Abstract
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Objective
To examine the validity of deterministic compared to probabilistic record linkage in the ascertainment of hospitalisations in two linked cohorts.
Study Design and Setting
HIV-negative (HIV-ve) (n=1325) and HIV-positive (HIV+ve) gay and bisexual men (n=557) recruited in Sydney, Australia were probabilistically and deterministically linked to a state-wide hospital registry (July 2000-June 2012).
Results
Using probabilistic linkage as the reference standard, deterministic linkage had higher specificity but much lower sensitivity [34.67% (95%CI 33.44-35.92)]. A disproportionate number of links missed were individuals with poorer socioeconomic and health indicators, including HIV status. Risk of hospitalisation compared to the general male population [HIV+ve SIR=1.45 (1.33-1.59); HIV-ve SIR=0.72 (0.67-0.78)] was significantly underestimated when deterministic linkage was used [HIV+ve SIR=0.46 (0.37-0.58); HIV-ve SIR=0.29 (0.24-0.35)]. The impact of linkage strategy on the calculation of incidence rate ratios (IRRs) was less, but a greater discrepancy in IRRs was seen for diagnostic categories where event rates were low or where the sensitivity of the deterministic linkage was differential between the two cohorts.
Conclusion
Linkage without proven high sensitivity and specificity should be carefully considered. In circumstances of undetermined sensitivity, SIRs should not be calculated as the extent of underestimation is unknown. The comparison of linked events within or between cohorts is more robust to linkage misclassification; however, selection bias does affect estimates and should be considered prior to linkage.
Keywords: Keywords: Probabilistic record linkage, Deterministic record linkage, Hospital discharge, Cohort study, Evaluation, Australia
1. Introduction
While randomised controlled trials (RCTs) remain the gold standard for assessing health intervention efficacy; they are not always feasible or adequately timely. With recent advances in information technology and related statistical methods there is increasing interest in leveraging ever-growing repositories of administrative data to support public health research. Furthermore, there has been a shift in medical research from processing internal data, which is extremely costly, to mining external data which is significantly more cost effective (1). Although administrative datasets have many assets, they also have certain weaknesses that must be taken into consideration. They are often collected for non-research purposes and can be limited in scope. Many of these limitations, however, can at least in part be overcome through linking data from multiple sources. Linking administrative datasets with one another can improve case and control group identification, measurement of risk factors and outcomes and allows for a more cost effective way of following study participants than longitudinal cohort studies. However while there are an increasing number of data linkage centres globally (2), data linkage particularly in countries where registries do not have a unique person identifier common to all administrative datasets or where errors exist in the identifiers that are used is not without difficulty.
There are two main approaches to data linkage: deterministic linkage and probabilistic linkage. Both have been utilised widely in previous research studies (3-8). The deterministic linkage process works by determining whether two data records agree or disagree given a set of patient identifiers (first name, last name, date of birth (dob), etc.). In deterministic linkage, a match will only be given if the two records agree, character for character, on all specified identifiers and the record pair is uniquely linked. In some circumstances deterministic linkage may be undertaken in multiple rounds whereby if records do not match on the first round (social security number (SSNs), first and last name, for example) then they may go through another round for matching (7 digits of SSNs, first and last name and date of birth, for example), this is termed “iterative or approximate deterministic linkage”(9). While deterministic linkage puts equal weight on all identifiers, probabilistic linkage, by contrast, is based on the notion that some identifiers have more discriminatory power than others. For example a match on a rare last name like ‘Wojciechowicz’ is less likely to occur by chance and is therefore assumed more likely to be true, than a match on a more common last name such as ‘Moore’. Phonetic equivalence, spelling distance and linkage algorithms are also widely used in probabilistic linkage to deal with common recording errors. Probabilistic linkage strategies work by assigning a weight to two records pairs based on the probability they are a “true match” which is determined from the strength of the linkage identifiers which have matched. If the sum of weights for a match is above a certain threshold value, the pair is considered a link (10, 11).
As a result of their differing linkage strategies, deterministic linkage tends to have lower sensitivity but higher specificity than probabilistic linkage (12). Thus it is recommended that, where possible some quantitative estimates of the sensitivity and specificity of the linkage process are reported, allowing the effect of these quantities on observed results to be assessed (13). Often in practice, however, analysis and linkage are undertaken separately to preserve pseudonymisation and maintain the separation principle (14) and it can be difficult for researchers to obtain specific information regarding how data quality was assessed and how linkage algorithms and clerical review were evaluated. In this study we undertook both a deterministic and probabilistic linkage of our cohorts, HIV-negative (HIV-ve) and HIV-positive (HIV+ve) gay and bisexual men (GBM) recruited in Sydney, Australia, to a state-wide administrative hospital registry. The factors associated with missing linkages are reported and the impact of the differing linkage strategies on outcomes are investigated. The purpose of this paper is to provide further assistance to researchers who are attempting to appraise the possible impact of errors in the linkage process.
