Summary
To evaluate a new therapy versus a control via a randomized, comparative clinical study or a series of trials, due to heterogeneity of the study patient population, a pre-specified, predictive enrichment procedure may be implemented to identify an “enrichable” subpopulation. For patients in this subpopulation, the therapy is expected to have a desirable overall risk-benefit profile. To develop and validate such a “therapy-diagnostic co-development” strategy, a three-step procedure may be conducted with three independent data sets from a series of similar studies or a single trial. At the first stage, we create various candidate scoring systems based on the baseline information of the patients via, for example, parametric models using the first data set. Each individual score reflects an anticipated average treatment difference for future patients who share similar baseline profiles. A large score indicates that these patients tend to benefit from the new therapy. At the second step, a potentially promising, enrichable subgroup is identified using the totality of evidence from these scoring systems. At the final stage, we validate such a selection via two-sample inference procedures for assessing the treatment effectiveness statistically and clinically with the third data set, the so-called holdout sample. When the study size is not large, one may combine the first two steps using a “cross-training-evaluation” process. Comprehensive numerical studies are conducted to investigate the operational characteristics of the proposed method. The entire enrichment procedure is illustrated with the data from a cardiovascular trial to evaluate a beta-blocker versus a placebo for treating chronic heart failure patients.
Keywords: Cox model, Cross-validation, Stratified medicine, Survival analysis, Therapy-diagnostic co-development
1. Introduction
In a typical randomized clinical trial, generally the assessment of a new therapy versus a control with respect to the risk-benefit profile is made for the entire study patient population. Due to heterogeneity of the study population, the conclusion of a positive (or negative) study on an average sense does not guarantee that the new therapy benefits (or does not benefit) uniformly all patients in the study population (Rothwell, 1995; Rothwell et al., 2005; Kent and Hayward, 2007). Recently various enrichment strategies were suggested and implemented in comparative trials (Freidlin and Simon, 2005; Jiang et al., 2007; Wang et al., 2007; Simon, 2008; Karuri and Simon, 2012; U.S. Food and Drug Administration, 2012). For predictive enrichment, the idea is to apply a systematic, pre-specified procedure to identify and validate a subpopulation whose patients would significantly benefit from the new therapy both clinically and statistically. This procedure may be utilized on data obtained from a single study or a series of trials conducted under similar settings.
A properly executed predictive enrichment procedure generally consists of three stages: at the first stage using data set 𝒜, we utilize all the relevant baseline information to fit the data with various prediction models to create several competing candidate scoring systems for stratifying future patients. The score from an individual patient is an estimated average treatment difference for future patients with similar baseline profiles. A larger score indicates that the patient has a higher chance to benefit from the new therapy. At the second stage with data set ℬ, we evaluate and compare these scoring systems and then develop a rule for identifying an enrichable subpopulation. At the last step with data set 𝒞, the holdout sample, we validate such a selection rule via proper two-sample inference procedures for comparing the two treatments for the enrichable subset of patients in 𝒞. Ideally each step would be conducted via an independent data set from the same underlying study population and interventions. With the data from a single comparative study, one may split the entire data set into two independent parts and use the first part as 𝒜 & ℬ for scoring system building and evaluation, as well as the identification of an enrichable subpopulation via a conventional “cross-training-evaluation” scheme. We then use the second part of the data as 𝒞 to examine if there is a strong evidence that patients in the enrichable subpopulation would respond favorably to the new therapy compared with the control.
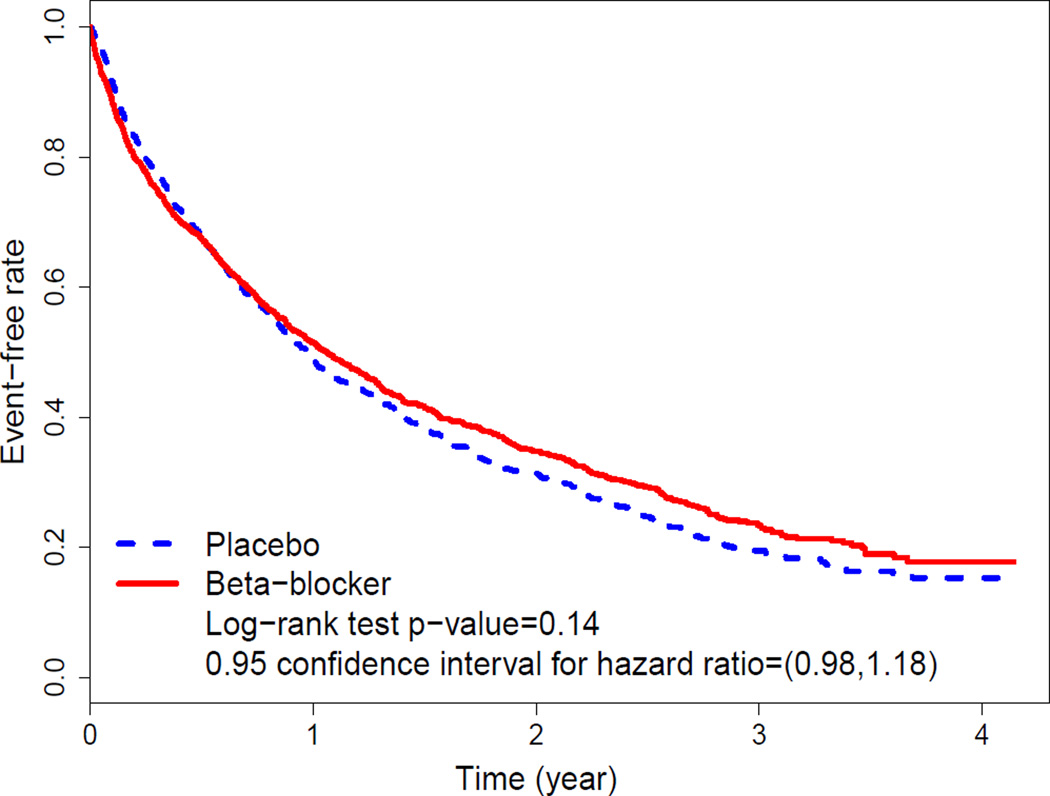
As an illustrative example, consider the data from a clinical trial “Beta-Blocker Evaluation of Survival Trial (BEST)” to investigate if bucindolol, a beta-blocker, would benefit patients with advanced chronic heart failure compared with a standard placebo (BEST, 2001). There were 2708 patients enrolled and followed for an average of two years. One of the primary goals of the study was to examine if the beta-blocker could reduce the overall hospitalization and mortality rates of the patients. With the time to either death or the first hospitalization as the endpoint, the Kaplan-Meier curves are given in Figure 1. The p-value based on the standard two-sample log-rank test is 0.14, with a hazard ratio (HR) estimate of 1.07 (placebo vs. beta-blocker) and its corresponding 0.95 confidence interval of (0.98, 1.18). For this outcome variable, the evidence that the beta-blocker was better than the placebo for the entire study population is not strong.
Figure 1.
Kaplan-Meier curves for the time to death or the first hospitalization with the data from the BEST study. This figure appears in color in the electronic version of this article.
