part enamel of tooth (fma:fma55629). An anonymous class is created representing the intersection of all conditions present in the reified frames instance (via OWL restrictions). This anonymous class is then linked to tooth via an existential restriction on the primary property (attributed_part).
The above were chosen as reasonable defaults, are based only on datatype, and are used when no other conversion procedure is specified in the converter's configuration file, as described in the next section. In the first two cases these defaults were chosen as they represent what we thought would be the most common conversions. In the reified relation case the default was chosen not because it is the best or most likely conversion, but because it was the only strategy that could be performed without additional information (as is supplied in the configuration file for some of the alternatives in the next section).
2.4 Alternatives to the default slot conversions
We provide several alternatives to the above default strategies for converting slots and their values into OWL. These alternatives allow us to tune the program, via the configuration file, to best match the meaning intended by the content authors.
Annotation properties, in OWL, are (loosely) those that are used for tagging/bookkeeping/provenance and have no logical consequences. However, in Frames it is not possible to automatically discover the slots that should be converted to annotation properties. The converter therefore allows us to specify, in the configuration file, which properties should be annotations. For example, we chose to convert to annotations our various FMA label properties, such as preferred_name, synonym, and FMAID. These are all without logical consequences, like the standard label property rdfs:label. We also use this strategy for attaching extra-ontology identifiers to FMA classes (i.e. Tailarach_id, UMLS_id, etc.). Any other attribute whose value describes the class itself (rather than individuals within the class) can optionally be converted to an annotation property.
Some of the slots in Frames that we would like to convert into annotation properties are actually reified (attributed). For example, values for the slot “preferred name” are instances of a class “Concept name”. Concept names have slots like name, authority, abbreviation, etc. In the conversion, we make values for the sub-property “name” direct values of the annotation property fma:preferred_name and all of the other slots and values are converted as annotations on the fma:preferred_name annotation (i.e., annotations on annotations). Synonym and non-English equivalent slots are converted in this same manner, as are other reified label properties (we have several other slots that are reified so as to assign both a term and an id to the class).
As an example, the following is a partial result of the conversion of a synonym:
<owl:Axiom>
<owl:annotatedSource rdf:resource=“http://.../fma/fma10420”/>
<owl:annotatedProperty rdf:resource=“http://.../fma/synonym”/>
<owl:annotatedTarget rdf:datatype=“...#string”>Stenon duct
</owl:annotatedTarget>
<authority rdf:datatype=“...#string”>Rosse MD</authority>
<authority rdf:datatype=“...#string”>Terminologia Anatomica 1998</authority>
...
</owl:Axiom>
The above conversion is governed by the following entry in the configuration file:
<slot_conv_class
slot_name= “Synonym”
primary_slot=“name”
conv_cls_name= “...ReifiedAnnotationPropertyConverter” />
The conversion of non-reified slots to annotation properties is simpler. FMAID, for example, is converted like this:
<owl:AnnotationProperty rdf:about=“http://.../fma/FMAID”>...
Values look like this:
<FMAID rdf:datatype=“...#string”>85821</FMAID>
The configuration looks like this:
<slot_conv_class
slot_name= “FMAID”
conv_cls_name= “...AnnotationPropertyConverter” />
Annotations on annotations are just one of the methods provided for the conversion of reified relations from Frames. The converter presently supports the following methods:
• Annotations on annotations as above.
• The default strategy (8,15), in which slots with value type class are converted to existential (“some”) restrictions and slots with values that are strings, integers, etc. are converted to DataHasValue expressions. The intersection of these becomes the value that corresponds to the reified slot.
• Property chaining improves upon the previous strategy, where appropriate, by adding a direct connection between the ‘primary’ value and the referring class. For example, consider the FMA slot “attributed part”. This slot is intended to connect a class like Heart to another class like Right atrium. But, it is not a simple binary relationship. We wish to add additional attributes to the relationship (e.g. when a whole object has multiple ways of partitioning we want to indicate which parts collectively form a non-overlapping whole). We say that the “primary” slot is “related part”, which is where the value Right atrium is found. We then create a chain attributed_part.related_part and make that chain a subproperty, in this case of a new property direct_part. By doing so, we retain a direct connection (i.e. direct_part) between the Heart and the Right atrium. But, the reification retains the other attributes as well.
<slot_conv_class slot_name=“attributed part”
conv_cls_name=“...ChainReifiedPropertyConverter”
primary_slot=“related part”
direct_property_name=“direct_part”
excluded_slots=“anatomical/arbitrary,shared/unshared.”/>
• Splitting a reified relation (easy case): In some cases a slot did not really need to be reified in the first place. For example, in the FMA the attributed property “chromosome pair number” has values that are instances of “Chromosome pair number value”. The latter, in turn, has slots “number of pairs per nucleus” and “ploidy”. Neither of these slots is an attribute on the other. Thus, it can be split without loss of information as follows:
Primordial_germ_cell number_of_pairs_per_nucleus “46”.
Primordial_germ_cell ploidy “diploid”.