2. Methods
2.1 Study Population and databases
Our study population included male participants recruited to the Health in Men (HIM) (HIV-ve) and Positive Health (pH) (HIV+ve) studies who provided informed consent for their study data to be used for data linkage. Both studies have been described in detail elsewhere (15, 16). Briefly, men were recruited from Sydney, New South Wales (NSW), Australia using similar community-based methods. Enrolment in HIV-ve cohort occurred from 2001 to 2004 and active follow-up ceased in 2007. Enrolment in HIV+ve occurred from 1998 to 2006 and follow up ceased in 2007. Participants in both studies either had sexual contact with at least one man during the previous 5 years or self-identified as gay, homosexual, queer or bisexual. In both studies the majority of participants identified as gay, homosexual or queer (35, 37). All participants provided demographic and personal identifying information such as first and last name, address, and date of birth in addition to answering annual questionnaires on sexual and drug use behaviour, STIs and STI testing, gay community involvement, general self-reported health and use of health care services.
Hospitalisation data for the study cohorts were obtained from the New South Wales Admitted Patient Data Collection (APDC). The APDC includes records for all hospital separations (discharges, transfers and deaths) from all public, private, psychiatric and repatriation hospitals in NSW as well as public multi-purpose services, private day procedure centres and public nursing homes. The APDC records include a range of demographic data items (e.g. date of birth, residential address, language spoken at home and country of birth), administrative items (e.g. admission and separation dates) and coded information (e.g. reason for admission, significant co-morbidities and complications and procedures performed during the admission). Diagnosis fields are coded according to the 10th revision of the International Classification of Disease-Australian Modification (ICD-10-AM). Patient name has only been recorded since 1 July 2000, so we restricted analysis to admissions from 1 July 2000 to the most recent available data at time of analysis (30 June 2012). Fact of death for the study participants was also determined from linkage to the Registry of Births, Deaths and Marriages (RBDM) for the same period and used to censor person-years observation.
2.2 Linkage Procedures
Record linkage was carried out by the Centre for Health Record Linkage in New South Wales, Australia (CHeReL). CHeReL provides a mechanism for researchers to linked health data while maintaining the pseudonymisation of health records.
2.2.1 Probabilistic
CHeReL used open source probabilistic record linkage software ChoiceMaker (ChoiceMaker Technologies Inc) to link participants in the HIV-ve and HIV+ve cohorts to their respective hospital admission records and death records. First name, last name, date of birth, address, and postcode were used as the personal identifiers to match records. Standardization and parsing techniques are used which allows comparison of common fields and assist matching. At the first stage an automated blocking algorithm was used which attempts to find all possible matches to an individual's personal identifiers. There are two types of blocking: i) an exact blocking algorithm which requires records to have the same set of valid fields and the same values for these fields, and ii) an automated blocking algorithm which builds a set of conditions that are used to find as many as possible records that potentially match each other. At this stage, blocking is used to determine all possible links rather than aiming for precision. This increases the efficiency and accuracy of the second stage which is a more detailed matching using machine learning technique for ‘scoring’ or assigning weights. Weights are assigned on the basis of matches on a series of weighted clues. An example of a clue is that the date of birth doesn't match or there is a match on the phonetic code for the first name. The weight for each clue is based on previously matched data and a machine learning process called Maximum Entropy Modelling.
ChoiceMaker also includes a “transitivity engine” which allows for a user-specified action in the case of transitive linkage problems. An example of this would be the circumstance where record A is a high probability match to both B and C but B and C are a low probability matches to each other the user can specify whether in this circumstance B and C should be linked, should not be linked, or should be held for clerical review. Another example would be in the case where a database, let say a registry of deaths, is presumed free of duplicates but record A (e.g. an individual's personal identifiers) links to two death records, again the user could pre-specify that in this circumstance the records be held for clerical review. Further through its utilisation of information contain within other linkages, the transitivity engine used by the CHeReL can account for some missing identifiers.
ChoiceMaker converts linkage weights to probabilities in the range of 0 to 1, with 0 representing a definite non-match and 1 representing a definite match. An upper and lower cut-off weight of 0.75 and 0.25 are used to determine which matches are true and false matches by CHeReL respectively. This is then followed by a clerical review of record pairs with linkage weights above and below the upper and lower cut-offs, respectively. A random sample of 1,000 groups of matched records with probabilities that lie above the upper cut-off are reviewed by hand, the cut-off point is adjusted and this process is repeated until the false positive rate is below 5 per 1,000. Similarly 1,000 records with probabilities close to the lower cut-off are reviewed. The cut off is adjusted and the process is repeated until the false negative rate is below 5 per 1,000O. Thus the decisions documented during the clerical review are used as a reference standard against which the decisions made by the probabilistic linkage algorithm are compared, allowing for the calculation of sensitivity and specificity of the linkage algorithm. For this project, the CHeReL reported the linkage quality as <4/1,000 false positive links and <5/1000 false negative links (17).