Now, suppose that we had a pre-specified, aforementioned enrichment strategy at the beginning of the study with respect to this endpoint. We first split the entire data set into two parts, using the data from the first 900 patients on the data listing for BEST provided by US National Institutes of Health as 𝒜 & ℬ for building and evaluating scoring systems to identify a potentially enrichable subpopulation. Then, we use the data from the remaining 1807 patients as the holdout sample 𝒞 to examine if the beta-blocker would benefit the patients of the selected subpopulation by reducing the rate of death or the first hospitalization compared with the standard placebo. For this study, there are 16 clinically relevant baseline covariates: age, sex, left ventricular ejection fraction (LVEF), systolic blood pressure (SBP), class of heart failure (Class III versus Class IV), obesity (Body mass index > 30 versus ≤ 30), resting heart rate, smoking status (ever versus never), history of hypertension, history of diabetes, ischemic heart failure etiology, presence of atrial fibrillation, race (white versus non-white) and estimated glomerular filtration rate (eGFR) adjusted for body surface area, which is categorized with cut-points of 45, 60, and 75.
We will use this example to illustrate the proposed procedure step by step. In Section 2, with data set 𝒜 from the BEST study as described above, we use a set of regression models, which relate the event time to its 16 baseline covariates, to estimate an average treatment difference (e.g., the HR of placebo versus beta-blocker) for patients who share similar baseline covariate profiles. This creates a continuous scoring system. If the model is a reasonably good approximation to the truth, a future patient with a large score is expected to benefit from the new therapy. An enrichable subpopulation would consist of patients whose scores are greater than an appropriately chosen threshold value. To avoid the problem of potential over-fitting, we discuss methods to choose this threshold value with an independent data set ℬ in Section 3. Since there is only one study for evaluating the beta-blocker in our illustrative example, the size of the data set at each step may not be large enough for obtaining a reliable scoring system. In Section 4, we present a “cross-training-evaluation” process by implementing the above two steps iteratively with various competing models and regularized estimation procedures to create the scoring systems. The final recommendation for an enrichable subgroup is then obtained by considering the totality of evidence from all of the candidate scoring systems and the related threshold values. In Section 5, we validate our selection by applying two-sample inference procedures to the enrichable subgroup in the holdout sample. Note that the primary criteria utilized for identifying a threshold value in Section 4 should be closely related to those for the final validation with the holdout sample. For instance, if the final primary assessment of the enrichable subgroup is based on the standard two-sample interval estimation procedure for the HR with the holdout sample, the choice of the threshold value in Section 4 would be made via the corresponding predicted interval estimate for the HR. That is, we choose the threshold value such that the predicted interval estimate suggests that using the data from the potential enrichable subgroup in the holdout sample, the beta-blocker can be demonstrated to have a clinically meaningful advantage over the placebo via the confidence interval estimate. In Section 6, we conduct a simulation study to compare the performance of our proposed procedure with two existing methods in the literature. Further remarks about the procedure are given in Section 7.
There are numerous novel proposals in the literature for identifying future patients to be treated by the new therapy (Foster et al., 2011, Qian and Murphy, 2011, Zhang et al., 2012, Zhang et al., 2012, Zhao et al., 2013). For example, using data from the BEST study, one may recommend treating future patients whose HR scores are greater than one with placebo vs. beta-blocker. However, such a selection procedure may not be applied directly to our problem. For instance, we may not be able to validate the superiority of the beta-blocker over the placebo using the data from the holdout sample (especially if the enrichable subgroup includes a sizable subset of patients for whom the benefit of beta-blocker is not clinically meaningful).
2. Patient-specific scoring systems for quantifying the between-group differences
In this section, we present the predictive enrichment scheme under the survival analysis setting. Let T be a time-to-event outcome, U be a p × 1 vector of baseline covariates and G denote the group indicator with 1 for the new therapy group and 0 for control. Furthermore, it is assumed that T may be censored by a censoring variable C, which is independent of T and U. For each patient, we observe (X, Δ, G, U), where X = min(T, C) and Δ denotes the censoring indicator with Δ = 1 if T is observed and 0 otherwise. The observations, {(Xi, Δi, Gi, Ui), i = 1, …, n1}, from data set 𝒜 consist of n1 independent and identical copies of (X, Δ, G, U). Conditional on U, let the population parameter for the group contrast, i.e. the treatment difference, be denoted by D(U) and a large value of D(U) indicates that the new therapy is better than the control. For example, given a patient population with a covariate vector of U, D(U) may be the difference of two event time medians or the model-based “constant” HR for the control versus the new therapy (Cox, 1972).
For U = u, we first consider D(u) being the HR for patients with baseline vector u. To estimate D(u) with the data set 𝒜, one may fit the data from each treatment group with a Cox model to obtain D̂(u) to approximate D(u). Specifically, for patients in Group G = k (k = 0, 1), consider the Cox model:
| (2.1) |
where Λk(·) is the group-specific underlying cumulative baseline hazard function and βk denotes the vector of unknown coefficients.
To obtain an estimate of βk from (2.1) when the dimension of U is high, we may utilize a regularized estimation procedure, for example, via lasso, adaptive lasso, elastic net or ridge regression (Tibshirani, 1997; Zou, 2006; Zhang and Lu, 2007; Friedman et al., 2010). Taking the elastic net procedure as an example, for k = 1, 2, β̂k is a minimizer of
where L(βk) is the partial likelihood function, and ‖βk‖p denotes the Lp– norm of βk, p = 1, 2. Here, λ1k and λ2k are the non-negative regularization parameters, which can be selected by cross-validation (Tibshirani, 1996). Note that the estimation procedure becomes the standard lasso method by setting λ2k ≡ 0. On the other hand, when λ1k ≡ 0, the resulting procedure is the standard ridge regression.
Now, assuming that the underlying hazard functions for two treatment groups are proportional to each other, one may summarize the treatment difference up to a constant based on . It is important to note that (2.1) is simply a working model, which is an approximation to the truth. Therefore, D̂(·) may not be able to completely capture the patient-specific treatment differences. In fact, even if the two Cox models are correctly specified, D̂(·) can only estimate D(·) up to a positive constant. On the other hand, this scoring system may work reasonably well for ranking the future patients with respect to the benefit from the new therapy. As an example, with the data from the first 900 patients in the BEST study discussed in Section 1, we randomly choose a subset of 450 patients and fit the corresponding data with the above procedure using the ridge regression. The resulting β̂0 and β̂1 are then used to obtain D̂(u) for each given covariate vector u. With such a scoring system, we can rank patients in a given data set. Generally there is a one-to-one correspondence between the scores and their ranks. Thus, we can use relative ranks as scores to identify an enrichable subgroup.
