Summary
Studies in mice and humans have revealed intriguing associations between host genetics and the microbiome. Here we report a 16S rRNA-based analysis of the gut microbiome in 1,126 twin pairs, a subset of which was previously reported. Tripling the sample narrowed the confidence intervals around heritability estimates and uncovered additional heritable taxa, some of which are validated in other studies. Repeat sampling of subjects showed heritable taxa to be temporally stable. A candidate gene approach uncovered associations between heritable taxa and genes related to diet, metabolism and olfaction. We replicate an association between Bifidobacterium and the lactase (LCT) gene locus and identify an association between the host gene ALDH1L1 and SHA-98 bacteria, suggesting a link between formate production and blood pressure. Additional genes detected are involved in barrier defense and self/non-self recognition. Our results indicate that diet-sensing, metabolism, and immune defense are important drivers of human-microbiome co-evolution.
Graphical Abstract

eTOCBlurb
Does host gentoype shape the microbiome? Goodrich et al. present a gut microbiome analysis of 1,126 twin pairs, which extends the association between host genetics and select bacterial taxa. Lactase nonpersistence was linked to higher levels of Bifidobacteria. Other gene/microbe links relate to diet and barrier defense.
Introduction
The gut microbiome is acquired from the environment starting at birth. Diversity builds up over the first few years of life, and thereafter is largely shaped by environmental factors such as age, diet, lifestyle, hygiene, and disease state. Patterns of microbiome diversity across human populations can be pronounced, with stark contrasts in membership and structure observed in stool collected from people living on different continents (De Filippo et al., 2010; Martínez et al., 2015; Yatsunenko et al., 2012). Such population differences in microbiomes may be largely driven by diet and lifestyle, but genetic ancestry cannot at present be excluded as an important shaper of microbial diversity. Within a population, genetically encoded differences between hosts, such as those that dictate food preferences, aspects of immunity or gut physiology, in principle could impact diversity and structure of the microbiome.
For several years twin studies have provided the basis for asking whether genetic variation in the host associates with genetic variation in the microbiome. The basic premise is the following: If the genetics of the host influence a phenotype, measures of the phenotype will be more similar within monozygotic (MZ) twin pairs compared to within dizygotic (DZ) twin pairs, assuming that since they are reared together, twins within a pair experience similar environments. Given a large enough twin population, these differences can be modeled to yield estimates of heritability, or the proportion of variance in the phenotype that can be attributed to genetic differences between hosts.
Early twin studies that used culture-based methods (Van de Merwe et al., 1983) and fingerprinting of fecal 16S rRNA gene amplicons (Stewart et al., 2005; Zoetendal et al., 2001) lacked the sample sizes required to estimate heritabilities of gut microbiota (e.g., relative abundances of taxa), but their results did suggest that some part of the gut microbiome was heritable. Two subsequent larger twin studies cast some ambiguity on this conclusion, however (Turnbaugh et al., 2009; Yatsunenko et al., 2012). Using a larger number of twins (~50 pairs of the Missouri twin registry) and 16S rRNA gene sequencing, both studies reported that MZ twin pairs had slightly more similar microbiomes compared to DZ twin pairs, though the differences were not statistically significant. These studies underscored that environmental factors are paramount in shaping the microbiome, yet they also hinted at a small host genetic effect.
In Goodrich et al. (2014), we increased the power to detect heritable microbiota with a 16S rRNA gene-based analysis of 416 twin pairs. As observed for the Missouri twin studies, the UniFrac distances for MZ twins were slightly less than for DZ twins, but due to the greater sample size, the difference reached statistical significance. Importantly, the greater number of subjects allowed us to detect taxa with significant heritabilities. Among common taxa (those found in at least 50% of samples) the most heritable was the family Christensenellaceae (phylum Firmicutes), which forms the hub of a co-occurrence network composed of other heritable taxa and is enriched in lean subjects. Experiments using fecal transplants from twin donors into germfree mice demonstrated that Christensenella minuta addition to a microbiome deficient in Christensenellaceae limited adiposity gain in the recipient mice. Together, these observations supported the notion that heritable microbes could drive the human phenotypes with which they associate.
A few studies have used quantitative measures derived from the gut microbiome as phenotypes in genetic association studies. Blekhman et al. (2015) performed an analysis of human genetic data generated as a byproduct of the Human Microbiome Project (HMP) for 93 subjects (Consortium, 2012). Without accounting for population structure, ethnicity, or geographic stratification, Blekhman reported correlations between the first principal component of the host genetic variation and the first principal coordinate of the stool UniFrac distances (Blekhman et al., 2015). A second study using the HMP’s human DNA data showed that mitochondrial haplotypes correlated with abundances of specific microbiota in stool (Ma et al., 2014). Both results reflect ancestry effects on the composition of the microbiome.
Diet differs among subjects and can rapidly alter microbial community composition (David et al., 2014), casting some doubt over the use of relative abundance data in genetic association studies. Davenport et al. (2015) circumvented this issue with a genome-wide analysis of the fecal microbiome composition in the Hutterites, a founder population that lives and eats communally (Davenport et al., 2015, 2014). These analyses revealed a suite of heritable taxa and highlighted links between microbial taxa and genes involved in barrier defense and immunity.
Studies in mice further reduce the environmental influences on the microbiome. Benson et al. (2010) performed the first quantitative trait loci (QTL) mapping study, using 645 mice from an advanced intercross line (Benson et al., 2010). QTL analysis detected thirteen genetic loci that were significantly associated with microbial abundances, and five additional suggestive loci. Many of the QTL were in regions with genes that play roles in the immune system. Subsequent studies in mice have confirmed some of these results (Leamy et al., 2014; McKnite et al., 2012). Comparisons across mouse and human studies highlight some common themes: genetic control of certain bacterial taxa (e.g., Turicibacter) and associations with immune-related genes.
Here, we report heritability and gene-association analyses for the TwinsUK cohort. We tripled the size of our initial dataset of Goodrich et al. (2014), now including 3,261 fecal samples from 2,731 individuals. This includes 489 dizygotic (DZ) twin pairs, 637 monozygotic (MZ) twin pairs and 530 samples collected at a second time-point. The goals of this study are to (i) calculate the heritabilities of specific components of the gut microbiota and (ii) associate abundances of microbes with host gene alleles through candidate gene analysis and genome-wide association. Using this expanded dataset we improve heritability estimates for taxa previously identified as heritable, expand the list of heritable taxa, and identify associations between a subset of the heritable taxa and candidate genes related to diet and immunity.
Results
Effect of an expanded dataset on heritability results
In our initial study of 416 twin pairs we estimated heritability for 909 widely shared taxa (Operational taxonomic units (OTU) and taxonomy bins). Heritability analysis revealed 5.3% of taxa had a heritability (A, see methods) greater than 0.2 with 95% confidence intervals that did not overlap zero. In the expanded sample set, we examined 945 taxa and this percentage was increased to 8.8%.
