Abstract
Premise of the study:
The One Thousand Plant Transcriptomes Project (1KP, 1000+ assembled plant transcriptomes) provides an enormous resource for developing microsatellite loci across the plant tree of life. We developed loci from these transcriptomes and tested their utility.
Methods and Results:
Using software packages and custom scripts, we identified microsatellite loci in 1KP transcriptomes. We assessed the potential for cross-amplification and whether loci were biased toward exons, as compared to markers derived from genomic DNA. We characterized over 5.7 million simple sequence repeat (SSR) loci from 1334 plant transcriptomes. Eighteen percent of loci substantially overlapped with open reading frames (ORFs), and electronic PCR revealed that over half the loci would amplify successfully in conspecific taxa. Transcriptomic SSRs were approximately three times more likely to map to translated regions than genomic SSRs.
Conclusions:
We believe microsatellites still have a place in the genomic age—they remain effective and cost-efficient markers. The loci presented here are a valuable resource for researchers.
Keywords: 1KP, microsatellite development, neutral markers, next-generation sequencing (NGS), non-neutral markers, simple sequence repeat (SSR), transcriptomes
Microsatellites have been used to answer a host of research questions over the past several decades, including studies involving forensics, population and conservation genetics, phylogeography, genomic mapping, and determining parentage (Ellegren, 2000; Esselink et al., 2004; Kalia et al., 2011). For many research projects using microsatellites, developing simple sequence repeat (SSR) loci can be the most expensive portion of the budget (Wei et al., 2014; Hodel et al., 2016a). Typically, one would generate next-generation sequence data to mine for SSRs; the initial sequencing cost would likely exceed US$1000, unless multiplexing is used (Jennings et al., 2011). In some cases, primers may already be developed for well-studied taxa, or existing primers may be able to amplify loci in closely related species (Kalia et al., 2011). As microsatellites and their flanking regions evolve relatively rapidly, loci often must be developed by the researcher specifically for the taxon of interest, either using that taxon, or a closely related one (e.g., a congener; Guichoux et al., 2011). As DNA sequencing costs decrease, online resources such as the Sequence Read Archive (SRA), curated by the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/sra), have grown substantially in the past decade. If researchers can develop SSR loci from existing, publicly available data sets, they may be able to eliminate one of the largest costs associated with using microsatellites. As of February 2016, the SRA had sequence data from 2623 species in Embryophyta, which could potentially be used to develop SSR loci. Another online resource, the One Thousand Plant Transcriptomes Project (1KP; www.onekp.com; Matasci et al., 2014), has 1334 sequenced and assembled transcriptomes available from over 1000 plant species. This has the potential to be an enormous resource for researchers.
In this paper, we take advantage of the availability of the 1KP transcriptomes to develop, and make available, over 5 million SSR loci for the green plant science community to use in their research. Botanists with limited budgets and/or research questions best addressed using microsatellites will be able to exploit this huge set of loci across the plant tree of life. Although recently developed techniques (e.g., RAD-Seq) can generate many more loci than are typically used in SSR studies, our companion review article in this issue indicates that microsatellites are still a valuable tool for researchers and can be more cost-effective than newer methods, especially if loci are already developed (Hodel et al., 2016a). In particular, microsatellites remain a vital tool for researchers with limited budgets (e.g., conservation scientists, students, researchers in developing countries). Our objectives in this article are to: (1) develop SSR loci from 1KP data, and (2) assess the utility of these loci using several approaches.
