Abstract
Most adaptive immune responses require the activation of specific T cells through the T cell antigen receptor–CD3 complex (TCR). Here we show that cholesterol sulfate (CS), a naturally occurring analog of cholesterol, inhibits CD3 ITAM phosphorylation, a crucial first step in T cell activation. Biochemical studies show that CS disrupted TCR multimers, apparently by displacing cholesterol, known to bind TCRβ. Moreover, CS-deficient mice displayed a heightened sensitivity to a self-antigen, whereas increasing CS content by intrathymic injection inhibited thymic selection, indicating that this molecule is an intrinsic regulator of thymocyte development. These results reveal a regulatory role for CS in TCR signaling and thymic selection, highlighting the importance of the membrane microenvironment in modulating cell surface receptor activation.
The major function of T lymphocytes is to mediate, directly or indirectly, immune responses against foreign antigens, while maintaining tolerance to endogenous (self) antigens. Specificity is achieved through the T cell antigen receptor (TCR), which can bind to peptide antigens bound to major histocompatibility complex molecules (MHC) on other cells. The TCR is expressed as a multi-subunit membrane receptor, including an antigen-recognizing heterodimer, TCRαβ (or TCRγδ), and a signaling module typically composed of three CD3 dimers: CD3εγ, CD3εδ and CD3ζζ1. All eight subunits in the TCR complex are type 1 transmembrane proteins2. Engagement of the TCR with specific peptide-MHC ligands initiates a transmembrane signal to trigger the phosphorylation of intracellular immunoreceptor tyrosine-based activation motifs (ITAMs) in CD3 subunits3. This initial phosphorylation then induces a series of signaling events in the T cell, including adaptor protein phosphorylation, formation of signaling complexes, intracellular calcium flux, immunological synapse formation, cytokine secretion and cell proliferation4, 5.
Despite the typically weak affinity of the TCR for its peptide-MHC ligand (1–150 μM)1, a given TCR is highly specific and sensitive for a particular peptide-MHC complex. It has been shown that even a single antigenic peptide together with endogenous peptides presented on MHC is able to trigger cytokine secretion in CD4+ T cells6. These paradoxical observations of the low affinity and high sensitivity are thought to be at least partially reconciled by the antigen-independent clustering of TCRs on the surface of T cells, reflecting both an intrinsic tendency of TCRs to self-associate, referred to as nanoclusters2, 7, 8, and their confinement on specific cytoskeleton-connected structures known as protein islands9, 10. Both phenomena result in a higher local concentration of TCRs on the T cell membrane and may be important for T cell sensitivity by enhancing the avidity to multimeric peptide-MHC11, 12, allowing for cooperativity between TCRs2, 13 and allowing often rare agonist peptide-MHC to rebind quickly, in order to signal continuously9, 14, 15, 16.
T cell sensitivity to a specific ligand is also tightly regulated during different developmental stages. Weak antigenic peptides that are unable to active mature effector T cells can efficiently induce selection in immature double-positive (DP, CD4+CD8+) thymocytes. The resulting thymocyte selection is driven by a successful triggering of TCR signal transduction17, 18, 19. Moreover, TCR nanoclustering increases from naïve T cells to effector and memory T cells, which contributes to the enhanced TCR sensitivity20. Thus, compared with naïve T cells, effector and memory T cells are more efficient at TCR signaling and more responsive to peptide-MHC21, 22, 23. These results highlight that intrinsic regulators of TCR signaling are involved in modulating T cell responsiveness at different developmental stages24.
Although several hundred different lipid species are known to exist in cell membranes25, and ~ 30% of the eukaryotic genome encodes for transmembrane proteins26, little is known about the relationship between these membrane lipids and proteins. However, recently it has been shown that the transition of the epidermal growth factor receptor from a low activity monomeric to a high activity dimeric state, can be inhibited by an interaction with the ganglioside GM3 (ref. 31). Importantly, one of our laboratories (W.W.S.) found that cholesterol specifically associates with the TCRβ chain and is necessary for TCR nanoclustering and a high avidity to multivalent peptide-MHC13. Other studies have shown that negatively charged phospholipids are involved in regulation of the TCR-CD3 conformation. Interactions between the CD3ε cytoplasmic domain and phospholipids might decrease the accessibility of phosphorylation sites in ITAMs27, 28 and also that the rise in intracellular Ca2+ that accompanies T cell activation dissociates the CD3ε cytoplasmic domain from the cell membrane to release the ITAM tyrosine residues to facilitate phosphorylation29.
In this report, we studied the role of cholesterol sulfate (CS) in modulating TCR signal transduction and its physiological implications on T cell development. We show that TCR signaling is repressed by CS, which is generally thought to function as a stabilizer of the cell membrane30. As a sulfated derivative of membrane cholesterol, we found here that CS disrupted cholesterol-driven TCR multimers by replacing cholesterol bound to TCRβ. Moreover, we found that the amount of CS was differentially regulated during thymocyte maturation, suggesting that the CS/cholesterol ratio may play a role in thymic selection. Increasing CS content leads to apoptosis in DP cells, apparently because it depressed the ability of those cells to be positively selected. In contrast, male mice deficient in the major enzyme responsible for sulfating cholesterol, SULT2B1, had enhanced thymocyte sensitivity to the HY self-ligand in a T cell autonomous manner. Thus CS plays an important role in modulating T cell signaling during thymocyte development.
RESULTS
Cholesterol sulfate inhibits TCR signaling
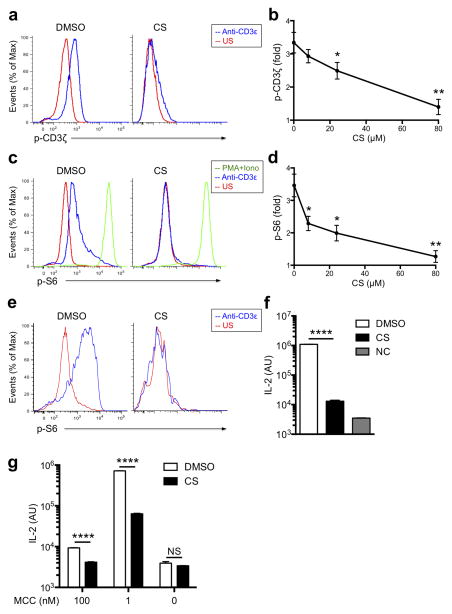
Among the different analogues derived from cholesterol, CS is the most abundant sterol sulfate in human plasma30. Previous studies have shown that the relative proportion of CS and cholesterol determine the phase separation and the domain curvature in model membranes31. To examine the possible influence of CS in T cell reactivity, we incubated 5C.C7 T cells (specific for a Moth Cytochrome C peptide, aa 88–103, in I-Ek) isolated from lymph nodes of transgenic mice with CS-supplemented media (Supplementary Fig. 1a), and examined CD3ζ phosphorylation upon anti-CD3ε TCR stimulation. Flow cytometry data showed that this CS pre-treatment resulted in a substantially attenuated response of anti-CD3-induced CD3ζ phosphorylation (Fig. 1a,b and Supplementary Fig. 1b). Furthermore, in both αβ T cells and γδ T cells, CS treatments blocked the TCR-mediated phosphorylation of the S6 ribosomal subunit (Fig. 1c–e), a key downstream component of the TCR signaling pathway that controls new protein synthesis32. Consistent with the phosphorylation data, TCR-induced up-regulation of the activation marker CD69 was also inhibited in CS-treated T cells (Supplementary Fig. 2). A similar inhibitory effect on S6 phosphorylation could also be observed after adenovirus-mediated overexpression of SULT2B1, the main sulfotransferase responsible for generating CS from cholesterol33 (Supplementary Fig. 3a,b). In contrast, CS did not affect phorbol ester (PMA) plus ionomycin-mediated S6 phosphorylation and CD69 up-regulation that bypasses TCR stimulation (Fig. 1c and Supplementary Fig. 2). Taken together these data suggest that CS specifically interacts with the TCR to inhibit transmembrane signaling without interfering with downstream components of this signaling pathway.