Alternative and model-free between-group contrast measures can also be considered, for example, the difference of two median or mean survival times. However, due to censoring, we may not be able to estimate median or mean survival time well with the observed data. We can modify the mean with so-called restricted mean survival time (RMST) as a summary to accommodate the study follow-up time, say, up to τ0 (Irwin, 1949; Karrison, 1987; Zucker, 1998; Murray and Tsiatis, 1999; Chen and Tsiatis, 2001; Andersen et al., 2004; Zhang and Schaubel, 2011; Royston and Parmar, 2011; Tian et al., 2012; Zhao et al., 2012). The RMST is simply the population average of the event-free times for τ0-year follow up. This quantity can be easily estimated by the area under the Kaplan-Meier curve up to τ0. For example, in BEST study, if we let τ0 = 3.5 (years), the estimated RMSTs for beta-blocker and the placebo are 1.51 and 1.42 (years), respectively. The p-value for testing the equality of the underlying two RMSTs based on their empirical counterparts is 0.19 and a 0.95 confidence interval for the difference of the RMSTs is (−0.11, 0.30). Note that unlike the HR, this between-group difference measure is model-free and readily interpretable clinically.
To construct a scoring system using the difference of RMSTs, one may use the regression models studied by Andersen et al. (2004) and Tian et al. (2014). Alternatively, we may use the Cox models above to estimate D(u), the difference of two RMSTs given U = u. The resulting estimate
where Λ̂k(t) is the Breslow’s estimator for the baseline cumulative hazard function.
Note that instead of fitting the data from each treatment group with a Cox model, respectively, one may use a single model including the treatment indicator, main covariate effects, and treatment by covariate interactions with regularized estimation procedures discussed above to fit the entire data set 𝒜 to create the scoring system. In Section 4, we will provide more details about other models for building the scoring systems.
3. Choosing an enrichable subpopulation with a scoring system
From data set 𝒜, we obtain a scoring system based on D̂(·) and define its corresponding rank score Q̂(·) = F̂{D̂(·)}, where F̂(·) is the empirical distribution function of D̂(U). If the working models utilized for establishing the individual scores are reasonably good approximations to the truth, we expect that a patient whose rank score is larger would be more likely to benefit from the new therapy. Therefore, it seems natural to consider an enrichable subgroup of patients in data set ℬ, whose Q̂(·) ≥ 1 − q0, where q0 is an appropriately selected threshold value representing the fraction of the enrichable subpopulation with respect to the entire study population. That is, given 0 < q0 < 1, the selected enrichable subgroup based on q0 consist of patients whose rank scores exceed the 100(1 − q0)th percentile.
Now, the question is how to choose this threshold value q0 at this stage. To answer this question, we need to know how we will validate the potentially promising subgroup using a threshold value q0, with the data from the holdout sample 𝒞. As an example, let us consider the Q̂(·) based on the estimated HR as the score created by two Cox models with the ridge regression method discussed in Section 2. We apply the above binary rule to identify a set ℰ of patients in the holdout sample-whose ranks of the HRs are greater than n3(1 − q0), where n3 is the sample size of the holdout sample. With the observations from ℰ, we test the null hypothesis that there is no difference between two arms via the logarithm of the two-sample HR estimator . If the resulting standardized test statistic Z is significantly large, then one claims that this selected subgroup is successfully validated.
Now for each given q, let HRq be the limit of and be the asymptotic variance of . With the observations from data set ℬ, the Z-test statistic for the holdout sample may be predicted via
| (3.1) |
where is the estimate for the HR from observations whose Q̂(·) ≥ 1 − q in data set ℬ and σ̂q is the corresponding estimate for σq.
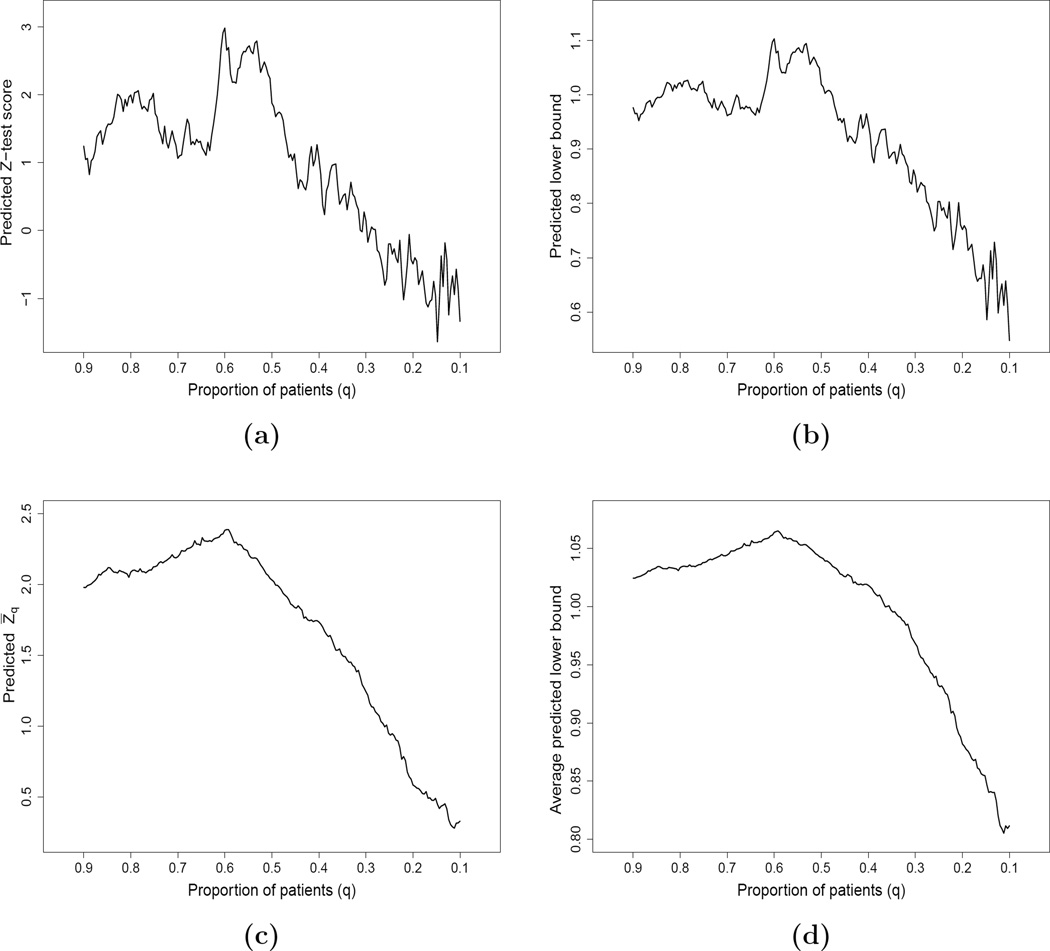
Utilizing the data from the first 900 patients in the BEST study, we considered using the HR scores created from Model (2.1) with ridge regression procedure and the data from 450 patients not used for fitting the models discussed in the previous section to obtain the predicted Z-test scores. Figure 2(a) gives the curve of such predicted Z-test scores over q. Although the right tail of this curve is not stable due to the small sample size, this predicted Z-test score curve is quite informative for choosing an appropriate threshold value q0. For example, the Z-test score achieves its maximal value of 2.98 when q = 0.6. This means that if we apply q0 = 0.6 as the threshold value to the holdout sample, this would be a good approximation to the observed two-sample standardized test statistic resulting from the final validation stage. If one would like to choose a larger enrichable subgroup, we may select a larger threshold value q0 but with a compromise of a smaller Z-test score.