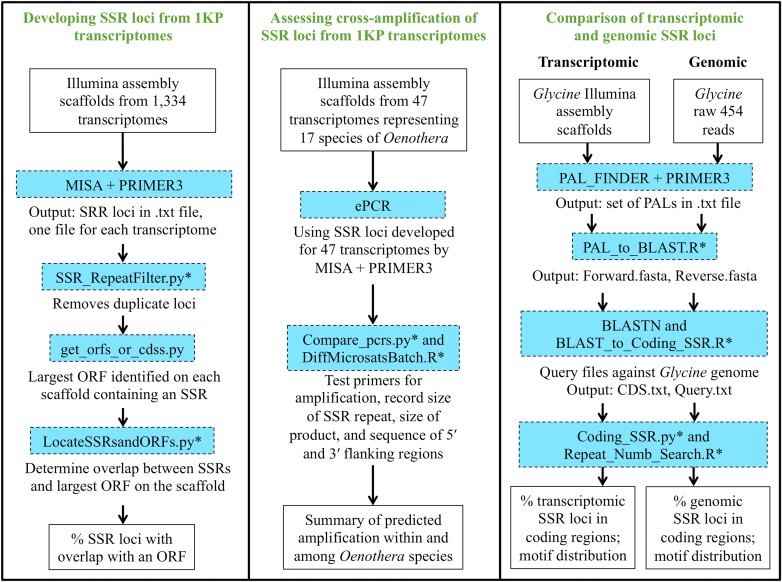
The growing prevalence in the literature of the development of microsatellite loci from transcriptome data sets (e.g., Aggarwal et al., 2007; Triwitayakorn et al., 2011; Zhang et al., 2012; Zheng et al., 2013) prompted us to characterize the microsatellite loci in the 1334 transcriptome samples from across green plants from the 1KP project. These data have the potential to be an enormous resource for developing large numbers of microsatellite markers inexpensively. However, generating markers from transcriptomes could favor microsatellites in translated regions of the genome, which may be less desirable than those from noncoding regions. To assess the utility of the markers generated from 1KP data, we use several approaches to ascertain if there are any biases associated with transcriptome-developed markers (i.e., if these loci are non-neutral; Fig. 1). We use Oenothera L. (Onagraceae) as a test case to assess how successfully transcriptome-derived SSRs can be used among congeners. Transcriptomes are available from 17 Oenothera species in the 1KP data, and some of the Oenothera species are represented by more than one individual, enabling us to report how well SSR loci will amplify in conspecific or congeneric taxa via simulated in silico PCR. Glycine Willd. (Fabaceae) was used in a separate test case to compare the number of loci, size distribution of loci, and genomic location of the loci between transcriptome-derived SSRs and genome-derived SSRs. This investigation was designed to assess whether microsatellite loci developed from transcriptomic data would be more likely to be found in translated regions of the genome. Figure 1 shows the workflow followed in each approach.
Fig. 1.
The workflows for developing SSR loci from 1KP transcriptomes, assessing cross-amplification of SSR loci from 1KP transcriptomes, and comparing transcriptomic and genomic SSR loci. Turquoise boxes indicate software packages or scripts; custom scripts (available at https://github.com/soltislab/transcriptome_microsats) designed by the authors are designated by asterisks.
METHODS AND RESULTS
Developing microsatellites from 1KP transcriptomes
We used commonly employed software packages for extracting microsatellite primers from transcriptome data from over 1000 green plant species using data from the 1KP Project. Microsatellite loci were identified using the packages MISA (version 1.0; Thiel et al., 2003) and Primer3 (version 2.3.4; Koressaar and Remm, 2007; Untergasser et al., 2012). The MISA script was modified slightly to format the output for the newer version of Primer3. MISA was run on 1334 transcriptomes, using the following minimum number of repeat settings: mononucleotide, 10 repeats; dinucleotide, six repeats; and tri- through hexanucleotide, five repeats. The maximum size for interruptions in compound loci was set to 25 bp. MISA identifies microsatellite loci in the transcriptome scaffolds and generates an input file with settings for Primer3. A set of custom scripts was developed to process further and summarize the microsatellites in these samples. These annotated scripts are available on GitHub at: https://github.com/soltislab/transcriptome_microsats, and the data files are available from the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.rb7h0; Hodel et al., 2016b). There is a file in the GitHub repository explaining the function of each script, and there is a directory named “Tutorial” containing a README file that will guide the user through the steps we took to develop loci and assess their utility.