2.2.2 Deterministic
A deterministic linkage of the HIV-ve and HIV+ve cohorts to their respective hospital admissions was also undertaken by CHeReL. Deterministic linkage was performed which required an exact match on all of the following fields: first name, last name, date of birth and sex.
2.3 Ethics Approval
Individual consent for data linkage was optional in the HIM and pH studies and was collected in addition to consent to participate in the study. Only data from participants who consented to data linkage were included in this analysis (93% of HIM and 74% of pH participants). We found no meaningul differences in examined cohort characteristics between those that consented compared with those that declined linkage. Examined cohort characteristics included ethnicity, education, employment, income, sexual identity, previous exposure to an STI or hepatitis C, self-reported health, Kessler 6 score of psychological distress, relationship status and HIV-serostatus, sexual risk taking behaviour, frequency of exercise, experiences of discrimination and alcohol use. Ethics approval this data linkage was granted by the University of New South Wales (NSW) and the NSW Population and Health Services Research Ethics Committee.
2.4 Statistical Analysis
The ICD-10-AM chapter heading for the primary diagnosis field was used to describe the principal reason for hospital admission. The sensitivity and specificity of deterministically linkage compared to probabilistic linkage of the cohorts to the APDC was calculated by ICD-10-AM chapter heading and 95% confidence intervals (CIs) estimated. We considered probabilistic linkage to be the reference standard here in view of the high sensitivity and specificity (99.5%).
Random effects Poisson regression analysis was performed to examine person-level characteristics associated with false-negative linkages i.e. probabilistic linkages that were missed via deterministic linkage. The person-level characteristics which were examined were age at time of admission (calculated from self-reported date of birth), year of hospital admission, year of cohort entry, whether the participant subsequently died (from linkage to the RBDM) and self-reported ethnicity, country of birth, highest level of education, employment, income, residential post code, experiences of discrimination and harassment, Kessler 6 score of psychological distress, smoking, injecting drug use, and hepatitis C status (all reported at entry into the cohort). Incidence rate ratios (IRRs) and corresponding 95%CIs were calculated. A p-value for homogeneity was reported. Analyses were performed using STATA (version 13; StataCorp LP, College Station, Texas, USA) and SAS (version 9.3; SAS Institute INC., North Carolina, USA).
Time at risk commenced at entry into the study cohort or opening of database for hospital admissions (1 July 2000), whichever was latest, with data right censored at death or the close of database (30 June 2012). The incidence of hospital admissions in the HIV-ve cohort and the HIV+ve cohort were compared with the incidence of hospital admissions in the general NSW male population (18) by calculating standardised incidence ratios (SIRs) . SIRs which had previously been calculated in the probabilistically linked cohort by Moore et al. (19) were compared to those calculated in the deterministically linked cohort. 95% confidence intervals (CIs) for the SIRs were calculated using the method by Stukel et al. (20). The incidence of hospital admissions in the HIV+ve cohort were comparted with the incidence of hospital admission in the HIV-ve cohort by calculating IRRs using random-effects Poisson methods. Further risk factors for all-cause hospitalisation in two cohorts were estimated by linkage strategy also using random-effects Poisson methods.
3. Results
3.1 Deterministic linkage accuracy
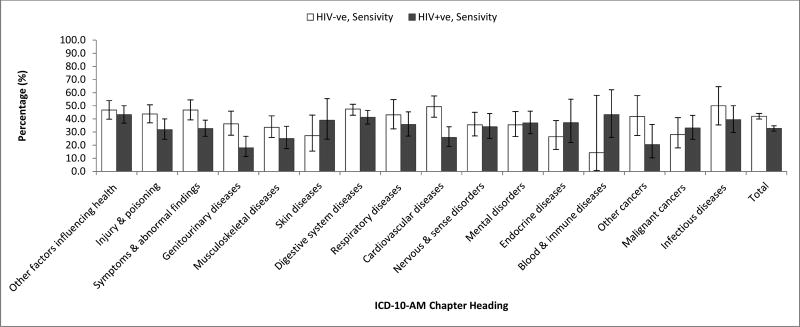
In a cohort of 1,325 HIV-ve GBM and 557 HIV+ve GBM there were 5,728 probabilistically linked hospital records of which a further 1,986 could be linked deterministically. The probabilistic linkage of the cohorts had extremely high sensitivity (99.5%, 95%CI: 98.8-99.8%) and specificity (99.6%, 95%CI: 98.9-99.9%), as reported by CHeReL which was determined by clerical review of matched and non-matched records (17). All records deterministically linked also linked probabilistically and were assumed to be true links thus specificity for the deterministic linkage using probabilistic linkage as the reference standard was assumed to be 100% for all diagnostic categories. Using probabilistic linkage as the reference standard, deterministic linkage had a sensitivity of 34.67% (95%CI: 33.44-35.92). The linkage sensitivity was significantly lower for the HIV+ve cohort (32.77%, 95%CI: 30.80-34.80) compared to the HIV-ve cohort (42.03%, 95%CI: 39.90-44.20). The sensitivity of the deterministic linkage varied by diagnostic category in both cohorts (Figure 1). The sensitivity of the linkages for cardiovascular diseases were significantly lower in the HIV+ve compared with the HIV-ve group.
