Abstract
Tropomodulins (Tmods) cap F-actin pointed ends and have altered expression in the brain in neurological diseases. The function of Tmods in neurons has been poorly studied and their role in neurological diseases is entirely unknown. In this paper we show that Tmod1 and Tmod2, but not Tmod3, are positive regulators of dendritic complexity and dendritic spine morphology. Tmod1 increases dendritic branching distal from the cell body and the number of filopodia/thin spines. Tmod2 increases dendritic branching proximal to the cell body and the number of mature dendritic spines. Tmods utilize two actin-binding sites and two tropomyosin (Tpm)-binding sites to cap F-actin. Overexpression of Tmods with disrupted Tpm-binding sites indicates that Tmod1 and Tmod2 differentially utilize their Tpm- and actin-binding sites to affect morphology. Disruption of Tmod1’s Tpm-binding sites abolished the overexpression phenotype. In contrast, overexpression of the mutated Tmod2 caused the same phenotype as wild type overexpression. Proximity ligation assays indicate that the mutated Tmods are shuttled similarly to wild type Tmods. Our data begins to uncover the roles of Tmods in neural development and the mechanism by which Tmods alter neural morphology. These observations in combination with altered Tmod expression found in several neurological diseases also suggest that dysregulation of Tmod expression may be involved in the pathology of these diseases.
Introduction
Actin regulation is an integral part of neuronal development (da Silva and Dotti, 2002; Gomez and Letourneau, 2014; Hur et al., 2012; Vitriol and Zheng, 2012). Monomeric actin (G-actin) can polymerize into long filaments (F-actin). Assembly and disassembly of F-actin is the mechanical step that translates many signaling pathways into directional growth in dendrites and axons. Actin polymerization is also the driving force in dendritic spine formation (Bellot et al., 2014). F-actin has two characteristic ends, a slow growing (pointed) and a fast growing (barbed) end. Actin filaments push on the membrane when monomers are added to the barbed ends near the membrane (Doherty and McMahon, 2008). Actin monomers dissociate from filaments at the pointed end, which is often rate-limiting in actin dynamics (Carlier et al., 1997). There are numerous proteins and signaling pathways that converge to alter actin dynamics. Actin dynamics is modulated by diverse processes, such as monomer sequestration, ADP-ATP exchange, and filament stabilization, destabilization, severing or capping (for reviews see (Bernstein and Bamburg, 2010; Birbach, 2008; Colpan et al., 2013; dos Remedios et al., 2003; Gunning et al., 2008; Silacci et al., 2004; Xue and Robinson, 2013).
Tropomodulins (Tmods) are a family of actin pointed end-capping proteins (Weber et al., 1994). Tmods are known to influence many processes, including muscle contraction, cellular motility and shape, and vesicle-membrane fusion (Fischer et al., 2003; Lim et al., 2015; Weber et al., 2007). Tmod isoforms are widely expressed throughout the body; three of the four mammalian Tmod isoforms, Tmod1, Tmod2, and Tmod3, are expressed in the brain (Almenar-Queralt et al., 1999; Cox and Zoghbi, 2000; Watakabe et al., 1996). Knockout of Tmod2 in mice results in reduced sensorimotor gating, hyperactivity, and impaired learning and memory (Cox et al., 2003). Knockout of Tmod2 also results in a 2-fold increase of Tmod1 expression in mice. This indicates a compensatory expression mechanism for Tmods in neurons and confounds the apparent consequences of Tmod2 knockout with Tmod1 overexpression. A similar effect was observed in N2a neuroblastoma cells after siRNA knockdown of Tmod2 (Fath et al., 2011).
Tmods form a pointed end cap by concerted action of two actin-binding sites (ABS1 and ABS2) and two tropomyosin (Tpm)-binding sites (Fowler et al., 2003; Greenfield et al., 2005; Kostyukova et al., 2006; Kostyukova et al., 2005). Tpms are a family of coiled-coil proteins that associate in a head-to-tail fashion along actin filaments (for reviews see (Gunning et al., 2015a; Gunning et al., 2015b)). Tmods have isoform specific affinities to different Tpms (Colpan et al., 2016; Kostyukova, 2007; Uversky et al., 2011). ABS1 and both Tpm-binding sites are all within the N-terminal half of Tmod and enable the majority of its capping ability of Tpm decorated filaments; this half of Tmods is unstructured and flexible until it binds onto Tpm and the pointed end of an actin filament (Greenfield et al., 2005; Kostyukova et al., 2000; Kostyukova et al., 2006; Kostyukova et al., 2005). ABS2 is made up by the entire C-terminal half which forms a highly structured leucine rich repeat (LRR) domain (Krieger et al., 2002; Rao et al., 2014). Tmods have a greater affinity for actin filaments decorated with Tpm than for bare filaments (Gregorio et al., 1995; Weber et al., 1994).
There are no known neurological disorders caused by mutations in Tmod; however, Tmod2 has altered expression in fetal Down syndrome, (Sun et al., 2011) mesial temporal lobe epilepsy (Yang et al., 2006), post-seizure (Sussman et al., 1994), post-stroke (Chen et al., 2007) and post-methamphetamine exposure (Iwazaki et al., 2006). The purpose and consequence of the altered expression is entirely unknown. In this report, we demonstrate changes caused by overexpression of Tmod1 and Tmod2 in both dendritic branching and spine morphology. Interestingly, no morphological changes are observed when Tmod3 is overexpressed. Furthermore, we report that, unlike Tmod2, Tmod1 requires its Tpm-binding ability to cause these changes. From these experiments we can begin to infer the relevant roles of Tmods in neuronal morphology as well as the reason for their altered expression in disease states.
Results
Tmod1 and Tmod2, but not Tmod3, increase complexity of dendritic arbor at different radial distances from the cell body and require different binding sites to alter the dendritic arbor
Tmod1 and Tmod2 overexpression increases the complexity of the dendritic arbor
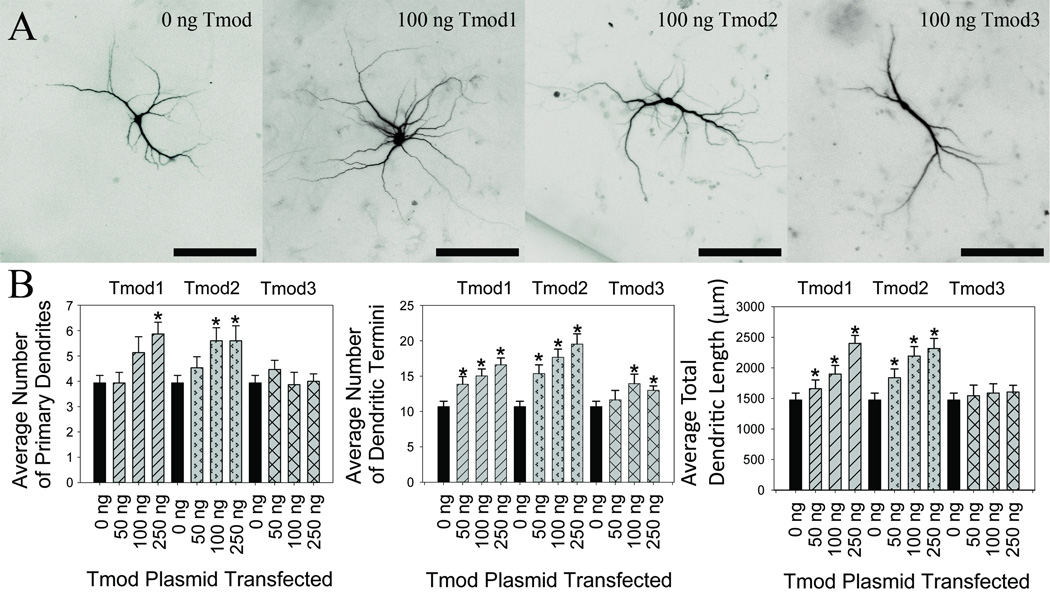
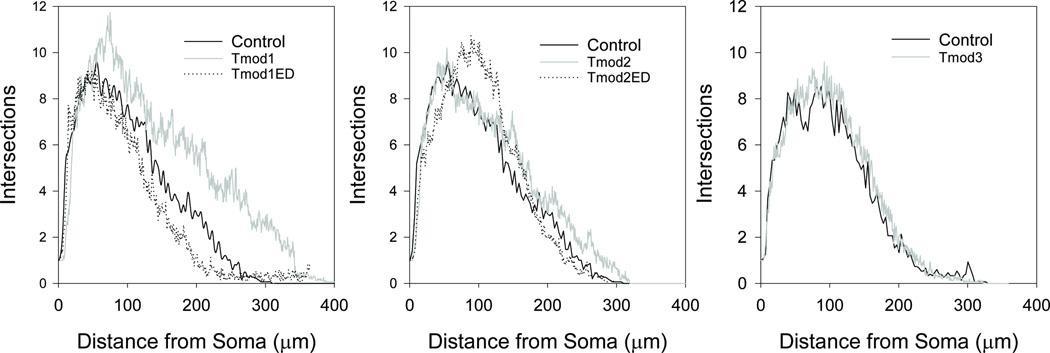
Tmod1 and Tmod2 alter length and number of neurite-like processes in PC12 and N2a cell lines in an isoform-specific manner (Fath et al., 2011; Guillaud et al., 2014; Moroz et al., 2013) but no data has been published to demonstrate effects of Tmods on neurite formation in neurons. We tested our hypothesis that Tmods have isoform-specific roles in dendritic morphology by overexpressing Clover Fluorescent Protein (ClFP) -tagged Tmod1, Tmod2 and Tmod3 in primary hippocampal neurons in a dose-response experiment. Dendrites were visualized using an mRFP-MAP2B which is restricted to dendrites and soma. Neurons were analyzed for number of primary dendrites, dendritic termini, total dendritic length and dendritic complexity. Primary dendrites are the number of dendritic processes from the cell body and dendritic termini are ends of branches from dendrites. The control groups for this experiment were neurons transfected with an empty vector. Overexpression of either Tmod1 or Tmod2 both caused an increase in the number of primary dendrites, dendritic termini and total dendritic length in a dose-dependent manner (Figure 1). The highest level of Tmod1 overexpression increased the average number of primary dendrites, dendritic termini and total dendritic length by 51 %, 55 % and 63 %, respectively (P < 0.05). Tmod2 overexpression increased the same measures by 44 %, 82 %, and 57 %, respectively (P < 0.05). Tmod3 overexpression had marginal impacts on the number of dendritic termini at the higher transfection amounts; there was no statistically significant change in the number of primary dendrites or total dendritic length. Interestingly, neither Tmod1 nor Tmod2 overexpression resulted in changes to the average length of dendritic branches (data not shown). We suspected that Tmod1 and Tmod2 may alter dendritic branching at different positions from the cell body. Sholl Analysis (Sholl, 1953) was conducted to test if Tmod1 and Tmod2 overexpression impacted the complexity of dendritic arbor at different distances from the soma. Tmod1 overexpression caused an increase in complexity after 150 µm distal from soma (Figure 2, P < 0.1); however, Tmod2 and Tmod3 overexpression had no observed effect. This suggests that Tmod1 positively impacts dendritic branching distal to the cell body.
