Abstract
Aging results in a significant decline in aerobic capacity and impaired mitochondrial function. We have tested the effects of moderate physical activity on aerobic capacity and a single bout of exercise on the expression profile of mitochondrial biogenesis, and fusion and fission related genes in skeletal muscle of human subjects. Physical activity attenuated the aging-associated decline in VO2 max (p<0.05). Aging increased and a single exercise bout decreased the expression of nuclear respiratory factor-1 (NRF1), while the transcription factor A (TFAM) expression showed a strong relationship with VO2max and increased significantly in the young physically active group. Mitochondrial fission representing FIS1 was induced by regular physical activity, while a bout of exercise decreased fusion-associated gene expression. The expression of polynucleotide phosphorylase (PNPase) changed inversely in young and old groups and decreased with aging. The A2 subunit of cyclic AMP-activated protein kinase (AMPK) was induced by a single bout of exercise in skeletal muscle samples of both young and old subjects (p<0.05). Our data suggest that moderate levels of regular physical activity increases a larger number of mitochondrial biogenesis-related gene expressions in young individuals than in aged subjects. Mitochondrial fission is impaired by aging and could be one of the most sensitive markers of the age-associated decline in the adaptive response to physical activity.
Keywords: Aging, Exercise, Skeletal muscle, Mitochondrial biogenesis, Fusion and fission
1. Introduction
Mitochondrial networks have a fundamental importance in energy generation, and play key roles during myogenesis, fiber maturation, the advancement of neuromuscular pathologies, apoptosis and natural fiber aging in skeletal muscle. Studies on human and animal muscle models have demonstrated that mitochondrial dysfunctions are linked to excessive generation of reactive oxygen species (ROS); metabolic alterations, including changes in energy metabolism; excitation–contraction coupling; ion homeostasis; and muscle fiber contraction/relaxation. Proteomic analyses have yielded over 900 distinct mitochondrial proteins in human muscle and all, except the 13 encoded by mitochondrial DNA, are imprinted on nuclear DNA (Pagliarini et al., 2008).
In skeletal muscle fibers, the mass of mitochondria is dependent on the muscle subtype. The number and functions of mitochondria required to maintain fiber homeostasis are determined by regulatory circuits/programs that are under synchronized regulation between the mitochondrial and nucleic genes. This regulation is required in order to maintain respiratory electron transport complexes and proteins that are required for physiological functions of mitochondrial networks, such as oxidative phosphorylation, ionic homeostasis, and mitochondrial uncoupling (thermogenesis). Most reports suggest that aging alters the mitochondrial efficiency and thus cellular metabolism (Kelly, 2011; Sahin et al., 2011). However, the detailed mechanisms are not completely understood (Bratic and Trifunovic, 2010).
We have recently documented an increased oxidative challenge in human muscle fibers in response to exercise (Radak et al., 2011). Specifically, we found a sustained increase in levels of ROS byproducts, lipid peroxidation and intrahelical 8-oxoguanine, and expression of anti-oxidant enzymes including Cu,ZnSOD and MnSOD in muscles of sedentary young and old subjects in response to acute physical activity. We also showed an increased activity, by post-translational modification of 8-oxoguanine DNA glycosylases and apurinic/apyrimidinic endonuclease-1, key repair proteins for excision and repair of 8-oxoguanine, in the DNA base excision repair pathways (Radak et al., 2011). Moreover, the SIRT3 expression is increased with physical fitness levels as found in human muscle biopsies of young but not old subjects. SIRT3 is a downstream target of PGC-1 and one of the regulators of mitochondrial ROS production (Kong et al., 2010; Qiu et al., 2010) and apoptosis (Bell et al., 2011). These observations and those by others suggest an oxidative stress response by mitochondria to acute physical exercise (Davies et al., 1982). Mitochondrial biogenesis is a very complex multifactorial process that involves nuclear and mitochondrial DNA-encoded proteins, and requires activation of mitochondrial transcription factor A (TFAM) (Chow et al., 2007; Scarpulla, 2008); nuclear respiratory factors (Nrf1, Nrf2) (Cartoni et al., 2005; Chow et al., 2007; Scarpulla, 2008) cyclic AMP-activated protein kinase (AMPK) (Sriwijitkamol et al., 2007) peroxisome proliferator-activated receptors (PPARs) and peroxisome proliferator activated receptor gamma coactivator 1-alpha (PGC-1α) (Arany, 2008; Scarpulla, 2008). Regular physical activity has been suggested to attenuate the progress of aging, including mitochondrial biogenesis (Safdar et al., 2011). However, data from human studies are limited. Moreover, age and physical activity dependent activation of the most important gene expressions in mitochondrial biogenesis need to be investigated, in order to properly evaluate role pre- and post translation processes. Human studies have a number of limitations, including the small size of biopsy samples, but the level of gene expression is important to study the efficiency of signaling pathways and protein synthesis.
The mitochondrial network is a dynamic system which can be altered by outer membrane mitofusions 1 and 2 proteins (Mfn1 and Mfn2) (Westermann, 2010) and mitochondrial fission proteins (FIS1 and 2) (Ding et al., 2010; Lee et al., 2007; Romanello et al., 2010; Twig et al., 2008). Moreover, the activity of uncoupling proteins (UCPs) and polynucleotide phosphorylase (PNPase) (Wang et al., 2010) also significantly impacts mitochondrial function. The effect of aging and/or physical activity on the organization of the mitochondrial network in human tissue is largely unknown. A single bout of exercise brings about oxidative and metabolic challenge in skeletal muscle, while regular physical activity result in a well characterized adaption to these challenges (Radak et al., 2009). Mitochondria are highly dynamic organelles which fuse and divide, and translocate through microtubules to more efficiently serve crucial cellular processes, such as respiration, heme biogenesis and apoptosis among others (Otera and Mihara, 2011).
Therefore, in the present study we tested the hypothesis that most of the genes that are involved in mitochondrial biogenesis and quality control would show decreased expression with aging. We have postulated that regular physical activity attenuates the age-associated impairments. Moreover, we suggested that a single bout of exercise would be a useful tool to test the adaptive capability, which is believed to decrease with aging.
2. Methods
Twenty four apparently healthy men, volunteered to participate in the present study. A written informed consent was signed by all participants regarding their participation after being informed of all risks, discomforts and benefits involved in the study. Procedures were in accordance with the Helsinki Declaration of 1975 and were approved by the ethical committee of the University of Thessaly. Participants were assigned to one of four groups, six subjects per group: a) young sedentary (YS, 26.0±4.5 yrs), b) young physically active (YA, 30.2±7.9 yrs), c) old sedentary (OS, 63.4±4.7 yrs), and d) old physically active (OA, 62.4±2.9 yrs).