Figure 2.
Predicted curves based on HR estimates with the first 900 patients by fitting two Cox models with ridge regression in BEST study: (a) predicted Z-test score with a random subset of 450 patients; (b) predicted lower bound of one-sided 0.95-CI with a random subset of 450 patients; (c) predicted Z̄q based on 100 random cross-validation; (d) predicted average lower bound of one-sided 0.95-CI based on 100 random cross-validation.
Alternatively, we may utilize a 0.95 confidence interval estimate for the two-sample HR as the inferential tool for decision making with the holdout sample. As in the aforementioned hypothesis testing case, one can estimate the expected bounds of the confidence interval based on the data in ℬ. For example, the expected value for the lower bound of a one-sided 0.95 confidence interval can be predicted as
| (3.2) |
Figure 2(b) gives the corresponding curve based on (3.2), which provides information about the size of the treatment effect over q. If we choose q0 = 0.6, the predicted lower bound would be about 1.1. Thus, we expect that for the holdout sample, it is likely that the observed lower bound of 0.95 one-sided interval for HRq0 would be around 1.1. Similarly, one may obtain the lower bound curve based on other treatment effect summaries such as the RMST.
4. Cross-training-evaluation
For each given scoring system, with two independent data sets 𝒜 and ℬ, the predicted Z-test score or the confidence interval lower bound curve as in Figure 2(a) or Figure 2(b) may not be stable due to small sample sizes. An alternative is to use the conventional “cross-training-evaluation” procedure iteratively for model fitting and evaluation. Specifically, we randomly divide the combined data set from the first and second stages into two independent pieces, 𝒜 and ℬ evenly, as the training and the evaluation sets. As before, we use the data from 𝒜 to build the scoring system and the data from ℬ to obtain the predicted Z-test score curve as a function of q. By repeating such random cross-validation process M times, the final curve Z̄q can be obtained by averaging those predicted Z-test score curves.
With the data from the first 900 patients in BEST, using this random cross-validation procedure with two Cox working models and ridge regression setting as discussed in the previous sections, the resulting Z̄q curve with M = 100 is given in Figure 2(c). Note that this curve is much more smooth than that in Figure 2(a) using a single training and an evaluation data set. The Z̄q has its highest value of 2.3 at q = 0.6.
Now, we may consider various working models and regularized estimation procedures to create candidate scoring systems using data set 𝒜 and then construct, for example, the Z̄q curves using data set ℬ iteratively via the above random cross-validation scheme. We use the data from BEST to illustrate how to implement this process. In the following, we listed different candidate scoring systems including two Cox-type models with four regularized estimation criteria (lasso, ridge, adaptive lasso and elastic net) for analyzing survival data: (1)Two.Cox_Lasso; (2)Two.Cox_Ridge; (3)Two.Cox_ALasso; (4) Two.Cox_ElaNet; (5)One.Cox_Lasso; (6)One.Cox_Ridge; (7)One.Cox_ALasso; (8)One.Cox_ElaNet. For instance, a procedure labeled as “Two.Cox_Ridge” means that we fit the data from each treatment group via a Cox model additively with baseline covariates using the L2 penalty as described in the previous section. On the other hand, a procedure labeled as “One.Cox_Lasso” means that we fit the data from both groups together via a single Cox model with treatment indicator, main covariate effects, and the treatment by covariate interactions using the L1 penalty. Note that “ElaNet” stands for elastic net and “ALasso” stands for adaptive lasso, respectively.
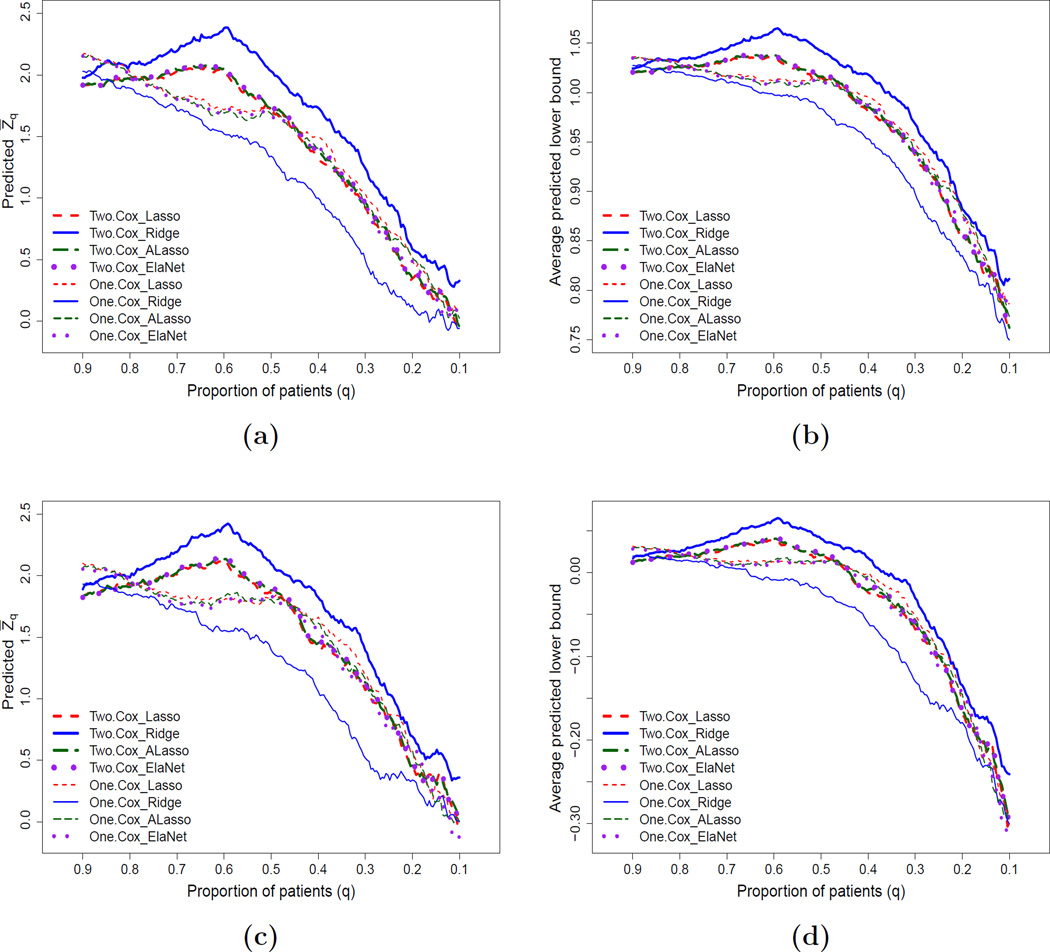
For the data from the first 900 patients in BEST, we use the eight procedures described above to implement the scoring system building and evaluation iteratively. The resulting predicted Z̄q curves for all these candidate scoring systems are given in Figure 3(a). Note that the largest Z̄q score is with the thicker solid curve evaluated at q = 0.6. If the size of the enrichable subgroup and the significance level are acceptable, we then choose the scoring system which generated the thicker solid curve to identify the enriched subgroup. For this case, it is based on two Cox models with the ridge regression. It is interesting to observe that scores built with two separate models appear to perform uniformly better compared to those constructed by fitting an overall interaction model in this data set. This suggests that fitting a more flexible model could potentially lead to better performance. We then use the two Cox models with the ridge regression procedure on the entire data set of the first 900 patients in BEST to obtain the final scoring system for ranking the future patients with respect to the benefit of beta-blocker. The resulting β̂0 and β̂1 are given in the Table 1. The enrichable subgroup would consist of patients whose scores are among the top 60% of the entire study population. This final selected enriched subgroup will then be formally validated using the data from the holdout sample.
