Abstract
The RNA Integrity Number (RIN) is often considered to be a critical measure of the quality of postmortem human brains. However, it has been suggested that RINs do not necessarily reflect the availability of intact mRNA. Using the Agilent bioanalyzer and qRT-PCR, we explored whether RINs provide a meaningful way of assessing mRNA degradation and integrity in human brain samples by evaluating the expression of 3’-5’ mRNA sequences of the cytochrome C-1 (CYC1) gene. Analysis of electropherograms showed that RINs were not consistently correlated with RNA or cDNA profiles and appeared to be poor predictors of overall cDNA quality. Cycle thresholds (CT) from qRT-PCR analysis to quantify the amount of CYC1 mRNA revealed positive correlations of RINs with amplification of full-length transcripts, despite the variable degree of linear degradation along the 3’-5’ sequence. These data demonstrate that in postmortem human brain tissue the RIN is an indicator of mRNA quantity independent of degradation, but does not predict mRNA integrity, suggesting that RINs provide an incomplete measure of brain tissue quality.
Keywords: RNA integrity number RIN, postmortem human brains, Agilent bioanalyzer, quantitative real-time polymerase chain reaction qRT-PCR, cytochrome C-1 CYC1
Graphical Abstract
Quality assessment of postmortem human brains by RNA integrity numbers (RINs) may be misleading, as they do not measure intact mRNAs. We show that the RIN is an indicator of mRNA quantity independent of degradation, but does not predict mRNA integrity, suggesting that RINs provide an incomplete measure of brain tissue quality. Our results resolve controversial assumption on interpreting quality assessments of human postmortem brains by RINs.
Introduction
Studies of gene expression in postmortem human brain tissues require well-characterized cases with minimal RNA degradation related to age, postmortem interval (PMI), disease states and medication effects. In particular, novel technology developments, such as whole genome or RNA sequencing (RNA-Seq) and laser-assisted microdissection (LMD), have resulted in an increase in studies to analyze the molecular composition of human brains (Benes 2006, Benes et al. 2007). The success of these labor-intensive efforts relies on the availability of well-characterized, high-quality postmortem brain tissues, which are usually provided by brain/tissue resource centers that determine brain quality by measuring tissue pH and RNA integrity (RIN) expressed either as 18S-28S ratios obtained with the Agilent bioanalyzer platform (Benes 2006, Konradi et al. 2004) or later, using RINs generated with additional software packages applied to Agilent bioanalyzer electropherogram data (Schroeder et al. 2006, Imbeaud et al. 2005). The RIN ranges between 1 and 10 indicating low or high RNA integrity, respectively; a RIN of 6 is typically considered by some to be a “quality threshold” in postmortem human brains, although these numbers are not consistent throughout the literature and across brain repositories.
In postmortem human brain materials, it is often generally assumed that determination of RINs, which measure the integrity of total RNA, is a surrogate for the characterization of mRNA quality. Extensive experience in this area, however, has suggested that RINs do not necessarily reflect the availability of intact mRNAs, and that this parameter is thought to be potentially misleading, particularly when used as a measure of the overall quality of postmortem brain tissue (Benes 2006). Consistent with this, in our studies using RNA derived from postmortem human brains (Benes et al. 2009, Konopaske et al. 2015, Pietersen et al. 2011, Pietersen et al. 2014a, Pietersen et al. 2014b, Ruzicka et al. 2015, Simunovic et al. 2009, Simunovic et al. 2010, Benes et al. 2007), we observed that 18S-28S ratios or the RIN were not necessarily reliable predictors of mRNA quality, as we were able to obtain high-quality expression profiling data from brain samples with both high or low RINs. In order to experimentally determine whether the RIN is in fact a meaningful and reliable predictor of mRNA integrity in human postmortem brain tissue, we systemically evaluated the relationship between RINs and the quantity and degradation of the cytochrome C-1 (CYC1) mRNA, as determined by quantitative real-time polymerase chain reaction (qRT-PCR), using RNA extracts available through the Harvard Brain and Tissue Resource Center (HBTRC).