Figure 1. Accuracy of deterministic linkage compared to probabilistic linkage (reference standard) by primary diagnosis in HIV-ve and HIV+ve cohorts.
3.2 Risk factors for missed deterministic linkages
Hospitalisations missed by deterministic linkage were more common in later years of admission (IRR >2006: 2.19 (2.01-2.39)), in participants recruited to the cohort in the first half of the recruitment period (IRR <2002: 1.77 (1.43-2.19)) and in those who subsequently died (IRR 13.09 (8.04-21.30) (Table 1). Missed linkages were more likely in Anglo-Australian/Anglo-Celtic participants (IRR 1.49 (1.15-1.93)), those who were younger at time of admission (IRR <40: 1.18 (0.99-1.41) and in those did not have a university or postdoctoral education (IRR tertiary: 1.88 (1.40-2.39), high school: 1.33(0.99-1.79), <high school: 3.16 (2.33-4.28). Those who were unemployed (IRR 3.99 (3.07-5.20), had experience discrimination (IRR 3.98 (2.87-5.51), had moderate or high levels of psychological distress (IRR 1.62 (1.23-2.13, 3.60 (2.76-4.70), respectively) and who were daily smokers (IRR 1.44 (1.15-1.79) were also more likely to have missed deterministic linkages.
Table 1. Association between cohort and registry characteristics and risk of missed record links.
| IRR (95%CI) | ||
|---|---|---|
|
|
||
| HIV Status | HIV-ve | 1 |
| HIV+ve | 4.03 (3.26-4.99) | |
| Age, years | >55 | 1 |
| 40-55 | 0.87 (0.77-0.99) | |
| <40 | 1.18 (0.99-1.41) | |
| Year of admission | <2006 | 1 |
| >2006 | 2.19 (2.01-2.39) | |
| Year of cohort entry | > 2002 | 1 |
| <2002 | 1.77 (1.43-2.19) | |
| Death record | No | 1 |
| Yes | 13.09 (8.04-21.30) | |
| Ethnicity | Other | 1 |
| Anglo-Australian/Anglo-Celtic | 1.49 (1.15-1.93) | |
| Country of Birth | Other | 1 |
| Australia | 1.46 (1.16-1.84) | |
| Education | University or Postgraduate | 1 |
| Tertiary education | 1.83 (1.40-2.39) | |
| Completed high school | 1.33 (0.99-1.79) | |
| 10 years of high-school or less | 3.16 (2.33-4.28) | |
| Employment | Employed | 1 |
| Unemployed | 3.99 (3.07-5.20) | |
| Income per week, $AUD | >1500 | 1 |
| 1000-1499 | 0.36 (0.28-0.46) | |
| 500-999 | 0.39 (0.29-0.52) | |
| <500 | 0.36 (0.27-0.49) | |
| Residential postcode area | Inner Regional | 1 |
| Major City | 0.86 (0.45-1.64) | |
| Outer Regional | 1.50 (0.19-11.71) | |
| Prior experience of discrimination | No | 1 |
| Yes | 3.98 (2.87-5.51) | |
| Kessler 6 Scale (Psychological Distress) | Low | 1 |
| Moderate | 1.62 (1.23-2.13) | |
| High | 3.60 (2.76-4.70) | |
| Daily Smoker | No | |
| Yes | 1.44 (1.15-1.79) | |
| Injecting drug use | No | 1 |
| Yes | 1.17 (0.78-1.75) | |
| Hepatitis C Positive (self-report) | No | 1 |
| Yes | 1.25 (0.82-1.91) |
Abbreviations: IRR- Incidence rate ratio; 95% CI- 95% confidence intervals;
3.3 Impact of deterministic linkage on effect sizes
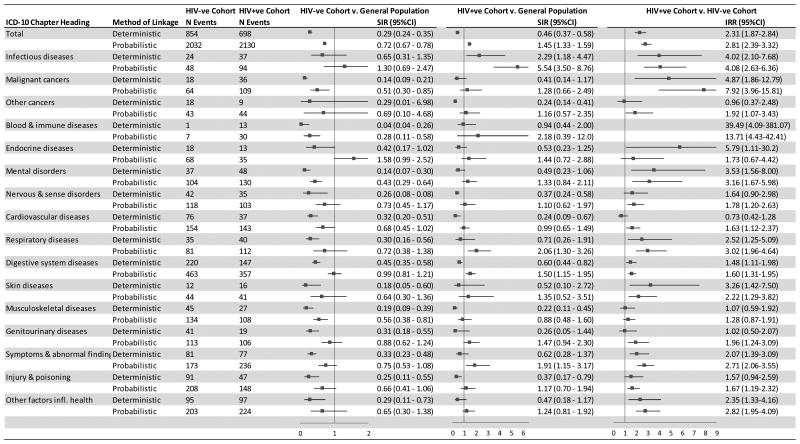
The probabilistic linkage overall found a lower risk of hospitalisation in the HIV-ve cohort compared to the general population (SIR 0.72, 95%CI: 0.67-0.78) (Figure 2). Similarly the deterministic linkage also found a lower risk of all-cause hospitalisation in the HIV-ve cohort compared to the general population (SIR 0.29 (95% CI 0.24-0.35)) albeit the magnitude of decreased risk was much larger in the deterministically linked cohort. A significantly lower risk of hospitalisation was seen for nervous and sense disorders, cardiovascular diseases, respiratory diseases, digestive system diseases, skin diseases, genitourinary diseases, symptoms and abnormal findings, injuries and poisonings and other factors influencing health using the deterministic but not the probabilistic linkage.
Figure 2. All-cause and cause-specific hospital risk among HIV-ve and HIV+ve cohorts compared with general population and HIV-ve compared with HIV+ve cohort (2000-2012) by record linkage method.