Figure 1. Effect of Tmod1, Tmod2 and Tmod3 overexpression on number and length of dendrites.
Primary hippocampal neurons were transfected with varying amounts of pCAGGs plasmid encoding a ClFP-tagged wild type Tmods. A. From left to right, representative images of transfected neurons. Shown images are RFP signals from mRFP-MAP2b used for morphology analysis (scale bars = 150 µm). B. Imaged neurons were analyzed for number of primary dendrites (left), dendritic termini (center) and total dendritic length (right). 25–30 neurons were analyzed for each condition. Error bars indicate SEM. Asterisks indicated statistically significant difference from 0 ng condition (ANOVA, P < 0.05).
Figure 2. Impact of Tmod overexpression on dendritic arbor.
The impact of overexpression of Tmod1, Tmod1ED, Tmod2, Tmod2ED and Tmod3 (100 ng DNA condition) on the dendritic arbor was investigated by Sholl Analysis. 15 neurons were analyzed per condition. Tmod1 was found to be statistically different from the control after 150 µm (ANOVA, P < 0.05).
L29E and L134D mutations in Tmod2 disrupt Tpm binding but not actin nucleation
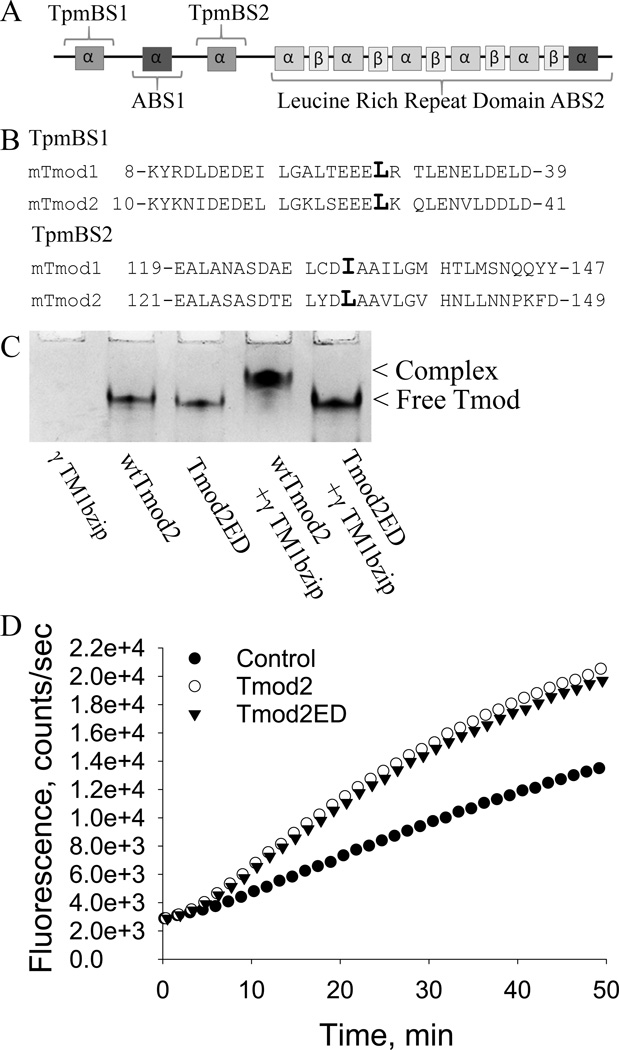
Tmod’s capping ability significantly increases in the presence of Tpm (Gregorio et al., 1995; Weber et al., 1994). The N-terminal domain contains the majority of the capping ability when Tpm is present (Greenfield et al., 2005; Kostyukova et al., 2006; Kostyukova et al., 2005). Overexpression of the N-terminal domains of either Tmod1 or Tmod2 in PC12 cells, which are used as a model for early neurite formation and extension (Das et al., 2004; Ohuchi et al., 2002; Schimmelpfeng et al., 2004), reduces the number and length of neurite-like processes (Guillaud et al., 2014). From these data, we expected that the Tpm-binding abilities of Tmod1 and Tmod2 may be necessary for the observed increase in branching caused by overexpression. The mutations L27E and I131D were shown to disrupt Tpm-binding ability of Tmod1 (Kong and Kedes, 2006; Kostyukova et al., 2006; Kostyukova et al., 2005). Alignment of Tmod1 and Tmod2 sequences indicated that L29 and L134 in Tmod2 correspond to L27 and I131 of Tmod1 (Figure 3A, B).
Figure 3. Tmod structure and effect of mutations in the Tpm-binding sites.
A. A schematic of the structural and functional domains of Tmods. B. Sequence alignments of Tmod1 and Tmod2 in the regions of the mutations used in this paper. The mutated leucines are indicated by a larger font size. C. Mutations in Tmod2’s Tpm binding sites disrupt Tpm binding. 190 pmol of wild type Tmod2 or 190 pmol Tmod2ED were loaded on a native gel either with or without 380 pmol of γTM1bzip. γTM1bzip does not enter the gel because it is positively charged. D. Influence of Tmod2 on the rate of actin polymerization. 0.2 µM of Tmod2 or Tmod2ED was added to 1 µM of G-actin (10% pyrene-labeled). Time-courses of actin polymerization was measured by the change in pyrene-actin fluorescence. Control: spontaneous actin polymerization.
We introduced the mutations L29E and L134D in Tmod2 (that will be referred to as Tmod2ED for simplicity) and tested Tpm- and actin-binding abilities of the mutated protein using the native gel electrophoresis and the actin nucleation assay. We confirmed that these mutations disrupted binding of Tmod2 with γTM1bzip, a chimeric peptide which contains the Tmod-binding site of TM5NM1 and TM5NM2 (Tpm3.1 and Tpm3.2, respectively (Geeves et al., 2015)) encoded by exon 1b of the Tpm3 gene (Kostyukova et al., 2007; Moroz et al., 2013). While there was a shift of the wild type Tmod2 band when the γTM1bzip was added, no shift was observed for the mutated Tmod2 (Fig. 3C). This indicated that the mutations disrupted Tmod2’s ability to bind Tpm3.1. The mutations did not affect actin-binding ability of Tmod2 because the mutated Tmod2 was able to nucleate actin polymerization similar to the wild type Tmod2 (Figure 3D).