Material and methods
Human tissue collection and RNA preparation
Postmortem human brains were collected using standard procedures at HBTRC that were approved by the Partners Human Research Committee. Written informed consent for the use of brains for research from male and female donors has been obtained by the legal next-of-kin. RNA was extracted from 50-100 mg occipital lobe tissue (Brodmann area 17) removed from snap frozen coronal slices that were immediately processed upon arrival at the HBTRC, placed in 1.0 ml TRIzol Reagent (Life Technologies, Grand Island, NY), and stored at −80 °C. To extract total RNA, the tissue was homogenized with motorized disposable pellet pestles (Sigma-Aldrich, St. Louis, MO), or using a Bead Ruptor 4 Mill Homogenizer (Omni International, Kennesaw, GA), mixed with 0.2 ml Chloroform, and centrifuged. The RNA-containing aqueous phase was collected, mixed with an equal amount of 75 % ethanol, and RNA was purified using the RNeasy Mini Kit (Qiagen, Valencia, CA) that included a DNase digestion step with RNase-free DNase I (Quiagen) for 15 min to eliminate genomic DNA contamination.
Brain quality measurement by Agilent bioanalyzer and qRT-PCR
RNA and cDNA samples were analyzed on an Agilent 2100 bioanalyzer using the 6000 Nano or Pico Kit, respectively, according to manufacturer's instructions (Agilent Technologies, Waldbronn, Germany). 1.2 μg of total RNA was converted to cDNA using the SuperScript III First-Strand Synthesis System with poly-dT primer extension and final RNase H treatment (Life Technologies). For detecting CYC1 transcripts, cDNA samples were subjected to PCR using the following primers: 5′-region: Forward (FW): 5′-GTCGTCGAAGTCTGGCCTTT-3′, Reverse (RV): 5′-CACGGTGAGACCACGGATAG-3′; Central-region: FW: 5′-GCCTCCTCTCTTCCTTGGAC-3′, RV: 5′-TCTTCATTGGGGCCGTCTTG-3′; 3′-region: FW: 5′-GGCATGGTGGTGAGGACTAC-3′, RV: 5′-CCCATGCGTTTTCGATGGTC-3′. For each sample, 1 μl of the cDNA solution (57 ng total RNA equivalent) was serially diluted 1:10, 1:100, 1:1,000, and 1:10,000, and PCR amplified in a Bio-Rad CFX Connect Real-Time PCR cycler using 0.2 μM primers and the iTaq Universal SYBR Green reagent (Bio-Rad, Hercules, CA).
Statistics
Pearson's correlation coefficients and p values were calculated using the Social Science Statistics program (http://www.socscistatistics.com/tests/pearson/Default2.aspx). Correlations were considered statistically significant when p values were less than 0.05 (p < 0.05).
Results
We determined the RINs of 179 brain samples (86 females, 93 males) and two control human skin fibroblasts from cell culture. Brains were from healthy individuals (n=33; 10 females, 23 males) or patients with neurodegenerative (n=101; 54 females, 47 males) or psychiatric disorders (n=45, 22 females, 23 males) with an average age of 69.6 +/−13.5 years (range 16 - 100). The average postmortem interval (PMI) was 20.1 +/−6.3 hrs (range 4.5 - 34.5) and average brain pH was 6.23 +/−0.27 (range 5.37 - 7.09) with consistent values across groups and sexes. Demographic analysis did not reveal any significant correlations of RIN with age, PMI, or diagnosis in all groups and sexes. However, we observed significant correlations of RINs with pH in all group and sex comparisons (r values between 0.2 and 0.55, p values < 0.05), except for healthy females (n=10; r = 0.546, p = 0.11) and males with neurodegenerative diseases (n=47; r = −0.0852, p = 0.57).