Many of the scaffolds in the SOAPdenovo-Trans (Xie et al., 2014) transcriptomes produced by the 1KP Project are inferred isoforms of the same gene. This leads to multiple microsatellites being identified for the same locus. Thus, we wrote a script (SSR_RepeatFilter.py) to filter redundant loci, discarding loci that used the same forward or reverse primers as another locus. After filtering, the largest open reading frame (ORF) was identified on each scaffold with a microsatellite locus using the get_orfs_or_cdss.py script (version 0.0.3; Cock et al., 2009). The location of the locus compared to the largest ORF was used to determine if the repeat was located in an exon using the script LocateSSRsandORFs.py. Although the largest ORF on a scaffold may not be the only one, or the true gene product, this was a reasonable way to characterize the location of microsatellites relative to translated regions. Microsatellite loci were considered to have substantial overlap with the ORF if 15 bp (five amino acids) of the repetitive element overlapped with the longest ORF on the scaffold.
To assess the cross-amplification and variability characteristics of the microsatellite loci developed from transcriptomes, we took advantage of a set of multiple Oenothera transcriptomes included in the 1KP Project. Electronic PCR was simulated with e-PCR (version 2.3.12; Schuler, 1997) using the primers for the microsatellite loci developed from 47 transcriptomes from 17 species of Oenothera in a pairwise all-by-all design, including self-amplification. In some cases, multiple equally good products are reported, mostly from inferred isoforms. These may bias the results slightly, likely increasing differences where isoform difference is in the microsatellite locus and increasing similarity where it is elsewhere on the scaffold. Either way, the effect is likely to be small (on the order of ±5%). We emphasize that this is an approximate test of cross-amplification, as transcriptome data are inherently incomplete due to both assembly issues and true differences of gene expression. It is entirely possible that a locus that fails to amplify is present in the genomic DNA of a sample, but was not expressed in the tissue used, or failed to be assembled. e-PCR settings allowed two mismatches and one indel for successful amplification, and the best e-PCR product was reported. In addition to amplification, we looked at the inferred PCR product and compared the following: size of microsatellite repeat, size of product, and sequence identity of 5′ and 3′ flanking regions between the source species and the target species (using the scripts compare_pcrs.py and DiffMicrosatsBatch.R). Genetic identities of the different species were taken from the average plastid genome sequence identity based on an alignment of 10 plastid genes also derived from the transcriptomes (atpA, atpB, matK, ndhH, psbA, psbB, rbcL, rpl33, rps11, and rps19). These 10 genes were chosen based on number of samples having data for these genes, as well as variability of the loci.
Comparing genomic and transcriptomic microsatellites
We used Glycine to compare the number of loci developed and the distribution of repeat size between genome- and transcriptome-derived microsatellites. We implemented the “Seq-to-SSR” approach (Castoe et al., 2012) to test two data sets of Glycine. We developed SSR loci from Glycine soja Siebold & Zucc. transcriptomes (using data from the 1KP Project) and from randomly sheared genomic G. max (L.) Merr. DNA using reads generated from 454 pyrosequencing (Swaminathan et al., 2007). We downloaded the raw DNA sequence reads from the 454 data set from NCBI Trace Archive (TI 1732557604–1733276192; Swaminathan et al., 2007). For both the transcriptomic and genomic data sets, we identified loci using the program PAL_FINDER (version 0.02.04; Castoe et al., 2012), which designs and characterizes PCR amplification primers flanking identified SSR loci (potentially amplifiable loci [PALs]) using the program Primer3 (version 2.0.0; Rozen and Skaletsky, 1999). We used the same minimum repeat number criteria for identifying a locus as in the MISA analysis in the previous Oenothera section: dinucleotide, six repeats; tri- through hexanucleotide, five repeats. We used a custom R script (PAL_to_BLAST.R) to remove duplicated loci, extract forward and reverse primer sequences, and write two FASTA files for the forward and reverse primers, respectively. The BLASTN search algorithm was used to query every primer against the annotated genome of G. max (NCBI assembly accession GCA_000004515.2). We then ran a script (BLAST_to_Coding_SSR.R) to process the results of the BLAST search and remove duplicates, which was necessary for the reasons mentioned above in the Oenothera section. Another script (Coding_SSR.py) used the processed results of the BLAST search and the annotated G. max genome to identify primers that occur within regions annotated as coding sequences (CDS). For each data set (genomic, transcriptomic), the percentage of loci in coding regions was averaged between the forward and reverse primers, and the two data sets were compared. We then filtered the output of Coding_SSR.py and PAL_FINDER to determine the distribution of repeat motifs using the script Repeat_Numb_Search.R.