Abbreviations: IRR- Incidence rate ratios; SIR- Standardised incidence ratios, N- Number of; 95%CI- 95% confidence intervals. Note: Incidence rate ratios greater than 9 and their respective confidence intervals have been left out of figure 2
The probabilistic linkage overall found a higher risk of all-cause hospitalisation in the HIV+ve cohort compared to the general population (SIR 1.45, 95%CI: 1.33-1.59), whereas the deterministic linkage found an overall reduce hospitalisation risk (SIR 0.46, 95%CI: 0.37-0.58). An increased risk of hospitalisation was seen for respiratory disorders, digestive system diseases, symptoms and abnormal findings in the probabilistically linked but not deterministically linked cohort. There was a statistically significant decreased risk of injuries and poisonings, musculoskeletal diseases, cardiovascular diseases, nervous and sense disorders and other cancers using deterministic linkage but not probabilistic linkage.
The impact of deterministic linkage compared to probabilistic linkage on the calculation of IRRs (comparing the HIV+ve to the HIV-ve cohort) was smaller compared to the impact on SIRs (Figure 2). There was a greater propensity for the IRRs using deterministically linked data to move towards the null as a result of the lower sensitivity generally in all diagnostic categories for the HIV+ve cohort. A greater discrepancy in IRRs was seen for diagnostic categories where event rates were low (e.g. blood and immune diseases, endocrine diseases, skin diseases) or where the sensitivity of the deterministic linkage was differential between the two cohorts (e.g. cardiovascular disease).
The ascertainment of risk factors for all-cause hospitalisation was also similar between the two linkage methods (Appendix A). The greatest discrepancies in IRRs were seen for risk factors with a lower linkage rate in the deterministic compared to probabilistic linkage method (e.g. education, employment and experiences of discrimination).
4. Discussion
This study aimed to examine the effects of differing linkage mechanisms, deterministic versus probabilistic, on ascertainment of outcomes. Event rates of hospitalisations for all diagnostic categories were significantly underestimated when deterministic linkage was used. While deterministic linkage is known to have lower sensitivity and higher specificity than probabilistic linkage, few studies have previously compared the application of different linkage algorithms on outcomes. Baldwin et al. (21) compared deterministic and probabilistic linkage algorithms for linking mothers and infants within electronic health records and found a much higher sensitivity of their deterministic linkage (74.5%, 95%CI: 73.8-75.2) compared to ours. Baldwin et al. deterministically linked mothers and infants on surname, address and infant's birthdate. It is surprising that our deterministic linkage, matching on first and surname and date of birth, performed so poorly comparatively. It is possible that our cohort, being a cohort of gay and bisexual men, who are more likely to experience discrimination from health care workers (22) are less likely to give correct self-identifying information in the health care setting thereby diminishing deterministic linkage sensitivity compared to other linked cohorts. It is known that other marginalized populations within Australia, such as Indigenous populations, are poorly identified within hospital data (23). It is also likely that iterative deterministic linkage, whereby linkage is performed in multiple rounds using different degrees of matching, may perform better than basic deterministic linkage as undertaken in this study.