Tmod1 requires its Tpm-binding sites to influence the dendritic arbor
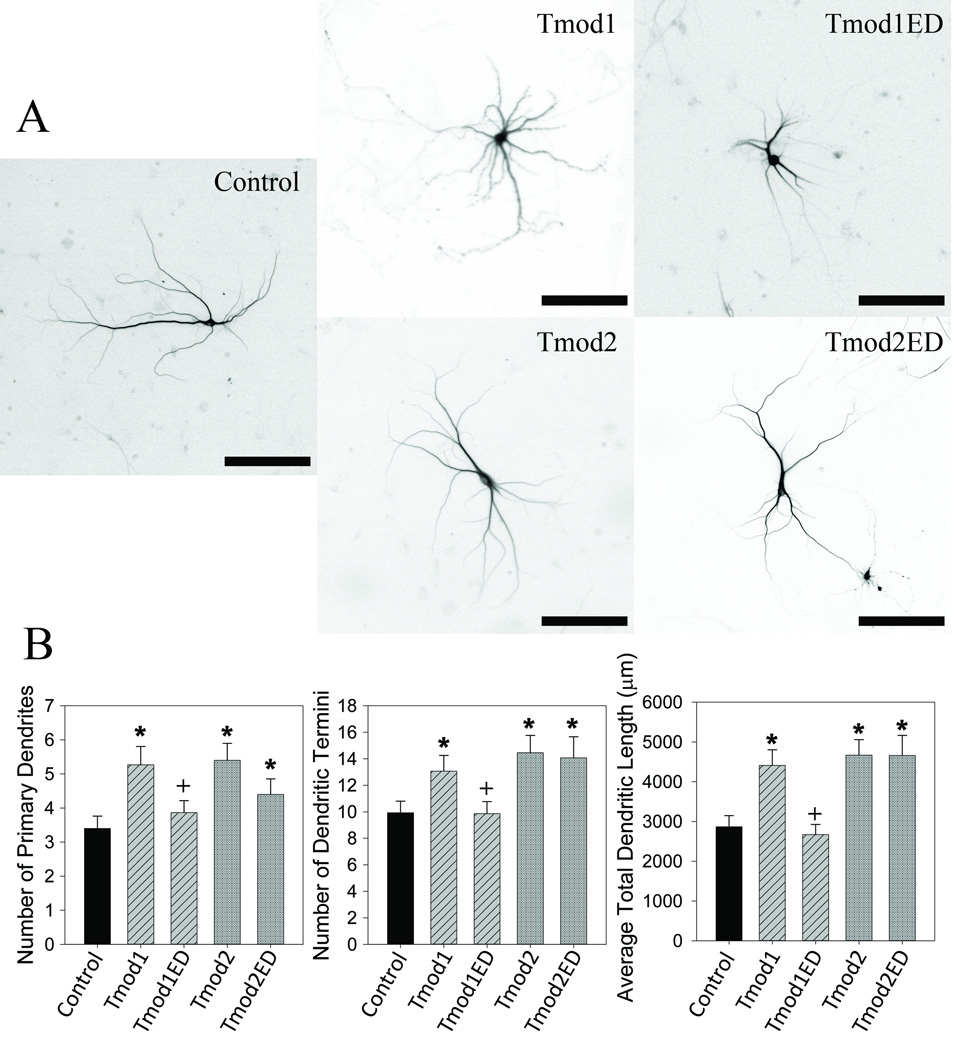
For expression in neurons we introduced L27E/I131D and L29E/L134D mutations in ClFP-Tmod1 and ClFP-Tmod2, respectively, which will be referred to as Tmod1ED and Tmod2ED. In our experiments with wild type Tmods, the 100 ng/ well transfection condition gave a good phenotypic response, so it was used in the following experiments. We overexpressed Tmod1ED and Tmod2ED in primary hippocampal neurons. We found that disruption of Tpm binding of Tmod1 abolished the overexpression phenotype observed with Tmod1 (Figure 4). Tmod1ED overexpressing neurons had a reduced number of primary dendrites, dendritic termini and significantly less total dendritic length, 27 %, 25 %, 40 % respectively (P < 0.05), compared to wild type overexpressing neurons. Tmod1ED overexpression resulted in morphology similar to the control (Figure 4). In contrast, overexpression of Tmod2ED caused no statistically significant differences compared to Tmod2 overexpression for primary dendrites, dendritic termini or total dendritic length. We compared the impact of the Tmods with disrupted Tpm binding by Sholl analysis and found that Tmod1ED had a profile similar to the control (Figure 2). Tmod2ED caused no statistically significant changes compared to both the control and Tmod2 overexpression.
Figure 4. Tpm binding is necessary for Tmod1’s but not for Tmod2’s impact on dendritic morphology.
Primary hippocampal neurons were transfected with pCAGGs plasmid encoding ClFP-tagged Tmod, wild type or with disrupted Tpm-binding sites. A. Representative images of neurons analyzed in this experiment. Shown images are RFP signals from RFP-MAP2b used for morphology analysis (scale bars = 150 µm). B. Imaged neurons were analyzed for number of primary dendrites (left), dendritic termini (center) and total dendritic length (right). 15–22 neurons were analyzed for each condition. Error bars indicate SEM. Asterisks and plus symbols indicate statistically significant difference from controls and wild type overexpression, respectively (ANOVA, P < 0.05).
Tmod1ED and Tmod2ED are found in sub-cellular compartments that are enriched with Tpm3.1 and Tpm3.2 (Tpm3.1/2), similar to wild type proteins
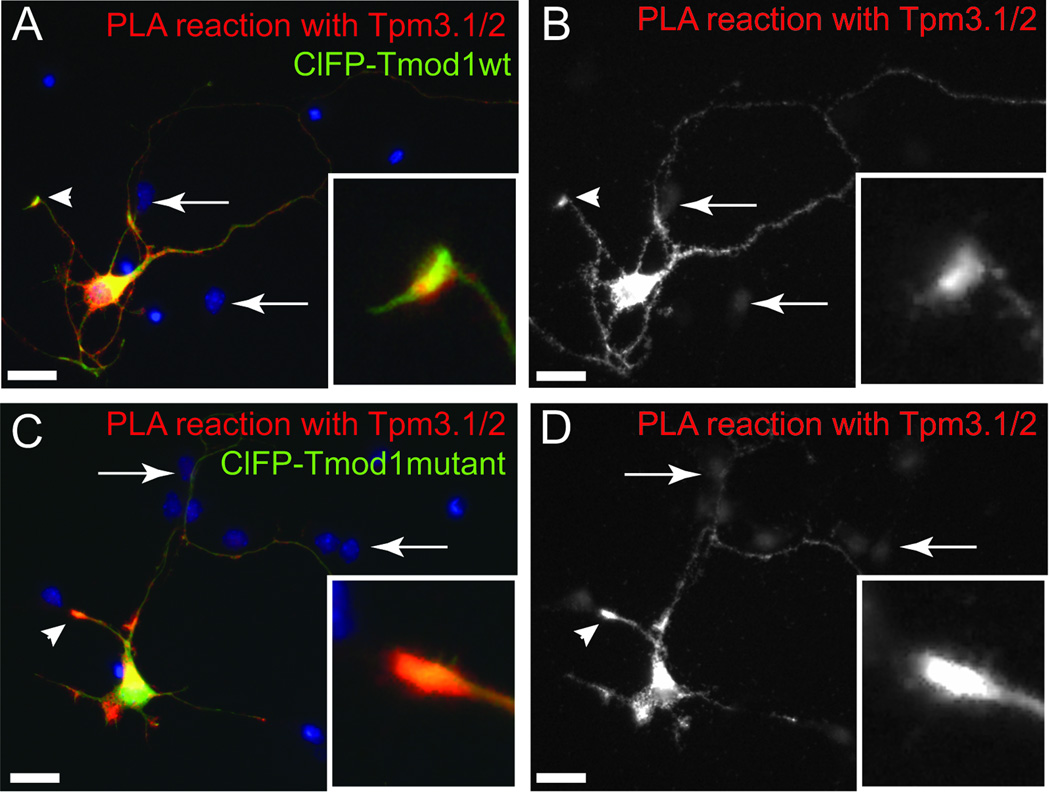
Tpm-binding is necessary for Tmod1’s ability to bind onto pointed ends in sarcomeres (Tsukada et al., 2011). We hypothesized that Tpm binding would be similarly necessary for Tmod co-localizing with actin filament pointed ends in neurons. We tested if mutations in Tmod1 and Tmod2 have an impact on the subcellular localization of Tmods by proximity ligation assays (PLA) (Figure 5) with endogenous Tpm3.1/2, which are highly expressed in cultured neurons (Fath et al., 2010; Schevzov et al., 2005). For this, ClFP-tagged Tmod1 or Tmod2, mutated or wild type, were overexpressed in hippocampal neurons. ClFP-Tmod1 (Figures 5 and S1) and ClFP-Tmod2 (not shown) were found in areas enriched with Tpm3.1/2. Interestingly, even though our mutations in Tmod1 impair direct binding with the Tpm (Kong and Kedes, 2006; Kostyukova et al., 2006; Kostyukova et al., 2005), the mutated Tmod1 is still found enriched in the growth cone compartment (Figure S1). Both mutated Tmods are found in close proximity to Tpm3.1/2 in the soma and neurites of the transfected neurons with a high PLA signal intensity at the tips of the neurites (Figure 5). No PLA signal was detected in non-transfected neurons (arrows, Figure 5).