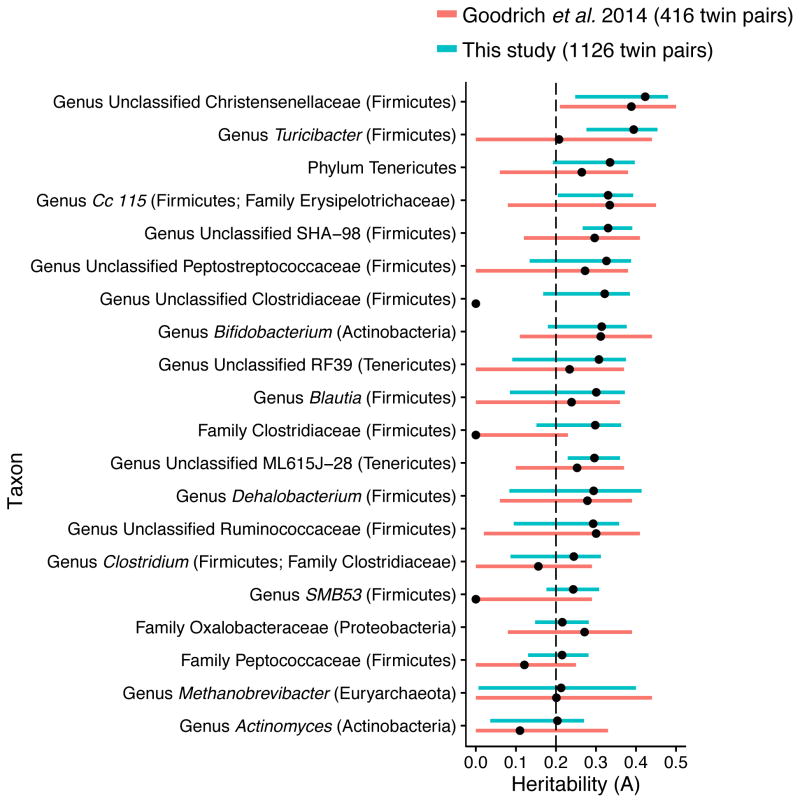
We found a significant correlation between the heritability estimates from the tripled sample set compared to the initial set (r = 0.57, P-value < 2.2 × 10−16; Figure 1 and Figure S1 and Table S1). Members of the Bacteroidetes phylum were generally not heritable, while taxa with heritability estimates A>0.2 belong to the Firmicutes, Actinobacteria, Tenericutes, and Euryarchaeota (Figure 1). The most heritable taxon in the expanded dataset was again the bacterial family Christensenellaceae (A = 0.42, 95% CI = 0.25 – 0.48). Most of the taxa with a heritability greater that 0.2 in the initial set maintained a high heritability. The main difference between sample sets is a decrease in the size of the confidence intervals. The ranking of heritabilities was slightly altered, notably, the genus Turicibacter jumped in the ranked heritability list from A = 0.21 (with a 95% confidence interval that overlapped zero) to A = 0.39 (95% CI = 0.28 – 0.45; Figure 1).
Figure 1. Expansion of the TwinsUK dataset results in similar microbial abundance heritability estimates with narrower confidence intervals.
Each point represents the heritability estimated as the proportion of variance in the microbial abundances (OTU abundances collapsed by taxonomic classification) that can be attributed to genetic effects (A). The bars show the 95% confidence intervals around the heritability estimates. Bars are colored by the dataset: Pink indicates heritability estimates reported in Goodrich et al. 2014 (171 MZ, 245 DZ) and Blue indicates heritability estimates using an additional 710 twin pairs (Blue; 637 MZ, 489 DZ twin pairs). The figure includes only taxa that are present in at least 50% of the TwinsUK participants and have A > 0.2 in the increased dataset. We also excluded any taxon that was highly correlated (r > 0.9) with another taxon at a lower taxonomic level. The Clostridium genus is known to be polyphyletic, however further examination revealed that the genus Clostridium within the Clostridiaceae family consists of mostly one Greengenes OTU 4465124 that is shared by 82.6% of the samples. See also Figure S1 and Table S1.
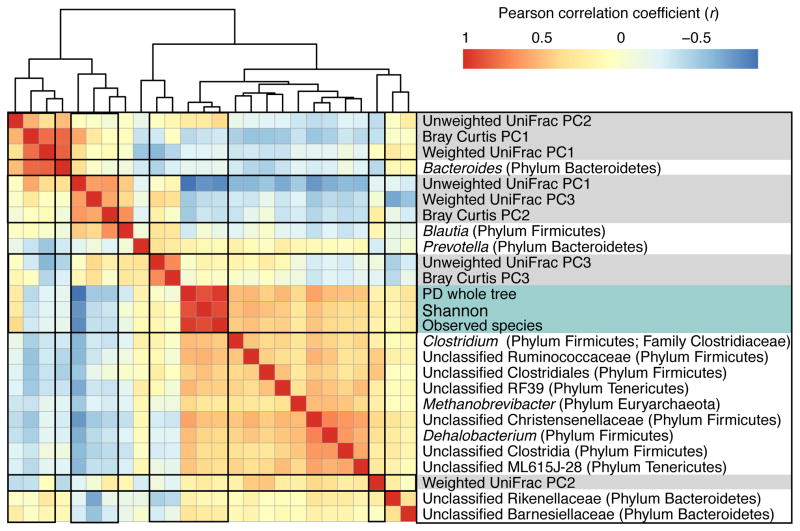
Additional taxa reached a heritability threshold of 0.2 when we tripled the size of the dataset (Figure 1). Importantly, the genus Methanobrevibacter, the predominant human gut archaeon, reached a significant level of heritability. Other taxa included the genera SMB53 and Actinomyces, and the families Clostridiaceae and Peptococcaceae. Measures of alpha-diversity were also heritable in the expanded dataset (Table S2; PD whole tree: A = 0.37, 95% CI = 0.17 – 0.44). Alpha-diversity is positively correlated with the relative abundances of the genus Methanobrevibacter (r = −0.37, BH adjusted P-value < 1 × 10−10), the family Christensenellaceae (r = −0.59, BH adjusted P-value < 1 × 10−10), and other members of its co-occurrence consortium (Figure 2). Alpha-diversity is also highly correlated with the first principal coordinate (PC) of principal coordinates analysis (PCoA) of the unweighted UniFrac distances (r = −0.90, Benjamini-Hochberg (BH) adjusted P-value < 1 × 10−10; Figure 2), most likely because its associated taxa drive differentiation of samples along PC1.
Figure 2. Alpha- and Beta- diversity are correlated with several microbiota that have a heritability estimate > 0.2.
Heatmap showing the correlation structure between the alpha-diversity metrics, the first three principal coordinates of the beta-diversity distance matrices, and the taxa that are correlated at |r| > 0.5 with one of the alpha- or beta- diversity metrics in the heatmap. See also Table S2.
Heritable microbes and temporal stability
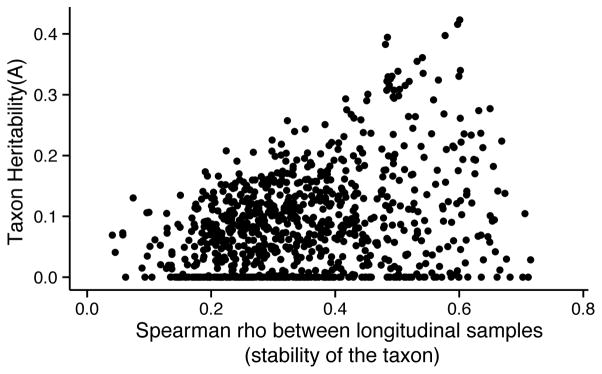
We collected repeat samples for a subset of the twins (530 samples were spaced a mean 946 +/− s.e.m. 15 days; Figure 3). Although non-heritable taxa displayed a wide range of stability, highly heritable taxa were associated with higher levels of stability, and absent from the low-end of the stability gradient.