The consequences of physical inactivity were determined by: a) maximal oxygen uptake (VO2max) values. According to the American College of Sports Medicine Guidelines these values are at the 20th percentile for their respective population (ACSM, 2005), b) physical activity scores (<9.0 in the Modified Baecke Questionnaire) (Voorrips et al., 1991), c) which further include a lower muscle capillarization rate, and d) a lower muscle cytochrome C protein content. Chronic physical activity was evidenced by: a) VO2max>35 ml/kg/min for old participants and >45 ml/kg/min for young participants — according to the American College of Sports Medicine Guidelines these values are at the 70th percentile of their respective populations (ACSM, 2005), b) physical activity scores (>over 10.0 in the Modified Baecke Questionnaire) (Voorrips et al., 1991), c) elevated muscle capillarization rate compared to control levels, and d) and higher muscle cytochrome C protein content compared to controls. The value of VO2max is in a certain degree dependent on genetics (Levine, 2008) and therefore, cannot definitively show the exact level of physical activity/inactivity. However, it is one of the most accepted markers of aerobic physical fitness (Abbott, 1994). Moreover, the inverse relationship between VO2max and mortality caused by specific pathologies such as cardiovascular diseases and cancer in aging populations, reflects the impact of VO2max on the quality of life and mortality in aged individuals (Ferreira et al., 2003; Morss et al., 2004; Sawada et al., 2003; Timmons et al., 2010).
The daily physical activity score was measured by the “International Physical Activity Questionnaire” (IPAQ) described by Craig et al. (2003). The daily step number of the subjects was evaluated by pedometry (Omron pedometer HJ-720IT-E2 or HJ-720).
All participants were non-smokers, free of musculoskeletal problems and orthopedic/neuromuscular limitations, had resting blood pressures below 140/85 mm Hg, received no antihypertensive medications, had no signs of cardiovascular/respiratory complications at rest and during a progressive diagnostic graded exercise test (GXT), had normal dietary habits, and did not consume aspirin, antioxidant compounds (including vitamins and minerals), certain medications (i.e., probucol, nebivolol, and anti-inflammatory agents) and alcoholic beverages, at least two weeks prior to the study.
Participants visited the laboratory on three occasions. During the first visit, participants were examined by a trained physician for limiting health complications. Percentage body fat was calculated from seven skinfold measures (chest, midaxillary, triceps, subscapular, abdomen, supraliac, and thigh), using a Harpenden caliper (Harpenden, HSK-BI, British Indicators, UK) on the right side of the body (ACSM, 2005). Subcutaneous skinfold thickness was measured sequentially, in triplicate, by the same investigator, using a standard technique (ACSM, 2005). If readings differed by more than 0.2 mm, a third measure was taken and the mean was recorded.
2.1. Exercise protocol
On the second visit, participants had their body height/weight and skin-folds measured and also underwent a GXT to evaluate VO2max.
VO2peak was determined during a GXT on a treadmill as previously described (Fatouros et al., 2004). All subjects completed the protocol. For most old subjects the exercise was equivalent to intense walking and in some cases light jogging. These subjects were free to RETIRE if they were tired or if there were signs of cardiac distress (based on EKG recordings).
On the third visit, a single bout of exercise WAS included, initially, 45-min of running on the treadmill at 70–75% of VO2max. After 45 min, speed was increased to PRODUCE 90% of VO2max and exercise was terminated at exhaustion (Michailidis et al., 2007). A single bout of exercise to exhaustion was applied to induce significant oxidative and metabolic challenges, which would allow for the testing of possible changes in the adaptive capacity of aging and a sedentary life-style.
All subjects were restricted from exercise two days prior to the single bout of exercise test.
2.2. Biopsy sampling
During the third visit, participants underwent a submaximal treadmill exercise bout to exhaustion. Muscle biopsies were collected before and immediately (about 10–15 min) following exercise. Two muscle specimens (pre- and post-exercise), of approximately 100–120 mg each were obtained from the Vastus lateralis of the same leg of each participant using the needle biopsy technique (Bergström, 1962). The biopsies were obtained approximately 20 cm from the mid patella of the right (dominant) leg with the application of suction (Terzis et al., 2008a).
2.3. Real time quantitative RT-PCR
The samples were immediately frozen in liquid nitrogen and stored at –70 °C. RNA isolation was performed using the Nucleospin® RNA/Protein kit in accordance with the manufacturer’s instructions (Macherey-Nagel, Düren, Germany). RNA concentration, purity and integrity (RIN) were measured by Bioanalizer (Agilent Technologies, USA) and RNA samples were stored at –70 °C. cDNA was synthesized using a cDNA Synthesis kit (Bioline, Luckenwalde, Germany) in accordance with the manufactures’ instructions. Briefly, the reaction conditions were as follows: 1 μg of RNA, 1μl of random primers, 0.5 mM dNTP, 1 μl of RNase inhibitor, and 0.25 μl of 200 μ/μL reverse transcriptase in a final volume of 20 μl, incubated for 60 min at 42 °C. Based on the principle of the Sybr Green detection method, EvaGreen® dye (Biotium, Hayward, CA, USA) was used to detect PCR products.
The thermal cycler instrument used was a Rotor-Gene 6000 (Corbett Research, Australia). ImmoMix™ (Bioline, Luckenwalde, Germany) complete ready-to-use heat-activated 2× reaction mix was used to amplify the target sequences. Only water, template, EvaGreen® and primers were added (final volume of 50 μL) and then pre-heated to 95 °C for 10 min. The PCR conditions were as follows: 95 °C for 10 s, 60 °C for 15 s, and 72 °C for 20 s.