Figure 5. Proximity Ligation Assay testing for close proximity of exogenous ClFP-tagged Tmods and endogenous Tpm3.1/2.
Hippocampal neurons, expressing wild type (wt) Tmod1 (A, B) or mutated Tmod1 (C, D) were fixed at DIV3 and examined by Proximity Ligation Assay (PLA). PLA has a theoretical maximum distance of 30–40 nm between epitopes for detection. Note the enrichment of the PLA reaction at the tips of the processes (arrowheads and boxed inserts in A–D). Arrows indicate non-transfected cells which are negative for PLA signals. Scale bar = 20 µm.
Tmod1 and Tmod2, but not Tmod3, increase different dendritic spine populations and require different binding sites to modulate dendritic spine morphology
Tmod1 and Tmod2 overexpression increase the number of different populations of dendritic spines
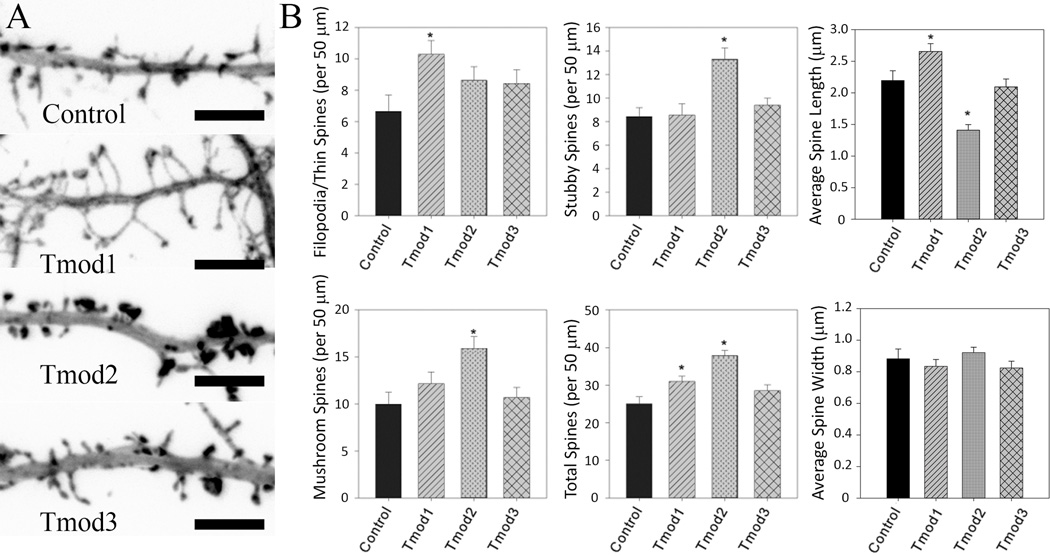
Dendritic spines are dynamic, actin-rich dendritic protrusions which are the postsynaptic side of the synapse. Our data confirmed isoform specific roles of Tmods in dendrite formation, which led us hypothesize that they also have specific roles in formation of dendritic spines. To visualize filopodia and spines, we co-expressed ClFP-Tmods and mRFP-actin in neurons. mRFP-actin is enriched in dendritic spines which enables easier visualization (Impey et al., 2010). Representative images of dendritic segments of neurons are shown on the left of Figure 6. Neurons overexpressing Tmod1 and Tmod2 were found to have increased number of dendritic spines and filopodia. Filopodia are rapidly forming and retracting dendritic protrusions that are precursors for dendritic spines (for review see (Cohen-Cory, 2002)). Glutamate is released from the axon terminal which signals to the postsynaptic compartment via N-methyl-D-aspartate receptors (NMDAR) which in turn promotes the development of a local filopodia into a dendritic spine (Kwon and Sabatini, 2011; Matus, 2000). It was difficult to distinguish between filopodia and thin spines in images, so they were counted as a single group. Tmod1 overexpression resulted in the increase of less mature protrusions (filopodia and thin spines), while Tmod2 overexpressing neurons had a greater number of more mature mushroom and stubby spines (Figure 6B). Tmod1 overexpression increased the number of filopodia/thin spines by 55 % and the total number of spines by 24 %. Tmod2 overexpressing neurons had 31 % more mushroom, 55 % more stubby and 22 % more total spines than the controls. Besides changing numbers of spines, we also observed changes in spine lengths. Tmod1 overexpressing neurons had spines that were 20 % longer than the controls. Tmod2 overexpressing neurons had spines that were 36 % shorter than the controls (Figure 6B). Previously, Tmod1 and Tmod2 were found to have distinct localization in neurons; Tmod1 localized to bundled actin while Tmod2 localized to more cytoplasmic domains (Fath et al., 2011). Tmods may have similar areas of influence in dendritic spines where Tmod1 influences spine necks and Tmod2 influences spine heads. Neither Tmod1 nor Tmod2 overexpression were found to alter the average head width of dendritic spines. Different from Tmod1 and Tmod2, overexpressed Tmod3 had no effect on the number, shape or average dimensions of dendritic spines (Figure 6).
Figure 6. The impact of Tmod overexpression on dendritic spine number and shape.
Primary hippocampal neurons were transfected with ClFP-tagged wild type Tmods. A. Representative images of dendritic fragments of analyzed neurons. Shown images are RFP signals from RFP-actin used for spine analysis (scale bar = 6 µm). B. Bar graphs show quantification of number of filopodia/thin spines, mushroom spines, stubby spines, total number of spines, and spine width and length. Error bars indicate SEM. Asterisks indicate statistically significant difference from controls (ANOVA, P < 0.05).
Tmod1 requires its Tpm-binding ability to alter number of dendritic spines
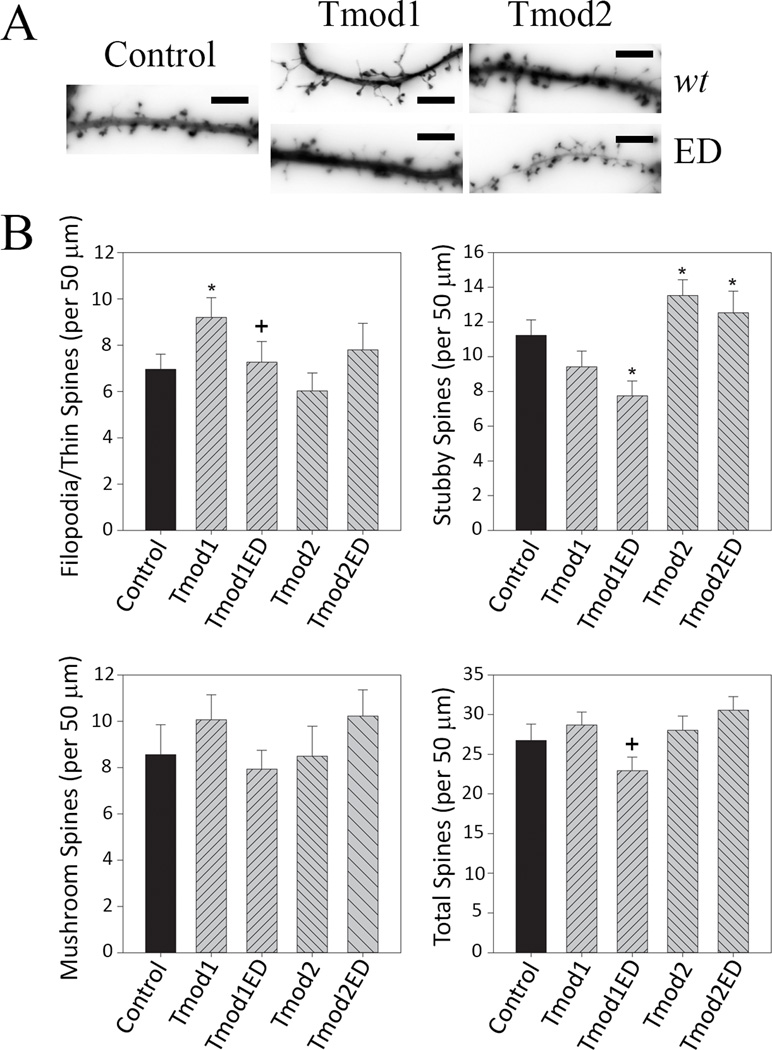
We tested if Tpm binding is the differentiating factor in Tmods’ modulation of dendritic spines by overexpressing Tmod1ED and Tmod2ED and analyzing for changes in dendritic spines (Figure 7). Similar to our dendrite experiments, disruption of Tpm-binding ability abolished the impact of Tmod1 overexpression on number and shape of dendritic spines. In contrast, Tmod2ED overexpression caused the same phenotype as Tmod2 overexpression. Tmod1ED overexpression did not cause filopodia/thin spines phenotype and also had 31 % fewer stubby spines than the controls (Figure 7). There were no statistically significant differences between Tmod2 and Tmod2ED overexpressing neurons with regards to the number and shape of dendritic spines.