Figure 1. Cholesterol sulfate blocks TCR signaling.
(a) Phospho-flow cytometry analyzing phosphorylated (p-) CD3ζ in cells treated with the vehicle DMSO (left) or 100 μM CS (right), then left unstimulated (US) or stimulated with anti-CD3ε. (b) The CS titration curve for fold changes of p-CD3ζ upon stimulation by anti-CD3ε. (c) Phospho-flow cytometry analyzing phosphorylated (p-) S6 in cells treated with the vehicle DMSO (left) or 100 μM CS (right), then left unstimulated (US) or stimulated with PMA and ionomycin (PMA + iono) or anti-CD3ε. (d) The CS titration curve for fold changes of p-S6 upon stimulation by anti-CD3ε. (e) Phospho-flow cytometry analyzing phosphorylated (p-) S6 in γδ T cells treated with the vehicle DMSO (left) or 100 μM CS (right), then left unstimulated (US) or stimulated with anti-CD3ε. (f, g) ELISA analysis of IL-2 from T cells treated with DMSO and CS, stimulated with pMCC-I-Ek loaded lipid bilayers (f) or MCC-pulsed CH27 APC cells as indicated concentrations (g). IL-2 abundance was presented in arbitrary units (AU) on a log scale. NC, non-stimulated control. *P < 0.05; **P < 0.01; ****P < 0.0001; NS, not significant, unpaired t-test mean and s.e.m. (b,d,f,g). Data are from one experiment representative of three independent experiments with similar results (a,c,e) or three independent experiments with biological duplicates in each (b,d; n = 2, f,g; n = 3).
We also measured the effect of CS on interleukin 2 (IL-2) secretion induced by a lipid bilayer loaded with pMCC-I-Ek and co-stimulatory proteins. We found that the secretion of IL-2 was decreased in CS-treated 5C.C7 T cells compared to DMSO-treated cells (Fig. 1f). Finally, we assessed T cell responses to CH27 antigen presenting cells (APC)-pulsed with MCC peptides. In line with the above data, a reduced secretion of IL-2 was observed in both CS-treated and SULT2B1 over-expressing T cells (Fig. 1g, Supplementary Fig. 3c), indicating that CS also inhibits T cell activation by antigen-pulsed APCs. These results show that CS inhibits T cell activation, and does so at a very early stage, where TCR engagement with its peptide-MHC ligand triggers CD3ζ phosphorylation.
Cholesterol sulfate decreases TCR avidity
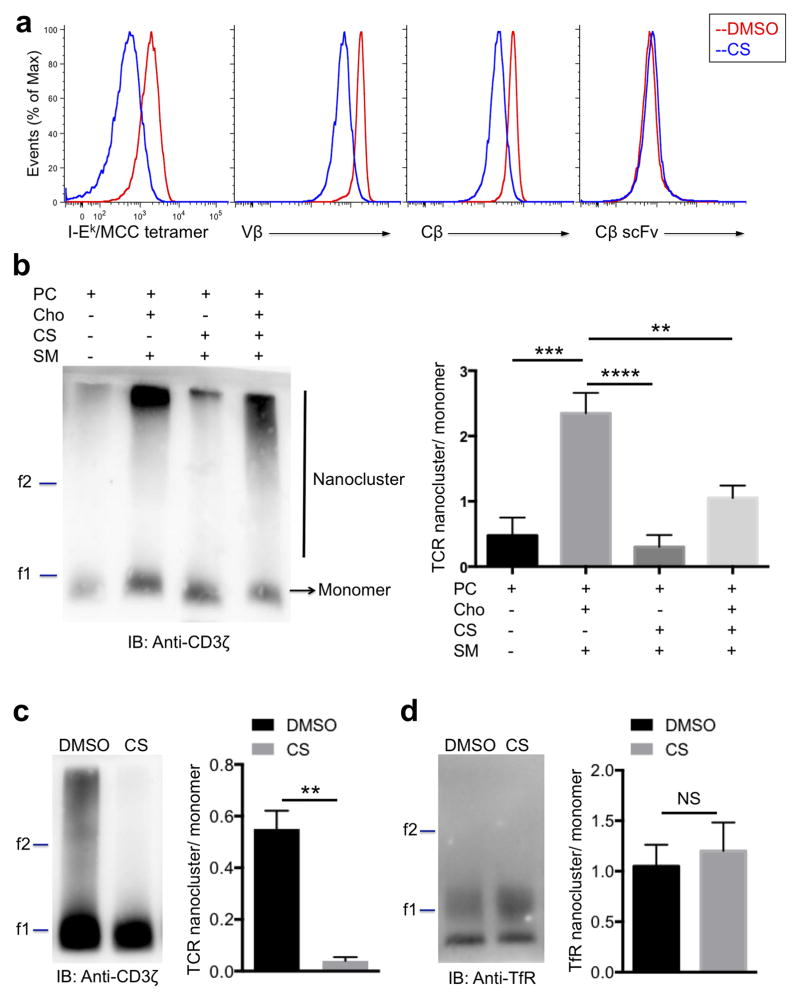
We then investigated whether CS influences the peptide-MHC binding avidity to the TCR by staining DMSO- or CS-treated 5C.C7 T cells with fluorescent pMCC-I-Ek tetramers. Our flow cytometry data showed that increasing CS abundance strongly decreased the binding of tetramers to the T cells (Fig. 2a, left), suggesting a reduced avidity between TCR and multivalent peptide-MHC. Consistent with the tetramer data, the binding of divalent antibodies directed against either the variable region or the constant region of TCRβ chain was also reduced by CS (Fig. 2a, middle). In contrast, CS did not affect the binding of monovalent single-chain variable fragment (scFv) to the same epitope of TCRβ chain (Fig. 2a, right). The fact that both peptide-MHC tetramer and divalent antibody binding, but not monovalent antibody binding, were impaired, indicated that CS could have an effect on the spatial organization of the TCR on the cell surface.
Figure 2. Cholesterol sulfate disrupts TCR nanoclusters.
(a) Flow cytometry analyzing 5C.C7 T cells treated with DMSO (red) or CS (blue), stained with pMCC-I-Ek tetramer, anti-Vβ3 antibody (KJ25), anti-Cβ antibody (H57-597) or anti-Cβ single chain Fv fragment (scFv) as indicated. (b) Blue-Native PAGE and immunoblot analysis of TCR nanoclusters from proteoliposomes with indicated lipid compositions. The ferritin markers, f1 and f2 corresponding to 440 kDa and 880 kDa, are shown. Band intensities of nanoclustered and monomeric TCRs were quantified with the Li-Cor Odyssey infrared imager and shown as a ratio of the two forms. (c, d) Blue-Native PAGE and immunoblot analysis of TCR and TfR nanoclusters from cells treated with DMSO or CS. Band intensities of nanoclustered and monomeric TCR or TfR were quantified with the Li-Cor Odyssey infrared imager and shown as a ratio of the two forms. **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant, unpaired t-test mean and s.e.m. (b,c,d). Data are from one experiment representative of three independent experiments with similar results (a,b,c,d) or three independent experiments with biological duplicates in each (b; n = 3, c,d; n = 2).