Human beta-actin gene served as an endogenous control gene. The validity of the signal was evaluated by melting analysis and agarose gel electrophoresis. To eliminate the differences in signal between different PCR runs, three control cDNAs were selected to validate the samples of each run. The integrity, purity and concentration of the obtained ribonucleic acids (RNAs) were determined with an Agilent 2100 Bioanalyzer. The absorbance ratio of the RNAs solution at wavelengths 260 nm/280 nm ranged from 2.01 to 2.27, and at wavelengths 260 nm/230 nm ranged from 1.89 to 2.1. The RNA integrity numbers (RINs) did not indicate degraded RNA; RINs ranged from 7.1 to 8.9. The average value of these samples served as a reference value to normalize all PCR runs. The following primers were used:
| Primers | Sequences |
|---|---|
| H-FIS1-F | ACCTGGCCGTGGGGAACTACC |
| H-FIS1-R | AGTTCCTTGGCCTGGTTGTTCTGG |
| H-LONP1-2-F | ATGGAGGACGTCAAGAAACG |
| H-LONP1-2-R | GATCTCAGCCACGTCAGTCA |
| H-MFN1-F | AGCTGGCTGTCTTGTACGTGTGT |
| H-MFN1-R | TGTAGTGACATCTGTGCCTGGACTGT |
| H-NRF1-F | CGCTCTGAGAACTTCATGGAGGAACAC |
| H-NRF1-R | GCCACATGGACCTGCTGCACTT |
| H-PNPT1-F | ACCGCTCTTTCCAGCTGGCTACT |
| H-PNPT1-R | GAGAGGGCTACGGAAGCGCCA |
| H-PPARGC1A(PGC-1a)-F | GTGAAGACCAGCCTCTTTGC |
| H-PPARGC1A(PGC-1a)-R | TCACGTCTCCATCTGTCAGC |
| H-PRKAA2-F | CGCCTCTAGTCCTCCATCTG |
| H-PRKAA2-R | AGCTCGGTAAACTTCAGCCA |
| H-PRKAB2-F | AAAGATTCAAATCCCCACCC |
| H-PRKAB2-R | TCAGCCTTCCAGTCTCAGGT |
| H-PRKAG1-F | CCACAAGCTCCAAATTGGTT |
| H-PRKAG1-R | GCAGACAAGCGGTTTAAAGG |
| H-PRKAG2-F | GAAGCAGAACCTGGATGAGC |
| H-PRKAG2-R | TGCTTCATTTACCACCACCA |
| H-PRKAG3-F | TTGTCATTTCAAGGCTGCTG |
| H-PRKAG3-R | GCACAAGTCTCTTGCCACAA |
| H-SIRT1-F | TGCGGGAATCCAAAGGATAATTCAGTGTC |
| H-SIRT1-R | CTTCATCTTTGTCATACTTCATGGCTCTATG |
| H-SIRT3-F | GTCGGGCATCCCTGCCTCAAAGC |
| H-SIRT3-R | GGAACCCTGTCTGCCATCACGTCAG |
| H-UCP3-F | GAGGAGGAGGGATTCTGGTC |
| H-UCP3-R | CAGGGCTTCTTTAGCACAGG |
| H-TFAM-F | TGCCTCATCCACCGGAGCGA |
| H-TFAM-R | CACAAAACTGAAGGGGGAGCGCA |
2.4. Activity assay
Cytochrome c (COX) oxidase activity was measured in muscle homogenates as previously described (Rooyackers et al., 1996). The activity of citrate synthase (CS) was assessed as reported earlier (Radak et al., 2000). Measurement of the capillary density (capillaries/mm2) was performed with an image analysis system (ImagePro; Media Cybernetics, Inc., Bethesda, Md.) at a known and calibrated magnification as previously described (Terzis et al., 2008b).
2.5. Statistical analyses
Statistical significance was assessed by the General Linear Model — Nested design ANOVA, followed by Duncan’s post hoc test. The significance level was set at p<0.05.
3. Results
3.1. Physiological data of the subjects
3.1.1. YS vs OS
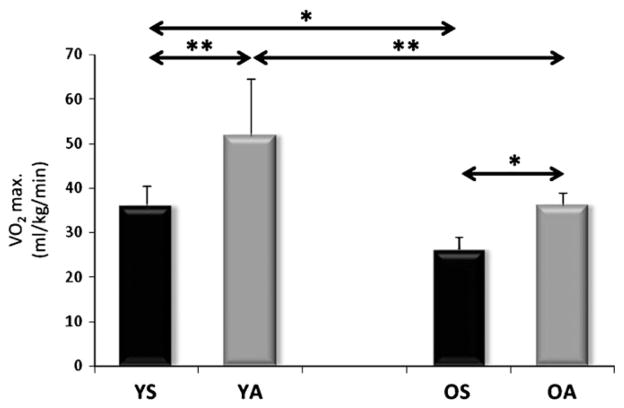
The body fat content of old sedentary individuals was higher than that of young subjects (p<0.05). The daily physical activity scores, judged by a questionnaire specifically designed for this purpose, showed that for aged individuals the rate of physical activity was decreased significantly (Table 1). Accordingly, the VO2max levels also showed (p<0.05) lower values for the aged subjects (Fig. 1).
Table 1.
| Young
|
Old
|
|||
|---|---|---|---|---|
| Sedentary | Active | Sedentary | Active | |
| VO2max (ml/kg/min) | 35.9±4.71,3 | 51.8±7.91,4 | 25.1±3.02,3 | 37.1±2.92,4 |
| Exercise time (min) | 47.8+2.01 | 53.2+1.01 | 45.8+2,72 | 48.2+2,42 |
| Capillarization (cap/fiber) | 1.5±0.21,3 | 1.8±0.061,4 | 1.2±0.12,3 | 1.5±0.22,4 |
| Citrate synthase(μmol/min/g) | 25.1±2.61,3 | 39.4±3.31,4 | 12.3±1.62,3 | 26.9±4.72,4 |
| Cytochrome C oxidase activity (μmol/min/g tissue) | 8.5±0.71,3 | 16.2±0.91,4 | 6.4±0.52,3 | 11.9±0.72,4 |
| Physical activity score (METS/day) | 893.5±173.61,3 | 2951.8±381.41,4 | 525.7±92.42,3 | 2509.8±315.12,4 |
| Steps/day | 7916.2±325.11,3 | 16.820.4±895.61,4 | 4855.9±414.72,3 | 12.365.8±649.12,4 |
Denotes significant difference between young sedentary and young active at p<0.05.
Denotes significant difference between old sedentary and old active at p<0.05
Denotes significant difference between the two sedentary groups at p<0.05.
Denotes significant difference between the two active groups at p<0.05.
Fig. 1.
VO2max data for the subjects. Regular physical activity increased the VO2max in both age groups, and aged subjects have significantly lower levels of VO2max. Values are means±SD for six subjects per group. * p<0.05, ** p<0.01. YS: young sedentary, YA: young physically active, OS: old sedentary, OA: old physically active.
3.1.2. YA vs OA
Regular physical activity increased VO2max levels to a larger degree in young (p<0.01) than in older age (p<0.05, Fig. 1). However, when the percentage differences between the sedentary and active groups were evaluated, the aged active individuals show greater differences from the sedentary values than young subjects. These differences are due to standard deviation in the groups. These data further suggest that even in older age VO2max can be maintained by regular physical activity.
The comparisons of YS to YA and OS to OA revealed that active individuals in both age groups had decreased body mass and body fat, and increased METS (Table 1). COX activity, CS activity and the rate of capillarization were all increased in both age groups as a result of regular physical activity (Table 1).