• Splitting a reified relation (more complicated case): In other cases, a reified slot in the Frames model could be divided into multiple non-reified slots, as in the case above, but where the properties chosen in the OWL model are based on the value (rather than the slot) used in Frames. For example, the FMA slot “attributed continuous with” in FMA Frames allows us to say not only that the esophagus is continuous with the stomach, but also that the latter lies in the inferior direction relative to the former. We could not split this into two statements like:
Esophagus continuous_with Stomach
Esophagus anatomical_coordinate “Inferior”
The above does not work because the esophagus is neither inferior nor superior in-and-of itself. Such directional information only makes sense relative to another object (the stomach). So, we instead support splitting like this:
Esophagus continuous_with Stomach
Esophagus superior_to Stomach
Note that the property in the second statement is based on the value “Inferior” in the original Frames relationship. The mappings between values and properties are configured in a mappings file, referenceable from the configuration file. Therefore this approach only works when a slot has a small finite set of possible values. The value to slot file contains entries like this:
Anterior=direct_posterior_to
Posterior=direct_anterior_to
...
The main configuration file entry looks like the following:
<slot_conv_class slot_name=“attributed continuous with”
conv_cls_name=“...SplitReifiedValPropertyConverter”
property_rename_map=“related object->direct_continuous_with”
config_map_delimiter=“->”
value_to_property_map=“anatomical coordinate->
resource/attrCoordMap.properties,
laterality->resource/attrCoordMap.properties”
value_slot=“related object”
excluded_slots=“surrounds,surrounded by,adjacent”/>
A no-op converter is also provided, which can be used to configure slots of any data type, to indicate to the converter that no corresponding OWL property should be generated for a given Frames slot. This is required because, if a slot is not mentioned in the configuration file, a default converter is used. Therefore, if we wish to ignore a slot, we must explicitly configure the conversion to do so.
2.5 Declaring the properties
Independent of property values, the properties themselves need to be declared. We state whether each new property will be a datatype, object, or annotation property based on the defaults and configurations stated above. We also define the domain and range of each property, as well as any facets on the range that can be gathered from the slot in Frames.
The domain of each property is class valued and is generated from the “Domain” facet in Frames. Note that this is a bit problematic for the FMA as often several classes are listed in the Frames model, but it would have been more appropriate to apply a common superclass of all. This happens frequently because authors do not typically edit the Domain facet directly on a slot. Rather they apply the slot at whatever classes they deem appropriate and Protégé automatically fills in the Domain. It is only upon later review of this facet that the authors see that they could have applied the property at a higher level (or lower in some cases).
When multiple classes are listed in the slot Domain facet, what does that mean and how should it be applied in OWL? As an example, the slot “arterial supply” has multiple Domain classes. The first two are “Subdivision of nervous system” and “Subdivision of genital system”. We do not want to say something like this:
arterial_supply rdfs:domain Subdivision_of_nervous_system
arterial_supply rdfs:domain Subdivision_of_genital_system
...
The above would suggest that any individual that has an arterial supply property is both a Subdivision of nervous system and a Subdivision of genital system (the intersection). What we want to say, to correspond to the Frames model, is that the individual must be a member of at least one of Subdivision of nervous system or Subdivision of genital system or ... (the union). So, the converter creates a class expression for this union and sets it as the property domain.
The range of each property, however, need not, in all cases, be a class. For a datatype property, we set the data type as its range. For object properties, we create a class union expression, as we did for domain, but this time based on the Frames facet “Allowed Superclasses”. A similar problem exists in Frames for this facet as was noted for the Domain facet, in that values are not entered at the facet directly but are auto-generated by Protégé based on slot usage. Often the authors would have preferred a higher-level class to best model their intension and to accommodate future values.
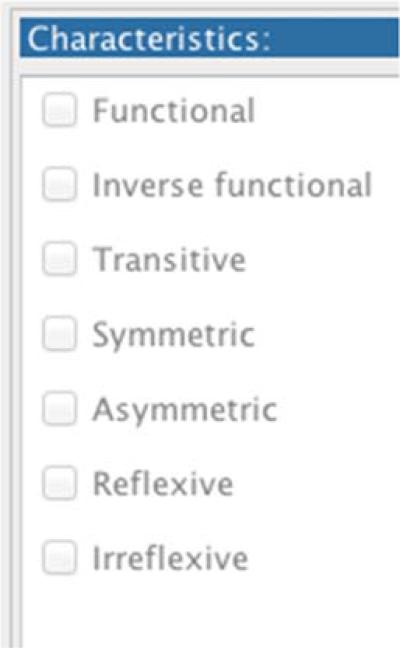
We looked at what other facets, beyond domain and range, could be carried over from Frames. The Protégé 4.× OWL editor has a properties characteristics view that shows the following:
If a slot in Protégé Frames has itself as its inverse, then we create a symmetric property axiom in OWL (for object properties). If a slot in Frames has max cardinality 1 then we create a functional property axiom in OWL. However, we cannot determine automatically from the Frames representation whether or not a slot has any of the other characteristics above (i.e. inverse functional, transitive, asymmetric, reflexive, or irreflexive).