Developing microsatellites from transcriptomes
Our methods identified 5,739,271 microsatellite loci from 1334 transcriptomes (supplementary files are available from the Dryad Digital Repository at http://dx.doi.org/10.5061/dryad.rb7h0; Hodel et al., 2016b). On average, each 1KP transcriptome yielded 4328 microsatellite loci (median: 3687; minimum: 181; maximum: 65,470; standard deviation: 3711). Of these loci, 96% were on scaffolds with an identifiable ORF, and 62% have some overlap with the ORF; however, on average, only 18% of the identified repeat motifs had a substantial (≥15 bp) overlap with the longest ORF on a scaffold (Table 1). The most prevalent microsatellite type was mononucleotide repeats (39% of total), and these were least likely to overlap with the ORF (3%). Trinucleotide repeats were the next most common type (33%), and a high percentage had substantial overlap with ORFs (33%).
Table 1.
Average number of SSR loci by repeat type and their location relative to identified open reading frames (ORFs). Note that percentages are based on unrounded values.
| SSR type | Avg. no. SSR loci (% of total) | Avg. no. with no overlap of ORF (% of type) | Avg. no. with any overlap of ORF (% of type) | Avg. no. with substantial (≥15 bp) overlap of ORF (% of type) |
| Compound | 176 (4) | 70 (40) | 106 (60) | 104 (59) |
| Complex compound | 7 (0) | 4 (49) | 4 (51) | 4 (50) |
| Mononucleotide | 1681 (39) | 833 (50) | 848 (50) | 46 (3) |
| Dinucleotide | 947 (22) | 436 (46) | 511 (54) | 99 (10) |
| Trinucleotide | 1427 (33) | 250 (18) | 1176 (82) | 472 (33) |
| Tetranucleotide | 55 (1) | 30 (54) | 25 (46) | 23 (41) |
| Pentanucleotide | 16 (0) | 9 (56) | 7 (44) | 6 (40) |
| Hexanucleotide | 20 (0) | 4 (21) | 16 (79) | 15 (79) |
| Total | 4328 (100) | 1635 (38) | 2693 (62) | 769 (18) |
Cross-amplification of microsatellites
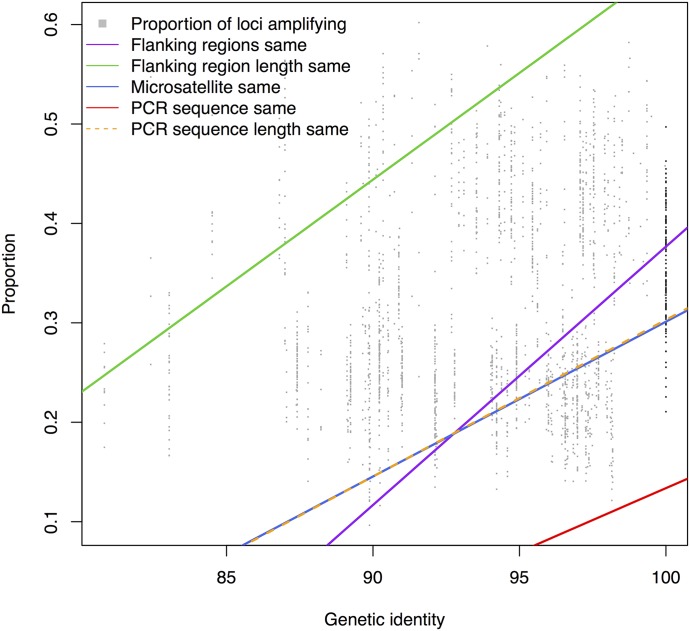
In the simulated PCR experiment using Oenothera, all of the loci amplified in the sample from which they were derived. However, only 53% of the loci amplified in other samples of the same species (Table 2, Fig. 2). Of those that amplified in multiple samples of the same species, the microsatellite repeat was of different length in 27% of the loci, the flanking region was of different length in 26% of the loci, and the flanking region sequence was different in 39% of the loci. In different species, between 18% and 24% of loci successfully amplified (Table 2). As the average plastid genome sequence identity is reduced, variation tends to increase in the microsatellite repeat, as well as the flanking region length and sequence (Fig. 2). However, amplification success values were quite similar across the distance classes selected (Table 2), with 76–79% of loci having variable repeat regions as well as variation in the length of the flanking region, and 94–96% of loci having sequence variation in the flanking region at all levels below amplification within the same species (Table 2). These values are even similar for genetic identities up to 98% (data not shown).