When comparing probabilistically linked versus deterministically linked datasets, the lower sensitivity of the deterministic linkage had a significant impact on the ascertainment of standardized incidence ratios, which compared rates of events within the linked cohort to rates in the known male population. As our results showed, linkage sensitivity could be so low at times as to invert the measure of association. This finding was driven by the fact that a large number of deterministically linked hospitalisations were missed while person-years of observation remained the same, thus leading to a comparative increase in the number of expected events compared to observed events and a significantly lower SIRs. We would recommend that when calculating SIRs on a linked cohort, investigators should report sensitivity and specificity of their data linkage which should be routinely provided by data providers and linkage centres. Further it is recommended that if linkage is shown to be poor as seen in our deterministic linkage an adjustment of SIRs be undertaken as previously described by Moore et al. (13). We would recommend that in circumstances of unknown quality of data linkage, SIRs not be calculated.
Our study also found that the sensitivity of deterministic linkage was discriminatory. In our cohort of HIV-negative and positive gay and bisexual men recruited in Sydney, Australia several factors were associated with the deterministic linkage accuracy. A disproportionate number of links missed by deterministic linkage (but captured by the probabilistic algorithm) were individuals with poorer socioeconomic and health indicators, including HIV status. Baldwin et al. (21) also found a selection bias in their deterministic linkage. Missed mother linkages by the deterministic algorithm were less likely to be married and Caucasian and more likely to be smokers with a low level of education and poorer prenatal care, suggesting that even when deterministic linkage sensitivity is significantly higher, selection bias in linkage remains. It is difficult to identify why certain person-level characteristics were associated with missed linkages, likely it is the result of a range of factors from data collection practices, linkage design and individual choices concerning disclosure of identifiers. If the failure to link records is a random error, then the effect on resulting estimates of effect is relatively straightforward and thus can be adjusted for (13, 24, 25) however, adjustment of outcomes when selection bias is present is difficult. Assessment of the characteristics of unlinked records could identify the presence of a strong selection bias in record linkage and should be performed more routinely by data linkage centres and reported by investigators.
The lower sensitivity and selection biases associated with deterministic linkage had a smaller impact on the ascertainment of HIV+ve/HIV-ve IRRs and risk factors for all-cause hospitalisation. However the direction of effect of the predictor hospital admission year (e.g. financial year) was reversed when deterministic linkage was use for the HIV+ve cohort. This is likely a result of later admission year being strongly associated with missing linkages and suggests the need for caution when selection bias in linkage may be present.
This study has several limitations. Linkage was undertaken in 2012 and hospitalisation data accessed was from 2000 to 2012, though to our knowledge CHeReL have not modified their linkage practices since 2012. We also could not identify the remaining incorrect linkages in the probabilistically linked dataset, due to privacy restrictions accessing the registry data. Further, we could not examine linkages identified by CHeReL as incorrect as these had already been removed. However CHeReL provided documentation to us that indicated these rates were low (<0.5%) (17). A further examination of probabilistic linkage and deterministic linkage using a reference standard dataset could shed further light in this regard, however reference standard datasets can be difficult to obtain. The generalisability of our findings may be affected by our cohort being predominantly those who identify as gay and bisexual men. Despite the inclusion criteria for both studies being open to men who engaged in same-sex sexual contact but who did not self-identify as gay, homosexual, queer or bisexual, few of these men were recruited to either study. Further, GBM in Australia recruited through non-online formats tend to be older, higher socioeconomic status and more likely to be of Caucasian ethnicity than other men who have sex with men cohorts. Previous studies have shown an association between non-Caucasian ethnicity and foreign born adults and poor linkage sensitivity which was not seen in our cohorts (21, 26). Sensitivity of linkage is dependent on characteristics of cohorts being linked and quality of identifiers used in linkage and may be higher in other settings. In addition, we were unable to undertake iterative deterministic linkage which would have likely performed better than the simple deterministic linkage used here. We believe future research comparing probabilistic linkage with more advanced deterministic linkage methods would be of value. Furthermore there is some evidence suggesting that possibly newer probabilistic linkage methods using imputation (27) may outperformed standard probabilistic linkage methods. This method uses imputation to account for missing data and has the added benefit in that it considers all the records (including unmatched records) compared with only retaining records with the highest weight as done in standard probabilistic linkage methods. Unfortunately probabilistic linkage using imputation is not yet routinely provided by linkage centres.