As expected, old physically active subjects had higher levels of body mass, body fat and body mass index, but lower levels of daily physical activity, as expressed by the metabolic equivalent of tasks ((METS) Table 1).
The assessment of VO2max levels showed that regular physical activity increased VO2max in both age groups (Fig. 1). However, the data also revealed that VO2max values are much lower than those found for athletes or master athletes, underscoring that the load of physical activity used in this study was moderate.
3.2. Gene expression levels
3.2.1. YS vs OS
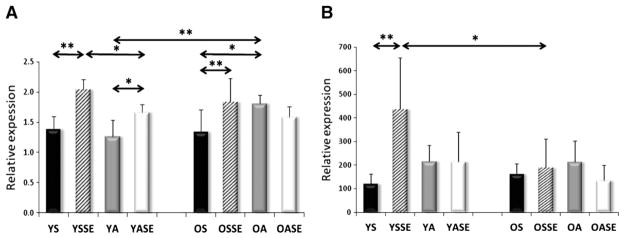
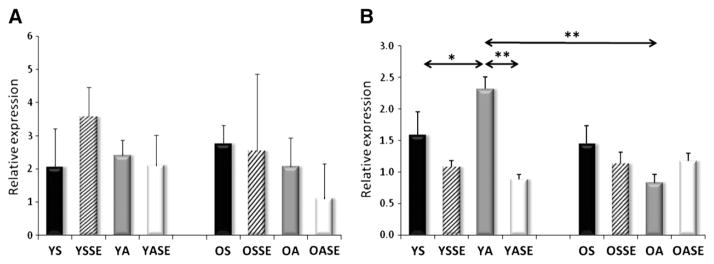
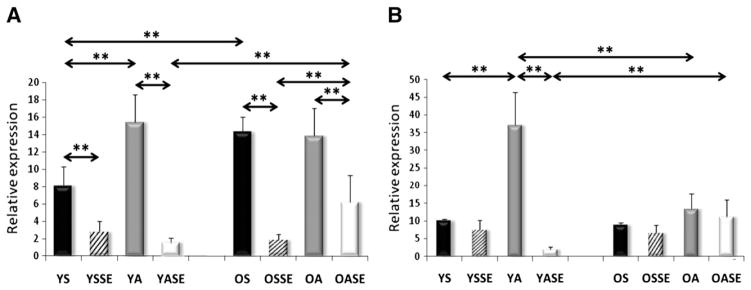
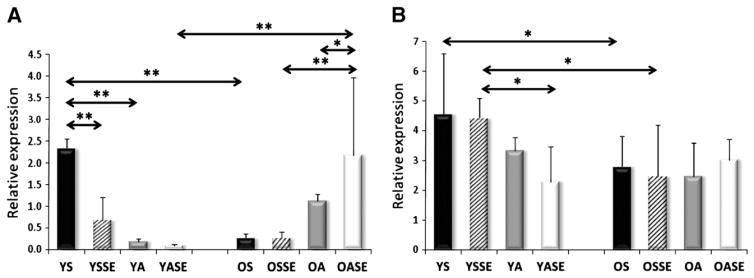
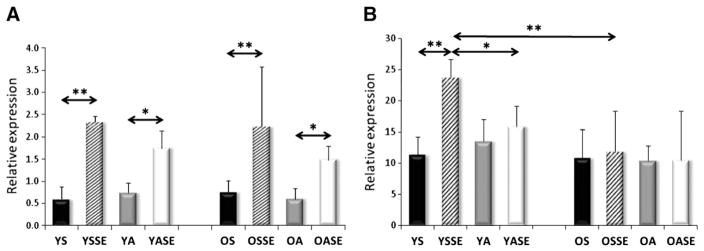
Significant differences were not observed in the mRNA levels of young and old sedentary individuals for SIRT1, PGC-1a, AMPK subunits, TFAM, Fis1, Mfn1, and Lon protease (Figs. 2–7). On the other hand, aged individuals showed elevated expression of NRF1 (p<0.01, Fig. 4A) and depressed levels of PNPase and UCP3 expression (p<0.01, Fig. 6).
Fig. 2.
mRNA expression of SIRT1 and PGC-1a. A single bout of exercise significantly increased the expression of SIRT1 in the young sedentary group and tended to show an increase in the active group. Similarly to SIRT1, PGC-1a only increased in YSSE group and differences were found between YSSE and OSSE groups. Values are means±SD for six subjects per group. * p<0.05, ** p<0.01. YS: young sedentary, YSSE: young sedentary with a single bout of exercise, YA: young physically active, YASE: young physically active with a single bout of exercise, OS: old sedentary, OSSE: old sedentary with a single bout of exercise, OA: old physically active, OSSE: old physically active with a single bout of exercise.
Fig. 7.
The levels of Lon protease (A) and SIRT3 (B) mRNA. The mitochondria localized Lon protease mRNA levels were not altered in the examined groups, while the mRNA levels of SIRT3 increased with regular physical activity in the young group. Values are means±SD for six subjects per group. * p<0.05, ** p<0.01. YS: young sedentary, YSSE: young sedentary with a single bout of exercise, YA: young physically active, YASE: young physically active with a single bout of exercise, OS: old sedentary, OSSE: old sedentary with a single bout of exercise, OA: old physically active, OSSE: old physically active with a single bout of exercise.
Fig. 4.
The mRNA level of NRF1 (A), TFAM (B). NRF1 expression was induced with aging, and physical activity in the young groups, and down-regulated by a single bout of exercise in both age groups. TFAM was induced by physical activity in the young group, but this change was not seen in the aged group. Values are means±SD for six subjects per group. * p<0.05, ** p<0.01. YS: young sedentary, YSSE: young sedentary with a single bout of exercise, YA: young physically active, YASE: young physically active with a single bout of exercise, OS: old sedentary, OSSE: old sedentary with a single bout of exercise, OA: old physically active, OSSE: old physically active with a single bout of exercise.
Fig. 6.
Expression of PNPase (A) and UCP3 (B) in human skeletal muscle. Aging significantly decreased the expression of PNPase. Moreover, both a single bout of exercise and regular physical activity decreased the mRNA content of PNPase in the young groups. On the other hand, the expression levels increased in OA and OASE groups. Significant differences were not observed in UCP expression among groups. Values are means±SD for six subjects per group. * p<0.05, ** p<0.01. YS: young sedentary, YSSE: young sedentary with a single bout of exercise, YA: young physically active, YASE: young physically active with a single bout of exercise, OS: old sedentary, OSSE: old sedentary with a single bout of exercise, OA: old physically active, OSSE: old physically active with a single bout of exercise.