Cardinality constraints beyond “1” are not presently handled by the converter. We did not try to anticipate all possible Frames knowledge construction patterns nor all OWL patterns. At present the converter only addresses constructs that we actually see in the FMA (or OCDM, section 2.7). It is, therefore, not (yet) a complete generic converter for any Frames ontology. Cardinality constraints greater than 1 are one such omission.
2.6 Converting the instances
In the FMA, the only instances are those representing reified relations. None of these are converted into OWL individuals. Those cases that still reify relations in OWL use class expressions.
2.7 Generality of the convertor: converting the Ontology of Craniofacial Development and Malformation
The convertor was primarily designed for converting the FMA. However, by allowing customizations to be coded as special Java classes in the convertor and activated in the XML configuration file we believe the convertor could be used for converting other Frames ontologies to OWL. We have partially verified this belief by using the same software to convert our Ontology of Craniofacial Development and Malformation (OCDM) from Frames to OWL. Each conversion has a unique configuration file.
Certain special cases (Java classes) and configuration directives needed to be added to the convertor so that it could handle the OCDM. Examples of these include:
Handling multiple component ontologies. Unlike the FMA the Frames version of the OCDM consists of multiple component ontologies, each implemented in a separate Frames file, and each included in the parent OCDM ontology by the Frames “Include” statement. In the conversion we thus created a parent OCDM.owl ontology, which then “Imports” separately converted OWL versions of each component ontology. Each component ontology was processed by a separate run of the convertor.
IRI generation. Unlike the FMA not all OCDM classes have an existing numerical identifier slot. We therefore used an auto-incrementing, 7 digit numerical IRI generator. To insure that the same id is chosen the next time the converter is run, all auto-generated ids are cached in a map that is written to persistent storage.
Mapping instances: Two ontologies in the OCDM serve to map concepts in one ontology to homologous concepts in another (human and mouse). These mappings are not applied directly to classes in either ontology. Rather, mapping classes exist whose individuals refer to both the source and target concepts (like an association table in a database). The choice to create mappings separate from the source and target concepts was to prevent polluting those classes as more and more mappings were made (there could conceptually be many, and they are outside of the domain of both the source and target ontologies). However, in OWL we have another way of keeping them separate. We do apply them as properties on the source class, but we do so in a separate OWL file, which imports the source. In this way the mappings are cleanly separable from the source domain.
External term instances: Many researchers in craniofacial development and abnormalities have their own set of commonly used terms (which may differ from those in the OCDM). There may be many such sets. We wanted the OCDM to capture these, but not necessarily set them as attributes of the class they refer to (per the same argument as above). So, rather than have classes refer to these terms, the terms refer to the classes as instances of the class External term. We handled these, during the OWL conversion, in the same way we handled mapping instances. The terms are now property values of the OCDM class, but in a separate OWL file.
These differences between the FMA and OCDM necessitated changes in the conversion code, which were implemented as Java classes that could be specified in the configuration code. It is likely that other Frames ontologies would lead to additional special cases that would in turn be activated for a specific ontology via directives in the configuration file. As more Frames ontologies are examined we believe the conversion Framework should allow these special cases to added and activated as needed by including directives in the configuration file, thus leading to a general purpose convertor of Frames ontologies to OWL. Validation of this assertion is beyond the scope of this paper, but since we are making the convertor code available as open source, others will be able to help validate the generality of the convertor by using and modifying the code to convert their own Frames ontologies.
3 Towards a More Reasonable FMA Ontology
The results of both the FMA and OCDM conversions are OWL files, transformations of the information from the Frames versions into a syntax that is compatible with other OWL ontologies. These conversions were initially released as versions 4.2.0 and 1.1 of the FMA and OCDM respectively, with the purpose of generating OWL representations as soon as possible so others could more easily interoperate with them than was possible in the Frames versions. At this time we also abandoned content development in Frames. We currently do all our content development on the OWL versions.
As mentioned in the previous section, many issues were left as post-conversion tasks to be done directly on the converted OWL files. In particular, in the interest of preserving all information, many assertions in Frames were kept during the conversion even when they resulted in unsatisfiable classes in OWL. A class is unsatisfiable if some combination of assertions (or inferences that follow from assertions) lead to a class that could not possibly have individuals (examples later in this section). It was not possible for the converter to know which assertions should be dropped or modified to fix such issues. Fixing unsatisfiable classes was therefore left as a post-conversion task.
We recognize that most OWL users see logical inference and the use of an OWL reasoner as an integral part of the processes of OWL development and use. We also recognize that reasoner identified problems, like unsatisfiable classes, are legitimate errors in the knowledge represented. Therefore we made it a post-conversion priority to clean up known (i.e. reasoner identified) logical issues in the FMA. In our future work we will perform a similar cleanup of the OCDM.