Figure 3.
Predicted curves for testing treatment difference with the first 900 patients based on 100 random cross-validation with various candidate scoring systems: (a) predicted Z̄q score based on two-sample HR estimates; (b) average predicted lower bound of 0.95 one-sided CI based on two-sample HR estimates; (c) predicted Z̄q score based on the difference of two estimated RMSTs; (d) average predicted lower bound of 0.95 one-sided CI based on the difference of two estimated RMSTs. This figure appears in color in the electronic version of this article.
Table 1.
Regression coefficient estimates by fitting two Cox models with ridge regression procedure using the first 900 patients in the BEST study.
| Covariate | β̂0 (Placebo) | β̂1 (Beta-blocker) |
|---|---|---|
| Age | 0.003 | −0.008 |
| Male | −0.003 | −0.091 |
| LVEF | −0.009 | −0.016 |
| I(45 < eGFR ≤ 60) | −0.087 | −0.460 |
| I(60 < eGFR ≤ 75) | −0.113 | −0.741 |
| I(eGFR > 75) | −0.171 | −1.072 |
| SBP | −0.002 | −0.005 |
| Class IV Heart Failure | 0.253 | 1.223 |
| I(BMI > 30) | −0.001 | 0.018 |
| Ever Smoker | −0.070 | −0.045 |
| Heart Rate | 0.003 | −0.007 |
| History of Hypertension | 0.040 | 0.001 |
| History of Diabetes | 0.059 | 0.125 |
| Ischemic Etiology | 0.041 | 0.261 |
| Atrial Fibrillation | 0.099 | 0.042 |
| White Race | 0.020 | −0.049 |
Now, if we are interested in using the size of the treatment difference for the final assessment of the treatment benefit, Figure 3(b) provides the corresponding average curves of the lower bounds of the 0.95 one-sided confidence interval estimates under the same setting. It appears that the above choice based on the thicker solid curve is appropriate.
If we are interested in utilizing the difference of RMSTs (up to τ0 = 3.5 years) for quantifying the treatment contrast, with the data from BEST and under the same setting discussed above, Figure 3(c) gives the average predicted Z̄q curves with various scoring systems listed above for testing the hypothesis that there is no treatment effect using the difference of two estimated RMSTs. It is interesting to note that the choice of the thicker solid curve with q0 = 0.6 would give us a reasonably sized enrichable subgroup and the largest predicted Z-test score. Similar conclusions can be made via the expected lower bound of the 0.95 one-sided confidence interval estimates, which is provided in Figure 3(d).
It is important to note that the selection rule for an enrichable subgroup at this stage may depend on the totality of evidence based on various criteria rather than a single one. For instance, if the primary analysis for the holdout sample is based on the test for the HR equal to one, one would use Figure 3(a) first to choose a set of subgroups with different sizes, whose predicted Z-test scores range from 2.0 to 2.4. Then among those subgroups, we can examine how these subgroups behave with respect to other features (such as the RMST difference) and make a final recommendation regarding the choice of the enrichable subpopulation. In addition to the Z-test score, the absolute magnitude of the treatment effect with respect to HR or RMST may also be considered. When selecting the scoring system, we may prefer to consider the model performance as well as the model complexity, which often results in the selection of a more parsimonious model. Although we do not force the selected candidate scoring system to be generated from a parsimonious model, when parsimony is of particular interest, one may restrict the selection to candidate procedures that generate sparse models for the building of candidate scoring systems.
5. Final validation of the selected enrichable subgroup
In Section 4, we choose a specific scoring system and a threshold value q0 for selecting a potentially promising subgroup. We apply this rule to the data in the holdout sample. For this selected subgroup of patients in data set 𝒞, one can then make inferences about the relative merits between two treatment groups.
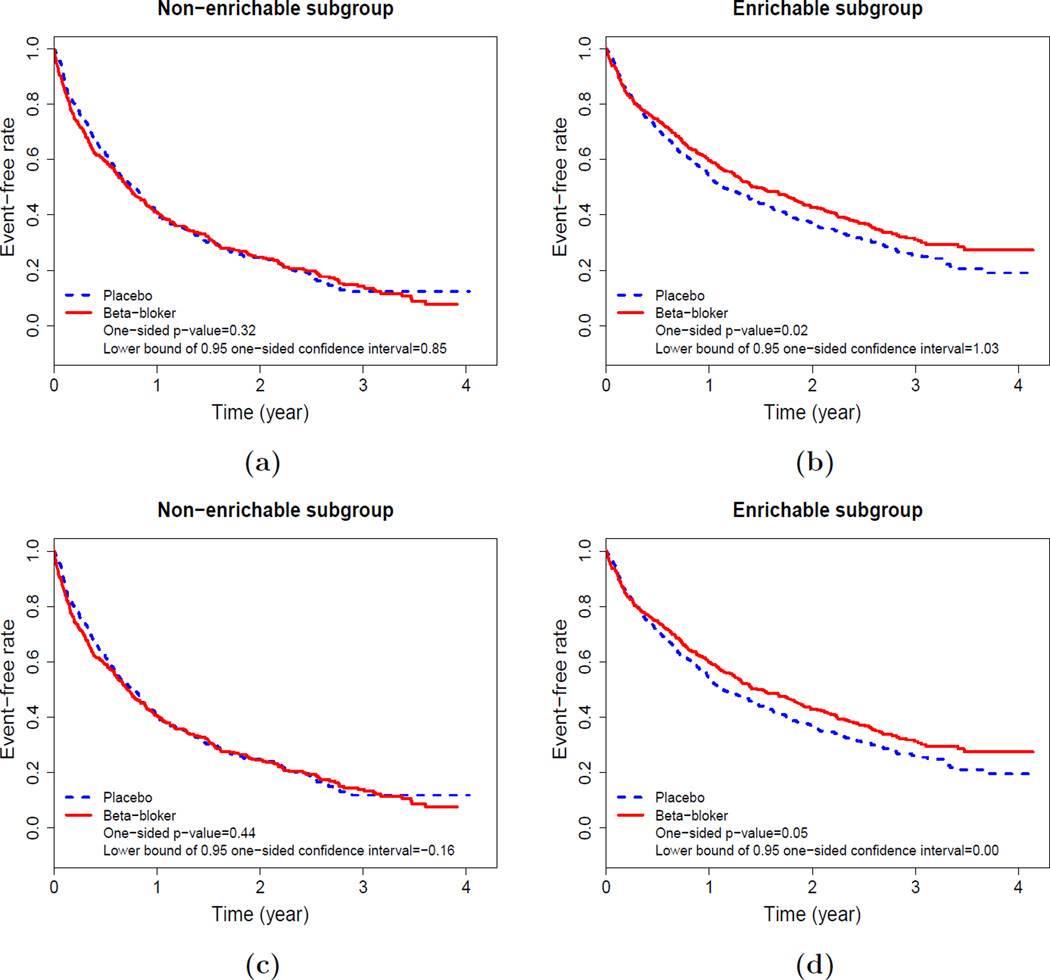
As an example, for the data from BEST, the holdout sample is obtained from the remaining 1807 patients. If we are interested in making inference about the “constant” HR, we use the scoring system generated from two independent Cox models with ridge regression and the threshold value q0 of 0.6. With the data from corresponding enrichable subgroup in the holdout sample, Figure 4(b) provides the Kaplan-Meier curves for two treatment groups, p-value of the two-sample test based on the HR estimates and the lower bound of the one-sided 0.95 confidence interval for the HR. The beta-blocker for patients in this enriched subgroup appears to prolong the event time compared with the placebo. Figure 4(a) also shows the non-significant inferential results for the non-enrichable subgroup.