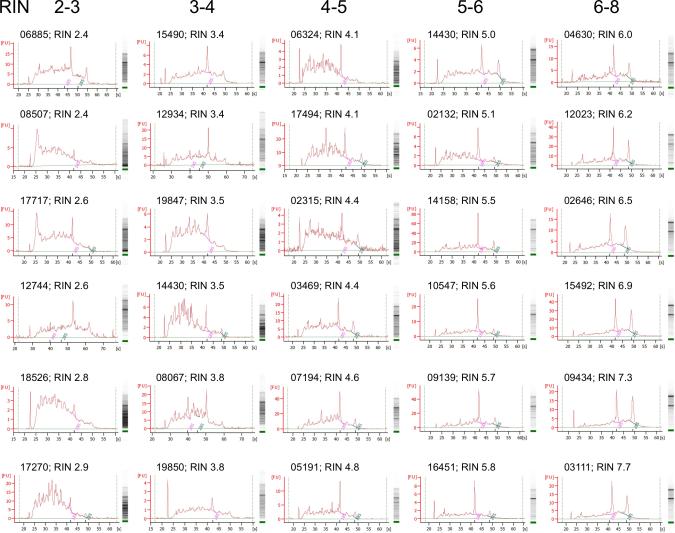
Consistent with observations from other studies (Birdsill et al. 2011, Chevyreva et al. 2008, Coulson et al. 2008, Koppelkamm et al. 2011, Lipska et al. 2006, Sheedy et al. 2012, Stan et al. 2006, Trabzuni et al. 2011, Webster 2006, Weis et al. 2007) we found a high variation of RINs in a range between 2.2 and 7.7, which prompted us to systematically evaluate the electrophoresis and electropherogram data obtained from the Agilent 2100 bioanalyzer in more detail. RNA electrophoretic traces consist of a series of different regions (pre-, 5S-, fast-, inter-, precursor-, and post-region), and peaks (marker, 18S and 28S ribosomal RNA) (Schroeder et al. 2006, Imbeaud et al. 2005, Weis et al. 2007), which change during RNA degradation. As expected, there was a good association of typical electropherogram traces (ribosomal peaks and baseline flatness (Schroeder et al. 2006, Imbeaud et al. 2005)) with calculated high or low RINs, e.g., 7.7 and 2.4, respectively (Fig. 1). However, there was also a substantial number of RINs that appeared to be inconsistent with the shape of the traces, and in most cases the differences were within a reading range of ±1.0. These inconsistencies were in particular observed with lower RINs (< 5), but also occurred at higher RINs (> 5) (Fig. 1), and could be attributed to reported detection of anomalies, such as additional or noise peaks between ribosomal peaks, and low molecular weight species (Schroeder et al. 2006, Imbeaud et al. 2005). In an attempt to quantify the results obtained from the Agilent bioanalyzer, we found no significant correlations between RINs and RNA concentration, signal area, or rRNA ratios (28S/18S), with the exception of RIN and rRNA ratio when RINs were greater than 6 (Table 1). Together with our observations on the electropherogram traces these data demonstrate that the RIN is an unreliable measure of RNA quality from postmortem brain tissue. Notably, these results did not depend on the methodology of RNA preparation, e.g., omitting the phenol (TriReagent) purification step, or using an Omni Bead Ruptor 4 homogenizer instead of manual pestle homogenization; however, variations in traces and corresponding RIN readings were also observed from the same RNA samples when repeatedly measured on separate chips or when analyzed by independent investigators on different Agilent bioanalyzers (data not shown).
Fig. 1.
RNA analysis by Agilent 2100 bioanalyzer. Electropherogram and electrophoresis assays from representative total RNA measurements with RINs between 2.4 and 7.7. Numbers indicate brain sample identification numbers.
Table 1.