Figure 3. Heritable taxa are among the most stable taxa in the TwinsUK dataset.
Heritability (A) of the microbial abundances (OTU abundances and abundances collapsed by taxonomic classification) are plotted against the Spearman correlation between longitudinal samples (530 individuals with samples collected at a second time-point spaced 946 +/− 15 days (mean +/− s.e.m)).
Candidate genes and single nucleotide polymorphisms (SNPs)
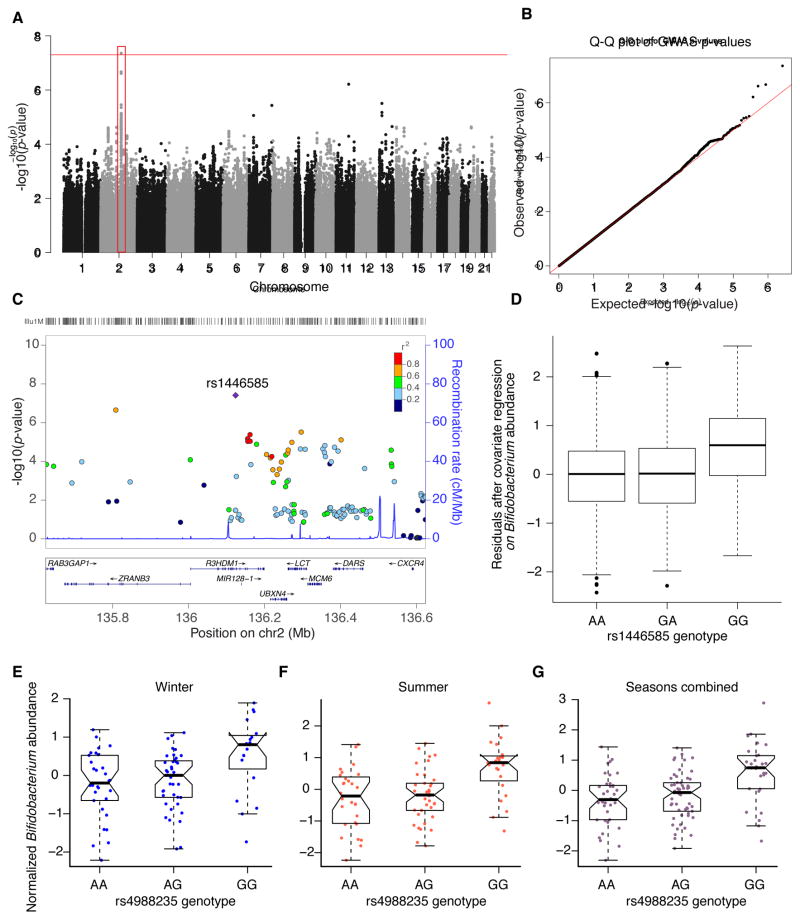
2,139 individuals (including 472 MZ twin pairs and 418 DZ twins pairs) had both genotype and 16S rRNA gene sequence data. We assembled a list of 37 sets of candidate genes or SNPs with suspected roles in shaping the microbiome (Table 2, S3 and S4). Included are genes implicated in previous mouse or human studies, genetic variants and genes associated with gut microbiome-related diseases, and genes involved in taste reception. To reduce the burden of multiple testing we performed association analysis on 20 heritable taxa (A > 0.2; Figure 1). Table 3 details the SNP-microbe associations for the significant candidate sets. We validated the association between Bifidobacterium and LCT reported by Blekhman et al. (LCT permutation P-value <0.001). Additionally, the candidate set analysis revealed an association between Bifidobacterium and a SNP within the gene RABGAP1, which is in linkage disequilibrium (LD) with variants in LCT (Figure 4). Given that these genes are in LD, it is not possible to know which of the genes is truly driving the association.
Figure 4. Relative abundances of the heritable genus Bifidobacterium are associated with genetic variants in the genomic locus containing the gene LCT.
(A) Genome-wide Manhattan plot: the x-axis represents the chromosome and position along the chromosome, and the y-axis is −log of the P-value for the association of the SNP (each dot) with the genus Bifidobacterium. The red box highlights the associated locus on chromosome 2 that contains the gene LCT. (B) Quantile-Quantile (Q-Q) plots of the P-values. The Q-Q plot measures deviation from the expected distribution of P-values. The diagonal (red) line represents the expected (null) distribution. (C) Close-up plots of a 1 Mb window around the SNP with the highest association, the coloring of the points represents r2 between a SNP and the SNP with the highest association in the locus (rs1446585, denoted by the purple diamond); r2 is calculated from the 1000 Genomes data on the CEU population. (D) Box-plot of Bifidobacterium normalized abundances within each genotype at the most strongly associated SNP (rs1446585; P-value = 4.38 × 10−8), the y-axis depicts the residuals from linear regression of the Box-Cox transformed abundances with the covariates (the number of 16S rRNA gene sequences per sample, age, gender, shipment date, collection method (postal or visit), ID of technician performing DNA extraction). (E–G). Normalized Bifidobacterium abundance within each genotype at the lactase persistence-associated SNP (rs4988235) in the Hutterite dataset. (E) Winter samples (P-value = 0.02). (F) Summer samples (P-value = 0.001). (G) Seasons combined (P-value = 4 × 10 −5). See also Table S5 and Table S6.
We observed a significant association between heritable taxa and the olfactory receptor gene OR6A2 (linked to cilantro soapy-taste (Eriksson et al., 2012); permutation P-value = 0.011) which was driven by the Erysipelotrichaceae Cc 115 genus. The gene CD36 had a significant association driven by the genus Blautia (permutation P-value = 0.009). CD36 is involved in a variety of functions, including long chain fatty acid tasting on the tongue (Silverstein and Febbraio, 2009). We also observed an association between the order SHA-98 (a component of the Christensenellaceae consortium) and the gene ALDH1L1 in the one-carbon metabolism gene set (permutation P-value = 0.006). Within a SNP set associated with IBD (Jostins et al., 2012), we detected a suggestive association (permutation P-value = 0.056) driven by the genus SMB53 and variants in the gene GNA12, involved in barrier defense and associated with ulcerative colitis (Lees et al., 2011).
GWAS with the expanded TwinsUK dataset
The above testing with candidate genes was sufficiently successful to suggest that genome-wide association analysis may reveal additional associations. When using the full dataset (945 taxa and 1,300,091 SNPs), no associations reached study-wide significance (correction for all tests; Table S5). The strongest GWA signal among the 20 heritable taxa used in the gene set analysis was the “unclassified Clostridiaceae” with SNP rs10055309 in the gene SLIT3 (P-value 1.20 × 10−8, BH adjusted P-value = 0.016). This taxon bin consists of several OTUs, but the OTU making up the majority of sequence counts is Greengenes OTU 4434334. The “unclassified Clostridiaceae” taxon bin is also highly correlated with the Clostridiaceae family (r = 0.89), which is also associated with the same SLIT3 SNP (P-value = 4.21 × 10−6, BH adjusted P-value = 0.14).