Figure 7. The impact of overexpression of mutated Tmods on dendritic spine numbers and shape.
Primary hippocampal neurons were transfected with ClFP-tagged Tmod, wild type or with disrupted Tpm-binding sites. A. Representative images of dendrite fragments analyzed in this experiment. Shown images are RFP signals from RFP-actin used for spine analysis (scale bar = 6 µm). B. Quantification of number of filopodia/thin spines, mushroom spines, stubby spines and total number of spines. 20–24 dendritic fragments were analyzed per condition. Error bars indicate S.E.M. Asterisks and plus symbols indicate statistically significant difference from controls and wild type overexpression, respectively (ANOVA, P < 0.05).
Discussion
The goal of this study was to explore the morphology-modulating effects of Tmods in the nervous system. Little is known about roles of different Tmod isoforms in neurons. There have been only a few publications studying Tmods in neurons. Knockout of Tmod2 in mice was found to cause reduced sensorimotor gating, hyperactivity and deficits in learning and memory (Cox et al., 2003). Brain slices of Tmod2 knockout mice also had increased long term potentiation indicating that Tmod2 has a role in synaptic plasticity. Tmod1 and Tmod2 were found to have distinct localization in neurons (Fath et al., 2011). Tmod1 localized to leading edges of extensions and Tmod2 had a more cytoplasmic localization. Tmod2 and an miRNA, which targets Tmod2 mRNA, were found to be required for induction of long term depression caused by NMDA treatment (Hu et al., 2014), this supports Cox and co-authors’ conclusion that Tmod2 is involved in spine plasticity (Cox et al., 2003).
Our data demonstrate that Tmod1 and Tmod2 are positive regulators of dendritic arbor complexity. Sholl Analysis shows that Tmod1 overexpression increases dendritic branching distal from the cell body but does not indicate any changes caused by Tmod2 overexpression. As Tmod2 does increase the number of dendrites, we expected to see an increase in complexity by Sholl Analysis. The program used to conduct Sholl Analysis had difficulty resolving dendrites that were very close or overlapping which more frequently occurs proximal to the cell body. We assume that the increase in branching caused by Tmod2 overexpression is proximal to the cell body, where detection is more difficult for analysis using this software. Interestingly, Tpm binding was found to be necessary for the dendritic phenotype caused by Tmod1 overexpression but not by Tmod2 overexpression. Immunostaining for endogenous Tpm3.1/2 and exogenous ClFP-Tmods showed that Tmods’ localization in primary neurons partially overlaps with localization of Tpm3.1/2. We did not expect precise co-localization because Tmods only bind at the pointed ends of actin filaments while Tpms bind along filaments. Our PLA experiments show that in primary neurons, mutations in Tmods that disrupt Tpm binding do not disrupt Tmods localization to areas with Tpm3.1/2. This unexpected result could be explained if Tmods do not require their Tpm-binding ability to be shuttled. If Tmods are still properly shuttled then they may still bind at the pointed ends of the same actin filaments as Tpm3.1/2 through direct interactions with actin filaments.
It is unclear how Tmods are shuttled in cells; removal of Tmod1’s LRR domain or mutations that disrupt conserved exposed hydrophobic clusters prevent localization of Tmod1 to filament ends in sarcomeres (Tsukada et al., 2011). Our studies in PC12 cells found that overexpression of Tmod2 reduced the extension of neurite-like processes but Tmod1 overexpression did not cause any observable changes (Guillaud et al., 2014). Overexpression of a chimeric Tmod with the N-terminal domain of Tmod1 and LRR domain of Tmod2 caused normal extension of neurite-like processes, which led us to conclude that the LRR domains contain the distinguishing feature between Tmod1 and Tmod2 for determining their role in morphology. We assume that the LRR domains of Tmods are influential in the differences in localization of Tmod1 and Tmod2 observed in neurons (Fath et al., 2011). The LRR domain, but not Tpm-binding ability, could be directing Tmods to their specific actin sub-populations before they can exert their influence on morphology.
While there is little data about Tmods’ effect on neural plasticity, there is no previous data about Tmod’s effect on dendritic spines. There is evidence that Tmod2 and miRNA-191, which targets Tmod2 mRNA, are necessary for dendritic spine remodeling in long term depression (Hu et al., 2014). Glutamate released from an axon terminal activates NMDA receptor (NMDAR) on a local filopodia which in turn begins specification as a dendritic spine (Kwon and Sabatini, 2011; Matus, 2000). NMDA treatment causes downregulation of miRNA-191 which in turn upregulates Tmod2 expression. In our study, Tmod2 overexpression increases the number of mature dendritic spines. This suggest that upregulation of Tmod2 may be a means by which neurons increase the number and strength of dendritic spines. This is an apparent contradiction with Cox et al’s observation that Tmod2 knockout increases potentiation (Cox et al., 2003); however, they note an increase in Tmod1 expression after knockout. It may be the case that other proteins, including Tmod3, could have altered expression and localization thereby obfuscating the exact role of Tmod2 in neural connectivity. For example, Tmod3 is known to have increased expression in red blood cells (Moyer et al., 2010) and aberrant localization in muscle cells after Tmod1 knockout (Gokhin and Fowler, 2011).
Our experiments show that overexpression of Tmod1 and Tmod2 but not Tmod3 increase the number of specific dendritic spine populations. Tmod1 overexpression increased filopodia and thin spines, while Tmod2 overexpression increased mushroom and stubby spines. This leads us to conclude that Tmod1 is a positive regulator of early stages of spinogenesis while Tmod2 is a positive regulator of spine maturation. The expression profiles of Tmods are not known in neurons; however, in N2a cells, Tmod1 protein levels increase with differentiation while Tmod2 expression levels are nearly constant (Fath et al., 2011). Disruption of Tpm binding of Tmod1 abolished the observed phenotype from overexpression. These data indicate that Tmod1 requires its Tpm-binding ability to modulate dendritic spine morphology but it is not required in Tmod2. Tmods could alter spine morphology through two different mechanisms: (1) by directly altering actin dynamics through their actin-binding sites or (2) by altering recruitment of actin-associated proteins through stabilizing specific Tpm-decorated filaments.
Decoration of actin filaments can alter recruitment of actin-associated proteins such as myosin motors. Tpm4.2 can recruit myosin II onto F-actin (Tojkander et al., 2011). Neurons overexpressing Tpm3.1 had greater number and length of neurites while neurons overexpressing Tpm1.7 had fewer neurites (Schevzov et al., 2005). In B35 neuroblastoma cells, overexpression of Tpm1.10, Tpm1.11, Tpm1.12 and Tpm4.2 alters branching and length of neurite-like processes (Curthoys et al., 2014) and elasticity of the cellular membrane (Jalilian et al., 2015). Tmods are known to have different affinities for Tpm isoforms (Kostyukova, 2007; Uversky et al., 2011). As Tmods preferentially bind to specific Tpm-decorated actin filaments, those filaments are stabilized and have slow turnover rates compared to other filaments. These preferentially stabilized filaments could accumulate thereby altering their relative distribution. Tpms have isoform-specific expression and localization in neurons (Guven et al., 2011; Had et al., 1994; Schevzov et al., 1997). TPM1 gene products (Tpm1.10, Tpm1.11 and Tpm1.12, previously TMBr1, TMBr2 and TMBr3 respectively) were found to localize to presynaptic compartments while TPM3 (Tpm3.1–9, previously TM5NM1–9) and TPM4 (Tpm4.2, previously TM4) gene products localize to dendritic spines. Decoration of F-actin with different Tpms changes recruitment of other actin-associated proteins onto the filament. Overexpression of Tmods could indirectly alter spine morphology through Tpms’ ability to recruit various actin-associated proteins.
Tmod3 was not found to impact the complexity of the dendritic arbor or the number and maturity of dendritic spines, which suggests that Tmod3 may be predominantly involved in other processes in neurons. Recently, Tmod3 was found to be a target for the kinase Akt2 in adipocytes which has implications for fusion of Glut4 carrying vesicles with the cellular membrane (Lim et al., 2015). A later study indicated that in adipocytes Tmod3 preferentially stabilizes actin subpopulations that are decorated with Tpm3.1 and thereby altering recruitment of different myosin isoforms, providing a mechanism for altered vesicle fusion (Kee et al., 2015). Future studies are needed to investigate if Tmod3 predominantly plays a role in vesicle-membrane fusion in the pre-synapse.