3.2.2. YA vs OA
The evaluation of mRNA levels of YA and OA subjects showed no differences in PGC1-a, AMPK isoforms, NRF1, Fis1, Mfn1, PNPase, Lon protease and UCP3 (Figs. 1–7). The expression of SIRT1 was elevated in OA compared to YA (Fig. 2A), whereas the expressions of TFAM (Fig. 4B), and SIRT3 were lower in OA (Fig. 7B).
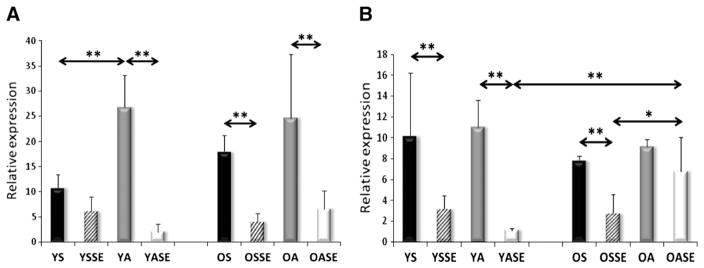
Young subjects who were involved in regular physical activity were found to have elevated NRF1, TFAM, FIS1 and SIRT3 mRNA levels compared to sedentary young individuals (Figs. 4,5A), while the PNPase expression was decreased in active subjects (Fig. 6A). No differences were found in the expression of SIRT1, PGC-1a, AMPK, Mfn1 and Lon protease mRNA levels (Figs. 2, 3, 5B and 7A).
Fig. 5.
mRNA levels of FIS1 and Mfn1 transcription factors. Regular physical activity increased the expression of FIS1 in young and old groups, while a single bout of exercise decreased the expression of FIS1 in young active and old sedentary and old active groups. The Mfn1 expressions were decreased by a single bout of exercise in young and old subjects. Values are means±SD for six subjects per group. * p<0.05, ** p<0.01. YS: young sedentary, YSSE: young sedentary with a single bout of exercise, YA: young physically active, YASE: young physically active with a single bout of exercise, OS: old sedentary, OSSE: old sedentary with a single bout of exercise, OA: old physically active, OSSE: old physically active with a single bout of exercise.
Fig. 3.
Expression of AMP-Kinase A2 (A), B2-subunit (B). Among the measured five isoforms of AMPK, significant alterations were found only in the expression of A2 and B2 subunits. * p<0.05, ** p<0.01. Values are means±SD for six subjects per group. YS: young sedentary, YSSE: young sedentary with a single bout of exercise, YA: young physically active, YASE: young physically active with a single bout of exercise, OS: old sedentary, OSSE: old sedentary with a single bout of exercise, OA: old physically active, OSSE: old physically active with a single bout of exercise.
3.2.3. YS vs YSSE
For young sedentary subjects, a single bout of exercise increased the mRNA levels of SIRT1and AMPK subunit2 (Figs. 2A, 3A), had no effect on the levels of PGC-1a, AMPK subunitB2, PNPase, UCP3 and Lon protease (Figs. 2B, 3B, 6, 7A), but significantly decreased NRF1, TFAM, Fis1, Mfn1, and SIRT3 expression levels compared to young physically active control values (Figs. 4, 5, 7B).
3.2.4. YA vs YASE
For young active individuals, a single bout of exercise significantly increased the expressions of SIRT1, PGC-1a, AMPK subunit A2, B2, maintained the expressions of TFAM, Fis1, UCP3, Lon protease, SIRT3, and decreased the mRNA levels in NRF1, Mfn1 and PNPase compared to young sedentary controls (Figs. 1–7).
3.2.5. OS vs OSSE
The mRNA levels of old sedentary subjects, after a single bout of exercise, were found to increase expressions of SIRT1 and AMPK subunit A2 (Figs. 2, 3A), maintain the levels of PGC-1a, AMPK subunit B2, TFAM, PNPase, UCP3, Lon protease, SIRT3 (Figs. 2, 3B, 4B, 6, 7) and decrease expression of NRF1, Fis1 and Mfn1 mRNA levels (Figs. 4A, 5), compared to those found for old sedentary control subjects.
3.2.6. OA vs OASE
mRNA expression levels for old physically active subjects, after an acute exercise bout, demonstrated an increase in AMPK subunit A2 and PNPase expressions (Figs. 3A, 6A), maintained levels of SIRT1, PGC-1a, AMPK subunit B2, TFAM, Mfn1, UCP3, Lon protease and SIRT3 (Figs. 2, 3B, 4B, 5B, 6B, 7), but decreased NRF1 and Fis1 expression levels compared to control values (Figs. 4A, 5A).
4. Discussion
One of objectives of this investigation was to study the effects of regular physical activity on young and old individuals and to ascertain whether moderate levels of physical activity could affect physiological parameters and the expression of mitochondrial biogenesis related genes. The data revealed that regular physical activity decreases body fat, BMI when compared to sedentary age-matched groups, and increases the level of VO2max in both age groups. Mitochondrial function closely affects VO2max (di Prampero, 2003), that is the level is inversely related to mortality in aged individuals (Radak et al., 2010; Timmons et al., 2010). In the present study a strong correlation between VO2max and TFAM (r=0.7397, p<0.01) was found and this fits well with previous evidence that elite athletes have a higher level of TFAM expression than moderately trained individuals (Norrbom et al., 2010), suggesting that TFAM expression levels could impact aerobic endurance. Interestingly enough, other measured genes did not show a relationship with VO2max.
Skeletal muscle readily responds to metabolic challenges (exercise/inactivity) and the plasticity of mitochondria is one of the key factors in this adaptability (Ljubicic et al., 2010). Increased numbers of mitochondria could result in an enhanced ATP supply, lower levels of ROS generation, and a more efficient signaling process (Holloszy, 2008; Koopman et al., 2010).
Therefore, we studied the expression profile of some key aspects of mitochondrial biogenesis involved genes as a result of aging. Our finding that aging increases the expression of NRF1 in human skeletal muscle is in accordance with earlier observations (Lezza et al., 2001). NRF1 is a proximal promoter of a number of mitochondrial genes involving some of the oxidative phosphorylation-related components, as well as the expression of TFAM (Scarpulla, 2008). NRF1, among other targets, activates antioxidant enzymes, and the Mn-SOD gene has been shown to be upregulated with aging (Radak et al., 2011). TFAM is also a downstream target of NRF1. However, the expression of TFAM did not change with aging in the present study, suggesting that this pathway could be attenuated by aging NRF-1 silencing suppresses target genes to mitochondrial biogenesis (Cam et al., 2004), although NRF-1 alone is not sufficient to stimulate mitochondrial biogenesis. Therefore, the decreased activities of COX and CS demonstrate that, with aging, mitochondrial function is decreased, and this is supported by the lack of simultaneous induction of NRF1 and TFAM. However, the fact that regular physical activity decreased the gap in VO2max and mitochondrial density related protein, such as CS and COX activity, between young control and aged active groups, implies that regular physical activity could attenuate the age-associated decline in important physiological functions.