Cholesterol sulfate disrupts TCR nanoclusters
It has been suggested that cholesterol plays an important role in driving the formation of TCR nanoclusters and that TCR nanoclustering enhances the avidity towards peptide-MHC tetramers11. To investigate the effect of CS on TCR nanoclustering, we isolated native, monomeric TCRs from lysates of M.mζ-SBP hybridoma T cells (expressing SBP-tagged mouse 2B4 TCRs) by using the detergent digitonin7, and reconstituted the TCRs in large unilamellar vesicles (LUV) with defined lipid components, as was done previously11. The proteoliposomes of different lipid composition were lysed with 0.5% Brij 96V to maintain the TCR nanoclusters, in case they would have formed in the liposomes. We then analyzed the TCR monomers and nanoclusters by Blue Native-PAGE. We found that high-molecular-weight TCR nanoclusters can form in the proteoliposomes containing the ternary lipid mixture of phosphatidylcholine (PC), cholesterol and sphingomyelin (SM), but not in proteoliposomes containing only PC (Fig. 2b), as reported before11. When cholesterol was substituted by CS in the ternary mixture, TCR nanoclusters were not detectable above background (Fig. 2b), demonstrating that CS does not support nanocluster formation. Compared with TCRs in cholesterol-containing proteoliposomes, fewer TCR nanoclusters with a lower molecular weight were formed in the proteoliposomes containing both cholesterol and CS together with PC and SM, suggesting that CS can affect TCR nanocluster formation even in the presence of cholesterol (Fig. 2b).
To test whether CS can disrupt existing TCR multimers in living cells, we incubated T cells with DMSO or CS, and purified TCRs from the treated T cells to analyze TCR nanoclustering by Blue Native-PAGE. Consistent with our in vitro proteoliposome data, we found a strong reduction of the TCR nanoclusters when extracted from CS treated-cells (Fig. 2c). In contrast, high-molecular-weight complexes of the transferrin receptor (TfR) were not sensitive to CS treatment (Fig. 2d). We conclude that CS can disrupt TCR-CD3 nanoclusters, that this is at least somewhat specific to the TCR, and this may be the major mechanism by which CS disrupted signaling.
Cholesterol sulfate displaces cholesterol from TCRβ
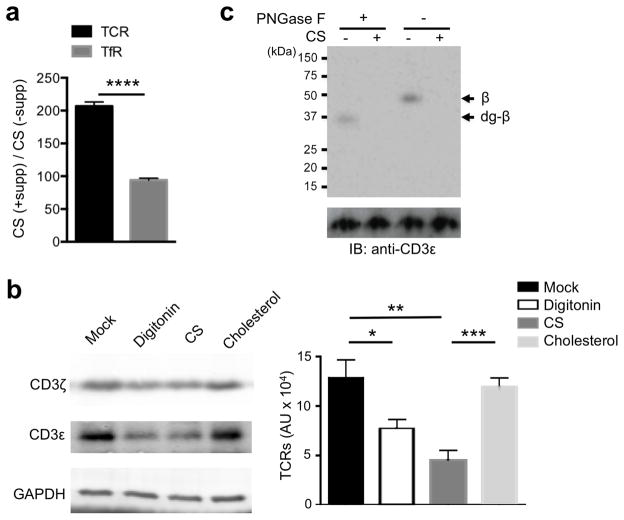
To investigate whether there is an interaction between CS and the TCR, we affinity-purified TCR-lipid and TfR-lipid complexes from DMSO- or CS-treated cells, extracted the protein-associated lipids with organic solutions, and quantified the CS amount with mass spectrometry. We calculated the ratio of CS concentration with and without CS supplementation. Notably, we found a 200-fold accumulation of CS that co-purified with the TCR, while less than a 100-fold accumulation of CS co-purified with the TfR as determined by mass spectrometry (Fig. 3a and Supplementary Table 1). Taken together, the higher than two-fold relative enrichment indicates that CS preferentially associates with the TCR, but not the TfR, in the cell membrane.
Figure 3. Cholesterol sulfate associates with TCR and disrupts cholesterol-TCR interaction.
(a) Mass spectrometry quantification of TCR- and TfR- associated CS with, CS (+supp), and without, CS (-supp), pre-incubation of the cells with CS. Data are presented as the ratio of CS amounts. (b) Immunoblot analysis of TCR in cholesterol beads treated with vehicle control, digitonin, CS or cholesterol. Band intensities of CD3ε were quantified with the Li-Cor Odyssey infrared imager and shown as arbitrary units (AU) in the right panel. (c) Autoradiogram and immunoblot analysis of TCRβ-cholesterol in the absence or presence of CS, then left untreated or subjected to PNGase F treatment as indicated. Proteins were separated by SDS-PAGE. The autoradiogram and the anti-CD3ε immunoblot are shown. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant, unpaired t-test mean and s.e.m. (a,b). Data are from one experiment representative of three independent experiments with similar results (c) or three independent experiments with biological duplicates in each (a,b; n = 3).
CS is much less abundant than cholesterol under normal physiological conditions34, 35. Because CS is able to reduce TCR nanoclusters despite an excess of cholesterol in the plasma membrane, we asked whether it could compete with cholesterol for TCR binding in vitro by using a lipid-protein pull down method36. T cell lysates were incubated with cholesterol-conjugated beads, then various free lipids were added to compete with the pre-formed interaction between cholesterol and TCR. Indeed, less TCR remained on the cholesterol-conjugated beads in the presence of free CS than free cholesterol, suggesting that CS can disrupt the binding of cholesterol to the TCR-CD3 complex (Fig. 3b). The efficiency with which CS displaced cholesterol was similar to that of digitonin that can break down TCR nanoclusters into monomers7, 8 (Supplementary Fig. 4).
We next used photoactivatable cholesterol-crosslinking11, 37 to assess whether CS affects the cholesterol-TCR interaction in living cells. T cells were grown with photoactivatable tritiated cholesterol, then CS was added to compete with the pre-formed binding of cholesterol and TCR. Upon activation by UV light, photocholesterol is covalently cross-linked to proteins in close proximity. TCRs were then purified from cellular lysates, and TCR subunits were separated by SDS-PAGE, and analyzed by autoradiography and immunoblotting (Fig. 3c). As reported previously11, we found that cholesterol binds to the TCRβ subunit, based on the molecular weight and N-glycosylation, as verified by the PNGase F treatment (Fig. 3c). The binding of cholesterol to TCRβ was decreased in the presence of CS (Fig. 3c), showing that CS can directly disrupt cholesterol binding to TCRβ in living T cells. Taken together, the data in this section shows that The TCR-CD3 complex involves CS preferentially compare to the Transferrin Receptor, and that CS can efficiently prevent cholesterol binding to the TCR, probably through direct competition.
Regulation of cholesterol sulfate contents in thymocytes
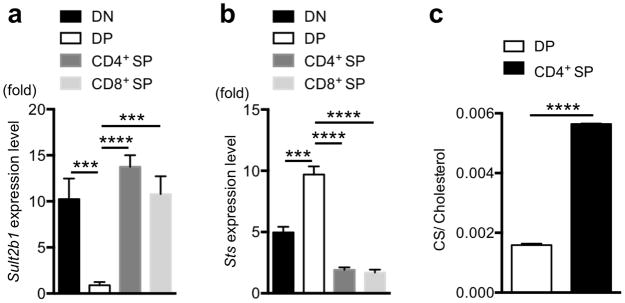
As thymic selection involves markedly different sensitivities to ligands as thymocytes mature38, we asked whether the abundance of CS is modulated in the different developmental stages. We obtained >99% pure thymocyte DP and single-positive (SP) subpopulations by flow cytometry based on CD4 and CD8 staining. We then measured the mRNA abundance of the CS-generating sulfotransferase gene, Sult2b1, and the sulfatase gene, Sts, responsible for metabolizing CS back to cholesterol by quantitative PCR (Fig. 4a,b). We found that the DP thymocytes have the lowest level of Sult2b1 expression and the highest expression level of Sts than other thymocyte subsets (Fig. 4a,b).