There is still considerable work to be done to characterize the role of Tmods in neural development. Tmods have other functions that are not explored in this paper but will be investigated in further studies. Beyond capping, Tmod1, Tmod2 and Tmod3 can also directly bind G-actin and either sequester or nucleate actin (Yamashiro et al., 2010). Tmod2 has the strongest nucleation ability of the three Tmods in this paper (Yamashiro et al., 2010). The cellular roles of these functions are largely unexplored since Tmods are generally thought of as capping proteins, not nucleating or sequestering proteins. Our data begin to clarify the roles of Tmod isoforms in neural development and Tmods’ functions that give rise to those roles. It also suggests the consequences of altered Tmod expression in various disease states. We demonstrated that increasing Tmod expression increases the complexity of the dendritic arbor and number of dendritic spines. Increased expression of Tmods in disease states may be an attempt by the brain to strengthen neural connections or increase their number. It is unclear if this increase in connections is a beneficial response or exacerbates the condition; further studies are needed to address these questions.
Methods
Plasmid construction
Wild type Tmod1 and wild type Tmod2 were previously developed in pEGFP-C1 and pReciever-M55, respectively (Guillaud et al., 2014). The Tmod sequences were sub-cloned into a Gateway pCAGGs destination vector containing an N-terminal clover fluorescent protein (ClFP) tag. ClFP is a derivative of green fluorescent protein (Shaner et al., 2013). Plasmids were sequenced to confirm Tmod sequences had been properly inserted (GENEWIZ, Inc). Plasmids for transfection experiments were purified using HiPure MidiPrep Plasmid purification Kits (Invitrogen) according to the manufacturer’s protocol.
The L27E/I131D and L29E/L134D mutations were introduced in Tmod1 and Tmod2 respectively, using the constructs containing wild type Tmods as templates. For this, the plasmids encoding Tmods were amplified by PCR using a set of two complementary oligonucleotides with codons changed for the desired mutations by Pfu Turbo DNA polymerase (Agilent Technologies, USA). The original template plasmids were digested using DpnI (New England Biolabs), and the mutated plasmids were transformed into Escherichia coli (max efficiency DH5α). All designed oligonucleotides were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). Introduced mutations were confirmed by DNA sequencing at GENEWIZ, Inc. (South Plainfield, NJ). Tmod1[L27E/I131D] and Tmod2[L29E/L134D] were sub-cloned into the pCAGGs vector as described for wild type Tmods.
PC12 cells were used as a vehicle to test that constructs expressed properly. PC12 cells were cultured as described previously (Guillaud et al., 2014). They were transfected using Lipofectamine 2000 according to the manufacturer’s protocol (Invitrogen). After transfection, PC12 cells were lysed in RIPA buffer (Santa Cruz Biotechnology). Tmod1, Tmod2, Tmod3, wild type and mutated, were then probed for by western blotting. The primary antibodies included polyclonal rabbit antibodies against Tmod1 (custom made by Thermo Fisher Scientific, Waltham, MA), Tmod2 and Tmod3 (custom made by Pacific Immunology, Ramona, CA). Optimal dilutions for each antibody were determined prior to experiments. The secondary antibodies used in the experiments were Peroxidase-conjugated AffiniPure goat anti-Rabbit IgG (H+L) (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). For each construct we saw only single bands corresponding to ClFP-Tmod fusion expression.
For expression in E.coli pReciever-B01 with a His-tagged Tmod2 was used as a template to introduce L29E/L134D mutations using the same primers (Guillaud et al., 2014).
Protein preparation and in vitro assays
Wild type Tmod2 and Tmod2ED were purified using the protocol for His-tagged Tmod2 purification described previously (Moroz et al., 2013). Tmod concentrations were measured using the difference method as described in (Guillaud et al., 2014; Kostyukova et al., 2007). G-actin and pyrene-iodoacetamide labeled G-actin were prepared as in (Kostyukova and Hitchcock-DeGregori, 2004). γTM1bzip was designed and synthesized as described in (Kostyukova et al., 2007). Binding assays were conducted using native gel electrophoresis as described previously (Moroz et al., 2013). Actin polymerization was measured by the change in pyrene-actin fluorescence using a PTI fluorometer (Lawrenceville, NJ) (excitation, 366 nm, and emission, 387 nm, with 2 nm slit). 1 µM G-actin (10% pyrenyl-actin) was mixed with 0.2 µM Tmod2 or Tmod2ED in G-buffer (2 mM Tris-HCl pH 8.0, 0.2 mM CaCl2, 0.01% (v/v) NaN3, 0.5 mM DTT, 0.2 mM ATP) and incubated for 1 minute. Polymerization reactions were started by adding 20X polymerization buffer to a final concentration 25 mM Imidazole, pH 7.0, 100 mM KCl, 2 mM MgCl2, 1 mM EGTA at room temperature. Spontaneous actin nucleation in the absence of Tmod was measured as a control.
Cell culture and transfection for morphological studies
Hippocampi of Sprague-Dawley rats (Charles River Laboratories) of both sexes were harvested and dissociated on postnatal day 0–1 (P1-P2). The dissociated cells were then plated at a density of 3.4×104 cells per cm2 on glass coverslips precoated with poly-L-lysine (Sigma; molecular weight 30,000) in 24-well plates with 1.9 cm2 bottom surface area per well. Cells were kept in Neurobasal A medium (Invitrogen) supplemented with B27 (Invitrogen) as described previously [Brewer, 1997]. Cultures were fixed with 4% paraformaldehyde in 60mM PIPES, 25mM HEPES, 5mM EGTA, 1mM MgCl2, and 87.6 mM sucrose at room temperature for 20 minutes. Fixed neurons were washed with PBS. Transfected and fixed neurons were permeablized using 0.1% Triton X-100 detergent (Bio-Rad Laboratories) in PBS for 20 minutes before washing with PBS. Cover slips were then mounted onto microscope slides using Elvanol. Cells used for morphological experiments were transfected with Lipofectamine 2000 (Invitrogen) using manufacturer’s protocol. Dissociated hippocampal neurons were transfected on the 6th day in vitro (DIV6) for all dendrite and dendritic spine experiments. Neurons were fixed on DIV9 for dendrite experiments and DIV12 for dendritic spine experiments. For co-localisation experiments of Tmod1 or Tmod1ED with Tpm3.1 and Tpm3.2, hippocampal neurons were transfected by Lipofectamine 3000 (Thermofisher scientific) on DIV1. The cells were fixed at DIV2 and immunostained using an antibody directed against Tpm3.1/2 [clone 2G10.2] (1:100; a kind gift from Prof P. Gunning, University of New South Wales, Sydney, Australia) (Schevzov et al., 2011).
Fluorescent microscopy and image quantification
Fluorescent images for dendrite and dendritic spine experiments were taken using an Olympus IX81 inverted confocal microscope (Olympus Optical) with a 60× oil-immersion lens, numerical aperture 1.4 and resolution 0.280 µm using Slidebook 5.5 Digital Microscopy Software. Dendritic spine z-stack images were processed using MetaMorph software from Molecular Devices (Sunnyvale, CA). A minimum of 15 neurons were traced for each experiment. A minimum of 20 dendritic fragments were counted for dendritic spine experiments. Neurons within each experimental condition were selected with similar levels of fluorescence corresponding to Tmod overexpression. Dendrites and dendritic spines were traced using the NeuronJ plugin (Meijering, 2010; Meijering et al., 2004; Schneider et al., 2012) for ImageJ (Schneider et al., 2012). Spines were counted and sorted manually using previously described criteria (Harris et al., 1992; Lesiak et al., 2014). Sholl Analyses were conducted using the Sholl Analysis (Ferreira et al., 2014) plugin for ImageJ. A series of concentric circles were drawn around representative neurons and the number of dendrites that intersected with each circle was counted.
Proximity ligation assay
Primary mouse hippocampal neurons were plated at a density of 70 × 103 cells on 12 mm glass coverslips (Menzel) as previously described (Fath et al., 2009). Cells were transfected 48 hours after plating with ClFP-Tmod1, ClFP-Tmod1ED, ClFP-Tmod2 or ClFP-Tmod2ED using Lipofectamine 3000. Cells were fixed 24 hours post transfection with 4% PFA in PBS for 15 min and then permeabilized with 0.1% Triton X-100 in PBS for 5 mins. The assay was carried out using the Duolink kit (Sigma Aldrich) in accordance to the manufacturer’s protocol. The primary antibodies used were anti Tpm3.1/2 [clone 2G10.2] and rabbit polyclonal anti-GFP (1:750) (Abcam). No PLA signal was detected in controls in which only 2G10.2 or only anti-GFP antibodies were used. For better visualization of the localization of ClFP-tagged Tmods, signal intensity of the green channel was adjusted (Figure 5). The localization of the PLA signal was examined in a total of 30 cells per experimental group from four independent experiments.
Statistical analysis
Data are presented as mean values ± standard error about the mean (SEM). One way ANOVA was used to test for statistical significance.
Supplementary Material
Acknowledgments
The authors thank Christopher J. Keller for technical assistance in purifying proteins for in vitro assays and Katherine Tyson for help with fluorescence microscopy.