3.1 Scope of clean-up
The FMA is a large and complex ontology that is, for all practical purposes, intractable to full OWL2 reasoning. A subset of OWL, the EL profile, limits axiom types to those over which ontology consistency, class expression subsumption, and instance checking can be decided in polynomial time (16). This makes EL well suited for large ontologies for which inference is desired. While the FMA is more expressive than the EL profile, it is still useful to identify logical issues that arise from considering only the subset of EL allowed axioms.
For this exercise, we looked specifically at cleaning up a large number of unsatisfiable classes present in the post-conversion FMA. It is worth noting that, if a class were unsatisfiable based on the axioms allowed under EL, it would still be unsatisfiable if we were to add back in the dropped axioms that made reasoning intractable. So, while EL reasoning might not identify all unsatisfiable classes in the ontology, the ones that it does identify are legitimate problems.
For this work we used the ELK reasoner version 0.4.2 (17) from within the Protégé editing environment. At present ELK does not yet support all axiom types from the EL profile. When reasoning on the FMA, ELK alerts the user that reasoning may be incomplete as it:
Only partially supports DataHasValue
Does not support ObjectPropertyRange
Does not support positive occurrences of ObjectUnionOf
We chose ELK because it was able to classify the FMA both successfully and very quickly (on the order of seconds). We used the “explanations” feature in Protégé to help us understand the sources of logical conflict.
3.2 Clean-up, error types and numbers
Unsatisfiable classes were observed to stem from 6 general types of representational errors:
1. Class X has an existential restriction on a property whose domain is disjoint with X or a superclass of X. X is unsatisfiable.
Class: X
SubClassOf:
P1 some V1
DisjointWith:
C1 (or superclass of C1)
(Object|Data)Property: P1
Domain:
C1
2. Class X is asserted to be a subclass of a class that it is also disjoint with (asserted or inferred). X is unsatisfiable.
Class: X
SubClassOf:
C1
DisjointWith:
C1 (or superclass of C1)
3. Class X has an existential restriction on 2 properties whose domains are disjoint. X is usatisfiable.
Class: X
SubClassOf:
P1 some V1,
P2 some V2
(Object|Data)Property: P1
Domain:
C1
(Object|Data)Property: P2
Domain:
C2
DisjointClasses: C1, C2
4. Class X is asserted to be a subclass of two classes C1 and C2 where C2 is a descendent of C1. Due to the normalization pattern introduced by converter, which makes all primitive sibling classes disjoint, X is asserted to be disjoint from C2 or a superclass of C2. X is unsatisfiable.
Class: X
SubClassOf:
C1,
C2
Class: C2
SubClassOf:
C1
DisjointClasses: X, C2
5. Class X is asserted to be a subclass of two classes C1 and C2. C1 and C2 have a common ancestor. Because the children of that ancestor are asserted to be disjoint (again the normalization pattern), so too must C1 and C2 be disjoint. X is unsatisfiable.
Class: X
SubClassOf:
C1,
C2
Class: C1
SubClassOf:
C3 (or subclass of C3)
Class: C2
SubClassOf:
C4 (or subclass of C4)
DisjointClasses: C3, C4
6. An anonymous class expression, created to accommodate an n-ary or attributed relationship from the Frames model, is asserted to be a subclass of a class representing a complex relationship-type (a mirror of the reification in Frames). While the anonymous class expression is a potential collection of physical anatomical entities (things where has_mass=True), the relationship-type class is a non-physical anatomical entity (things where has_mass=False). Therefore the class expression is unsatisfiable.
This error types differs from the others in that it is an error introduced by the converter itself (inappropriate superclass assignment). The previous error types are errors that already existed in Frames, but were unidentified. This illustrates the curatorial potential to be gained by maintaining the FMA in OWL and utilizing an OWL reasoner.
Not discussed above, but related to some of the errors seen, was an issue with disjoint classes. When the FMA was converted, all primitive sibling classes were made disjoint. In the interim between the conversion and the clean-up of unsatisfiable classes, some classes were moved in the isa hierarchy (i.e. superclass assertion changed). Changing the asserted superclass in Protégé does not change any disjoint classes axioms it is referenced in. Changes such as re-assigning a class from its current parent to a more specific descendent introduces an error (type #4 above). During the clean-up we wrote a script to re-assert the normalization pattern.
Table 1 shows the progress of the clean-up. Numbers contained in Table 1 pertain to counts of unsatisfiable classes in the FMA. However, they may not be complete counts, rather they refer to counts of unsatisfiable classes as identified by ELK. Errors were identified and fixed using the Protégé explanations workbench. Though we've categorized the types of errors discovered, these cannot be selected on nor counted via the explanations tool. Therefore, we were unable to fix the errors one type at a time, but proceeded one explanation at a time.
Table 1.