Recent evidence suggests that PNPase plays an important role in mitochondrial biogenesis by regulating the transport of nuclear DNA coded RNAs into the mitochondrial matrix (Wang et al., 2010). PNPase is important for the homeostasis of mitochondria (Chen et al., 2006) and can induce cellular senescence (Sarkar et al., 2005). Ablation of PNPase leads to a significant drop in enzymatic activities of respiratory complexes and decreased ATP production, leading to activation of AMPK phosphorylation (Chen et al., 2006). PNPase knockdown results in impaired mitochondrial membrane potential, which can readily affect fusion and fission (Chen et al., 2005). Interestingly, over expression of PNPase could also be harmful to cells, since it can cause increased production of ROS and inflammation (Sarkar et al., 2004). Thus, both up and down regulation of PNPase could impair cell function (Hayakawa and Sekiguchi, 2006). In this study we have found significantly lower expression levels of PNPase in aged individuals. This could be due to the reduction of membrane potential that occurs with aging (Sastre et al., 2003). However, the very different response of young and old individuals, in terms of PNPase expression, to exercise bouts or regular physical activity, complicates the evaluation of these data.
Recent findings suggest that SIRT3 is important to reduce oxidative stress (Kong et al., 2010; Qiu et al., 2010) and the induction of SIRT3 in young subjects could indicate an adequate response to exercise-associated oxidative challenge, while the lack of response in aged groups could indicate that SIRT3 related adaptive response is decreased as a result of aging. This observation could be in accordance with the fact that SIRT3 gene is implicated in longevity of humans (Rose et al., 2003).
Besides measuring important genes in the biogenesis of mitochondria we also measured genes which are involved in the quality control of the mitochondrial network.
Fis1 is one of the key proteins required for mitochondrial fission (Romanello et al., 2010). Fis1 silencing has been shown to result in increased levels of oxidized proteins in the mitochondria, due to decreased mitochondrial autophagy (Twig et al., 2008). Fis1 mRNA and protein levels increase readily in skeletal muscle of rats (Ding et al., 2010), in response to a single bout of exercise. Moreover, it appears that mitochondrial fission could play a preventive role in cellular senescence by down-regulating mitochondrial elongation (Lee et al., 2007). Therefore, the present finding that regular physical activity increases Fis1 expression in young but not in old subjects, is important, and points out that aging could impair the adaptive response of mitochondrial fission to regular physical activity.
Accordingly to our knowledge, this is the first study on the effect of exercise on mitochondrial fission and fusion in human skeletal muscle. It is important to clarify this finding by making additional measurements of protein levels.
Aging might impair adaptive capability. Therefore, we monitored how aging and chronic physical activity influence the adaptive response to mild oxidative challenge as induced by a single bout of exercise. In young groups, a single bout of exercise increased the expression of four measured genes (SIRT1, PGC-1a, AMPK A2 and B2 subunits) while in aged subjects only SIRT1 and AMPK A2 subunits were induced. This finding strongly indicates that in young individuals the mitochondrial biogenesis-related genes were readily activated, while with aging, this response was attenuated. NRF1 and Mfn1 genes were decreased in both age groups by the acute exercise, and in old groups the Fis1 expression also decreased. Fusion enables mitochondria to mix their contents within interconnected mitochondrial reticula in order to minimize abnormalities. Fission on the other hand segregates mitochondria from the network, especially abnormal and damaged units, to facilitate their removal by autophagy (Benard and Karbowski, 2009). Well-coordinated fusion and fission minimize the formation of giant mitochondria and dysfunctional mitochondria. Indeed, cells which are unable to remove damaged mitochondria, due to the ablation of Fis1, show senescence-associated phenotypic changes (Lee et al., 2007). The different responses of Fis1 in young and old individuals to regular physical activity and acute exercise might be important, and could indicate that the mitochondrial fission-associated quality control is impaired with aging. Besides the elimination of damaged mitochondrial particles, fission is important to the adequate distribution of mitochondria, based on the local ATP need (Otera and Mihara, 2011).
The other striking difference to the response of acute exercise was observed for the PNPase gene. It has been suggested that oxidative stress decreases the expression of PNPase (Hayakawa and Sekiguchi, 2006). However, there is a lack of relationship between 8-oxoguanine levels in DNA (Radak et al., 2011) and PNPase activity expression for these subjects. Noting the important role of PNPase in mitochondrial function (Wang et al., 2010) and the enigma of the results for this enzyme, suggests a need for further study to appropriately define the role of PNPase in aging and its possible link to the quality control regulators of mitochondrial biogenesis and fusion/fission.
The data from this study suggest that aging slightly affects the gene expression profile of mitochondrial biogenesis and quality control genes. Moderate levels of regular physical activity improve VO2max, decrease body fat and BMI, and increase a larger number of mitochondrial biogenesis-related gene expressions in young individuals than in aged subjects.
However, physically active individuals, show similar levels of VO2max, and mitochondrial density, as judged from the activity of CS and COX, than young inactive subjects, emphasizing the importance of regular physical activity to decelerate the aging process. Aging decreases the adaptive response of mitochondrial biogenesis-related gene expression to regular physical activity and to a single bout of exercise. Mitochondrial fission appears to be impaired by aging, at least at the level of gene expression, and to be one of the most sensitive markers of the age-associated decline in adaptive responses to physical activity.
Acknowledgments
We are grateful to the support of Prof. A.W. Taylor in the preparation of this paper. The present work was supported by Hungarian grants from ETT 38388, OTKA (K75702) TAMOP/Magiszter awarded to Z. Radák.