FUNDING SOURCES: This project was supported by startup funds to ASK, NIH/NIMH grant to GW (MH086032), the Australian National Health and Medical Research Council Grant APP1083209 to TF, and partially by Anne and Russ Fuller Interdisciplinary Research Fellowship to KTG. KTG was supported by a NIH/NIGMS-funded predoctoral fellowship (T32 GM008336).
References
- Almenar-Queralt A, Lee A, Conley CA, Ribas de Pouplana L, Fowler VM. Identification of a novel tropomodulin isoform, skeletal tropomodulin, that caps actin filament pointed ends in fast skeletal muscle. The Journal of biological chemistry. 1999;274:28466–28475. doi: 10.1074/jbc.274.40.28466. [DOI] [PubMed] [Google Scholar]
- Bellot A, Guivernau B, Tajes M, Bosch-Morato M, Valls-Comamala V, Munoz FJ. The structure and function of actin cytoskeleton in mature glutamatergic dendritic spines. Brain research. 2014;1573:1–16. doi: 10.1016/j.brainres.2014.05.024. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends in cell biology. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birbach A. Profilin, a multi-modal regulator of neuronal plasticity. BioEssays : news and reviews in molecular, cellular and developmental biology. 2008;30:994–1002. doi: 10.1002/bies.20822. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. The Journal of cell biology. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Liao WP, Lu Q, Wong WS, Wong PT. Upregulation of dihydropyrimidinase-related protein 2, spectrin alpha II chain, heat shock cognate protein 70 pseudogene 1 and tropomodulin 2 after focal cerebral ischemia in rats--a proteomics approach. Neurochemistry international. 2007;50:1078–1086. doi: 10.1016/j.neuint.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Cohen-Cory S. The developing synapse: construction and modulation of synaptic structures and circuits. Science. 2002;298:770–776. doi: 10.1126/science.1075510. [DOI] [PubMed] [Google Scholar]
- Colpan M, Moroz NA, Gray KT, Cooper DA, Diaz CA, Kostyukova AS. Tropomyosin-binding properties modulate competition between tropomodulin isoforms. Archives of biochemistry and biophysics. 2016 doi: 10.1016/j.abb.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpan M, Moroz NA, Kostyukova AS. Tropomodulins and tropomyosins: working as a team. Journal of muscle research and cell motility. 2013;34:247–260. doi: 10.1007/s10974-013-9349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox PR, Fowler V, Xu B, Sweatt JD, Paylor R, Zoghbi HY. Mice lacking Tropomodulin-2 show enhanced long-term potentiation, hyperactivity, and deficits in learning and memory. Molecular and cellular neurosciences. 2003;23:1–12. doi: 10.1016/s1044-7431(03)00025-3. [DOI] [PubMed] [Google Scholar]
- Cox PR, Zoghbi HY. Sequencing, expression analysis, and mapping of three unique human tropomodulin genes and their mouse orthologs. Genomics. 2000;63:97–107. doi: 10.1006/geno.1999.6061. [DOI] [PubMed] [Google Scholar]
- Curthoys NM, Freittag H, Connor A, Desouza M, Brettle M, Poljak A, Hall A, Hardeman E, Schevzov G, Gunning PW, Fath T. Tropomyosins induce neuritogenesis and determine neurite branching patterns in B35 neuroblastoma cells. Molecular and cellular neurosciences. 2014;58:11–21. doi: 10.1016/j.mcn.2013.10.011. [DOI] [PubMed] [Google Scholar]
- da Silva JS, Dotti CG. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nature reviews. NeuroScience. 2002;3:694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- Das KP, Freudenrich TM, Mundy WR. Assessment of PC12 cell differentiation and neurite growth: a comparison of morphological and neurochemical measures. Neurotoxicology and teratology. 2004;26:397–406. doi: 10.1016/j.ntt.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annual review of biophysics. 2008;37:65–95. doi: 10.1146/annurev.biophys.37.032807.125912. [DOI] [PubMed] [Google Scholar]
- dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiological reviews. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- Fath T, Agnes Chan YK, Vrhovski B, Clarke H, Curthoys N, Hook J, Lemckert F, Schevzov G, Tam P, Watson CM, Khoo PL, Gunning P. New aspects of tropomyosin-regulated neuritogenesis revealed by the deletion of Tm5NM1 and 2. European journal of cell biology. 2010;89:489–498. doi: 10.1016/j.ejcb.2009.11.028. [DOI] [PubMed] [Google Scholar]
- Fath T, Fischer RS, Dehmelt L, Halpain S, Fowler VM. Tropomodulins are negative regulators of neurite outgrowth. European journal of cell biology. 2011;90:291–300. doi: 10.1016/j.ejcb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath T, Ke YD, Gunning P, Gotz J, Ittner LM. Primary support cultures of hippocampal and substantia nigra neurons. Nature protocols. 2009;4:78–85. doi: 10.1038/nprot.2008.199. [DOI] [PubMed] [Google Scholar]
- Ferreira TA, Blackman AV, Oyrer J, Jayabal S, Chung AJ, Watt AJ, Sjostrom PJ, van Meyel DJ. Neuronal morphometry directly from bitmap images. Nature methods. 2014;11:982–984. doi: 10.1038/nmeth.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RS, Fritz-Six KL, Fowler VM. Pointed-end capping by tropomodulin3 negatively regulates endothelial cell motility. The Journal of cell biology. 2003;161:371–380. doi: 10.1083/jcb.200209057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler VM, Greenfield NJ, Moyer J. Tropomodulin contains two actin filament pointed end-capping domains. The Journal of biological chemistry. 2003;278:40000–40009. doi: 10.1074/jbc.M306895200. [DOI] [PubMed] [Google Scholar]
- Geeves MA, Hitchcock-DeGregori SE, Gunning PW. A systematic nomenclature for mammalian tropomyosin isoforms. Journal of muscle research and cell motility. 2015;36:147–153. doi: 10.1007/s10974-014-9389-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhin DS, Fowler VM. Cytoplasmic gamma-actin and tropomodulin isoforms link to the sarcoplasmic reticulum in skeletal muscle fibers. The Journal of cell biology. 2011;194:105–120. doi: 10.1083/jcb.201011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TM, Letourneau PC. Actin dynamics in growth cone motility and navigation. Journal of neurochemistry. 2014;129:221–234. doi: 10.1111/jnc.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ, Kostyukova AS, Hitchcock-DeGregori SE. Structure and tropomyosin binding properties of the N-terminal capping domain of tropomodulin 1. Biophysical journal. 2005;88:372–383. doi: 10.1529/biophysj.104.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio CC, Weber A, Bondad M, Pennise CR, Fowler VM. Requirement of pointed-end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes. Nature. 1995;377:83–86. doi: 10.1038/377083a0. [DOI] [PubMed] [Google Scholar]
- Guillaud L, Gray KT, Moroz N, Pantazis C, Pate E, Kostyukova AS. Role of tropomodulin's leucine rich repeat domain in the formation of neurite-like processes. Biochemistry. 2014;53:2689–2700. doi: 10.1021/bi401431k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiological reviews. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC. The evolution of compositionally and functionally distinct actin filaments. Journal of cell Science. 2015a;128:2009–2019. doi: 10.1242/jcs.165563. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. Tropomyosin - master regulator of actin filament function in the cytoskeleton. Journal of cell Science. 2015b;128:2965–2974. doi: 10.1242/jcs.172502. [DOI] [PubMed] [Google Scholar]
- Guven K, Gunning P, Fath T. TPM3 and TPM4 gene products segregate to the postsynaptic region of central nervous system synapses. Bioarchitecture. 2011;1:284–289. doi: 10.4161/bioa.1.6.19336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Had L, Faivre-Sarrailh C, Legrand C, Mery J, Brugidou J, Rabie A. Tropomyosin isoforms in rat neurons: the different developmental profiles and distributions of TM-4 and TMBr-3 are consistent with different functions. Journal of cell Science. 1994;107(Pt 10):2961–2973. doi: 10.1242/jcs.107.10.2961. [DOI] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. The Journal of neuroscience : the official journal of the Society for NeuroScience. 1992;12:2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yu D, Gu QH, Yang Y, Tu K, Zhu J, Li Z. miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nature communications. 2014;5:3263. doi: 10.1038/ncomms4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur EM, Saijilafu, Zhou FQ. Growing the growth cone: remodeling the cytoskeleton to promote axon regeneration. Trends in neurosciences. 2012;35:164–174. doi: 10.1016/j.tins.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Davare M, Lesiak A, Fortin D, Ando H, Varlamova O, Obrietan K, Soderling TR, Goodman RH, Wayman GA. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Molecular and cellular neurosciences. 2010;43:146–156. doi: 10.1016/j.mcn.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwazaki T, McGregor IS, Matsumoto I. Protein expression profile in the striatum of acute methamphetamine-treated rats. Brain research. 2006;1097:19–25. doi: 10.1016/j.brainres.2006.04.052. [DOI] [PubMed] [Google Scholar]