Table 2.
Summary of the electronic cross-amplification among species of Oenothera. The first row is percentage of all loci that amplify in other samples of the same species. The other rows are percentage of amplifying loci that are polymorphic between samples. Genetic identity (GI) is based on nucleotide identity at 10 plastid loci.
| Same species (100% GI) | Very similar (95% to <100% GI) | Similar (90% to <95% GI) | Less similar (<90% GI) | |
| % Amplification | 53 | 24 | 20 | 18 |
| % Polymorphic repeat | 27 | 76 | 79 | 79 |
| % Flanking sequence length polymorphic | 26 | 76 | 79 | 79 |
| % Flanking sequence polymorphic | 39 | 94 | 96 | 95 |
Fig. 2.
The relationship between genetic identity and the proportion of loci that will successfully amplify, the proportion of flanking regions that are the same, the proportion of flanking regions with the same length, the proportion with the same microsatellite, the proportion with the same PCR sequence, and the proportion of PCR sequences that have the same length. The sequence is defined as the whole sequence between the primers, so the flanking regions are the sequence between the primer and the repeat, or either side of the repeat. The orange line is dashed to enable the reader to distinguish it from the blue line.
Comparing genomic and transcriptomic microsatellites
Our Glycine genomic data set contained 717,309 raw 454 reads, for a total of 84,930,732 bases (Table 3). Using the Seq-to-SSR method (Castoe et al., 2012) and custom scripts to remove duplicates, we identified 532 unique PALs in this data set. The Glycine transcriptomic data set was composed of 364,755 Illumina assembly scaffolds, with a total of 133,872,643 bases. The transcriptomic data set yielded 7186 unique PALs. In the genomic data set, most (53.8%) of the loci were dinucleotide repeats, and 39.8% of the loci were trinucleotide repeats. Most (56.4%) of the loci in the transcriptomic data set were trinucleotide repeats, and the second most common motif was dinucleotide repeats (40.3%). The BLAST search results revealed that for the genomic data set, 65 primers (12.2%) were in translated regions, whereas for the transcriptomic data set, 2552 primers (35.5%) were in translated regions (Table 3). In our Glycine analysis, excluding trinucleotide and hexanucleotide repeats still yields 3106 PALs, of which only 698.5 (22.5%) are in coding regions. Although this is a low proportion of the total loci available, it is double the percentage of translated loci found using genomic loci.
Table 3.