References
- Abbott PV. Analysis of a referral-based endodontic practice: Part 1. Demographic data and reasons for referral. J Endod. 1994;20:93–96. doi: 10.1016/S0099-2399(06)81190-8. [DOI] [PubMed] [Google Scholar]
- ACSM. Guidelines for Exercise Testing and Prescription. Lippincott Williams & Wilkins; Philadelphia: 2005. [Google Scholar]
- Arany Z. PGC-1 coactivators and skeletal muscle adaptations in health and disease. Curr Opin Genet Dev. 2008;18:426–434. doi: 10.1016/j.gde.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard G, Karbowski M. Mitochondrial fusion and division: regulation and role in cell viability. Semin Cell Dev Biol. 2009;20:365–374. doi: 10.1016/j.semcdb.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström J. Muscle electrolytes in man. Scand J Clin Lab Invest. 1962;14:1–110. [Google Scholar]
- Bratic I, Trifunovic A. Mitochondrial energy metabolism and ageing. Biochim Biophys Acta. 2010;1797:961–967. doi: 10.1016/j.bbabio.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Cartoni R, Leger B, Hock MB, Praz M, Crettenand A, Pich S, Ziltener JL, Luthi F, Deriaz O, Zorzano A, Gobelet C, Kralli A, Russell AP. Mitofusins 1/2 and ERRalpha expression are increased in human skeletal muscle after physical exercise. J Physiol. 2005;567:349–358. doi: 10.1113/jphysiol.2005.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, Hong JS, McBride HM, Koehler CM, Teitell MA, French SW. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol Cell Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LS, Greenlund LJ, Asmann YW, Short KR, McCrady SK, Levine JA, Nair KS. Impact of endurance training on murine spontaneous activity, muscle mitochondrial DNA abundance, gene transcripts, and function. J Appl Physiol. 2007;102:1078–1089. doi: 10.1152/japplphysiol.00791.2006. [DOI] [PubMed] [Google Scholar]
- Craig CL, Marshall AL, Sjostrom M, Bauman AE, Booth ML, Ainsworth BE, Pratt M, Ekelund U, Yngve A, Sallis JF, Oja P. International physical activity questionnaire: 12-country reliability and validity. Med Sci Sports Exerc. 2003;35:1381–1395. doi: 10.1249/01.MSS.0000078924.61453.FB. [DOI] [PubMed] [Google Scholar]
- Davies KJ, Quintanilha AT, Brooks GA, Packer L. Free radicals and tissue damage produced by exercise. Biochem Biophys Res Commun. 1982;107:1198–1205. doi: 10.1016/s0006-291x(82)80124-1. [DOI] [PubMed] [Google Scholar]
- di Prampero PE. Factors limiting maximal performance in humans. Eur J Appl Physiol. 2003;90:420–429. doi: 10.1007/s00421-003-0926-z. [DOI] [PubMed] [Google Scholar]
- Ding H, Jiang N, Liu H, Liu X, Liu D, Zhao F, Wen L, Liu S, Ji LL, Zhang Y. Response of mitochondrial fusion and fission protein gene expression to exercise in rat skeletal muscle. Biochim Biophys Acta. 2010;1800:250–256. doi: 10.1016/j.bbagen.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Fatouros IG, Jamurtas AZ, Villiotou V, Pouliopoulou S, Fotinakis P, Taxildaris K, Deliconstantinos G. Oxidative stress responses in older men during endurance training and detraining. Med Sci Sports Exerc. 2004;36:2065–2072. doi: 10.1249/01.mss.0000147632.17450.ff. [DOI] [PubMed] [Google Scholar]
- Ferreira I, Twisk JW, Stehouwer CD, van Mechelen W, Kemper HC. Longitudinal changes in VO2max: associations with carotid IMT and arterial stiffness. Med Sci Sports Exerc. 2003;35:1670–1678. doi: 10.1249/01.MSS.0000089247.37563.4B. [DOI] [PubMed] [Google Scholar]
- Hayakawa H, Sekiguchi M. Human polynucleotide phosphorylase protein in response to oxidative stress. Biochemistry. 2006;45:6749–6755. doi: 10.1021/bi052585l. [DOI] [PubMed] [Google Scholar]
- Holloszy JO. Regulation by exercise of skeletal muscle content of mitochondria and GLUT4. J Physiol Pharmacol. 2008;59(Suppl 7):5–18. [PubMed] [Google Scholar]
- Kelly DP. Cell biology: ageing theories unified. Nature. 2011;470:342–343. doi: 10.1038/nature09896. [DOI] [PubMed] [Google Scholar]
- Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman WJ, Nijtmans LG, Dieteren CE, Roestenberg P, Valsecchi F, Smeitink JA, Willems PH. Mammalian mitochondrial complex I: biogenesis, regulation, and reactive oxygen species generation. Antioxid Redox Signal. 2010;12:1431–1470. doi: 10.1089/ars.2009.2743. [DOI] [PubMed] [Google Scholar]
- Lee S, Jeong SY, Lim WC, Kim S, Park YY, Sun X, Youle RJ, Cho H. Mitochondrial fission and fusion mediators, hFis1 and OPA1, modulate cellular senescence. J Biol Chem. 2007;282:22977–22983. doi: 10.1074/jbc.M700679200. [DOI] [PubMed] [Google Scholar]
- Levine BD. VO2max: what do we know, and what do we still need to know? J Physiol. 2008;586:25–34. doi: 10.1113/jphysiol.2007.147629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezza AM, Pesce V, Cormio A, Fracasso F, Vecchiet J, Felzani G, Cantatore P, Gadaleta MN. Increased expression of mitochondrial transcription factor A and nuclear respiratory factor-1 in skeletal muscle from aged human subjects. FEBS Lett. 2001;501:74–78. doi: 10.1016/s0014-5793(01)02628-x. [DOI] [PubMed] [Google Scholar]
- Ljubicic V, Joseph AM, Saleem A, Uguccioni G, Collu-Marchese M, Lai RY, Nguyen LM, Hood DA. Transcriptional and post-transcriptional regulation of mitochondrial biogenesis in skeletal muscle: effects of exercise and aging. Biochim Biophys Acta. 2010;1800:223–234. doi: 10.1016/j.bbagen.2009.07.031. [DOI] [PubMed] [Google Scholar]
- Michailidis Y, Jamurtas AZ, Nikolaidis MG, Fatouros IG, Koutedakis Y, Papassotiriou I, Kouretas D. Sampling time is crucial for measurement of aerobic exercise-induced oxidative stress. Med Sci Sports Exerc. 2007;39:1107–1113. doi: 10.1249/01.mss.0b013e318053e7ba. [DOI] [PubMed] [Google Scholar]
- Morss GM, Jordan AN, Skinner JS, Dunn AL, Church TS, Earnest CP, Kampert JB, Jurca R, Blair SN. Dose response to exercise in women aged 45–75 yr (DREW): design and rationale. Med Sci Sports Exerc. 2004;36:336–344. doi: 10.1249/01.MSS.0000113738.06267.E5. [DOI] [PubMed] [Google Scholar]
- Norrbom J, Wallman SE, Gustafsson T, Rundqvist H, Jansson E, Sundberg CJ. Training response of mitochondrial transcription factors in human skeletal muscle. Acta Physiol (Oxf) 2010;198:71–79. doi: 10.1111/j.1748-1716.2009.02030.x. [DOI] [PubMed] [Google Scholar]