Figure 4. Developmental regulation of CS abundances in thymocytes.
(a, b) Quantitative PCR analysis of SULT2B1 and STS mRNA from sorted DN, DP, CD4+ SP and CD8+ SP thymocytes. β-actin was used for normalization. (c) Mass spectrometry quantification of CS and cholesterol from DP and CD4+ SP thymocytes. Total lipids were extracted from sorted thymocyte populations, and CS and cholesterol were quantified by small molecule mass spectrometry. The ratio of CS/cholesterol is shown. ***P < 0.001; ****P < 0.0001, unpaired t-test mean and s.e.m. (a,b,c). Pooled data are from three individual experiments (n = 4).
This reciprocal gene expression pattern of Sult2b1 and Sts suggested DP thymocytes may contain low amounts of CS. To test this, we extracted total lipids from sorted DP and CD4+ SP thymocytes and quantified the amounts of CS and cholesterol by mass spectrometry. In line with the gene expression data, we found that the ratio of CS/cholesterol is 3-fold lower in DP cells compared with CD4+ SP cells (Fig. 4c and Supplementary Table 2).
To assess the impact of CS in different thymocyte subsets that actively regulate their CS biosynthesis and metabolism, we next incubated thymocytes with various concentrations of CS, and monitored their TCR signaling by measuring calcium flux (Supplementary Fig. 5). Without CS, TCR cross-linking antibodies induced calcium fluxes in all three types of thymocytes (Supplementary Fig. 5). The calcium flux of DP thymocytes was measurably reduced by a 5 μM concentration of exogenous CS, while CD4+ and CD8+ SP thymocytes were not affected (Supplementary Fig. 5), demonstrating that DP thymocytes are more sensitive to CS than SP thymocytes. Similar to our signaling data on mature T cells. TCR-mediated calcium fluxes of all the thymocyte populations were impaired by 10 μM CS (Supplementary Fig. 5). These data show that CS abundance is regulated during thymocyte differentiation, together with other factors17, 24, and the lower amount of CS in DP thymocytes may contribute to their enhanced sensitivity.
Cholesterol sulfate induces apoptosis in DP thymocytes
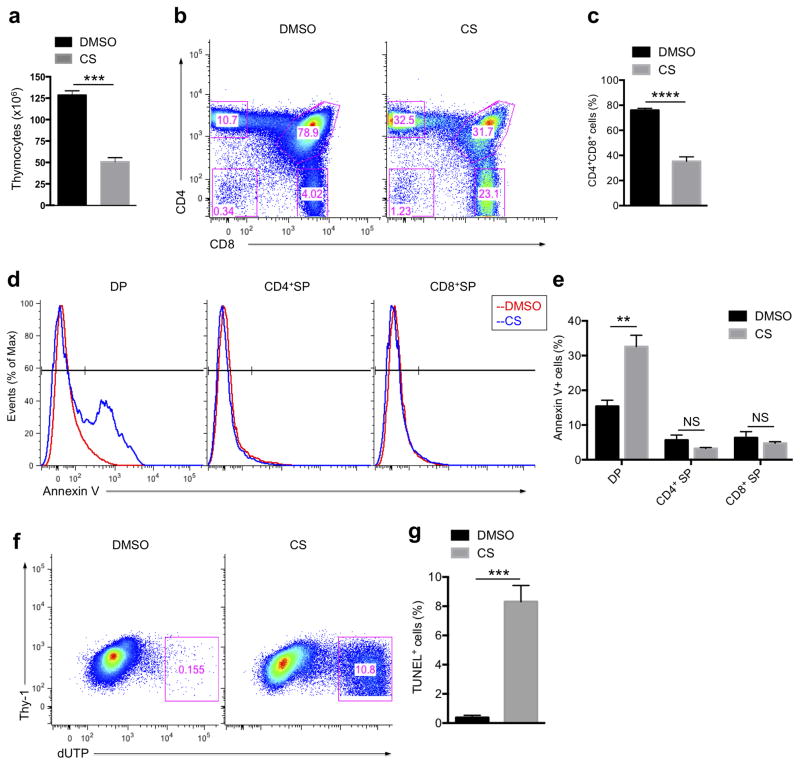
We next investigated the effects of CS modulation on thymic selection, where immature T cells (thymocytes) need to express TCRs that bind weakly to self-peptide–MHC complexes in order to mature, a process known as positive selection18, 39. At the same time, many, but not all T cells that have TCRs specific for self-peptide–MHCs are eliminated through a process termed negative selection40. We increased the amount of CS by an intrathymic injection and found that the number of total thymocytes was decreased with CS injection (Fig. 5a and Supplementary Fig. 1a), particularly DP thymocytes, which decreased by over half (75% vs 35%) as a percentage of the total number of cells (Fig. 5b,c). To test if the observed reduction of DP cells was due to the failure of positive selection, which occurs at this stage, we used Annexin V staining to measure the surface exposure of phosphatidylserine (PS), a marker of early apoptotic cells, as DP thymocytes that fail positive selection die by apoptosis41. CS specifically increased PS exposure on the surface of DP cells, but not on CD4+ or CD8+ SP cells (Fig. 5d,e), indicating that CS specifically leads to DP cell apoptosis. Moreover, our TUNEL assay showed that a massive number of late-stage apoptotic cells accumulated in the thymus with increasing CS concentration (Fig. 5f,g), confirming the failure of selection in these thymocytes. CS did not affect apoptosis in pre-selection DP thymocytes in OT-I TCR-transgenic Rag2−/− (KO) β2m-null mice42, 43, suggesting that intrathymic CS injection leads to increased cell death in DP cells by preventing positive selection, instead of non-specific toxicity, although we cannot rule out the possibility of an effect on thymic stroma, or other non thymocytes, which impairs their ability to mediate positive selection (Supplementary Fig. 6). We conclude that the increasing the concentration of CS in the thymus reduces the sensitivity of DP thymocytes to the signals that these cells normally get from self-peptide-MHC ligands, and thus more DP cells undergo apoptosis than usual.
Figure 5. Increasing cholesterol sulfate abundance induces apoptosis in double positive (DP) thymocytes.
(a) Cell number counting of total thymocytes after intrathymic injection of DMSO control or CS. The numbers were estimated using a hemocytometer. (b) Flow cytometry analysis of thymocyte populations. Mice were treated as in a. (c) Statistical analysis for the percentage of DP thymocytes. (d) Flow cytometry analysis of surface exposure of phosphatidylserine. Mice were treated as in a, then thymocytes were subjected to FITC-Annexin V staining. DP (left), CD4+ SP (middle) and CD8+ SP (right) populations were gated and Annexin V staining intensities were analyzed. (e) Statistical analysis for the percentage of Annexin V positive populations. (f) Flow cytometry analysis of TUNEL positive thymocyte population. Mice were treated as in a, then thymocytes were subjected to TUNEL staining and analyzed by flow cytometry. (g) Statistical analysis for the percentage of TUNEL positive populations. **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant, unpaired t-test mean and s.e.m. (a,c,e,g). Data are from one experiment representative of three independent experiments with similar results (b,d,f) or pooled three independent experiments (a,c,g; n = 4, e; n = 5).
SULT2B1 deficiency enhances negative selection
We next investigated the effect of decreasing CS abundance in the thymus by analyzing Sult2b1−/− mice44 that we extensively crossed onto the C57BL/6 background. These mice have a two-fold decrease in their CS/cholesterol ratio compared with their wild-type counterparts (Supplementary Fig. 7a). The percentages of DP and SP populations in both thymocytes and splenocytes are comparable between wild-type and Sult2b1−/− mice (Supplementary Fig. 7b).