Figure 4.
Two-sample inferences for the HR or two RMSTs up to 3.5 year follow-up with the holdout sample: (a)–(b): inference based on HR; (c)–(d): inference based on two RMSTs up to 3.5 year follow-up. This figure appears in color in the electronic version of this article.
Now, if we are interested in quantifying the treatment contrast using the difference of RMSTs up to τ0 = 3.5 years, Figure 4(d) provides the comparison results for the enrichable subgroup and Figure 4(c) gives the results for the non-enrichable subset. For the enrichable subgroup, the one-sided p-value is 0.05 with the lower bound of the confidence interval of 0. Note that for the entire study population, the corresponding one-sided p-value via the test statistic based on the difference of two estimated RMSTs is 0.10 and one-sided 0.95 confidence interval is (−0.08, +∞).
With the event time data from the BEST study, we created two sets of patient-specific scores using the HR and the RMST difference as the treatment effect summary measures. If we choose the “optimal” threshold values based on their average predicted Z-test scores from Figures 3(a) and 3(c), respectively, to define the enrichable subgroups, these two resulting subgroups in the holdout sample are almost identical to each other. This may suggest that the selection of the enrichable subgroup is quite robust to the choice of the summary measure for BEST study. Note that the inference procedure at this step is conditional on the enrichable subgroup selected based on the cross-training-evaluation data. The target of the inference is not the treatment effect of a unknown underlying subgroup but the treatment effect of the identified enrichable subgroup conditional on the cross-training-evaluation data, which will be used in the future after successful validation with the holdout sample.
6. Numerical Study
We conduct an extensive numerical study to examine whether the proposed procedure can maintain the nominal type I error rate under the null of absence of treatment effect in any subgroup of patients. We also compare the efficiency of the enrichment procedure with two existing competitors in identifying the enrichable subgroup when the true enrichable subgroup exists. The first alternative method fits a single lasso regularized Cox regression model with main treatment and covariate effects, as well as all treatment by covariate interactions. The second alternative method is adapted from the virtual twin approach originally proposed for binary outcomes in Foster et al. (2011). Specifically, we first fit boosted Cox regression models to the survival data with all covariates in the two arms separately. We use the R function gbm with all parameters being set as default, except that the maximum depth of variable interactions, the maximum number of trees, and the embedded bagging fraction are set as 3, 1000, and 0.5, respectively. The fitted models are then used to estimate the treatment effect for each individual. Subsequently, we use regression tree with the estimated individual treatment effect as the response variable and all covariates to further simplify the treatment effect estimates. Following Foster et al. (2011), we use the R function rpart with default settings for the regression tree, except that the minimum terminal node size and the complexity parameter are set as 20 and 0.02, respectively. For both methods, the treatment effect for individual patient is calculated as the difference in event rate at τ0 = 3 years based on fitted models, and individuals belong to the enrichable subgroup if their estimated subject-specific treatment effects are greater than the observed average treatment effect in the entire cohort by at least dc. Here, dc represents the minimum additional treatment benefit compared with an “average patient” for an individual to be considered “specially enrichable”. In practice, dc is subjectively chosen by users considering specific applications (Foster et al., 2011). Here, we consider 4 dc values: 0.05, 0.10, 0.15, and 0.20 in our comparisons.
Under the null and alternative hypotheses, we generate 1:1 randomized trials mimicking the BEST study with event time as the primary outcome. The total sample size is set as 1500 or 3000. We simulate the 16 covariates mentioned in the Introduction. Specifically, we first generate the discrete covariates based on their empirical distributions in the BEST study, and then generate the continuous covariates based on the fitted multivariate linear regression model by regressing the continuous covariates on the discrete covariates in the BEST study. For the null case of no treatment effect, the event times for both treatment and placebo groups are generated from the fitted additive log-normal model with 16 covariates based on the observed BEST data. For the alternative case with enrichable subgroup, the survival times for treatment group are generated by adding 0.1+θ0 × I(Male and LVEF ≥ 25) to the linear predictor. We consider θ0 = 0.5 and θ0 = 1.0 representing moderate and large treatment effects, respectively. Note that male participants with LVEF≥ 25 consist of 32% of the BEST cohort. We generate the censoring times from the observed nonparametric Kaplan-Meier estimate for the censoring distribution in the BEST study. The censoring proportion is approximately 30%. We simulate 500 data sets for each scenario. For each simulated data set, two thirds of the observations are used as the holdout samples for validation. To compare the efficiency of the proposed procedure with the two alternative methods, in addition to the enrichable subgroup finding rate, we also estimate the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). These estimates were derived by generating an independent validation data set of size 10,000, and then compare the true enrichable subgroup (i.e., males with LVEF ≥ 25) to the enrichable subgroups identified using the proposed procedure and two alternative methods.
The simulation results are summarized in Table 2. Note that the the proposed procedure has a nominal 5% false enrichable finding rate under the null scenario regardless of the sample size. It is not a surprise, since under the null, the true treatment effect in any subgroup identified at steps 1 and 2 is zero and there is only 5% random chance to observe a significant treatment effect at step 3 based on holdout sample at the 0.05 significance level. On the other hand, the false enrichable finding rates of the virtual twin and lasso regularized Cox regression methods are sensitive to dc and sample sizes and vary from 0% to about 100% in our numerical study. For example, the false enrichable finding rate of the virtual twin procedure is 7.6% with dc = 0.1 and a sample size of 1500, and reduces to 1.4% with the increased sample size of 3000. Similarly, the false finding rate of the lasso regularized Cox model is 5% with dc = 0.15 and a sample size of 3000, but is inflated to 13.8% with the reduced sample size of 1500. Neither of these two methods with dc selected a priori guarantees to control the false enrichable finding rate. Under the alternative scenarios, the proposed procedure has a higher enrichable finding rate than the two alternatives when they use dc value yielding approximately comparable false enrichable finding rates under the null scenario. In addition, the proposed procedure also has comparable or higher sensitivity, specificity, PPV and NPV.
Table 2.