Correlations of data obtained from Agilent 2100 bioanalyzer. r and p values of linear regression curves for RINs with RNA concentrations (ng/μl), signal area, and rRNA ratios (28s/18s). Significant values are in italic.
| RIN | 2-3 | 3-4 | 4-5 | 5-6 | 6-7 | 7-8 | |
|---|---|---|---|---|---|---|---|
| N | 20 | 37 | 45 | 39 | 31 | 7 | |
| RIN/conc. | r value | −0.075 | −0.146 | −0.043 | −0.064 | 0.039 | −0.663 |
| p value | 0.7565 | 0.3886 | 0.7791 | 0.6988 | 0.8329 | 0.1045 | |
| RIN/area | r value | −0.062 | −0.197 | −0.109 | 0.031 | 0.067 | −0.681 |
| p value | 0.7951 | 0.2425 | 0.4760 | 0.8523 | 0.7207 | 0.0928 | |
| RIN/ratio | r value | 0.239 | −0.158 | −0.007 | 0.154 | 0.443 | 0.746 |
| p value | 0.3094 | 0.3502 | 0.9636 | 0.3499 | 0.0126 | 0.0540 | |
The quality of the RIN system has been determined to compute assessment metrics for 91% of RNA profiles (Imbeaud et al. 2005). However, the observed variability in the electropherograms of our sample collection prompted us to question if RIN, indeed, represents a reliable measure of mRNA integrity and thus brain tissue quality; afterall, mRNAs constitute less than 5% of all RNA species. We, therefore, analyzed the cDNA from 24 selected RNA samples with lower RINs ranging from 2.4 to 6.5 and the two fibroblasts controls (RINs 9.6 and 9.8) by an Agilent bioanalyzer (Fig. 2). There was a typical bell-shape curve in the control samples reflecting the expected transcription of small- and large-sized mRNAs. In contrast, all cDNAs from the brain samples had a left shift of the curves indicating an increase in the smaller and a decrease in the larger transcripts, consistent with mRNA degradation. However, in all cases, there was no apparent association of the cDNA electropherograms with the RNA profiles and the corresponding RINs, indicating that in the postmortem brain tissue RIN is a poor predictor of overall cDNA integrity.
Fig. 2.
RNA and cDNA analysis by Agilent 2100. Representative electropherogram and electrophoresis assays from 10 RNA and corresponding cDNA measurements used in qRT-PCR analysis. Numbers indicate brain sample identification numbers.
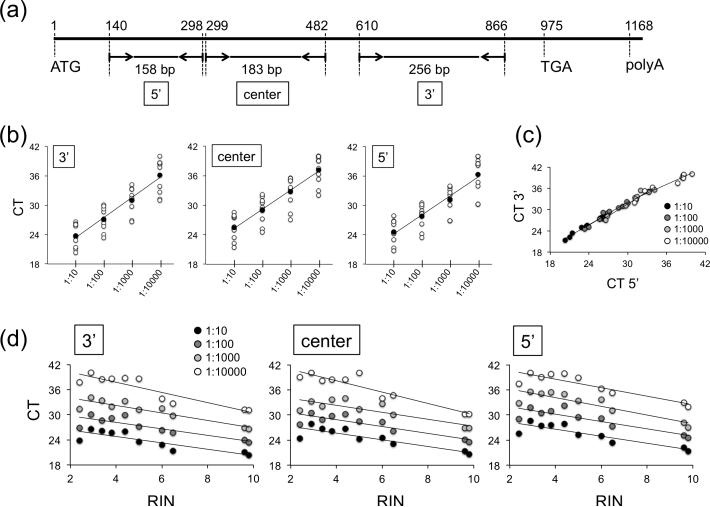
To determine the integrity of the mRNA, we analyzed serial dilutions of equal cDNA input material from eight representative brain samples and the two fibroblast controls by PCR, amplifying the 5’, center, and 3’ region of the CYC1 transcript (Fig. 3a) - CYC1 is a gene integrally associated with the electron transport chain and has been determined as a suitable housekeeping transcript in studies on postmortem brain material from patients that are affected with Alzheimer's disease (Penna et al. 2011) or glioma (Grube et al. 2015). In all samples we found linear amplification across the entire length of the cDNA (Fig. 3b), with significant cycle threshold (CT) correlations for the 3′ and 5′ region (Fig. 3c, Table 2). In addition, there were significant negative correlations between CT values and RINs (Fig. 3d, Table 2).