Comparison of genomic and transcriptomic SSRs developed in Glycine. The number of reads, total number of bases, reads containing an SSR, reads containing a potentially amplifiable locus (PAL), unique PALs, mean number of PALs in a coding region, and percentage of PALs found in a coding region are presented for both data sets. The analyses to determine which loci were in translated regions were run on both forward and reverse loci, and the results were averaged. Thus, some of the counts of motifs in coding regions are not integers.
| Statistic of data set | Genomic | Transcriptomic |
| Reads | 717,309 (raw 454 reads) | 364,755 (Illumina assembly scaffolds) |
| Total bases | 84,930,732 | 133,872,643 |
| Reads/scaffolds with SSR | 7368 | 13,667 |
| Reads/scaffolds with primers (PALs) | 624 | 8456 |
| Total unique PALs | 532 | 7186 |
| Dinucleotides, total | 286 | 2893 |
| Trinucleotides, total | 212 | 4050 |
| Tetranucleotides, total | 24 | 174 |
| Pentanucleotides, total | 9 | 39 |
| Hexanucleotides, total | 1 | 30 |
| Dinucleotides, % of total | 53.8 | 40.3 |
| Trinucleotides, % of total | 39.8 | 56.4 |
| Tetranucleotides, % of total | 4.5 | 2.4 |
| Pentanucleotides, % of total | 1.7 | 0.5 |
| Hexanucleotides, % of total | 0.2 | 0.4 |
| Mean PALs in coding region | 65 | 2552 |
| % of PALs in coding region | 12.2 | 35.5 |
| Dinucleotides, in coding region | 17.5 | 643 |
| Trinucleotides, in coding region | 46 | 1838 |
| Tetranucleotides, in coding region | 1.5 | 48 |
| Pentanucleotides, in coding region | 0 | 7.5 |
| Hexanucleotides, in coding region | 0 | 15 |
| Dinucleotides, % of total in coding region | 26.9 | 25.2 |
| Trinucleotides, % of total in coding region | 70.8 | 72.0 |
| Tetranucleotides, % of total in coding region | 2.3 | 1.9 |
| Pentanucleotides, % of total in coding region | 0.0 | 0.3 |
| Hexanucleotides, % of total in coding region | 0.0 | 0.6 |
| Dinucleotides, % in coding relative to total dinucleotides | 6.1 | 22.2 |
| Trinucleotides, % in coding relative to total trinucleotides | 21.7 | 45.4 |
| Tetranucleotides, % in coding relative to total tetranucleotides | 6.3 | 27.6 |
| Pentanucleotides, % in coding relative to total pentanucleotides | 0.0 | 19.2 |
| Hexanucleotides, % in coding relative to total hexanucleotides | 0.0 | 50.0 |
CONCLUSIONS
Developing microsatellites from transcriptomes
Based on our analyses, transcriptome data sets such as those generated by the 1KP Project can be a valuable source of microsatellite loci for researchers. To develop a community resource, we provide a list of 5,739,271 candidate microsatellite loci from 1334 transcriptomes across more than 1000 species of green plants. This collection of microsatellite loci should serve as a valuable resource for researchers across all clades of green plants wishing to start using microsatellites for related species. Because SSR loci can be used in species closely related to the species for which the markers were designed, the loci we developed could potentially be used in tens of thousands of plant species (assuming each set of loci is amplifiable in at least 10 closely related species). Although there are unexplored considerations for microsatellites linked to translated regions (Loire et al., 2013), only 18% of the microsatellite loci identified have 15 bp or more of the repeat within the largest inferred ORF on the scaffold (Table 1). Thus, while closely linked to translated regions, it appears that the majority of these loci are not themselves within a translated region of a gene.
Comparison of transcriptomic and genomic microsatellites
Ideally, SSR markers would be selectively neutral; researchers often have concerns that these markers are prone to selective pressures when they occur in or near coding regions (Morgante et al., 2002). SSR loci in Glycine are three times more likely to occur in translated regions when derived from transcriptomic data than genomic data (35.8% vs. 11.8%, Table 3). However, the vast majority of loci found in translated regions are trinucleotide repeats (75.0% of genomic loci and 72.3% of transcriptomic loci). Researchers who are concerned about selection affecting inferences made using transcriptomic SSR loci could exclude trinucleotide repeats (and, to be safe, hexanucleotide repeats) to avoid translated regions. As noted above, excluding trinucleotide and hexanucleotide repeats resulted in 22.1% of transcriptomic loci mapping to translated regions in a BLAST search. This would still leave a researcher with over 5000 potential SSR loci; he or she could then select primers to test from this subset of loci.