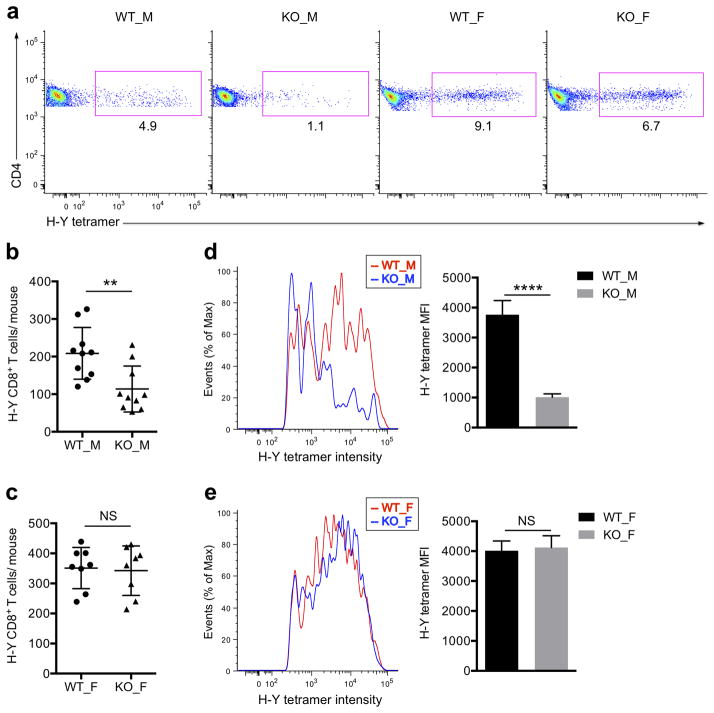
Whereas it has long been thought that self-specific T cells are very efficiently eliminated in the thymus, we have recently found that many are not, including HY-specific CD8+ T cells in mice and humans40. H-Y is a Y chromosome encoded antigen commonly used to monitor negative selection in thymocytes40, 45, 46, and so we measured the frequency of mature H-Y specific T cells in the periphery of male and female Sult2b1−/− mice in comparison to wild-type controls (Fig. 6a). There was a significant reduction (~2x) in the frequency of H-Y specific CD8+ T cells in male Sult2b1−/− mice compared to their wild-type counterparts (Fig. 6b,c). Consistently, the H-Y specific CD8 SP thymocytes also decreased in male Sult2b1−/− mice (Supplementary Fig. 8). Furthermore, we found the tetramer staining pattern of these male mice was noticeably skewed towards lower avidity H-Y specific cells, based on the intensity of tetramer staining (Fig. 6d,e). Our data show that the Sult2b1−/− mice display a heightened sensitivity to the negative selection to H-Y self-antigen.
Figure 6. Effects of Sult2b1 deficiency on mature T cells recognizing the HY self-antigen.
(a) FACS plots gated for H-Y Uty Db tetramer+ CD8+ T cells from wild-type male (WT_M), Sult2b1-deficient male (KO_M), wild-type female (WT_F), and Sult2b1-deficient female (KO_F) mice after magnetic enrichment. (b) The total numbers of H-Y tetramer positive CD8+ T cells in male WT and Sult2b1-deficient mice. (c) The total numbers of H-Y tetramer positive CD8+ T cells in female WT and Sult2b1-deficient mice. (d, e) Left, fluorescence intensity histogram of H-Y Uty:Db tetramer binding CD8+ T cells from a. Right, statistical analysis for the median fluorescence intensity (MFI) of HY tetramer positive populations. **P < 0.01; ****P < 0.0001; NS, not significant, unpaired t-test mean and s.d. (b,c,d,e). Data are from one experiment representative of three independent experiments with similar results (a,d,e) or pooled three independent experiments (b; n = 10, c; n = 8, d,e; n = 5).
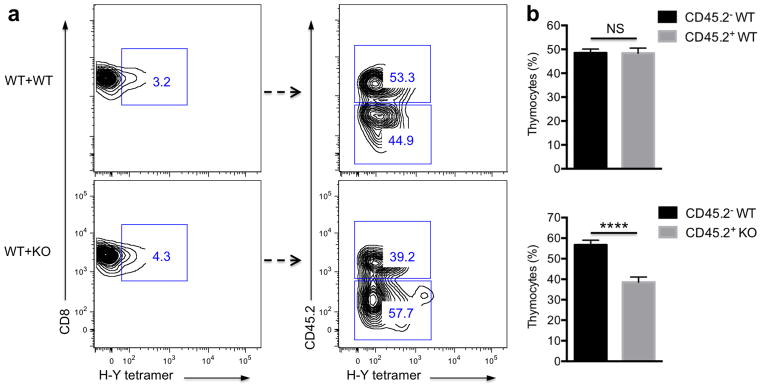
Because it is possible that the effect described above is mediated by some of the many other cell types that thymocytes encounter during their maturation, we also performed a mixed bone marrow chimera experiment to test this possibility. Lethally irradiated male B6.SJL mice were reconstituted with a 1:1 mixed bone marrow of either wild-type B6.SJL (CD45.2−) and wild-type B6 (CD45.2+) or wild-type B6.SJL (CD45.2−) and Sult2b1−/− B6 (CD45.2+). We then examined the H-Y specific thymocytes developed from wild-type and Sult2b1−/− bone marrow. Based on congenic marker gating of the tetramer-positive populations (Fig. 7a), we showed that even in the presence of wild-type cells, fewer CD8+ H-Y-specific thymocytes develop from Sult2b1−/− bone marrow compared to wild-type cells in the same animal (Fig. 7b), although the difference between wild-type and Sult2b1−/− was not as pronounced as in separated mice. These data suggest that the loss of Sult2b1 increases the sensitivity of H-Y specific thymocytes to negative selection, and that this effect is largely independent of wild-type cells and tissues in that same environment. Thus, the effect of the SULT2B1 deficiency is mostly intrinsic to cells carrying that defect.
Figure 7. Fewer H-Y specific thymocytes were developed from Sult2b1-deficient bone marrow.
(a) Representative FACS plots of CD8+ SP thymocytes gated on H-Y Uty Db tetramer+ populations (left). Tetramer+ populations were further gated based on the expression of CD45.2 congenic marker (right). (b) Statistical analysis for the percentage of CD45.2− and CD45.2+ populations. ****P < 0.0001; NS, not significant, unpaired t-test mean and s.e.m. (b). Data are from one experiment representative of three independent experiments with similar results (a) or pooled three independent experiments (b), n = 8 in WT+WT group, and n=12 in WT+KO group.
DISCUSSION
Although there are many specialized lipids that occur in nature, and as components of cell membranes, their biological roles are largely unknown. But an attractive and potentially general property may be in modulating the conformation and activity of membrane receptors involved in cell signaling, as seen with the EGF receptor and the GM3 lipid47 and cholesterol binding to the TCR-CD3 complex and its role, together with sphingomyelin in nanocluster formation11. Recent study shows that cholesterol enhances CD8+ T cell signaling to potentiate the antitumour response48.
In this study, we describe a new aspect to the cited TCR-CD3 data by our finding that a naturally occurring form of cholesterol, cholesterol sulfate, can act as a negative regulator of TCR-mediated cellular activation at the very earliest stage of signaling following ligand binding. We find that sufficient concentrations of exogenous CS disrupt TCR-CD3 nanocluster formation, which correlates with successful signal transduction, and that the mechanism of this is likely the fact that it interferes with cholesterol binding. Since the dimerization and multimerization of surface receptors is a common way to control signaling events shared with many cell types, our finding suggests a novel way in which a membrane lipid can modulate signaling.