Interestingly, GWA revealed that the SNP with the strongest association to Bifidobacterium lies within the gene R3HDM1 (P-value = 4.38 × 10−8, BH adjusted P-value = 0.057). However, several genes in this locus are in strong linkage disequilibrium (LD), including LCT and the gene RABGAP1 mentioned above (Figure 4). Although our genotype dataset does not contain the SNP (rs4988235) that correlates directly with lactase persistence, it is in LD with the SNP (rs1446585; 1000 Genomes Phase 3 GBR population r2 = 0.89), which in our dataset has the greatest association with Bifidobacterium. In our analysis, individuals who carry the rs1446585(G) allele associated with lactase nonpersistence have higher levels of Bifidobacteria in stool.
For validation we examined the association of the lactase persistence-associated SNP (rs4988235) with the relative abundance of Bifidobacterium using data from a GWAS on the fecal microbiome composition in the Hutterites in two seasons (Davenport et al., 2015). As observed in the TwinsUK dataset, nonpersisters have significantly higher levels of Bifidobacteria (winter P-value = 0.02, summer P-value = 0.001, seasons combined P-value = 4 × 10−5; Figure 4E–G)
GWAS for beta-diversity measures
To examine the association between genetic variation and beta-diversity metrics, including Bray Curtis dissimilarity, weighted UniFrac distance, and unweighted UniFrac distance, we applied microbiomeGWAS (Hua et al., 2015) to a subset of our dataset that included only unrelated individuals (n = 1,248; see Experimental Procedures). A SNP (rs563779) within the gene UHRF2 was associated with weighted UniFrac distance at a study-wide significance threshold (P-value= 9.77 × 10−9), while two SNPs in LD on chromosome 4 are associated with Bray Curtis dissimilarity at a relaxed threshold (rs9997915, P-value = 1.44 × 10−8; rs1593554, P-value = 1.16 × 10−8).
Associations with imputed gene expression
We utilized a gene expression based approach using the PrediXcan framework, in which SNPs are used to infer gene expression across a range of tissues (Gamazon et al., 2015). Association testing is then performed between taxon abundance and the imputed gene expression values for each tissue. In addition to reducing the multiple-testing burden, a strength of this approach is the interpretability of the results, as imputed gene expression is a biologically plausible intermediate phenotype through which inter-individual genetic variation may act to influence microbiome composition in the gut. We used PrediXcan to obtain imputed gene expression values for 40 tissues and performed association testing for each of those tissues with each of the taxa. While no genetic associations met study-wide significance, inferred expression of SIGLEC15 was associated with the abundance of Akkermansia at a tissue-wide significance level in transverse colon (P = 6.21 × 10−9). Additionally, several other taxa were associated to genes at a significance threshold of P = 5 × 10−8 (Table S6).
Discussion
We report heritability and genome-wide association analyses for fecal microbiome data obtained from 1,126 twin pairs from the TwinsUK registry. Heritability estimates were broadly similar when tripling the number of subjects from our previous report (Goodrich et al., 2014), and the confidence intervals were narrower with the expanded dataset, making the estimates more robust. The number of heritable taxa was increased, and alpha-diversity was also found to be heritable.
To put microbiome heritabilities in context, they are a little lower than those of other complex traits measured in the same population: Systolic Blood pressure (0.51; Menni et al., 2013), anxiety (0.44; Davies et al., 2015) and serum Vitamin D (0.43; Hunter et al., 2001). In a meta-analysis paper of twin studies, the average heritability for diseases of the digestive system is 0.31 (Polderman et al., 2015). Although on the low side, these results for 16S rRNA gene data from stool suggest that heritabilities for the microbiome may be refined and increased with sample types other than stool, and/or other data types.
We showed that heritable taxa are temporally stable over long periods suggesting a lesser effect of environmental factors on their relative abundances. This could be explained by a strong dependence of these heritable taxa on host physiology or metabolism. Deeper analysis of strains within these taxa may reveal adaptations to specific host genotypes, as has been shown for Helicobacter pylori strains in the stomach and host genotypes (Suerbaum and Josenhans, 2007). Several of the heritable and stable taxa have also been associated with genetic loci in mouse QTL studies. This suggests that the host-microbial connections are widespread in mammals, and may be ancient associations.
The twins in this study were genotyped, allowing for tests of association between SNPs and microbiome traits. Because there is a set of candidate genes whose function gives them a strong prior expectation to be relevant to gut function, we focused on these tests first. Candidate gene association tests uncovered associations between heritable taxa and genes related to diet, carbohydrate metabolism and olfaction, barrier defense and self/non-self recognition. Associations of genes involved in barrier defense and immunity have also been observed in mouse QTL studies (Benson et al., 2010, McKnite et al., 2012, Org et al., 2015) and plant microbiome GWAS (Horton et al., 2014).
Bifidobacterium and the LCT gene locus
The association for which we have the highest confidence is between Bifidobacterium and the LCT gene locus (Figure 4). Bifidobacterium is heritable in the TwinsUK population, the HMP, the Hutterites, and in mouse studies (Figure 5). LCT encodes lactase, the enzyme that hydrolyzes lactose in the upper gastrointestinal tract. In mammals, lactase production ceases at weaning, but lactase persistence has evolved independently several times in a subset of humans. Lactase nonpersisters may experience lactose intolerance when lactose reaches the large intestine and is fermented. The direction of the genetic association shows lactase nonpersisters to harbor higher levels of Bifidobacterium. Since Bifidobacteria are members of the large intestine and metabolize lactose, one can speculate that lactase persisters harbor lower levels of Bifidobacteria because of low lactose availability, whereas nonpersisters who ingest milk provide relatively more lactose to Bifidobacteria.
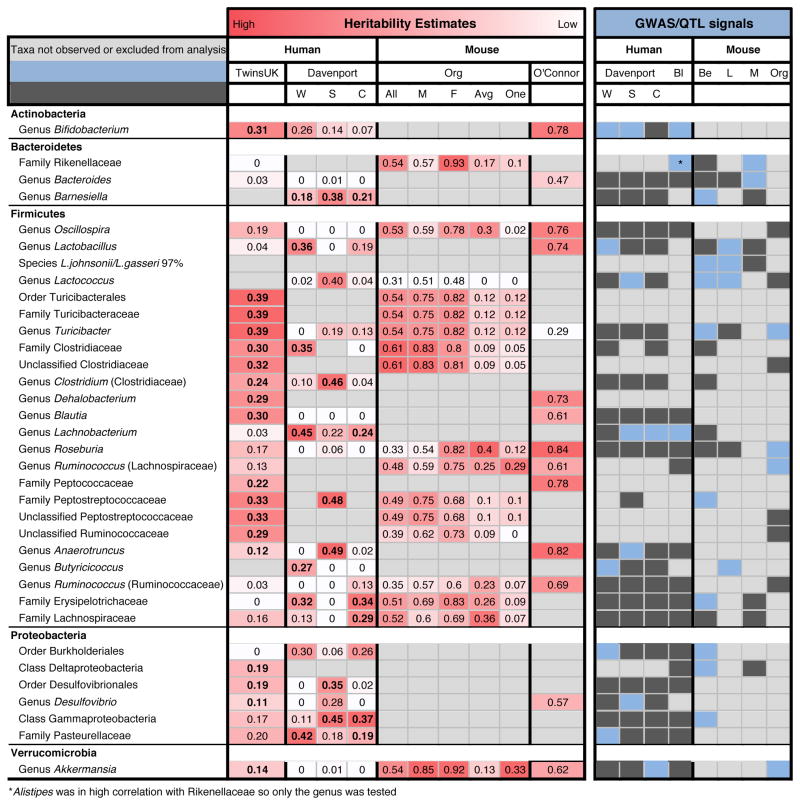
Figure 5. Comparison of taxa estimated as heritable or linked to genes in at least two human GWA or mouse QTL studies.