Unsatisfiable Class Clean-Up Progress
| Description | Unsatisfiable | Total | % Unsatisfiable |
|---|---|---|---|
| Initial conversion | 72659 | 100080 | 73% |
| Post clean-up of types 1, 2, and 3 errors | 73459 | 103847 | 71% |
| Post set primitive siblings disjoint correction | 73438 | 103847 | 71% |
| Post clean-up type 6 errors | 3 | 103857 | <1% |
| Post clean-up type 4 errors | 1 | 103857 | <1% |
| Post clean-up type 5 errors | 0 | 103857 | 0% |
Table 1 appears to suggest that little progress was made cleaning up types 1, 2, and 3 errors, or resetting sibling class disjointness. Rather it looks as though almost all of the clean-up involved fixing type 6 errors. This is not necessarily the case. The FMA is a network of classes highly connected via existential restrictions. For example, the class Heart is a subclass of (regional_part some Right_atrium). Therefore if Right_atrium is unsatisfiable then so is Heart (and anything similarly connected to Heart). So, one unsatisfiable class may cascade to many. It wasn't until we cleaned up all the errors in a large connected sub-network that we saw real changes in the numbers.
The result of the conversion and post-conversion of the FMA is a single OWL file that classifies quickly under the ELK reasoner in Protégé. Because of this fast classification we are able to keep the ELK reasoner active as we enter new knowledge in Protégé, which allows us to correct EL level logical errors at the time of content creation. We are thus already seeing the benefits of converting to OWL in our own work.
In our current workflow we use Protégé to edit and add content to the current OWL version of the FMA. New releases of the FMA are released via our website every 3 months.
4 Using the Converted FMA
Many groups have used the FMA in the past. In fact, a Google Scholar search for “biomedical informatics” lists the primary FMA paper (1) first, with 1038 citations as of this writing, surpassing even the number of citations to the primary textbook in the field. Because of this widespread use, and because the conversion of the FMA to OWL is a major change from the Frames version, we discuss in this section the changes that will likely need to be made in applications, illustrated by changes that we needed to make to our own applications.
Although we have not done a systematic review of the papers that cite the FMA, we have observed various types of uses based on samples of these papers, conversations and collaborations with users, and our own experience.
4.1 Controlled terminology
The primary use of the FMA is as a controlled terminology. FMA IDs are either used directly in other ontologies and terminologies, or are declared to be equivalent to existing IDs. FMA IDs are also often used for data annotation, as for example regions corresponding to anatomical structures in 2-D or 3-D images (18). Because the FMA was initially in Frames and not open source, many other ontologies and terminologies invented their own anatomy axes, with the result that these ontologies and data are annotated with IDs that are different than those provided by the FMA, yet are semantically equivalent to them. Now that we provide definitive FMA IRIs that are based on FMA IDs, existing ontologies and annotated data can be programmatically updated by either 1) replacing their existing FMA IDs with FMA IRIs, 2) providing external mappings for FMA IDs, or 3) in the case of ontologies, providing the FMA IRIs as annotations or as OWL Equivalents. These kinds of mappings will always be required as the semantic web evolves into a global knowledge base by linking together many local efforts.
4.2 Reasoning
Most uses of the FMA for reasoning (as for example, classification) have been done by informatics researchers. In fact one of the main reasons for efforts by others to convert the FMA to OWL was that the Frames version was not amenable to reasoning. Since the FMA is now natively in OWL these research efforts should be greatly facilitated without major changes.
4.3 Queries
Many ontologies and terminologies are made accessible to applications via query, in particular via SPARQL (19). For example, in our own earlier work we periodically converted the Frames version of the FMA to OWL Full, using a script developed by Noy (10). We then embedded the OWL Full version in an Apache Jena triple store, and provided a SPARQL query service that could in turn be accessed by our Query Integrator (20), which permits SPARQL queries over the ontology to be combined with XQuery queries over data annotated with FMA terms or IDs.
We have built several applications using this infrastructure, including our current FMA browser that accesses saved SPARQL queries over the FMA to create an interactive browser (21); a brain map integration system that provides “intelligent” querying of brain images annotated with neuro FMA terms (22); and an ontology-based scene generator that combines queries over the FMA with a library of 3-D models to construct interactive web based 3-D scenes for anatomy education and data visualization (23).
Adapting these applications to the converted FMA involved two main tasks: relating the annotated images to the new FMA IRIs, and adapting the queries. Since the images were annotated with FMA IDs or strings representing FMA concepts we did not try to change the annotations, but instead used saved queries to deconstruct the FMA IRIs to components that match the embedded annotations.
However, adapting the earlier SPARQL queries potentially would have required more effort since these queries took advantage of the fact that in OWL Full classes are directly related (as in Liver has_part Right_lobe_of_liver), whereas in our OWL 2 conversion classes are generally related via existential restrictions (e.g. Liver subclassOf [has_part some Right_lobe_of_liver]).
In order to minimize the changes to the existing queries we took advantage of the punning feature of OWL 2, in which a single identifier (IRI) can refer to both a class and an individual, and individuals can be directly related via properties such as has_part. Each week we therefore programmatically take the most recent version of the FMA in OWL2, compute a separate punning file, and use both to populate an Apache Jena triple store that is then available to our Query Integrator. Using these puns allowed us to make only minimal changes to our previous saved SPARQL queries.