That this is biologically meaningful in vivo is indicated by our data showing that the enzyme responsible for the production of CS, SULT2B1, is expressed less abundantly in thymic DP than in SP thymocytes (CD4+ or CD8+). DP thymocytes are the most sensitive cells in the maturation of T cells19, since this is the stage where positive selection for very weak binding to endogenous peptide-MHCs needs to occur for the cells to mature further. Mass spectroscopy confirms that this lower SULT2B1 is accompanied by lower content of CS compared to cholesterol in the membranes of these cells. Further indicating a role in T cell sensitivity for CS is our finding that intrathymic injection of CS results in considerably increased cell death, likely due to thymocytes treated in this way being less sensitive to endogenous ligands. Complementary to these data and further indicating a role for the CS/cholesterol ratio in thymic selection, we find that SULT2B1-deficient mice delete HY-specific T cells more efficiently at the SP stage. This is consistent with less CS in the cell membrane allowing a greater sensitivity to negative selection. But other cells in the thymus might be affected by the SULT2B1 deficiency and influence selection as well, and so it is important to note that in a mixed bone marrow chimera experiment, only the thymocytes carrying the SULT2B1 deficiency showed any additional deletion, and not the wild type cells they developed alongside. This shows that this is largely a cell autonomous effect, and CS production within the cell itself is the most important factor in this circumstance. In addition, one could ask why there is no obvious effect on positive selection of the SULT2B1 deficiency. Here we would suggest that the CS content in DP cells is already at a minimum with respect to thymocyte sensitivity, and that further lowering fails to result in any increase in the efficiency of positive selection.
In conclusion, we present evidence here that CS is a specific, negative regulator of T cell signaling through the TCR-CD3 complex, which, at the least is important in regulating the sensitivity of thymocytes during their maturation. It is also tempting to speculate that modulating CS abundance might also be important in other situations involving T cell reactivity, or that it could be useful in the treatment of excessive T cell activity. Future work will be needed to fully demonstrate the in vivo relevance of CS. Importantly, CS is the main circulating sulfated steroid in human plasma, with a concentration of ~6 μM, but elevations of CS concentrations have been observed under certain pathological conditions, including but not limited to, recessive X-linked ichthyosis, cirrhosis of the liver, hypothyroidism and hypercholesterolemia30, 49. Thus, it would be interesting to carefully examine T cell function in these patients. In conclusion, our work here adds to the increasing evidence that specialized membrane lipids can have important roles as modulators of cell surface receptors involved in cell signaling.
ONLINE METHODS
Cell culture and mice
5C.C7 T cells were harvested and cultured as described previously24. Briefly, T cells from lymph nodes of 5C.C7 mice were harvested and primed with 10 μM MCC peptides (ANERADLIAYLKQATK). Cells were stimulated on the second day of culture by 50 units/ml of recombinant mouse IL-2. After 7–9 days of culture, T cell blasts were used for various in vitro activation and staining assays. Before stimulation, T cell blasts were incubated with 100 μM cholesterol sulfate or DMSO control in RPMI-1640 media supplemented with 1% lipid-free BSA or 5% lipid-deficient FCS for 2 h at 37 °C. CH27 cells, a mouse B lymphoma cell line, were pre-loaded with MCC peptides overnight before stimulation experiments. M.m ζ-SBP and M.hTfR-SBP cells were cultured as described previously11. Sult2b1−/− mice were back-crossed onto C57BL/6 genetic background at least 10 times. Jackson Laboratory SNP analysis indicated that they were greater than 99% C57BL/6 background. For the experiment of intrathymic injection, mice were anesthetized and 20 μl DMSO or cholesterol sulfate (25 mM) was injected into thymus by a Hamilton syringe (10 μl each lobe). For the bone marrow chimera experiments, B6.SJL-Ptprca Pepcb/ BoyJ (Stock Number: 002014) mice were purchased from Jackson Laboratory. Male wild-type B6.SJL mice were subjected to lethal irradiation (950 Rads), then reconstituted with a 1:1 mixed bone marrow of wild-type B6.SJL (CD45.2−) and wild-type B6 (CD45.2+), or wild-type B6.SJL (CD45.2−) and Sult2b1−/− B6 (CD45.2+) mice. 5 × 106 bone marrow cells were injected into each irradiated recipient intravenously. Ten to twelve weeks after bone marrow transfer, thymocytes were isolated for tetramer staining and flow cytometry analysis. Mice were bred and maintained in the Research Animal Facility at Stanford University Department of Comparative Medicine Animal Facility in accordance with guidelines of the US National Institutes of Health.
IL-2 ELISA
The 5C.C7 T cell blasts were added onto a planar glass-supported bilayer (loaded with pMCC-I-Ek, B7 and ICAM proteins) or mixed with MCC pre-loaded CH27 APC. Cells were incubated 8 h at 37 °C. Secretion of IL-2 in the supernatant was assessed by ELISA with rat antibody to mouse IL-2 (JES6-1A12, BD Pharmingen), biotinylated rat antibody to mouse IL-2 (JES6-5H4, BD Pharmingen) and Europium-labeled streptavidin (Perkin Elmer).
Flow cytometry
Phospho-flow was used to detect phosphorylation of CD3ζ chain and S6. Briefly, T cells were stimulated with 10 μg/mL anti-CD3 ε antibody for 15 min at 37 °C. Cells were fixed with 1.5 % paraformaldehyde at 25 °C for 10 min. After washes with PBS, T cells were suspended in ice-cold methanol and kept at −20 °C for 30 min to allow for permeabilization. After washes with PBS, cells were stained with fluorescent phosphor-specific antibody, and analyzed by LSRII (BD Biosciences). For cell sorting, thymocytes were harvested from thymus of 6-week-old BALB/c mice. Cells were stained with fluorescent antibodies against cell surface markers CD4 (RM4-5), CD8 (53-6.7), F4/80 (BM8), Gr-1 (RB6-8C5), CD11b (M1/70), CD11c (N418), Ter119 (TER-119), CD19 (6D5) and Live/Dead Aqua amine. Labeled cells were sort into CD4+CD8+ DP cells, CD4+CD8−, CD4−CD8+ SP cells and CD4−CD8− double-negative cells populations using Aria cell sorter (BD Biosciences) at Stanford FACS facility. FACS data were analyzed with FlowJo software (Tree Star, Inc.).
Calcium flux measurement
Mouse thymocytes were stained with 8 μg/ml of Atto488-NHS Ester (Sigma) or Cy3-NHS Ester (GE Healthcare) for 20 min at 25 °C. Atto488-labeled and Cy3-labeled thymocytes were then incubated with DMSO and CS at various concentrations, respectively. Cells were then stained for surface markers for 10 min at 25 °C. After stained with Indo-1 AM (Life Technologies) at a final concentration of 2 μM at 37 °C for 30 min, cells were washed and resuspended with HBSS + 2% FBS. Anti-CD3ε (clone 145-2C11), a final concentration of 3 μg/ml, and anti-Hamster Ig (Jackson Immunoresearch), a final concentration of 8 μg/ml, were added to cells for crosslinking TCRs. Calcium flux data were acquired with LSR II UV (BD Biosciences). The data were analyzed with FlowJo software (Treestar) by comparing the ratio of emission of Indo-1 on the Indo-1 Blue (450/50 nm) channel to the Indo-I Green (525/50 nm) channel.