SSRs are often presumed to be neutral, but can be subject to both positive and negative selection for a variety of reasons (e.g., genetic hitchhiking) that are undetectable to a researcher without extensive genomic resources available for the taxon of interest (Haasl and Payseur, 2011). Additionally, in some cases, genic SSRs may be effectively neutral (e.g., trinucleotide-repeat loci in highly conserved genes). Furthermore, it is standard procedure in microsatellite development not to test loci for selection, as it is not feasible to do so before collecting genotype data. Many published SSR studies have included loci found in coding regions because they did not or could not verify the locations of loci within the genome (Gardner et al., 2011). Our Glycine results, combined with the results of the Oenothera data sets, should give researchers confidence that they can use transcriptomic microsatellites without concerns about neutrality that exceed those concerns in typical SSR studies—especially if trinucleotide repeats are avoided. Furthermore, some research questions can be answered with non-neutral microsatellites (e.g., Aberlenc-Bertossi et al., 2014). However, we stress that current and future microsatellite studies should use caution when making assumptions about neutrality of SSR loci, regardless of whether they are derived from genomic or transcriptomic data. We recommend that researchers who use the 1KP SSR loci test for selection in their own data sets, rather than assume that the loci are behaving in a neutral manner. In summary, we have demonstrated methods to identify loci with substantial overlap with an ORF, or that are found in translated regions. Although these methods require annotated genomic resources for the taxon of interest, researchers working with less well-studied groups may be able to identify and remove (or preferentially include, depending on the research questions) loci in translated regions by using a BLAST search on their PALs to identify closely related taxa with rich genomic resources.
LITERATURE CITED
- Aberlenc-Bertossi F., Castillo K., Tranchant-Dubreuil C., Chérif E., Ballardini M., Abdoulkader S., Gros-Balthazard M., et al. 2014. In silico mining of microsatellites in coding sequences of the date palm (Arecaceae) genome, characterization, and transferability. Applications in Plant Sciences 2: 1300058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal R. K., Hendre P. S., Varshney R. K., Bhat P. R., Krishnakumar V., Singh L. 2007. Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theoretical and Applied Genetics 114: 359–372. [DOI] [PubMed] [Google Scholar]
- Castoe T. A., Poole A. W., de Koning A. P. J., Jones K. L., Tomback D. F., Oyler-McCance S. J., Fike J. A., et al. 2012. Rapid microsatellite identification from Illumina paired-end genomic sequencing in two birds and a snake. PLoS ONE 7: e30953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P. J. A., Antao T., Chang J. T., Chapman B. A., Cox C. J., Dalke A., Friedberg I., et al. 2009. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25: 1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. 2000. Microsatellite mutations in the germline: Implications for evolutionary inference. Trends in Genetics 16: 551–558. [DOI] [PubMed] [Google Scholar]
- Esselink G. D., Nybom H., Vosman B. 2004. Assignment of allelic configuration in polyploids using the MAC-PR (microsatellite DNA allele counting–peak ratios) method. Theoretical and Applied Genetics 109: 402–408. [DOI] [PubMed] [Google Scholar]
- Gardner M. G., Fitch A. J., Bertozzi T., Lowe A. J. 2011. Rise of the machines—Recommendations for ecologists when using next generation sequencing for microsatellite development. Molecular Ecology Resources 11: 1093–1101. [DOI] [PubMed] [Google Scholar]
- Guichoux E., Lagache L., Wagner S., Chaumeil P., Léger P., Lepais O., Lepoittevin C., et al. 2011. Current trends in microsatellite genotyping. Molecular Ecology Resources 11: 591–611. [DOI] [PubMed] [Google Scholar]