- Otera H, Mihara K. Molecular mechanisms and physiologic functions of mitochondrial dynamics. J Biochem. 2011;149:241–251. doi: 10.1093/jb/mvr002. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Radak Z, Sasvari M, Nyakas C, Taylor AW, Ohno H, Nakamoto H, Goto S. Regular training modulates the accumulation of reactive carbonyl derivatives in mitochondrial and cytosolic fractions of rat skeletal muscle. Arch Biochem Biophys. 2000;383:114–118. doi: 10.1006/abbi.2000.2042. [DOI] [PubMed] [Google Scholar]
- Radak Z, Atalay M, Jakus J, Boldogh I, Davies K, Goto S. Exercise improves import of 8-oxoguanine DNA glycosylase into the mitochondrial matrix of skeletal muscle and enhances the relative activity. Free Radic Biol Med. 2009;46:238–243. doi: 10.1016/j.freeradbiomed.2008.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radak Z, Hart N, Sarga L, Koltai E, Atalay M, Ohno H, Boldogh I. Exercise plays a preventive role against Alzheimer’s disease. J Alzheimers Dis. 2010;20:777–783. doi: 10.3233/JAD-2010-091531. [DOI] [PubMed] [Google Scholar]
- Radak Z, Bori Z, Koltai E, Fatouros IG, Jamurtas AZ, Douroudos II, Terzis G, Nikolaidis MG, Chatzinikolaou A, Sovatzidis A, Kumagai S, Naito H, Boldogh I. Age-dependent changes in 8-oxoguanine-DNA glycosylase activity are modulated by adaptive responses to physical exercise in human skeletal muscle. Free Radic Biol Med. 2011;51:417–423. doi: 10.1016/j.freeradbiomed.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radak Z, Bori Z, Koltai E, Fatouros I, Jamurtas A, Douroudos I, Terzis G, Nikolaidis M, Chatzinikolaou A, Sovatzidis A, Kumagai S, Naito H, Boldogh I. Age-dependent changes in 8-oxoguanine-DNA-glycosylase activity is modulated by adaptive responses to physical exercise in human skeletal muscle. Free Radic Biol Med. 2011;51:417–423. doi: 10.1016/j.freeradbiomed.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, Milan G, Masiero E, Del Piccolo P, Foretz M, Scorrano L, Rudolf R, Sandri M. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 2010;29:1774–1785. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooyackers OE, Adey DB, Ades PA, Nair KS. Effect of age on in vivo rates of mitochondrial protein synthesis in human skeletal muscle. Proc Natl Acad Sci U S A. 1996;93:15364–15369. doi: 10.1073/pnas.93.26.15364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, BonaFe M, Franceschi C, Tan Q, Boiko S, Yashin AI, De Benedictis G. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38:1065–1070. doi: 10.1016/s0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- Safdar A, Bourgeois JM, Ogborn DI, Little JP, Hettinga BP, Akhtar M, Thompson JE, Melov S, Mocellin NJ, Kujoth GC, Prolla TA, Tarnopolsky MA. Endurance exercise rescues progeroid aging and induces systemic mitochondrial rejuvenation in mtDNA mutator mice. Proc Natl Acad Sci U S A. 2011;108:4135–4140. doi: 10.1073/pnas.1019581108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, Maser RS, Tonon G, Foerster F, Xiong R, Wang YA, Shukla SA, Jaskelioff M, Martin ES, Heffernan TP, Protopopov A, Ivanova E, Mahoney JE, Kost-Alimova M, Perry SR, Bronson R, Liao R, Mulligan R, Shirihai OS, Chin L, DePinho RA. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Lebedeva IV, Emdad L, Kang DC, Baldwin AS, Jr, Fisher PB. Human polynucleotide phosphorylase (hPNPaseold-35): a potential link between aging and inflammation. Cancer Res. 2004;64:7473–7478. doi: 10.1158/0008-5472.CAN-04-1772. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Park ES, Emdad L, Randolph A, Valerie K, Fisher PB. Defining the domains of human polynucleotide phosphorylase (hPNPaseOLD-35) mediating cellular senescence. Mol Cell Biol. 2005;25:7333–7343. doi: 10.1128/MCB.25.16.7333-7343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastre J, Pallardo FV, Vina J. The role of mitochondrial oxidative stress in aging. Free Radic Biol Med. 2003;35:1–8. doi: 10.1016/s0891-5849(03)00184-9. [DOI] [PubMed] [Google Scholar]
- Sawada SS, Muto T, Tanaka H, Lee IM, Paffenbarger RS, Jr, Shindo M, Blair SN. Cardiorespiratory fitness and cancer mortality in Japanese men: a prospective study. Med Sci Sports Exerc. 2003;35:1546–1550. doi: 10.1249/01.MSS.0000084525.06473.8E. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Sriwijitkamol A, Coletta DK, Wajcberg E, Balbontin GB, Reyna SM, Barrientes J, Eagan PA, Jenkinson CP, Cersosimo E, DeFronzo RA, Sakamoto K, Musi N. Effect of acute exercise on AMPK signaling in skeletal muscle of subjects with type 2 diabetes: a time-course and dose–response study. Diabetes. 2007;56:836–848. doi: 10.2337/db06-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzis G, Georgiadis G, Stratakos G, Vogiatzis I, Kavouras S, Manta P, Mascher H, Blomstrand E. Resistance exercise-induced increase in muscle mass correlates with p70S6 kinase phosphorylation in human subjects. Eur J Appl Physiol. 2008a;102:145–152. doi: 10.1007/s00421-007-0564-y. [DOI] [PubMed] [Google Scholar]
- Terzis G, Stratakos G, Manta P, Georgiadis G. Throwing performance after resistance training and detraining. J Strength Cond Res. 2008b;22:1198–1204. doi: 10.1519/JSC.0b013e31816d5c97. [DOI] [PubMed] [Google Scholar]
- Timmons JA, Knudsen S, Rankinen T, Koch LG, Sarzynski M, Jensen T, Keller P, Scheele C, Vollaard NB, Nielsen S, Akerstrom T, MacDougald OA, Jansson E, Greenhaff PL, Tarnopolsky MA, van Loon LJ, Pedersen BK, Sundberg CJ, Wahlestedt C, Britton SL, Bouchard C. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J Appl Physiol. 2010;108:1487–1496. doi: 10.1152/japplphysiol.01295.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips LE, Ravelli AC, Dongelmans PC, Deurenberg P, Van Staveren WA. A physical activity questionnaire for the elderly. Med Sci Sports Exerc. 1991;23:974–979. [PubMed] [Google Scholar]
- Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, Lightowlers RN, Morse HC, III, Koehler CM, Teitell MA. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]