The color gradient over the heritability estimates ranges from the lowest heritability estimate (White) to the highest heritability estimate (Red) in the given study. For the TwinsUK heritability estimates the bold values indicate heritability estimates with a 95% confidence interval not overlapping 0. The estimates for Davenport et al. (2015) are the proportion of variance explained (PVE) estimates (“chip heritability”). We report the Winter (W), Summer (S), and seasons Combined (C) datasets. For the Davenport study bold values indicate heritability estimates with a standard error not overlapping 0. For Org et al. (2015) we report results using all mice (All), just males (M), just females (F), using an average per strain (Avg), and using a single mouse per strain (One). No significance value was reported for the Org and O’Connor heritability estimates. The coloring over the QTL/GWAS studies indicates if each taxon had a significant association (Blue) or not (Dark Grey) in the given study. Light grey indicates that the taxon was not observed in the given study or was excluded from the studies analysis for other reasons. The QTL and GWAS studies shown in the figure are the top GWAS hits from Davenport et al. (2015) (Davenport), the top stool GWAS results from Blekhman et al. (2015) (Bl), QTL results form Benson et al. (2010) (B), QTL results from Leamy et al. (2014) (L), QTL results from McKnite et al. (2012) (M), and GWAS results from Org et al. (2015) (Org). See Experimental Procedures for more details.
Turicibacter and the Peptostreptococcaceae
The genus Turicibacter is the second most heritable taxon in our analysis. Turicibacter is the sole genus of the family Turicibacteriaceae (Firmicutes), and its relative abundance is correlated with Peptostreptococcaceae (r=0.66). The host genetic associations with these taxa are validated in humans and in mice (Figure 5). Benson et al. (2010) associated Turicibacter with a QTL on MMU7 and Peptostreptococcaceae with a QTL on MMU1, but so far we have not detected any allelic associations with Turicibacter by GWA, in the candidate gene sets tested, or by pathway enrichment.
Both taxa are active members of the small intestinal microbiome (Bouhnik et al., 1999; Kamada et al., 2013; Leimena et al., 2013; Looft et al., 2014; Oh et al., 2012). Turicibacter has been linked to host immunity (Kellermayer et al., 2011; Dimitriu et al., 2013; Rausch et al., 2015) and is positively associated with inflammation (Rausch et al., 2015). Turicibacter has been isolated from blood of human patients, and analysis of its genome reveals genes consistent with a pathobiont lifestyle (Cuív et al., 2011). Oddly, Turicibacter has also been noted as highly abundant at later stages of dead pig decomposition (Yang et al., 2012). Turicibacter thus appears to be a pathobiont with the genes necessary to exploit host inflammation and that can also subsist on the host after death.
Akkermansia
We noted an association between Akkermansia and predicted tissue-specific expression levels of the gene SIGLEC15. This gene encodes a sialic acid-binding immunoglobulin-like lectin that participates in the discrimination of self and non-self and is highly conserved across mammals (Angata et al., 2007; Macauley et al., 2014). The physical niche of the genus Akkermansia is the intestinal mucus where it uses mucins as a carbon source (Derrien et al., 2004). Sialic acid is one the outermost carbohydrate decorations of mucin. Akkermansia can cleave sialic acids: Its genome encodes four neuraminidase enzymes responsible for the removal of sialic acids from glycoconjugates (Huang et al., 2015). SIGLEC15 is expressed on macrophages and DCs in humans, and is reported to be expressed in ileum tips in mice (Sommer et al., 2015). Another gene reported by Sommer et al. (2015) to be expressed at the villus tips of the small intestine, PLD1, was linked genetically with Akkermansia in the Hutterites (Davenport et al., 2015). Given that both studies associate Akkermansia with genes expressed in its physical niche space, an investigation of host genetic interaction with this important bacterium is warranted.
Other gene-taxon associations
We provide here for future reference other gene-microbe associations from this study requiring validation. The genus Blautia is heritable in this dataset and in the mouse study of O’Connor (Figure 5). Blautia associated here with CD36, a gene involved in fat sensing on the tongue and the promotion of absorption of long-chain fatty acids in the gut (Silverstein and Febbraio, 2009). Interestingly, Omega-3 fatty acid feeding has been shown to increase levels of Blautia in mice (Myles et al., 2014), and mice associated with a simplified community including Blautia show upregulation of CD36 (Woting et al., 2014).
Our strongest signal in the GWAS was between unclassified Clostridiaceae and the SNP rs10055309 in the gene SLIT3. SLIT3 is a secreted protein with high expression in the glandular cells of the stomach, duodenum and small intestine (Sanz-Pamplona et al., 2014). SLIT3 was reported as frequently methylated in colorectal cancers (Dickinson et al., 2004) has been shown to exhibit dysregulated exons in colorectal adenomas (Pesson et al., 2014). Notably, SLIT3 expression was shown to be upregulated in colon crypts when germfree mice are conventionalized (Sommer et al., 2015). Until these gene-microbe links are validated they are essentially hypothetical, however, it is interesting that the gene products are expressed in the gut epithelium.
The Christensenellaceae consortium
The family Christensenellaceae remained the most highly heritable taxon. Taxa we reported as heritable and which had relative abundances correlated to those of the Christensenellaceae family (SHA-98, Dehalobacterium, RF39, ML615J–28) were again heritable in the larger dataset.
The genus Methanobrevibacter, a member of the Christensenellaceae consortium, also reached a significant level of heritability in this analysis. The abundances of taxa within the heritable consortium correlate positively with alpha-diversity, also heritable. Whether the methanogenesis and associated fermentative dynamics drive a more diverse microbiota, or higher levels of methanogens and species richness are both results of other factors, such as a high-fiber diet, remains to be ascertained.
Methanobrevibacter smithii is the dominant methanogen in the human gut and MZ twins have previously been shown to have greater concordance for the carriage of this archaeon than DZ twins (Hansen et al., 2011). Studies across mammal species (Hackstein and van Alen, 1996) and within bovine lines (Roehe et al., 2016) have also suggested that host genetics influence levels of methanogens. Methanogen carriage has been associated with leanness and with a better metabolic profile in obese humans (Le Chatelier et al., 2013). Given a fermentable diet, a genetic predisposition for high methanogen levels may lead to methane production and an overall metabolism that partly explain the leaner phenotype associated with the Christensenellaceae consortium.
We observed an association between the order SHA-98 (a member of the Christensenellaceae consortium) and ALDH1L1, which codes for an aldehyde dehydrogenase involved in formate oxidation. Formate is produced endogenously by the host and is also a fermentation product that acts as a major interspecies electron carrier between syntrophs (Boone et al., 1989; Pavlostathis et al., 1990). Production of formate by a Clostridium sp. has been shown to promote growth of a syntroph in a gnotobiotic mouse model (Rey et al., 2013). Methanobrevibacter smithii can use formate in methanogenesis (Miller et al., 1982). Formate concentration in urine has been shown to be significantly correlated with systolic and diastolic blood pressure in 4,630 subjects in Asia, USA and Europe (Holmes et al., 2008). Holmes et al. (2008) also reported that urinary formate and Na+ excretion were positively correlated, and given the importance of Na+ in blood pressure, suggested an unrecognized role for formate in its regulation. Hypertension is a major risk factor for stroke, and a SNP in ALDH1L1 has also been associated with ischemic stroke (Xie et al., 2013). Interestingly, the microbiomeGWAS tool, which uses beta-diversity distances in GWAS, also revealed an association between weighted UniFrac and a SNP in the gene UHRF2, which has been identified as linked to ischemic stroke as well. Our results suggest the hypothesis that the Christensenellaceae-methanogen consortium regulates the thermodynamics of fermentation in the gut, including formate production and consumption. This activity interacts with the host’s own enzymatic activities to impact formate levels, with repercussions for blood pressure.