Fig. 3.
PCR analysis of full-length CYC1 cDNA. (a) Schematic of primer design covering the 5′, center, and 3′ regions of the CYC1 mRNA between the ATG (start) and TGA (stop) codon. Numbers indicate nucleotide positions. (b) CT values for 10 samples (8 brains and 2 fibroblast controls (#205 and #231)) show linear amplification of PCR products in 4 dilutions along the entire cDNA. RINs for individual samples are 2.4 (#06885), 2.9 (#17270), 3.4 (#15490), 3.8 (#08067), 4.4 (#02315), 5.0 (#17717), 6.0 (#04630), 6.5 (#02646), 9.6 (#205), and 9.8 (#231) (see Fig. 2). The linear regression curves depict average CT values for all samples in each dilution. 3′: r = 0.99064, p = 0.0094; center: r = 0.99712, p = 0.0027; 5′: r = 0.98567, p = 0.0144. (c) Linear regression curve showing distribution of CT values for the 3′ and 5′ region demonstrate positive correlations in all dilutions. (d) RINs negatively correlate with CT values. The r and p values in panels (c) and (d) are summarized in Table 2.
Table 2.
r and p values of linear regression curves shown in Fig. 3 panels (c) and (d). Significant values are in italic.
| Distribution of CT values for 3′ and 5′ cDNA region |
Correlation of RINs with CT values |
|||||||
|---|---|---|---|---|---|---|---|---|
| 3′ region |
center region |
5′ region |
||||||
| Dilution | r value | p value | r value | p value | r value | p value | r value | p value |
| 1:10 | 0.990 | 0.00001 | −0.859 | 0.00145 | −0.880 | 0.00081 | −0.888 | 0.00062 |
| 1:100 | 0.982 | 0.00001 | −0.864 | 0.00127 | −0.884 | 0.00071 | −0.876 | 0.00089 |
| 1:1000 | 0.973 | 0.00001 | −0.910 | 0.00027 | −0.842 | 0.00230 | −0.899 | 0.00040 |
| 1:10000 | 0.968 | 0.00001 | −0.915 | 0.00022 | −0.937 | 0.00007 | −0.900 | 0.00040 |
Discussion
A number of studies, including our own, have shown that good-quality RNA can be derived from postmortem human brain tissue and that samples with varying RINs can successfully be assessed using biochemical and molecular assays, such as microarrays, qPCR, or RNA-Seq (Benes et al. 2009, Konopaske et al. 2015, Pietersen et al. 2011, Pietersen et al. 2014a, Pietersen et al. 2014b, Ruzicka et al. 2015, Simunovic et al. 2009, Simunovic et al. 2010, Trabzuni et al. 2011, Gilbert et al. 1981, Sajdel-Sulkowska et al. 1988). Here, we report inconsistencies between RINs and corresponding RNA electropherogram profiles, as well as a lack of correlations of RINs with RNA concentration, signal area, and rRNA ratios, that limit the interpretation of the RIN as a singular or absolute measure of RNA quality from postmortem human brains. In particular, variations of readings in the range of ±1.0 should be taken into account when interpreting RINs. It should be noted that our traces and RINs matched those reported by other investigators that analyzed RNA degradation (Stan et al. 2006, Imbeaud et al. 2005, Schroeder et al. 2006, Weis et al. 2007). However, in contrast to these published data, in which RNA degradation was controlled under laboratory conditions, RNA degradation in the postmortem brain tissue was uncontrolled, which could have been a contributing factor to the observed variability in trace patterns.