- Haasl R. J., Payseur B. A. 2011. Multi-locus inference of population structure: A comparison between single nucleotide polymorphisms and microsatellites. Heredity 106: 158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel R. G. J., Segovia-Salcedo M. C., Landis J. B., Crowl A. A., Sun M., Liu X., Gitzendanner M. A., et al. 2016a. The report of my death was an exaggeration: A review for researchers using microsatellites in the 21st century. Applications in Plant Sciences 4: 1600025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel R. G. J., Gitzendanner M. A., Germain-Aubrey C. C., Liu X., Crowl A. A., Sun M., Landis J. B., et al. 2016b. Data from: A new resource for the development of SSR markers: Millions of loci from a thousand plant transcriptomes. Dryad Digital Repository. http://dx.doi.org/10.5061/dryad.rb7h0 [DOI] [PMC free article] [PubMed]
- Jennings T. N., Knaus B. J., Mullins T. D., Haig S. M., Cronn R. C. 2011. Multiplexed microsatellite recovery using massively parallel sequencing. Molecular Ecology Resources 11: 1060–1067. [DOI] [PubMed] [Google Scholar]
- Kalia R. K., Rai M. K., Kalia S., Singh R., Dhawan A. K. 2011. Microsatellite markers: An overview of the recent progress in plants. Euphytica 177: 309–334. [Google Scholar]
- Koressaar T., Remm M. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23: 1289–1291. [DOI] [PubMed] [Google Scholar]
- Loire E., Higuet D., Netter P., Achaz G. 2013. Evolution of coding microsatellites in primate genomes. Genome Biology and Evolution 5: 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasci N., Hung L.-H., Yan Z., Carpenter E. J., Wickett N. J., Mirarab S., Nguyen N., et al. 2014. Data access for the 1,000 Plants (1KP) project. GigaScience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., Hanafey M., Powell W. 2002. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nature Genetics 30: 194–200. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. 1999. Primer3 on the WWW for general users and for biologist programmers. In S. Misener and S. A. Krawetz [eds.], Methods in molecular biology, vol. 132: Bioinformatics methods and protocols, 365–386. Humana Press, Totowa, New Jersey, USA. [DOI] [PubMed] [Google Scholar]
- Schuler G. D. 1997. Sequence mapping by electronic PCR. Genome Research 7: 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan K., Varala K., Hudson M. E. 2007. Global repeat discovery and estimation of genomic copy number in a large, complex genome using a high-throughput 454 sequence survey. BMC Genomics 8: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel T., Michalek W., Varshney R. K., Graner A. 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theoretical and Applied Genetics 106: 411–422. [DOI] [PubMed] [Google Scholar]
- Triwitayakorn K., Chatkulkawin P., Kanjanawattanawong S., Sraphet S., Yoocha T., Sangsrakru D., Chanprasert J., et al. 2011. Transcriptome sequencing of Hevea brasiliensis for development of microsatellite markers and construction of a genetic linkage map. DNA Research 18: 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., Remm M., Rozen S. G. 2012. Primer3—New capabilities and interfaces. Nucleic Acids Research 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N. A., Bemmels J. B., Dick C. W. 2014. The effects of read length, quality and quantity on microsatellite discovery and primer development: From Illumina to PacBio. Molecular Ecology Resources 14: 953–965. [DOI] [PubMed] [Google Scholar]
- Xie Y., Wu G., Tang J., Luo R., Patterson J., Liu S., Huang W., et al. 2014. SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30: 1660–1666. [DOI] [PubMed] [Google Scholar]
- Zhang J., Liang S., Duan J., Wang J., Chen S., Cheng Z., Zhang Q., et al. 2012. De novo assembly and characterization of the transcriptome during seed development, and generation of genic-SSR markers in peanut (Arachis hypogaea L.). BMC Genomics 13: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Pan C., Diao Y., You Y., Yang C., Hu Z. 2013. Development of microsatellite markers by transcriptome sequencing in two species of Amorphophallus (Araceae). BMC Genomics 14: 490. [DOI] [PMC free article] [PubMed] [Google Scholar]