Conclusion
Our results highlight gene-microbe interactions from recent evolutionary adaptation to diet, its sensing and metabolism in the gut. In the case of the Bifidobacterium-LCT link, host genetics most likely shape the microbiome through diet preference, which itself is heritable. These signals contrast with the immune-related genes uncovered in studies where diet is controlled. For links to immune genes to be detected in human populations where diet is unrestricted, very large numbers of subjects may be necessary.
Most microbiome-genetic studies use 16S rRNA gene diversity as the phenotype, and other data types, such as metagenomic data will focus on functions which can be shared across taxa. However, any sequence based results are limited by their nature as relative abundances. Ultimately, results of sequence-based studies require validation with quantitative measures such as qPCR. The gene-microbe associations uncovered here will require validation across multiple studies, but they support incorporating measures of diet and microbiome in studies seeking a genetic basis for disease risk susceptibility, particularly for diseases involving chronic conditions of over and under-nutrition.
Experimental Procedures
Sample collection
All work involving human subjects was approved by the Cornell University IRB (Protocol ID 1108002388). Fecal samples were collected as described previously (Goodrich et al., 2014; Jackson et al., 2015). The sample set consists of 3,261 fecal samples from 2,731 individuals (530 individuals sampled at a second time-point).
Sample processing and 16S rRNA gene sequencing
DNA extraction, amplification of the V4 hypervariable region of the 16S rRNA gene (primers 515F and 806R), purification and pooling was performed on all fecal samples as previously described (Goodrich et al., 2014). The pooled amplicons were sequenced using the Illumina Miseq platform with 2×250bp paired-end sequencing.
16S rRNA gene data analysis
Mate-pair merging, de-multiplexing, quality control and OTU picking were performed using QIIME version 1.8 (Quantitative Insights Into Microbial Ecology; Caporaso et al., 2010). Taxa filtering, covariate regression, and details on the correlations between diversity metrics and taxa are described in the Supplemental Information.
Heritability calculations
The ACE model (Eaves et al., 1978) was used to estimate heritability for 945 taxa, three alpha-diversity metrics, and the first three PCs from PCoA of each of the beta-diversity metrics (details provided in Supplemental Information). P-value adjustment to correct for all 945 taxa was done using the Benjamini-Hochberg algorithm.
Stability of microbiota
530 individuals (including 125 DZ twin pairs and 69 MZ twin pairs) supplied two or more serial fecal samples spaced 3 to 1632 days apart (946 +/− 15 days (mean +/− s.e.m.)). For the stability analysis two samples were randomly chosen from each individual. Spearman correlation coefficients between longitudinal samples were calculated for each taxon.
Host genetic association analyses
The participants in this study were previously genotyped and the genotype data imputed using IMPUTE version 2 (Howie et al., 2009), and quality checked as previously described (Moayyeri et al., 2013). GEMMA (v0.94) was used to perform SNP-microbe association tests (Zhou et al., 2012). Association analysis details for the candidate gene and SNP sets, taxon genome-wide analysis, and imputed gene expression are provided in Supplemental Information. The tool microbiomeGWAS (Hua et al., 2015) was used to perform a GWAS on the beta-diversity measures. Only one twin per family was included in this analysis, for a total sample size of 1,248 individuals.
Bifidobacterium validation in the Hutterite dataset
16S rRNA gene sequencing analysis and genotyping is described in the original study (Davenport et al., 2015). Briefly, the dataset consists of data from the Hutterites sampled during two seasons (summer n = 91; winter n = 93; seasons combined n = 127). GEMMA was used to perform the association analysis between Bifidobacterium and the SNP rs4988235.
Cross-study comparisons of taxa influenced by host genetics
We compiled heritability and QTL/GWAS results from published human and mouse studies and summarize these results in Figure 5. For the human studies this includes the current study, the Hutterites (Davenport et al., 2015) and the HMP (Blekhman et al., 2015). The mouse studies examined are the advanced intercross lines (Benson et al., 2010; Leamy et al., 2014), the Hybrid Mouse Diversity Panel (Org et al., 2015), collaborative cross/diversity outbred mapping panels (O’Connor et al., 2014), and recombinant inbred strains (McKnite et al., 2012). Details are provided in Supplemental Information.
Supplementary Material
Highlights.
16S rRNA-based analysis of the gut microbiome in 1,126 twin pairs
Heritable bacterial taxa are temporally stable
Bifidobacterium associates with lactase; formate production links to blood pressure
Gene-microbe links involve genes related to diet, metabolism, oflaction, defense
Acknowledgments
We thank the participants of the TwinsUK registry. We thank Jessica Sutter, Qiaojuan Shi, Jillian Waters, Gabriela Surdulescu and Ayrun Nessa. This work was funded by NIH RO1 DK093595, DP2 OD007444, a David and Lucile Packard Foundation Fellowship (R.E.L.), the Arnold and Mabel Beckman Foundation (R.E.L.), The Cornell Center for Comparative Population Genomics (R.E.L., J.K.G.), a National Science Foundation Graduate Fellowship (J.K.G.), The Wellcome Trust, the European Community’s Seventh Framework Programme (FP7/2007-2013). The study also receives support from the National Institute for Health Research (NIHR) BioResource Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London.
Footnotes
Supplemental Information includes Extended Experimental Procedures and data and can be found with this article online.
Author Contributions. R.E.L. and A.G.C. supervised the study, and J.T.B. and T.D.S. helped design study and provided comments and discussion. J.T.B. and T.D.S. oversaw collection of samples; J.K.G., R.E.L., and E.R.D. oversaw microbial data generation; J.K.G. and E.R.D performed the analysis with contributions from C.O., R. K., M.B. and M.A.J.; J.K.G., E.R.D. and R.E.L. prepared the manuscript, with comments from A.G.C, T.D.S, J.T.B, and M.A.J.