4.4 Summary
Because most informatics research using the FMA is already in OWL we expect that these researchers will find the converted FMA easier to use than the Frames version since they won't have to create their own OWL versions. Since many existing ontologies already provide FMA ID annotations, their developers can use the existing IDs to construct or provide mappings to the new FMA IRIs, which are based on the FMA IDs. Similarly, data providers can either construct or provide mappings to new FMA IRIs within their data based on the existing annotations. Applications that have previously used SPARQL to access the FMA Full translation of FMA Frames should not have to make extensive changes to their queries if they take advantage of the puns that are added to our FMA triple store each week. Thus, we believe that extensive changes will not be required for applications that are currently using the FMA.
5 Discussion
In this report we have described the methods we used to convert the FMA to OWL2, and then to clean up the converted FMA so that it classifies under an EL reasoner.
The methods are embodied in a configurable Java program with relation-centric conversion rules. As such the program has the potential to generalize to other Frames ontologies. In particular we have tested this generality by showing that the same program can be used to convert our OCDM from Frames to OWL2, with the only changes being new Java classes in the convertor that are activated via new directives in the configuration file. Further validation of the generality of the program will occur as others attempt to use it to convert their own Frames ontologies (of which there are still several). The conversion software is open source (see Section 7: Availability below).
5.1 Beyond conversion
We do not claim that the conversion methods lead to a complete, logically consistent OWL representation of a Frames ontology. Rather, our goal was to capture all the information specified in the original Frames ontology, in a syntax that permits use by the tools being developed for OWL ontologies, and in a manner that supports interoperability with other community ontologies. The converted ontology then is not only interoperable with the semantic web, it also provides a starting point for post-conversion cleanup of the type we describe in section 3.
Any post-conversion cleanup of a converted Frames ontology is likely to be highly idiosyncratic to the particular ontology content. However, in all cases use of a polynomial-time reasoner like ELK, plus the explanation capability of Protégé, should prove useful. Since we have not yet cleaned up the converted OCDM we will be able to test the utility of these tools when we apply the same methods to cleaning up the OCDM.
5.2 Some remaining issues
The following sections give examples of some of the issues that remain in the FMA following our post-conversion cleanup. There are undoubtedly others that will be discovered by the ontology community.
Classification under more powerful reasoners
As noted in section 3 we have only cleaned up the FMA to classify under the ELK reasoner because of the intractability of the FMA to full DL reasoning. Creating a full DL reasoner over the FMA that completes in tractable time should provide a challenge of interest to semantic web researchers.
External references
One of our stated goals was that the FMA and OCDM in OWL should contribute to the semantic web and a global knowledge base. To do so it must be able to interoperate with other ontologies, rather than recreating their content. This is particularly relevant for the OCDM as there are many overlapping resources already available. Referencing external OWL concepts from Frames was achieved by tagging FMA classes with unique identifiers for their corresponding OWL counterparts. However, that means that we have classes in the FMA and in the OCDM Frames models that are equivalent to classes defined elsewhere.
These duplicate classes are created in the OWL models as well, as it is a direct conversion from Frames. References to external classes are carried over as class annotations. However, there are preferred strategies for handling this in OWL. For example, we could remove a class definition and replace all references to it with direct references to the external class. Or we could keep the class and declare it as equivalent to or as a subclass of a class defined elsewhere. We will explore the best approaches to eliminating this redundancy and amend our OWL ontologies in future augmentations.
Some values from
One thing that cannot be determined from the Frames model is whether a connection between a class and another class represents exactly one (e.g. every heart has exactly one right atrium) or one or more. There are at least two cases where a class-to-class relationship might mean one or more:
Non-countable parts
Epithelium_proper_of_stomach_has_part Surface_mucous_cell_of_stomach. Clearly there isn't only one cell that is part of the epithelium of stomach. But equally clearly, we are not going to enumerate those cells as Epithelial_cell_of_stomach_1, ...
Generalized parts
Lung has part Lobe_of_lung. This statement is intended to indicate that lungs have lobes, not necessarily just one. But this isn't easily distinguishable, automatically, from the case of heart and right atrium.
We chose to convert these to existential restrictions (e.g. “some”, 1 or more). Though this isn't the strongest model we could have made in some cases (i.e. the case of Heart and Right atrium) it is still correct.
Property duplication
Several of the OCDM sub-ontologies use what are conceptually the same slots. But, rather than include common relations ontology, they each reintroduce the slot definitions. So the converter does the same thing, basing the base portion of the IRI on the defining ontology. Thus, the fully converted OCDM would have cho:regional_part and cmo:regional_part properties that should be the same but are not. It would be better for the sake of query and/or reasoning to use the same property. But, the converter is going off of the Frames model. Of course, this could be addressed post conversion by creating a common super-property of the two (preferably something from a community accepted relation ontology). But this is just an example of how Frames issues get duplicated in OWL.
Slots with no values
In Frames, there is a notion of a class having a slot, even when that slot has no value(s). What does this really mean? A class with a slot without value(s), suggests that the class does not stand in that relationship to anything. But this is the same interpretation as a class which simply lacks the slot altogether.