5. Conclusions
There is an increasing move to utilise record linkage both in Australia and internationally and the findings of this study expand the existing evidence base about the relative performance of probabilistic and deterministic linkage approaches for the linkage of cohorts with electronic health data. We hope our research highlights the extreme importance of determining the sensitivity and specificity of record linkage and in the future there is a move to make the provision of this information more common place. However we recognise that it is often difficult in practicality to access records which would define a ‘reference standard’ for comparison. In such circumstances, our results suggest that SIRs should not be calculated as underestimation may be considerable and threaten the scientific validity of the study. The comparisons of linked events within or between cohorts are more robust; however, selection bias does affect estimates and should be considered prior to linkage.
What is new?
While deterministic linkage is known for lower sensitivity and higher specificity than probabilistic linkage, few studies have previously compared the application of different linkage algorithms on outcomes. When comparing probabilistically linked versus deterministically linked datasets, the lower sensitivity of the deterministic linkage had a significant impact on the ascertainment of standardized incidence ratios, which compared rates of events within the linked cohort to rates in the known male population. Sensitivity could be so low as to invert the measure of associations. Poor sensitivity and selection bias of deterministic linkage had a smaller impact on the ascertainment of incidence rate ratios which compared event rates between deterministically linked cohorts. Our results suggest that in circumstances of undetermined sensitivity, SIRs should not be calculated. The comparisons of linked events within or between cohorts are more robust; however, selection bias does affect estimates and should be considered prior to linkage.
Acknowledgments
The authors would like to thank the participants, the dedicated pH and HIM study teams and the participating doctors and clinics for their contribution to the HIM and pH studies. The authors would also like to acknowledge the assistance of the New South Wales Centre for Health Record Linkage in the conduct of this study.
The Kirby Institute and the Centre for Social Research in Health are funded by the Australian Government Department of Health and Ageing. The Health in Men Cohort study was funded by the National Institutes of Health, a component of the USA Department of Health and Human Services (NIH/NIAID/DAIDS: HVDDT Award N01-AI-05395), the National Health and Medical Research Council in Australia (Project grant #400944), the Australian Government Department of Health and Ageing (Canberra) and the New South Wales Health Department (Sydney). The Positive Health Cohort study was funded by the Australian Government Department of Health and Ageing (Canberra) and the New South Wales Health Department (Sydney). HG is funded by an NHMRC research fellowship. The content of this publication is solely the responsibility of the authors and does not necessarily represent the view of any of the institutions mentioned above.
References
- 1.Martin-Sanchez F, Verspoor K. Big data in medicine is driving big changes. Yearb Med Inform. 2014;9:14–20. doi: 10.15265/IY-2014-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Population Data Linkage Network. Data linkage centres 2015 [04/08/2015] Available from: http://www.ihdln.org/data-linkage-centres.
- 3.Li Q, Glynn RJ, Dreyer NA, Liu J, Mogun H, Setoguchi S. Validity of claims-based definitions of left ventricular systolic dysfunction in Medicare patients. Pharmacoepidemiol Drug Saf. 2011;20(7):700–8. doi: 10.1002/pds.2146. [DOI] [PubMed] [Google Scholar]
- 4.Hammill BG, Hernandez AF, Peterson ED, Fonarow GC, Schulman KA, Curtis LH. Linking inpatient clinical registry data to Medicare claims data using indirect identifiers. Am Heart J. 2009;157(6):995–1000. doi: 10.1016/j.ahj.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swart A, Meagher NS, van Leeuwen MT, Zhao K, Grulich A, Mao L, et al. Examining the quality of name code record linkage: what is the impact on death and cancer risk estimates? A validation study. Australian and New Zealand journal of public health. 2015;39(2):141–7. doi: 10.1111/1753-6405.12287. [DOI] [PubMed] [Google Scholar]
- 6.Lawson EH, Ko CY, Louie R, Han L, Rapp M, Zingmond DS. Linkage of a clinical surgical registry with Medicare inpatient claims data using indirect identifiers. Surgery (United States) 2013;153(3):423–30. doi: 10.1016/j.surg.2012.08.065. [DOI] [PubMed] [Google Scholar]