Accession Numbers. The 16S rRNA gene sequences have been deposited in the European Nucleotide Archive (ENA), European Bioinformatics Institute, with accession number XX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angata T, Tabuchi Y, Nakamura K, Nakamura M. Siglec-15: an immune system Siglec conserved throughout vertebrate evolution. Glycobiology. 2007;17:838–846. doi: 10.1093/glycob/cwm049. [DOI] [PubMed] [Google Scholar]
- Bachmanov AA, Beauchamp GK. Taste receptor genes. Annu Rev Nutr. 2007;27:389–414. doi: 10.1146/annurev.nutr.26.061505.111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, Spector TD, Keinan A, Ley RE, Gevers D, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DR, Johnson RL, Liu Y. Diffusion of the Interspecies Electron Carriers H(2) and Formate in Methanogenic Ecosystems and Its Implications in the Measurement of K(m) for H(2) or Formate Uptake. Appl Environ Microbiol. 1989;55:1735–1741. doi: 10.1128/aem.55.7.1735-1741.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhnik Y, Alain S, Attar A, Flourié B, Raskine L, Sanson-Le Pors MJ, Rambaud JC. Bacterial populations contaminating the upper gut in patients with small intestinal bacterial overgrowth syndrome. Am J Gastroenterol. 1999;94:1327–1331. doi: 10.1111/j.1572-0241.1999.01016.x. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter D, Dhar S, Mitchell LM, Fu B, Tyson J, Shwan NAA, Yang F, Thomas MG, Armour JAL. Obesity, starch digestion and amylase: association between copy number variants at human salivary (AMY1) and pancreatic (AMY2) amylase genes. Hum Mol Genet. 2015;24:3472–3480. doi: 10.1093/hmg/ddv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium H. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuív PÓ, Klaassens ES, Durkin AS, Harkins DM, Foster L, McCorrison J, Torralba M, Nelson KE, Morrison M. Draft genome sequence of Turicibacter sanguinis PC909, isolated from human feces. J Bacteriol. 2011;193:1288–1289. doi: 10.1128/JB.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, Gilad Y. Genome-Wide Association Studies of the Human Gut Microbiota. PLoS One. 2015;10:e0140301. doi: 10.1371/journal.pone.0140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport ER, Mizrahi-Man O, Michelini K, Barreiro LB, Ober C, Gilad Y. Seasonal variation in human gut microbiome composition. PLoS One. 2014;9:e90731. doi: 10.1371/journal.pone.0090731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Verdi S, Burri A, Trzaskowski M, Lee M, Hettema JM, Jansen R, Boomsma DI, Spector TD. Generalised Anxiety Disorder--A Twin Study of Genetic Architecture, Genome-Wide Association and Differential Gene Expression. PLoS One. 2015;10:e0134865. doi: 10.1371/journal.pone.0134865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- Deschamps M, Laval G, Fagny M, Itan Y, Abel L, Casanova JL, Patin E, Quintana-Murci L. Genomic Signatures of Selective Pressures and Introgression from Archaic Hominins at Human Innate Immunity Genes. Am J Hum Genet. 2016;98:5–21. doi: 10.1016/j.ajhg.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RE, Dallol A, Bieche I, Krex D, Morton D, Maher ER, Latif F. Epigenetic inactivation of SLIT3 and SLIT1 genes in human cancers. Br J Cancer. 2004;91:2071–2078. doi: 10.1038/sj.bjc.6602222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriu PA, Boyce G, Samarakoon A, Hartmann M, Johnson P, Mohn WW. Temporal stability of the mouse gut microbiota in relation to innate and adaptive immunity. Environ Microbiol Rep. 2013;5:200–210. doi: 10.1111/j.1758-2229.2012.00393.x. [DOI] [PubMed] [Google Scholar]
- Eaves LJ, Last KA, Young PA, Martin NG. Model-fitting approaches to the analysis of human behaviour. Heredity. 1978;41:249–320. doi: 10.1038/hdy.1978.101. [DOI] [PubMed] [Google Scholar]
- Eriksson N, Wu S, Do CB, Kiefer AK, Tung JY, Mountain JL, Hinds DA, Francke U. A genetic variant near olfactory receptor genes influences cilantro preference. Flavour. 2012;1:1–7. [Google Scholar]
- Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, Nicolae DL, et al. Consortium GT. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstein JHP, van Alen TA. Fecal Methanogens and Vertebrate Evolution. Evolution. 1996;50:559–572. doi: 10.1111/j.1558-5646.1996.tb03868.x. [DOI] [PubMed] [Google Scholar]
- Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, et al. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfield GM, Chapman RW, Karlsen TH, Lammert F, Lazaridis KN, Mason AL. The genetics of complex cholestatic disorders. Gastroenterology. 2013;144:1357–1374. doi: 10.1053/j.gastro.2013.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, Loo RL, Stamler J, Bictash M, Yap IKS, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton MW, Bodenhausen N, Beilsmith K, Meng D, Muegge BD, Subramanian S, Vetter MM, Vilhjálmsson BJ, Nordborg M, Gordon JI, et al. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat Commun. 2014;5:5320. doi: 10.1038/ncomms6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Song L, Yu G, Goedert JJ, Abnet CC, Landi MT, Shi J. MicrobiomeGWAS: a tool for identifying host genetic variants associated with microbiome composition. 2015 doi: 10.3390/genes13071224. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Wang MM, Kulinich A, Yao HL, Ma HY, Martínez JER, Duan XC, Chen H, Cai ZP, Flitsch SL, et al. Biochemical characterisation of the neuraminidase pool of the human gut symbiont Akkermansia muciniphila. Carbohydr Res. 2015;415:60–65. doi: 10.1016/j.carres.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Hunter D, De Lange M, Snieder H, MacGregor AJ, Swaminathan R, Thakker RV, Spector TD. Genetic contribution to bone metabolism, calcium excretion, and vitamin D and parathyroid hormone regulation. J Bone Miner Res. 2001;16:371–378. doi: 10.1359/jbmr.2001.16.2.371. [DOI] [PubMed] [Google Scholar]
- Jackson MA, Goodrich JK, Maxan M-E, Freedberg DE, Abrams JA, Poole AC, Sutter JL, Welter D, Ley RE, Bell JT, et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut. 2015 doi: 10.1136/gutjnl-2015-310861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse AR, Da Costa LA, Campos H, El-Sohemy A. Associations between polymorphisms in the AHR and CYP1A1-CYP1A2 gene regions and habitual caffeine consumption. Am J Clin Nutr. 2012;96:665–671. doi: 10.3945/ajcn.112.038794. [DOI] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermayer R, Dowd SE, Harris RA, Balasa A, Schaible TD, Wolcott RD, Tatevian N, Szigeti R, Li Z, Versalovic J, et al. Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll-like receptor 2-knockout mice. FASEB J. 2011;25:1449–1460. doi: 10.1096/fj.10-172205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Leamy LJ, Kelly SA, Nietfeldt J, Legge RM, Ma F, Hua K, Sinha R, Peterson DA, Walter J, Benson AK, et al. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol. 2014;15:552. doi: 10.1186/s13059-014-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees CW, Barrett JC, Parkes M, Satsangi J. New IBD genetics: common pathways with other diseases. Gut. 2011;60:1739–1753. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- Leimena MM, Ramiro-Garcia J, Davids M, van den Bogert B, Smidt H, Smid EJ, Boekhorst J, Zoetendal EG, Schaap PJ, Kleerebezem M. A comprehensive metatranscriptome analysis pipeline and its validation using human small intestine microbiota datasets. BMC Genomics. 2013;14:530. doi: 10.1186/1471-2164-14-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim U, Wang SS, Hartge P, Cozen W, Kelemen LE, Chanock S, Davis S, Blair A, Schenk M, Rothman N, et al. Gene-nutrient interactions among determinants of folate and one-carbon metabolism on the risk of non-Hodgkin lymphoma: NCI-SEER case-control study. Blood. 2007;109:3050–3059. doi: 10.1182/blood-2006-07-034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looft T, Allen HK, Cantarel BL, Levine UY, Bayles DO, Alt DP, Henrissat B, Stanton TB. Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 2014;8:1566–1576. doi: 10.1038/ismej.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Coarfa C, Qin X, Bonnen PE, Milosavljevic A, Versalovic J, Aagaard K. mtDNA haplogroup and single nucleotide polymorphisms structure human microbiome communities. BMC Genomics. 2014;15:257. doi: 10.1186/1471-2164-15-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley MS, Crocker PR, Paulson JC. Siglec-mediated regulation of immune cell function in disease. Nat Rev Immunol. 2014;14:653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I, Stegen JC, Maldonado-Gómez MX, Eren AM, Siba PM, Greenhill AR, Walter J. The gut microbiota of rural papua new guineans: composition, diversity patterns, and ecological processes. Cell Rep. 2015;11:527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnite AM, Perez-Munoz ME, Lu L, Williams EG, Brewer S, Andreux PA, Bastiaansen JWM, Wang X, Kachman SD, Auwerx J, et al. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One. 2012;7:e39191. doi: 10.1371/journal.pone.0039191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menni C, Mangino M, Zhang F, Clement G, Snieder H, Padmanabhan S, Spector TD. Heritability analyses show visit-to-visit blood pressure variability reflects different pathological phenotypes in younger and older adults: evidence from UK twins. J Hypertens. 2013;31:2356–2361. doi: 10.1097/HJH.0b013e32836523c1. [DOI] [PubMed] [Google Scholar]
- Miller TL, Wolin MJ, Conway de Macario E, Macario AJ. Isolation of Methanobrevibacter smithii from human feces. Appl Environ Microbiol. 1982;43:227–232. doi: 10.1128/aem.43.1.227-232.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayyeri A, Hammond CJ, Valdes AM, Spector TD. Cohort Profile: TwinsUK and healthy ageing twin study. Int J Epidemiol. 2013;42:76–85. doi: 10.1093/ije/dyr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocellin S, Verdi D, Pooley KA, Nitti D. Genetic variation and gastric cancer risk: a field synopsis and meta-analysis. Gut. 2015;64:1209–1219. doi: 10.1136/gutjnl-2015-309168. [DOI] [PubMed] [Google Scholar]
- Myles IA, Pincus NB, Fontecilla NM, Datta SK. Effects of parental omega-3 fatty acid intake on offspring microbiome and immunity. PLoS One. 2014;9:e87181. doi: 10.1371/journal.pone.0087181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor A, Quizon PM, Albright JE, Lin FT, Bennett BJ. Responsiveness of cardiometabolic-related microbiota to diet is influenced by host genetics. Mamm Genome. 2014;25:583–599. doi: 10.1007/s00335-014-9540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh PL, Martínez I, Sun Y, Walter J, Peterson DA, Mercer DF. Characterization of the ileal microbiota in rejecting and nonrejecting recipients of small bowel transplants. Am J Transplant. 2012;12:753–762. doi: 10.1111/j.1600-6143.2011.03860.x. [DOI] [PubMed] [Google Scholar]
- Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Yoshida S, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org E, Parks BW, Joo JWJ, Emert B, Schwartzman W, Kang EY, Mehrabian M, Pan C, Knight R, Gunsalus R, et al. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 2015;25:1558–1569. doi: 10.1101/gr.194118.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlostathis SG, Miller TL, Wolin MJ. Cellulose fermentation by continuous cultures of Ruminococcus albus and Methanobrevibacter smithii. Appl Microbiol Biotechnol. 1990;33:109–116. [Google Scholar]
- Pesson M, Volant A, Uguen A, Trillet K, De La Grange P, Aubry M, Daoulas M, Robaszkiewicz M, Le Gac G, Morel A, et al. A gene expression and pre-mRNA splicing signature that marks the adenoma-adenocarcinoma progression in colorectal cancer. PLoS One. 2014;9:e87761. doi: 10.1371/journal.pone.0087761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polderman TJC, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, Posthuma D. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47:702–709. doi: 10.1038/ng.3285. [DOI] [PubMed] [Google Scholar]
- Rausch P, Steck N, Suwandi A, Seidel JA, Künzel S, Bhullar K, Basic M, Bleich A, Johnsen JM, Vallance BA, et al. Expression of the Blood-Group-Related Gene B4galnt2 Alters Susceptibility to Salmonella Infection. PLoS Pathog. 2015;11:e1005008. doi: 10.1371/journal.ppat.1005008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replication DIG, Meta-analysis C, Mahajan A, Go MJ, Zhang W, Below JE, et al. Asian Genetic Epidemiology Network Type 2 Diabetes C, South Asian Type 2 Diabetes C, Mexican American Type 2 Diabetes C, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples C. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FE, Gonzalez MD, Cheng J, Wu M, Ahern PP, Gordon JI. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc Natl Acad Sci USA. 2013;110:13582–13587. doi: 10.1073/pnas.1312524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehe R, Dewhurst RJ, Duthie CA, Rooke JA, McKain N, Ross DW, Hyslop JJ, Waterhouse A, Freeman TC, Watson M, et al. Bovine Host Genetic Variation Influences Rumen Microbial Methane Production with Best Selection Criterion for Low Methane Emitting and Efficiently Feed Converting Hosts Based on Metagenomic Gene Abundance. PLoS Genet. 2016;12:e1005846. doi: 10.1371/journal.pgen.1005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Pamplona R, Berenguer A, Cordero D, Molleví DG, Crous-Bou M, Sole X, Paré-Brunet L, Guino E, Salazar R, Santos C, et al. Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer. Mol Cancer. 2014;13:46. doi: 10.1186/1476-4598-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood T, Allayee H, Bennett BJ. Choline metabolites: gene by diet interactions. Curr Opin Lipidol. 2016;27:33–39. doi: 10.1097/MOL.0000000000000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Nookaew I, Sommer N, Fogelstrand P, Bäckhed F. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. 2015;16:62. doi: 10.1186/s13059-015-0614-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Stewart JA, Chadwick VS, Murray A. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J Med Microbiol. 2005;54:1239–1242. doi: 10.1099/jmm.0.46189-0. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5:441–452. doi: 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Merwe JP, Stegeman JH, Hazenberg MP. The resident faecal flora is determined by genetic characteristics of the host. Implications for Crohn’s disease? Antonie Van Leeuwenhoek. 1983;49:119–124. doi: 10.1007/BF00393669. [DOI] [PubMed] [Google Scholar]
- Woting A, Pfeiffer N, Loh G, Klaus S, Blaut M. Clostridium ramosum promotes high-fat diet-induced obesity in gnotobiotic mouse models. MBio. 2014;5:e01530–01514. doi: 10.1128/mBio.01530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Wood AR, Lyssenko V, Weedon MN, Knowles JW, Alkayyali S, Assimes TL, Quertermous T, Abbasi F, Paananen J, et al. Genetic variants associated with glycine metabolism and their role in insulin sensitivity and type 2 diabetes. Diabetes. 2013;62:2141–2150. doi: 10.2337/db12-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Palmer AA, de Wit H. Genetics of caffeine consumption and responses to caffeine. Psychopharmacology. 2010;211:245–257. doi: 10.1007/s00213-010-1900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Hong SH, Cho SB, Lim JS, Bae SE, Ahn H, Lee EY. Characterization of microbial community in the leachate associated with the decomposition of entombed pigs. J Microbiol Biotechnol. 2012;22:1330–1335. doi: 10.4014/jmb.1205.05006. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, de Visser JAGM, de Vos WM. The Host Genotype Affects the Bacterial Community in the Human Gastrointestinal Tract. Microb Ecol Health Dis. 2001:13. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.