Mandible does not have a ‘branch’ slot. This makes sense since mandibles are not the kind of things that have branches. But, for example, all arteries have a branch slot. These are the sorts of things that do have branches. But this isn't interpreted as all arteries “must have” a branch. Some arteries have defined branches, like the Facial artery, which has branch Tonsillar artery. But others do not (the Tonsillar artery has no defined branches in the FMA).
Thus, in Frames the semantics of a slot without values is not clearly defined. And indeed, through conversations with content authors, the semantics are to be interpreted in a context dependent manner. In closed world semantics, a model is deemed to be complete and anything unsaid is considered untrue. But, empty slots are often considered as tacit acknowledgement that the model is not yet complete, despite the semantics of interpretation. At other times, the slot is complete, and the assertion that a slot has no values is deliberate. This occurs when modeling a class that is the kind of thing that could have a relationship type (like Arteries can have branches), but that does not have that relationship type (though Tonsillar artery is an Artery, it is a terminal branch and has no defined branches of its own). At times it means that a slot was applied too broadly (this is a modeling error in Frames). At other times it occurs because the values differ by subclass. This generally occurs higher in the subclass hierarchy (i.e. the class Artery has the branch property, but with no values, as the branches differ by specific artery).
In any case, an empty slot cannot always be interpreted to mean that there is a value that has not yet been defined. Sometimes it means there are no values, and sometimes it means that the values differ by subclass (a common superclass of all branches could have been asserted, like Artery in this case).
In OWL, on the other hand, there is no notion of a class having a property potentially without values. If adding a slot to a class in Frames (independent of values) means that the class is the sort of thing that might have that slot, we don't really need to say anything in OWL to capture this. In the open world, any class may have any property, even if it has not been asserted. Now, we can prevent the assertion that a class must have a specific property (or more precisely, if the aforementioned assertion is stated, then the class become unsatisfiable). For example, we could say that the domain of the branch property is Artery. And we could say that the class Muscle_tissue is disjoint from Artery. If we further say Muscle_tissue subClassOf (branch some X), it would be inferred that Muscle_tissue is unsatisfiable. But, in OWL, we never list the properties that can hold for individuals in a class, only those that must hold.
We have found that this issue is a stumbling block for people moving from Frames to OWL. The first thing many want to do is list the properties that apply to a class, before saying what their values are. That is a Frames approach, not an OWL approach.
6 Conclusions
The primary result of the work described in this report is an OWL2 version of the FMA, which captures all or as much as possible of the information in the Frames model, which classifies under an EL reasoner, and which is represented in a syntax that is compatible with the global knowledge base evolving through the semantic web. Secondary results are an OWL2 version of the OCDM that we will clean up in the near future to classify under EL, and generalizable conversion code that can be applied to other Frames ontologies.
Although there are other OWL versions of the FMA, none of them were produced by the FMA authors themselves, so are not as complete or as faithful to our modeling goals. In addition none of the other developers was in a position to change the content of the FMA so it would classify under an EL reasoner. For these reasons we propose that the version of the FMA (and OCDM) that we have developed in this work should become the official release of the OWL FMA reference ontology, and should replace all others.
The availability of a single OWL version of the FMA that is continuously updated by its authors and that utilizes a single scheme for URI's should go a long way, not only towards providing a common and complex artifact for ontology research, but also towards achieving interoperability among anatomy representations.
Anatomy is fundamental to understanding in biomedicine, but even more importantly for informatics, it is fundamental as a framework for organizing most other biomedical information because most physiological processes and diseases involve or are manifestations of anatomical entities. Thus, by providing a consistent, standardized version of the most widely used anatomy reference, the evolution of a semantic-web enabled structural information framework for organizing and linking biomedical information should be greatly facilitated.
HIGHLIGHTS.
Foundational Model of Anatomy (FMA) converted to OWL2
Ontology for Craniofacial Development and Malformation (OCDM) converted to OWL2
Frames to OWL2 ontology converter is configurable and reusable
Conversion is non-lossy but resulting model will require further refinement
Acknowledgment
This work was supported by the National Institutes of Health grant number DE24417.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7 Availability
The FMA webpage is http://www.si.washington.edu/projects/fma, which in turn links to a Summary of FMA Resources http://www.si.washington.edu/content/summary-fma-resources.
Links provided on this page include various mailing lists, an issue tracker, the FMA Browser (updated weekly), the latest release (updated approximately every 3 months), and the creative commons license under which the FMA is released. The FMA is also available via BioPortal.
The OCDM webpage is http://www.si.washington.edu/projects/ocdm, which provides a link to the latest version. The OCDM is also available via the FaceBase Consortium Hub https://www.facebase.org/ocdm/.
The conversion code is available at http://trac.biostr.washington.edu/trac/wiki/framesToOwl.