TCR purification
TCR complexes were purified from M.mζ-SBP T cells as described previously11. Briefly, M.mζ-SBP cells were harvested and lysed with 20 mM Bis-Tris pH 7.0, 500 mMζ-aminocaproic acid, 20 mM NaCl, 2 mM EDTA, 10% glycerol and 1% Digitonin or 0.5% Brij 96V as indicated. SBP-tagged TCRs were affinity-purified using streptavidin-conjugated agarose (GE Healthcare). Proteins were eluted by 30 min incubation at 4 °C with buffer containing 4 mM free biotin. For quantifying TCR-associated lipids with mass spectrometry, Brij 96V was changed to 0.01% ProteaseMax™ Surfactant (Promega) during washing steps of the agarose beads. After elution of TCR-lipid complexes with biotin, formatic acid was added to the eluates to adjust pH to 2–3. The solution was incubated at 37 °C for 30 min to get rid of the ProteaseMax before lipid extraction.
Proteoliposome reconstitution
Large unilamellar vesicles (LUVs) with different membrane compositions including cholesterol (Sigma), cholesterol sulfate (Sigma), soybean phosphatidylcholine and egg sphingomyelin (Lipoid) were prepared as described previously11. Lipid content was determined using thin layer chromatography. The diameters of the vesicles were measured by dynamic light scattering (Zetamaster S, Malvern Instruments). Approximately 100 ng of the purified TCR in 100 μl 0.01% Triton X-100 containing buffer was mixed with 100 μl of 2 mM prepared liposomes, and 40 μl 0.1% Triton X-100 was added. Samples were agitated for 30 min at 4 °C, and the detergent was removed by adsorption to 2–3 mg of BioBeads SM-2 (BioRad) at 4 °C overnight. To analyze TCR nanoclusters, proteoliposomes were lysed by the detergent-supplemented buffers. Blue Native PAGE was performed as previously described50.
Lipid extraction and mass spectrometry quantification
Total lipids of T cells or eluted TCR-lipids complexes were extracted with the two-step Bligh and Dyer method. Briefly, mix and vortex T cells with chloroform/methanol (1/2) for 10 min. Then add chloroform with additional vortex, and water with final vortex. After centrifugation, transfer the lower phase to a new tube. Add chloroform to the upper phase to repeat the extraction procedure. Cholesterol-26,26,26,27,27,27-D6 and 5, 24-cholestadien-3β-ol sulfate sodium salt were added to the extraction as internal controls of cholesterol and cholesterol sulfate, respectively. Evaporate under nitrogen gas, the lipid extract was dissolved in 200 μl methanol. Cholesterol and cholesterol sulfate were separated by Accela 1250 HPLC system (Thermo Fisher Scientific) with BEH C18 column. The samples were quantified by TSQ Vantage triple quadrupole mass spectrometer (Thermo Fisher Scientific) at Stanford University mass spectrometry facility. The MS of cholesterol and cholesterol sulfate were operated in APCI and ESI mode, respectively.
Cholesterol-TCR interaction assay
The cholesterol pull down assay was performed as described36. In brief, 20 μl cholesterol-coupled beads and 1 ml T cell lysate containing 1% Brij 96V were incubated for 30 min at 4 °C with rotation. 5.6 mM digitonin, cholesterol sulfate, cholesterol or solvent control (DMSO or ethanol) was added to the suspensions to compete the TCR binding of cholesterol beads. The beads were rotated overnight, and then washed 4 times with 1 ml wash buffer (200 mM Tris-HCl pH 8.0, 137 mM NaCl, 10% Glycerol, 2 mM EDTA, 1% Brij 96V). Proteins on the beads were separated by 12% SDS-PAGE and subjected to immunoblot analysis.
Photoactive cholesterol binding experiments
The experiments were performed as described previously11, 37. M.m ζ-SBP cells were incubated for 18 h in lipid-free medium containing the [5,6-3H] 7-Azi-5a-cholestan-3b-ol photocholesterol-mβCD complex (10 μCi/ml). CS was added to the cell culture at 80 μM concentration for 5 h. Labeled Cells were washed and UV-irradiated for 8 min. After cell lysis, TCR complexes were affinity-purified with streptavidin-conjugated agarose, and PNGase F (NEB) treatments were performed as indicated. The samples were subjected to SDS-PAGE followed by tritium autoradiography and immunoblot.
Tetramer enumeration
Tetramer enrichment was performed similarly as previously described40, 46. Single-cell suspensions were prepared from the spleens or thymi harvested from wild-type littermate and Sult2b1-deficient mice. Red blood cells (RBCs) were then lysed with ACK buffer. With equal numbers of total cells from each groups, PE-conjugated HY Uty-WMHHNMDLI:Db tetramers (MBLI) were added to the cell suspension at a 1:10 dilution. Cells were incubated for 1 h at 4 °C co-stained with anti-CD8. After washing, cells were resuspended in PBS, 2 mM EDTA and 0.5% BSA, and anti-PE microbeads (Miltenyi Biotec) were added to the cell suspension. Samples were incubated at 4 °C for 15 min and then washed. Cells were applied to pre-washed MACS LS columns. Following three washes, the bound fraction was eluted and resuspended in FACS buffer (PBS, 2 mM EDTA and 5% FBS). Tetramer-bound fractions were stained with 1:100 dilution of fluorophore-conjugated antibodies to the following markers: F4/80 (BM8), Gr-1 (RB6-8C5), CD11b (M1/70), CD11c (N418), Ter119 (TER-119), CD19 (6D5), CD4 (RM4-5) and Live/Dead Aqua amine (Life Technologies). After staining, the washed samples were collected on a BD LSR II and analyzed using FlowJo (Tree Star).
Statistical analysis
The statistical significance of the two data sets was determined by Student’s t-test. Statistical analysis was done using Prism 6 software (GraphPad Software).
Supplementary Material
Acknowledgments
We thank Y.-H. Chien, A. Habtezion, Y. Wong, and M. Lozano for helpful discussions, and X. Zeng, E.W. Newell, W.O. Gorman, L. Wagar, J. Xie, K.H. Roh, A. Morath, M. Swamy, J. Huppa, and J. Huang for experimental assistance, and D. Russell for providing Sult2b1−/− mice embryos. This work was supported by grants from the US National Institutes of Health (RO1 AI022511 to M.M.D.) and the Howard Hughes Medical Institute (M.M.D.). W.W.S. received grants from the Deutsche Forschungsgemeinschaft through EXC294 and SCHA 976/2-1. This study was also supported in part by the Excellence Initiative of the German Research Foundation (GSC-4, Spemann Graduate School).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
F.W. and M.M.D. conceived the project. Experiments were designed and conducted by F.W. unless indicated otherwise. F.W., K.B.-G. and C.Z. worked together on TCR proteoliposome reconstitution and BN-PAGE. K.B.-G. and C.Z. performed cholesterol-beads pull down assay. W.W.S. contributed important ideas and technical support. F.W. and M.M.D. wrote the manuscript. W.W.S. and other authors edited the manuscript.