As for measuring mRNA integrity, our data show that RINs in the lower ranges appear to be a poor predictor of cDNA quality when analyzed by the Agilent platform. However, the qRT-PCR results demonstrated that samples with both low and high RINs had full-length cDNA and that descending RINs correlate with decreasing cDNA input material, indicating that RINs positively correlate with the amount of intact mRNA, regardless of the degree of linear degradation along the 3’-5’ sequence. These results are consistent with data from other studies using Exon arrays (Trabzuni et al. 2011) or qPCR on housekeeping genes to assay RNA qualities measured by RINs (Imbeaud et al. 2005, Koppelkamm et al. 2011), i.e., RINs were the highest for cell culture samples when compared with RINs from tissue, and RINs significantly correlated with relative gene expression (CT values) and RNA degradation under laboratory conditions. Importantly, the study by Imbeaud et al. demonstrated equal PCR amplification of the 3′ and 5′ cDNA regions of the glucoronidase β (GUSB) and transferrin receptor (TFRC) genes on samples with different RINs, corroborating our results on postmortem tissue that RINs are indicative of linear degradation of full-length mRNA.
Taken together, our data indicate that, in analyzing postmortem human brain tissue, the RIN reflects a decrease in full-length mRNA quantity, rather than integrity, suggesting that it is an incomplete measure of overall brain quality.
Acknowledgements
This study was supported by NIH HHSN271201300030C to HBTRC (FMB, TUWW). KCS: Study design and conception, data generation and analysis, manuscript preparation. TUWW: Study design and conception, data analysis, manuscript preparation. GT, SS: Data generation. SB, SS, FMB: Manuscript editing.
Abbreviations
- RIN
RNA integrity number
- qRT-PCR
real-time polymerase chain reaction
- CT
cycle threshold
Footnotes
The authors declare no conflict of interest.
References
- Benes FM. Strategies for improving sensitivity of gene expression profiling: regulation of apoptosis in the limbic lobe of schizophrenics and bipolars. Prog Brain Res. 2006;158:153–172. doi: 10.1016/S0079-6123(06)58008-2. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci U S A. 2007;104:10164–10169. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Lim B, Subburaju S. Site-specific regulation of cell cycle and DNA repair in post-mitotic GABA cells in schizophrenic versus bipolars. Proc Natl Acad Sci U S A. 2009;106:11731–11736. doi: 10.1073/pnas.0903066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsill AC, Walker DG, Lue L, Sue LI, Beach TG. Postmortem interval effect on RNA and gene expression in human brain tissue. Cell Tissue Bank. 2011;12:311–318. doi: 10.1007/s10561-010-9210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevyreva I, Faull RL, Green CR, Nicholson LF. Assessing RNA quality in postmortem human brain tissue. Exp Mol Pathol. 2008;84:71–77. doi: 10.1016/j.yexmp.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Coulson DT, Brockbank S, Quinn JG, Murphy S, Ravid R, Irvine GB, Johnston JA. Identification of valid reference genes for the normalization of RT qPCR gene expression data in human brain tissue. BMC Mol Biol. 2008;9:46. doi: 10.1186/1471-2199-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JM, Brown BA, Strocchi P, Bird ED, Marotta CA. The preparation of biologically active messenger RNA from human postmortem brain tissue. J Neurochem. 1981;36:976–984. doi: 10.1111/j.1471-4159.1981.tb01689.x. [DOI] [PubMed] [Google Scholar]
- Grube S, Gottig T, Freitag D, Ewald C, Kalff R, Walter J. Selection of suitable reference genes for expression analysis in human glioma using RT-qPCR. J Neurooncol. 2015;123:35–42. doi: 10.1007/s11060-015-1772-7. [DOI] [PubMed] [Google Scholar]
- Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, Mueller O, Schroeder A, Auffray C. Towards standardization of RNA quality assessment using user- independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005;33:e56. doi: 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopaske GT, Subburaju S, Coyle JT, Benes FM. Altered prefrontal cortical MARCKS and PPP1R9A mRNA expression in schizophrenia and bipolar disorder. Schizophr Res. 2015;164:100–108. doi: 10.1016/j.schres.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- Koppelkamm A, Vennemann B, Lutz-Bonengel S, Fracasso T, Vennemann M. RNA integrity in post-mortem samples: influencing parameters and implications on RT-qPCR assays. Int J Legal Med. 2011;125:573–580. doi: 10.1007/s00414-011-0578-1. [DOI] [PubMed] [Google Scholar]
- Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, Kleinman JE. Critical factors in gene expression in postmortem human brain: Focus on studies in schizophrenia. Biol Psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Penna I, Vella S, Gigoni A, Russo C, Cancedda R, Pagano A. Selection of candidate housekeeping genes for normalization in human postmortem brain samples. Int J Mol Sci. 2011;12:5461–5470. doi: 10.3390/ijms12095461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen CY, Lim MP, Macey L, Woo TU, Sonntag KC. Neuronal type-specific gene expression profiling and laser-capture microdissection. Methods Mol Biol. 2011;755:327–343. doi: 10.1007/978-1-61779-163-5_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen CY, Mauney SA, Kim SS, et al. Molecular profiles of pyramidal neurons in the superior temporal cortex in schizophrenia. Journal of neurogenetics. 2014a;28:53–69. doi: 10.3109/01677063.2014.882918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen CY, Mauney SA, Kim SS, et al. Molecular profiles of parvalbumin immunoreactive neurons in the superior temporal cortex in schizophrenia. Journal of neurogenetics. 2014b;28:70–85. doi: 10.3109/01677063.2013.878339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka WB, Subburaju S, Benes FM. Circuit- and Diagnosis-Specific DNA Methylation Changes at gamma-Aminobutyric Acid-Related Genes in Postmortem Human Hippocampus in Schizophrenia and Bipolar Disorder. JAMA Psychiatry. 2015;72:541–551. doi: 10.1001/jamapsychiatry.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajdel-Sulkowska EM, Majocha RE, Salim M, Zain SB, Marotta CA. The postmortem Alzheimer brain is a source of structurally and functionally intact astrocytic messenger RNA. J Neurosci Methods. 1988;23:173–179. doi: 10.1016/0165-0270(88)90189-6. [DOI] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy D, Harding A, Say M, Stevens J, Kril JJ. Histological assessment of cerebellar granule cell layer in postmortem brain; a useful marker of tissue integrity? Cell Tissue Bank. 2012;13:521–527. doi: 10.1007/s10561-011-9265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic F, Yi M, Wang Y, et al. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson's disease pathology. Brain : a journal of neurology. 2009;132:1795–1809. doi: 10.1093/brain/awn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic F, Yi M, Wang Y, Stephens R, Sonntag KC. Evidence for gender-specific transcriptional profiles of nigral dopamine neurons in Parkinson disease. PloS one. 2010;5:e8856. doi: 10.1371/journal.pone.0008856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan AD, Ghose S, Gao XM, Roberts RC, Lewis-Amezcua K, Hatanpaa KJ, Tamminga CA. Human postmortem tissue: what quality markers matter? Brain Res. 2006;1123:1–11. doi: 10.1016/j.brainres.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D, Ryten M, Walker R, Smith C, Imran S, Ramasamy A, Weale ME, Hardy J. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem. 2011;119:275–282. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MJ. Tissue preparation and banking. Prog Brain Res. 2006;158:3–14. doi: 10.1016/S0079-6123(06)58001-X. [DOI] [PubMed] [Google Scholar]
- Weis S, Llenos IC, Dulay JR, Elashoff M, Martinez-Murillo F, Miller CL. Quality control for microarray analysis of human brain samples: The impact of postmortem factors, RNA characteristics, and histopathology. J Neurosci Methods. 2007;165:198–209. doi: 10.1016/j.jneumeth.2007.06.001. [DOI] [PubMed] [Google Scholar]