References
- 1.Rosse C, Mejino JLV. A reference ontology for bioinformatics: the Foundational Model of Anatomy. J Biomed Inform. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Rosse C, Mejino JLV. The Foundational Model of Anatomy Ontology. In: Burger A, Davidson D, Baldock R, editors. Anatomy Ontologies for Bioinformatics: Principles and Practice. Springer; 2007. pp. 59–117. [Google Scholar]
- 3.Noy N, Fergerson R, Musen M. The Knowledge Model of Protégé-2000: Combining Interoperability and Flexibility. In: Dieng R, Corby O, editors. Knowledge Engineering and Knowledge Management Methods, Models, and Tools [Internet] Springer; Berlin Heidelberg: 2000. pp. 17–32. (Lecture Notes in Computer Science; vol. 1937). Available from: http://dx.doi.org/10.1007/3-540-39967-4_2. [Google Scholar]
- 4.OKBC Working Group Open Knowledge Base Connectivity [Internet] 2005 Available from: http://www.ai.sri.com/~okbc/
- 5.World Wide Web Consortium [2016 Jan 12];OWL 2 Web Ontology Language Primer (Second Edition) [Internet] Available from: http://www.w3.org/TR/owl2-primer/
- 6.Berners-Lee T, Hendler J, Lassila O. The semantic web. Sci Am. 2001;284:34–43. [Google Scholar]
- 7.Golbreich C, Grosjean J, Darmoni SJ. The Foundational Model of Anatomy in OWL2 and its use. Artif Intell Med. 2013;57(2):119–32. doi: 10.1016/j.artmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Golbreich C, Grosjean J, Darmoni SJ. Proceedings of the 13th Conference on Artificial Intelligence in Medicine [Internet] Springer-Verlag; AIME'11; Berlin, Heidelberg: 2011. The FMA in OWL 2. pp. 204–214. Available from: http://dl.acm.org/citation.cfm?id=2040981.2041012. [Google Scholar]
- 9.Golbreich C, Zhang S, Bodenreider O. The foundational model of anatomy in OWL: Experience and perspectives. Web Semant Online. 2006;4(3):181–95. doi: 10.1016/j.websem.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noy NF, Rubin DL. Translating the Foundational Model of Anatomy into OWL. J Web Semant. 2008;6:133–6. doi: 10.1016/j.websem.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dameron O, Rubin DL, Musen MA. Proceeding, American Medical Informatics Association Fall Symposium [Internet] Washington, DC: 2005. Challenges in converting frame-based ontology into OWL: the Foundational Model of Anatomy case-study. pp. 181–5. Available from: http://sig.biostr.washington.edu/share/sigweb/pubs/DameronAMIA2005.pdf. [PMC free article] [PubMed] [Google Scholar]
- 12.Brinkley JF, Borromeo C, Clarkson M, Cox TC, Cunningham ML, Detwiler LT, et al. The Ontology of Craniofacial Development and Malformation for translational craniofacial research. Am J Med Genet Part C Semin Med Genet. 2013;164(4):232–45. doi: 10.1002/ajmg.c.31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shadbolt N, Hall W, Berners-Lee T. The semantic web revisited. IEEE Intell Syst. 2006:96–101. [Google Scholar]
- 14.World Wide Web Consortium [2016 Jan 12];SKOS Simple Knowledge Organization System Reference [Internet] Available from: http://www.w3.org/TR/skos-reference/
- 15.World Wide Web Consortium [2016 Jan 12];Defining N-ary Relations on the Semantic Web [Internet] Available from: http://www.w3.org/TR/swbpn-aryRelations/
- 16.World Wide Web Consortium [2016 Jan 13];OWL 2 Web Ontology Language Profiles (Second Edition) [Internet] Available from: http://www.w3.org/TR/owl2-profiles/
- 17.Kazakov Y, Krotzsch M, Simancik F. The incredible ELK. J Autom Reason. 53(1):1–61. [Google Scholar]
- 18.Wang KC, Salunkhe A, Morrison J, Lee P, Mejino JLV, Detwiler LT, et al. Ontology-based image navigation: exploring 3T MR neurography of the brachial plexus using AIM and RadLex. Radiographics. 2015;35(1):142–51. doi: 10.1148/rg.351130072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Wide Web Consortium SparQL query language for RDF [Internet] 2010 Available from: http://www.w3.org/TR/rdf-sparql-query/
- 20.Brinkley JF, Detwiler LT. A query integrator and manager for the Query Web. J Biomed Inform. 2012;45:975–91. doi: 10.1016/j.jbi.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarkson M. [2016 Jan 15];Foundational Model Browser [Internet] Available from: http://www.si.washington.edu/content/foundational-modelbrowser.
- 22.Detwiler LT, Suciu D, Franklin JD, Moore EB, Poliakov AV, Lee ES, et al. Distributed XQuery-based integration and visualization of multimodality data: Application to brain mapping. Front Neuroinformatics [Internet] 2009:3. doi: 10.3389/neuro.11.002.2009. Available from: http://sigpubs.biostr.washington.edu/archive/00000234/ [DOI] [PMC free article] [PubMed]
- 23.Nilsen T. Ontology-driven education: Teaching anatomy with intelligent 3D grames on the web [Internet] [PhD] University of Washington; 2015. Available from: http://hdl.handle.net/1773/33983. [Google Scholar]