Comparing the performance of the proposed procedure and two alternative methods. The numbers are in percent.
| Proposed | Virtual twins (dc) | Lasso (dc) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.05 | 0.10 | 0.15 | 0.20 | 0.05 | 0.10 | 0.15 | 0.20 | ||
| Sample size 1500 | |||||||||
| Null scenario | |||||||||
| Enrichable finding | 4.8 | 67.6 | 7.6 | 0.2 | 0 | 76 | 35.4 | 13.8 | 4.4 |
| Alternative scenario: θ0 = 0.5 | |||||||||
| Enrichable finding | 30.6 | 80.6 | 15.4 | 2.2 | 0 | 92 | 61.6 | 32.6 | 14.4 |
| Sensitivity | 55.4 | 25.3 | 1.1 | 0.1 | 0 | 28.1 | 7 | 1.5 | 0.3 |
| Specificity | 53.8 | 94.2 | 99.7 | 100 | 100 | 86.9 | 97.7 | 99.7 | 100 |
| PPV | 36.9 | 48.5 | 8.9 | 1.5 | 0 | 46.6 | 39.1 | 26.2 | 14.1 |
| NPV | 76.1 | 76.1 | 71.2 | 71.1 | 71.1 | 75.1 | 72.1 | 71.3 | 71.1 |
| Alternative scenario: θ0 = 1.0 | |||||||||
| Enrichable finding | 61.2 | 99.6 | 40.2 | 7.8 | 1 | 98.8 | 83.6 | 59 | 36.6 |
| Sensitivity | 65.2 | 78.2 | 15.9 | 0.5 | 0.1 | 46.8 | 16.9 | 5.4 | 1.4 |
| Specificity | 49.7 | 95.1 | 99.2 | 100 | 100 | 86.5 | 96.8 | 99.3 | 99.9 |
| PPV | 41.6 | 86.7 | 35 | 6.3 | 0.9 | 61.1 | 62.5 | 50.4 | 37.1 |
| NPV | 82.1 | 92.2 | 75 | 71.1 | 71.1 | 80.7 | 74.3 | 72.1 | 71.3 |
| Sample size 3000 | |||||||||
| Null scenario | |||||||||
| Enrichable finding | 4.4 | 39.2 | 1.4 | 0 | 0 | 66 | 22.6 | 5.0 | 1.0 |
| Alternative scenario: θ0 = 0.5 | |||||||||
| Enrichable finding | 54 | 67.4 | 10.8 | 1.4 | 0 | 95.2 | 59.8 | 27.6 | 9.6 |
| Sensitivity | 70.6 | 24.9 | 0.6 | 0.1 | 0 | 26.5 | 4.3 | 0.6 | 0.1 |
| Specificity | 39.3 | 96.7 | 99.9 | 100 | 100 | 91.7 | 99.2 | 99.9 | 100 |
| PPV | 36.2 | 48.8 | 8.3 | 1.1 | 0 | 58.4 | 47.3 | 25.2 | 10.7 |
| NPV | 79.9 | 76.7 | 71.2 | 71.1 | 71.1 | 75.7 | 71.8 | 71.2 | 71.1 |
| Alternative scenario: θ0 = 1.0 | |||||||||
| Enrichable finding | 93.6 | 99.4 | 46.4 | 12.6 | 3.8 | 99.8 | 93.6 | 75.4 | 49.4 |
| Sensitivity | 78.7 | 93.2 | 23.9 | 0.7 | 0.2 | 56.6 | 20.5 | 5.2 | 1 |
| Specificity | 41.9 | 98.2 | 99.7 | 100 | 100 | 89.3 | 98.2 | 99.7 | 100 |
| PPV | 41.9 | 95.2 | 44.4 | 11.9 | 3.7 | 70.1 | 79.8 | 72.1 | 53 |
| NPV | 87.6 | 97.5 | 77.3 | 71.2 | 71.1 | 84 | 75.4 | 72.1 | 71.3 |
In our simulation study reported above, we compared the proposed 3-stage procedure as a package with two existing competing procedures with respect to identifying the enrichable subgroup with a single data set. Note that the comparisons do not suggest the inferiority of the competing procedures in identifying the potential enrichable subgroups, since one can also reserve a holdout sample to control the type I error as we proposed. In fact, as a powerful discovery tool, the virtual twin approach can be added on top of the eight candidate procedures utilized in the BEST data for illustration to construct additional scoring systems, which might further improve the quality of the selected enrichable subgroup to be validated in the holdout sample.
7. Remarks
If there are two clinical trials conducted under the same setting, the data from one study can be used for building and evaluating the scoring systems via the “cross-training-evaluation” procedure described in the paper. Then we use the data from the second study for the assessment of the treatment effectiveness with pre-specified two-sample inference procedures to the sample of the selected enrichable population. It is important to note that the criteria for evaluating scoring systems and selection of the potentially promising subgroup should be consistent with those utilized for the holdout sample. Like the conventional cross-validation procedure for model building and selection, it is not clear how to choose the size of the data set at each stage of the process. It is generally recommended that the final validation set size should be relatively large compared to that for the training and evaluation to obtain greater precision of the inference procedures conducted for the holdout sample (Shao, 1993). The sample size required to construct the enrichable subgroup depends on many factors such as the ability of the covariates in capturing the differential treatment response. In practice, one may perform simulation study to investigate the power of the proposed method under different scenarios. Such a practice can be useful in study planning. However, the results need to be interpreted with caution due to their dependence on the models used in the simulation.
It is interesting to note that for our analysis of the data from BEST, the statistical significance of the two-sample test with the holdout sample for the enrichable subgroup is not as impressive as its predicted counterpart obtained at the training-evaluation stage. This may be partially due to the phenomenon of over-optimism even with cross-validation which does not correct for the overfitting due to the selection of an optimal q or an optimal model among all candidate models for building the scoring system. It is also important to keep in mind that the Z score in the holdout sample is a random variable by nature and would be difficult to predict with great precision.
The choice of a summary measure for the between-group difference is crucial. The HR is a heavily model-based treatment contrast measure. Although the proportional hazards assumption seems to be reasonable in the BEST study example, it is likely to be violated in general. On the other hand, a model-free and clinically meaningful two-sample contrast measure such as the difference of two RMSTs or medians seems more appropriate when there is no strong evidence to support a model-based summary at the beginning of the study. It is important to note that the selection rule for an enrichable subgroup at this stage may depend on the totality of evidence based on various criteria. For instance, we may consider both the Z-test score value and the absolute magnitude of the treatment difference among the enrichable subgroup. Both the HR and RMST may be used to quantify the treatment effect. The employment of multiple criteria reflects the multidimensional requirements for the final enrichable subgroup. Furthermore, the evidences such as the stability of the performance of a candidate choice in the “cross-training-evaluation” also helps to reduce the chance that an enrichable rule is selected because of its superior performance based on a single criterion by random chance, since we may choose an enrichable subgroup having slightly inferior average performance but based on a simple working model with more stable performance.