References
- 1.Davis MM, et al. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 2.Schamel WW, Alarcon B. Organization of the resting TCR in nanoscale oligomers. Immunol Rev. 2013;251:13–20. doi: 10.1111/imr.12019. [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty AK, Weiss A. Insights into the initiation of TCR signaling. Nat Immunol. 2014;15:798–807. doi: 10.1038/ni.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malissen B, Gregoire C, Malissen M, Roncagalli R. Integrative biology of T cell activation. Nat Immunol. 2014;15:790–797. doi: 10.1038/ni.2959. [DOI] [PubMed] [Google Scholar]
- 5.Davis MM, et al. T cells as a self-referential, sensory organ. Annu Rev Immunol. 2007;25:681–695. doi: 10.1146/annurev.immunol.24.021605.090600. [DOI] [PubMed] [Google Scholar]
- 6.Huang J, et al. A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4(+) T cells. Immunity. 2013;39:846–857. doi: 10.1016/j.immuni.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schamel WW, et al. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J Exp Med. 2005;202:493–503. doi: 10.1084/jem.20042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schamel WW, Risueno RM, Minguet S, Ortiz AR, Alarcon B. A conformation- and avidity-based proofreading mechanism for the TCR-CD3 complex. Trends Immunol. 2006;27:176–182. doi: 10.1016/j.it.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Lillemeier BF, et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci U S A. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molnar E, et al. Cholesterol and sphingomyelin drive ligand-independent T-cell antigen receptor nanoclustering. J Biol Chem. 2012 doi: 10.1074/jbc.M112.386045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alarcon B, Swamy M, van Santen HM, Schamel WW. T-cell antigen-receptor stoichiometry: pre-clustering for sensitivity. EMBO Rep. 2006;7:490–495. doi: 10.1038/sj.embor.7400682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Martin N, et al. Cooperativity between T cell receptor complexes revealed by conformational mutants of CD3epsilon. Sci Signal. 2009;2:ra43. doi: 10.1126/scisignal.2000402. [DOI] [PubMed] [Google Scholar]
- 14.Huppa JB, et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 2010;463:963–967. doi: 10.1038/nature08746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman E, et al. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity. 2011;35:705–720. doi: 10.1016/j.immuni.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature. 1995;375:148–151. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- 17.Davey GM, et al. Preselection thymocytes are more sensitive to T cell receptor stimulation than mature T cells. J Exp Med. 1998;188:1867–1874. doi: 10.1084/jem.188.10.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 19.Pircher H, Rohrer UH, Moskophidis D, Zinkernagel RM, Hengartner H. Lower receptor avidity required for thymic clonal deletion than for effector T-cell function. Nature. 1991;351:482–485. doi: 10.1038/351482a0. [DOI] [PubMed] [Google Scholar]
- 20.Kumar R, et al. Increased sensitivity of antigen-experienced T cells through the enrichment of oligomeric T cell receptor complexes. Immunity. 2011;35:375–387. doi: 10.1016/j.immuni.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 22.Kersh EN, et al. TCR signal transduction in antigen-specific memory CD8 T cells. J Immunol. 2003;170:5455–5463. doi: 10.4049/jimmunol.170.11.5455. [DOI] [PubMed] [Google Scholar]
- 23.Fahmy TM, Bieler JG, Edidin M, Schneck JP. Increased TCR avidity after T cell activation: a mechanism for sensing low-density antigen. Immunity. 2001;14:135–143. [PubMed] [Google Scholar]
- 24.Li QJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Rost B. Comparing function and structure between entire proteomes. Protein Sci. 2001;10:1970–1979. doi: 10.1110/ps.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu C, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell. 2008;135:702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gagnon E, Schubert DA, Gordo S, Chu HH, Wucherpfennig KW. Local changes in lipid environment of TCR microclusters regulate membrane binding by the CD3{varepsilon} cytoplasmic domain. J Exp Med. 2012 doi: 10.1084/jem.20120790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi X, et al. Ca(2+) regulates T-cell receptor activation by modulating the charge property of lipids. Nature. 2012 doi: 10.1038/nature11699. [DOI] [PubMed] [Google Scholar]
- 30.Strott CA, Higashi Y. Cholesterol sulfate in human physiology: what’s it all about? J Lipid Res. 2003;44:1268–1278. doi: 10.1194/jlr.R300005-JLR200. [DOI] [PubMed] [Google Scholar]
- 31.Bacia K, Schwille P, Kurzchalia T. Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proc Natl Acad Sci U S A. 2005;102:3272–3277. doi: 10.1073/pnas.0408215102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalland ME, Oberprieler NG, Vang T, Tasken K, Torgersen KM. T cell-signaling network analysis reveals distinct differences between CD28 and CD2 costimulation responses in various subsets and in the MAPK pathway between resting and activated regulatory T cells. J Immunol. 2011;187:5233–5245. doi: 10.4049/jimmunol.1101804. [DOI] [PubMed] [Google Scholar]
- 33.Fuda H, Lee YC, Shimizu C, Javitt NB, Strott CA. Mutational analysis of human hydroxysteroid sulfotransferase SULT2B1 isoforms reveals that exon 1B of the SULT2B1 gene produces cholesterol sulfotransferase, whereas exon 1A yields pregnenolone sulfotransferase. J Biol Chem. 2002;277:36161–36166. doi: 10.1074/jbc.M207165200. [DOI] [PubMed] [Google Scholar]
- 34.Bergner EA, Shapiro LJ. Increased cholesterol sulfate in plasma and red blood cell membranes of steroid sulfatase deficient patients. J Clin Endocrinol Metab. 1981;53:221–223. doi: 10.1210/jcem-53-1-220. [DOI] [PubMed] [Google Scholar]
- 35.Epstein EH, Jr, Krauss RM, Shackleton CH. X-linked ichthyosis: increased blood cholesterol sulfate and electrophoretic mobility of low-density lipoprotein. Science. 1981;214:659–660. doi: 10.1126/science.6945674. [DOI] [PubMed] [Google Scholar]
- 36.Beck-Garcia E, Beck-Garcia K, Schlosser A, Schamel WW. Analysis of interactions between proteins and fatty acids or cholesterol using a fatty acid/cholesterol pull-down assay. Anal Biochem. 2013;436:75–77. doi: 10.1016/j.ab.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 37.Thiele C, Hannah MJ, Fahrenholz F, Huttner WB. Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat Cell Biol. 2000;2:42–49. doi: 10.1038/71366. [DOI] [PubMed] [Google Scholar]
- 38.Hogquist KA, Jameson SC. The self-obsession of T cells: how TCR signaling thresholds affect fate ‘decisions’ and effector function. Nat Immunol. 2014;15:815–823. doi: 10.1038/ni.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hogquist KA, Jameson SC, Bevan MJ. The ligand for positive selection of T lymphocytes in the thymus. Curr Opin Immunol. 1994;6:273–278. doi: 10.1016/0952-7915(94)90101-5. [DOI] [PubMed] [Google Scholar]
- 40.Yu W, et al. Clonal Deletion Prunes but Does Not Eliminate Self-Specific alphabeta CD8(+) T Lymphocytes. Immunity. 2015;42:929–941. doi: 10.1016/j.immuni.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surh CD, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–103. doi: 10.1038/372100a0. [DOI] [PubMed] [Google Scholar]
- 42.Daniels MA, et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 43.Au-Yeung BB, et al. Quantitative and temporal requirements revealed for Zap70 catalytic activity during T cell development. Nat Immunol. 2014;15:687–694. doi: 10.1038/ni.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong B, et al. Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc Natl Acad Sci U S A. 2009;106:18831–18836. doi: 10.1073/pnas.0909731106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moon JJ, et al. Tracking epitope-specific T cells. Nat Protoc. 2009;4:565–581. doi: 10.1038/nprot.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaffert SA, et al. mir-181a-1/b-1 Modulates Tolerance through Opposing Activities in Selection and Peripheral T Cell Function. J Immunol. 2015;195:1470–1479. doi: 10.4049/jimmunol.1401587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coskun U, Grzybek M, Drechsel D, Simons K. Regulation of human EGF receptor by lipids. Proc Natl Acad Sci U S A. 2011;108:9044–9048. doi: 10.1073/pnas.1105666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang W, et al. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature. 2016;531:651–655. doi: 10.1038/nature17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams ML, Elias PM. Stratum corneum lipids in disorders of cornification: increased cholesterol sulfate content of stratum corneum in recessive x-linked ichthyosis. J Clin Invest. 1981;68:1404–1410. doi: 10.1172/JCI110391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swamy M, Siegers GM, Minguet S, Wollscheid B, Schamel WW. Blue native polyacrylamide gel electrophoresis (BN-PAGE) for the identification and analysis of multiprotein complexes. Sci STKE. 2006;2006:pl4. doi: 10.1126/stke.3452006pl4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.