- 7.Herman AA, McCarthy BJ, Bakewell JM, Ward RH, Mueller BA, Maconochie NE, et al. Data linkage methods used in maternally-linked birth and infant death surveillance data sets from the United States (Georgia, Missouri, Utah and Washington state), Israel, Norway, Scotland and Western Australia. Paediatric and Perinatal Epidemiology. 1997;11(SUPPL. 1):5–22. doi: 10.1046/j.1365-3016.11.s1.11.x. [DOI] [PubMed] [Google Scholar]
- 8.Kilkenny MF, Dewey HM, Sundararajan V, Andrew NE, Lannin N, Anderson CS, et al. Readmissions after stroke: Linked data from the australian stroke clinical registry and hospital databases. Medical Journal of Australia. 2015;203(2):102–6. doi: 10.5694/mja15.00021. [DOI] [PubMed] [Google Scholar]
- 9.Dusetzina SB, Tyree S, Meyer AM, Meyer A, Green L, Carpenter WR. Linking Data for Health Services Research: A Framework and Instructional Guide. Rockville, MD: Agency for Healthcare Research and Quality; 2014. Sep, [PubMed] [Google Scholar]
- 10.Campbell KM. Impact of record-linkage methodology on performance indicators and multivariate relationships. Journal of substance abuse treatment. 2009;36(1):110–7. doi: 10.1016/j.jsat.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Zhu VJ, Overhage MJ, Egg J, Downs SM, Grannis SJ. An Empiric Modification to the Probabilistic Record Linkage Algorithm Using Frequency-Based Weight Scaling. Journal of the American Medical Informatics Association : JAMIA. 2009;16(5):738–45. doi: 10.1197/jamia.M3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tromp M, Ravelli AC, Bonsel GJ, Hasman A, Reitsma JB. Results from simulated data sets: probabilistic record linkage outperforms deterministic record linkage. Journal of clinical epidemiology. 2011;64(5):565–72. doi: 10.1016/j.jclinepi.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Moore CL, Amin J, Gidding HF, Law MG. A New Method for Assessing How Sensitivity and Specificity of Linkage Studies Affects Estimation. PloS one. 2014;9(7):e103690. doi: 10.1371/journal.pone.0103690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelman CW, Bass AJ, Holman CDJ. Research use of linked health data — a best practice protocol. Australian and New Zealand journal of public health. 2002;26(3):251–5. doi: 10.1111/j.1467-842x.2002.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 15.Jin F, Prestage GP, Mao L, Kippax SC, Pell CM, Donovan B, et al. Transmission of herpes simplex virus types 1 and 2 in a prospective cohort of HIV-negative gay men: the health in men study. The Journal of infectious diseases. 2006;194(5):561–70. doi: 10.1086/506455. [DOI] [PubMed] [Google Scholar]
- 16.Prestage G, Mao L, Kippax S, Jin F, Hurley M, Grulich A, et al. Use of Viral Load to Negotiate Condom Use Among Gay Men in Sydney, Australia. AIDS and behavior. 2009;13(4):645–51. doi: 10.1007/s10461-009-9527-0. [DOI] [PubMed] [Google Scholar]
- 17.Centre for Health Record Linkage (CHeReL) Quality Assurance 2015 [cited 2015 27th August] Available from: http://www.cherel.org.au/quality-assurance.
- 18.Centre for Epidemiology and Evidence. HealthStats NSW Sydney2014 [cited 2014 22/05] Available from: www.healthstats.nsw.gov.au.
- 19.Moore CL, Grulich AE, Prestage G, Gidding HF, Jin F, Mao L, et al. Hospitalisation rates and associated factors in community-based cohorts of HIV-infected and -uninfected gay and bisexual men. HIV Medicine. 2015:n/a–n/a. doi: 10.1111/hiv.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stukel TA, Glynn RJ, Fisher ES, Sharp SM, Lu-Yao G, Wennberg JE. Standardized rates of recurrent outcomes. Statistics in medicine. 1994;13(17):1781–91. doi: 10.1002/sim.4780131709. [DOI] [PubMed] [Google Scholar]
- 21.Baldwin E, Johnson K, Berthoud H, Dublin S. Linking mothers and infants within electronic health records: a comparison of deterministic and probabilistic algorithms. Pharmacoepidemiology and Drug Safety. 2015;24(1):45–51. doi: 10.1002/pds.3728. [DOI] [PubMed] [Google Scholar]
- 22.Hughes M, Kentlyn S. Older Lesbians and Work in the Australian Health and Aged Care Sector. Journal of Lesbian Studies. 2015;19(1):62–72. doi: 10.1080/10894160.2015.959875. [DOI] [PubMed] [Google Scholar]
- 23.Australian Institute of Health and Welfare. Improving the quality of Indigenous identification in hospital separations data. Canberra, Australia: 2005. [Google Scholar]
- 24.Lahiri P, Larsen M. Regression analysis with linked data. Journal of the American Statistical Association. 2005;100:222–30. [Google Scholar]
- 25.Krewski D, Dewanji A, Wang Y, Bartlett S, Zielinski J, Mallick R. The effect of record linkage errors on risk estimates in cohort mortality studies. Survey Methodology. 2005;31:13–21. [Google Scholar]
- 26.Lariscy JT. Differential record linkage by Hispanic ethnicity and age in linked mortality studies: implications for the epidemiologic paradox. J Aging Health. 2011;23(8):1263–84. doi: 10.1177/0898264311421369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldstein H, Harron K, Wade A. The analysis of record-linked data using multiple imputation with data value priors. Statistics in medicine. 2012;31(28):3481–93. doi: 10.1002/sim.5508. [DOI] [PubMed] [Google Scholar]