Supplementary Material
Acknowledgements
This manuscript was prepared using BEST Research Materials obtained from the NHLBI Biologic Specimen and Data Repository Information Coordinating Center and does not necessarily reflect the opinions or views of the BEST investigators or the NHLBI. We are grateful to the Editor, Associate Editor, and two referees for their constructive comments on the article. The research is partially supported by the US NIH grants and contracts (R01-HL089778, R01-GM079330, R01-AI024643, R21-AG049385, UM1AI068616 and UM1AI068634).
Footnotes
The R code implementing the proposed method is available with this paper at the Biometrics website on Wiley Online Library.
References
- Andersen PK, Hansen MG, Klein JP. Regression analysis of restricted mean survival time based on pseudo-observations. Lifetime Data Analysis. 2004;10:335–350. doi: 10.1007/s10985-004-4771-0. [DOI] [PubMed] [Google Scholar]
- BEST. A trial of the beta-blocker bucindolol in patients with advanced chronic heart failure. The New England Journal of Medicine. 2001;344:1659–1667. doi: 10.1056/NEJM200105313442202. [DOI] [PubMed] [Google Scholar]
- Chen P-Y, Tsiatis AA. Causal inference on the difference of the restricted mean lifetime between two groups. Biometrics. 2001;57:1030–1038. doi: 10.1111/j.0006-341x.2001.01030.x. [DOI] [PubMed] [Google Scholar]
- Cox DR. Regression models and life-tables. Journal of the Royal Statistical Society. Series B (Methodological) 1972;34:187–220. [Google Scholar]
- Foster JC, Taylor JMG, Ruberg SJ. Subgroup identification from randomized clinical trial data. Statistics in Medicine. 2011;30:2867–2880. doi: 10.1002/sim.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freidlin B, Simon R. Adaptive signature design: an adaptive clinical trial design for generating and prospectively testing a gene expression signature for sensitive patients. Clinical Cancer Research. 2005;11:7872–7878. doi: 10.1158/1078-0432.CCR-05-0605. [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. Journal of Statistical Software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Irwin J. The standard error of an estimate of expectation of life, with special reference to expectation of tumourless life in experiment with mice. Journal of Hygiene. 1949;47:188–189. doi: 10.1017/s0022172400014443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Freidlin B, Simon R. Biomarker-adaptive threshold design: a procedure for evaluating treatment with possible biomarker-defined subset effect. Journal of the National Cancer Institute. 2007;99:1036–1043. doi: 10.1093/jnci/djm022. [DOI] [PubMed] [Google Scholar]
- Karrison T. Restricted mean life with adjustment for covariates. Journal of the American Statistical Association. 1987;82:1169–1176. [Google Scholar]
- Karuri SW, Simon R. A two-stage bayesian design for co-development of new drugs and companion diagnostics. Statistics in Medicine. 2012;31:901–914. doi: 10.1002/sim.4462. [DOI] [PubMed] [Google Scholar]
- Kent DM, Hayward RA. Limitations of applying summary results of clinical trials to individual patients: the need for risk stratification. The Journal of the American Medical Association. 2007;298:1209–1212. doi: 10.1001/jama.298.10.1209. [DOI] [PubMed] [Google Scholar]
- Murray S, Tsiatis AA. Sequential methods for comparing years of life saved in the two-sample censored data problem. Biometrics. 1999;55:1085–1092. doi: 10.1111/j.0006-341x.1999.01085.x. [DOI] [PubMed] [Google Scholar]
- Qian M, Murphy S. Performance guarantees for individualized treatment rules. Annals of statistics. 2011;39:1180–1210. doi: 10.1214/10-AOS864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell PM. Can overall results of clinical trials be applied to all patients? The Lancet. 1995;345:1616–1619. doi: 10.1016/s0140-6736(95)90120-5. [DOI] [PubMed] [Google Scholar]
- Rothwell PM, Mehta Z, Howard SC, Gutnikov SA, Warlow CP. From subgroups to individuals: general principles and the example of carotid endarterectomy. The Lancet. 2005;365:256–265. doi: 10.1016/S0140-6736(05)17746-0. [DOI] [PubMed] [Google Scholar]
- Royston P, Parmar MK. The use of restricted mean survival time to estimate the treatment effect in randomized clinical trials when the proportional hazards assumption is in doubt. Statistics in Medicine. 2011;30:2409–2421. doi: 10.1002/sim.4274. [DOI] [PubMed] [Google Scholar]
- Shao J. Linear model selection by cross-validation. Journal of the American statistical Association. 1993;88:486–494. [Google Scholar]
- Simon R. Designs and adaptive analysis plans for pivotal clinical trials of therapeutics and companion diagnostics. Expert Opinion on Medical Diagnostics. 2008;2:721–729. doi: 10.1517/17530059.2.6.721. [DOI] [PubMed] [Google Scholar]
- Tian L, Cai T, Zhao L, Wei L-J. On the covariate-adjusted estimation for an overall treatment difference with data from a randomized comparative clinical trial. Biostatistics. 2012;13:256–273. doi: 10.1093/biostatistics/kxr050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Zhao L, Wei L-J. Predicting the restricted mean event time with the subject’s baseline covariates in survival analysis. Biostatistics. 2014;15:222–233. doi: 10.1093/biostatistics/kxt050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological) 1996;58:267–288. [Google Scholar]
- Tibshirani R. The lasso method for variable selection in the cox model. Statistics in Medicine. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration. Guidance for industry: Enrichment strategies for clinical trials to support approval of human drugs and biological products. 2012 http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM332181.htm.
- Wang S-J, O’Neill RT, Hung H. Approaches to evaluation of treatment effect in randomized clinical trials with genomic subset. Pharmaceutical Statistics. 2007;6:227–244. doi: 10.1002/pst.300. [DOI] [PubMed] [Google Scholar]
- Zhang B, Tsiatis AA, Davidian M, Zhang M, Laber E. Estimating optimal treatment regimes from a classification perspective. Stat. 2012;1:103–114. doi: 10.1002/sta.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Tsiatis AA, Laber EB, Davidian M. A robust method for estimating optimal treatment regimes. Biometrics. 2012;68:1010–1018. doi: 10.1111/j.1541-0420.2012.01763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HH, Lu W. Adaptive lasso for cox’s proportional hazards model. Biometrika. 2007;94:691–703. [Google Scholar]
- Zhang M, Schaubel DE. Estimating differences in restricted mean lifetime using observational data subject to dependent censoring. Biometrics. 2011;67:740–749. doi: 10.1111/j.1541-0420.2010.01503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Tian L, Cai T, Claggett B, Wei L-J. Effectively selecting a target population for a future comparative study. Journal of the American Statistical Association. 2013;108:527–539. doi: 10.1080/01621459.2013.770705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Tian L, Uno H, Solomon SD, Pfeffer MA, Schindler JS, Wei LJ. Utilizing the integrated difference of two survival functions to quantify the treatment contrast for designing, monitoring, and analyzing a comparative clinical study. Clinical Trials. 2012;9:570–577. doi: 10.1177/1740774512455464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H. The adaptive lasso and its oracle properties. Journal of the American Statistical Association. 2006;101:1418–1429. [Google Scholar]
- Zucker DM. Restricted mean life with covariates: modification and extension of a useful survival analysis method. Journal of the American Statistical Association. 1998;93:702–709. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.