- Jalilian I, Heu C, Cheng H, Freittag H, Desouza M, Stehn JR, Bryce NS, Whan RM, Hardeman EC, Fath T, Schevzov G, Gunning PW. Cell elasticity is regulated by the tropomyosin isoform composition of the actin cytoskeleton. PloS one. 2015;10:e0126214. doi: 10.1371/journal.pone.0126214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee AJ, Yang L, Lucas CA, Greenberg MJ, Martel N, Leong GM, Hughes WE, Cooney GJ, James DE, Ostap EM, Han W, Gunning PW, Hardeman EC. An actin filament population defined by the tropomyosin Tpm3.1 regulates glucose uptake. Traffic. 2015;16:691–711. doi: 10.1111/tra.12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong KY, Kedes L. Leucine 135 of tropomodulin-1 regulates its association with tropomyosin, its cellular localization, and the integrity of sarcomeres. The Journal of biological chemistry. 2006;281:9589–9599. doi: 10.1074/jbc.M512064200. [DOI] [PubMed] [Google Scholar]
- Kostyukova A, Maeda K, Yamauchi E, Krieger I, Maeda Y. Domain structure of tropomodulin: distinct properties of the N-terminal and C-terminal halves. European journal of biochemistry / FEBs. 2000;267:6470–6475. doi: 10.1046/j.1432-1327.2000.01738.x. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS. Leiomodin/tropomyosin interactions are isoform specific. Archives of biochemistry and biophysics. 2007;465:227–230. doi: 10.1016/j.abb.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS, Choy A, Rapp BA. Tropomodulin binds two tropomyosins: a novel model for actin filament capping. Biochemistry. 2006;45:12068–12075. doi: 10.1021/bi060899i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyukova AS, Hitchcock-DeGregori SE. Effect of the structure of the N terminus of tropomyosin on tropomodulin function. The Journal of biological chemistry. 2004;279:5066–5071. doi: 10.1074/jbc.M311186200. [DOI] [PubMed] [Google Scholar]
- Kostyukova AS, Hitchcock-Degregori SE, Greenfield NJ. Molecular basis of tropomyosin binding to tropomodulin, an actin-capping protein. Journal of molecular biology. 2007;372:608–618. doi: 10.1016/j.jmb.2007.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyukova AS, Rapp BA, Choy A, Greenfield NJ, Hitchcock-DeGregori SE. Structural requirements of tropomodulin for tropomyosin binding and actin filament capping. Biochemistry. 2005;44:4905–4910. doi: 10.1021/bi047468p. [DOI] [PubMed] [Google Scholar]
- Krieger I, Kostyukova A, Yamashita A, Nitanai Y, Maeda Y. Crystal structure of the C-terminal half of tropomodulin and structural basis of actin filament pointed-end capping. Biophysical journal. 2002;83:2716–2725. doi: 10.1016/S0006-3495(02)75281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HB, Sabatini BL. Glutamate induces de novo growth of functional spines in developing cortex. Nature. 2011;474:100–104. doi: 10.1038/nature09986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesiak A, Zhu M, Chen H, Appleyard SM, Impey S, Lein PJ, Wayman GA. The environmental neurotoxicant PCB 95 promotes synaptogenesis via ryanodine receptor-dependent miR132 upregulation. The Journal of neuroscience : the official journal of the Society for NeuroScience. 2014;34:717–725. doi: 10.1523/JNEUROSCI.2884-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CY, Bi X, Wu D, Kim JB, Gunning PW, Hong W, Han W. Tropomodulin3 is a novel Akt2 effector regulating insulin-stimulated GLUT4 exocytosis through cortical actin remodeling. Nature communications. 2015;6:5951. doi: 10.1038/ncomms6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A. Actin-based plasticity in dendritic spines. Science. 2000;290:754–758. doi: 10.1126/science.290.5492.754. [DOI] [PubMed] [Google Scholar]
- Meijering E. Neuron tracing in perspective. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 2010;77:693–704. doi: 10.1002/cyto.a.20895. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Moroz N, Guillaud L, Desai B, Kostyukova AS. Mutations changing tropomodulin affinity for tropomyosin alter neurite formation and extension. PeerJ. 2013;1:e7. doi: 10.7717/peerj.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer JD, Nowak RB, Kim NE, Larkin SK, Peters LL, Hartwig J, Kuypers FA, Fowler VM. Tropomodulin 1-null mice have a mild spherocytic elliptocytosis with appearance of tropomodulin 3 in red blood cells and disruption of the membrane skeleton. Blood. 2010;116:2590–2599. doi: 10.1182/blood-2010-02-268458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi T, Maruoka S, Sakudo A, Arai T. Assay-based quantitative analysis of PC12 cell differentiation. Journal of neuroscience methods. 2002;118:1–8. doi: 10.1016/s0165-0270(02)00116-4. [DOI] [PubMed] [Google Scholar]
- Rao JN, Madasu Y, Dominguez R. Mechanism of actin filament pointed-end capping by tropomodulin. Science. 2014;345:463–467. doi: 10.1126/science.1256159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevzov G, Bryce NS, Almonte-Baldonado R, Joya J, Lin JJ, Hardeman E, Weinberger R, Gunning P. Specific features of neuronal size and shape are regulated by tropomyosin isoforms. Molecular biology of the cell. 2005;16:3425–3437. doi: 10.1091/mbc.E04-10-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevzov G, Gunning P, Jeffrey PL, Temm-Grove C, Helfman DM, Lin JJ, Weinberger RP. Tropomyosin localization reveals distinct populations of microfilaments in neurites and growth cones. Molecular and cellular neurosciences. 1997;8:439–454. doi: 10.1006/mcne.1997.0599. [DOI] [PubMed] [Google Scholar]
- Schevzov G, Whittaker SP, Fath T, Lin JJ, Gunning PW. Tropomyosin isoforms and reagents. Bioarchitecture. 2011;1:135–164. doi: 10.4161/bioa.1.4.17897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmelpfeng J, Weibezahn KF, Dertinger H. Quantification of NGF-dependent neuronal differentiation of PC-12 cells by means of neurofilament-L mRNA expression and neuronal outgrowth. Journal of neuroscience methods. 2004;139:299–306. doi: 10.1016/j.jneumeth.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, Davidson MW, Wang J. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nature methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. Journal of anatomy. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Silacci P, Mazzolai L, Gauci C, Stergiopulos N, Yin HL, Hayoz D. Gelsolin superfamily proteins: key regulators of cellular functions. Cellular and molecular life sciences : CMLs. 2004;61:2614–2623. doi: 10.1007/s00018-004-4225-6. [DOI] [PubMed] [Google Scholar]
- Sun Y, Dierssen M, Toran N, Pollak DD, Chen WQ, Lubec G. A gel-based proteomic method reveals several protein pathway abnormalities in fetal Down syndrome brain. Journal of proteomics. 2011;74:547–557. doi: 10.1016/j.jprot.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Sussman MA, Sakhi S, Tocco G, Najm I, Baudry M, Kedes L, Schreiber SS. Neural tropomodulin: developmental expression and effect of seizure activity. Brain research. Developmental brain research. 1994;80:45–53. doi: 10.1016/0165-3806(94)90088-4. [DOI] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Schevzov G, Hotulainen P, Naumanen P, Martin C, Gunning PW, Lappalainen P. A molecular pathway for myosin II recruitment to stress fibers. Current biology : CB. 2011;21:539–550. doi: 10.1016/j.cub.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Tsukada T, Kotlyanskaya L, Huynh R, Desai B, Novak SM, Kajava AV, Gregorio CC, Kostyukova AS. Identification of residues within tropomodulin-1 responsible for its localization at the pointed ends of the actin filaments in cardiac myocytes. The Journal of biological chemistry. 2011;286:2194–2204. doi: 10.1074/jbc.M110.186924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Shah SP, Gritsyna Y, Hitchcock-DeGregori SE, Kostyukova AS. Systematic analysis of tropomodulin/tropomyosin interactions uncovers fine-tuned binding specificity of intrinsically disordered proteins. Journal of molecular recognition : JMR. 2011;24:647–655. doi: 10.1002/jmr.1093. [DOI] [PubMed] [Google Scholar]
- Vitriol EA, Zheng JQ. Growth cone travel in space and time: the cellular ensemble of cytoskeleton, adhesion, and membrane. Neuron. 2012;73:1068–1081. doi: 10.1016/j.neuron.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Kobayashi R, Helfman DM. N-tropomodulin: a novel isoform of tropomodulin identified as the major binding protein to brain tropomyosin. Journal of cell Science. 1996;109(Pt 9):2299–2310. doi: 10.1242/jcs.109.9.2299. [DOI] [PubMed] [Google Scholar]
- Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin caps the pointed ends of actin filaments. The Journal of cell biology. 1994;127:1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber KL, Fischer RS, Fowler VM. Tmod3 regulates polarized epithelial cell morphology. Journal of cell Science. 2007;120:3625–3632. doi: 10.1242/jcs.011445. [DOI] [PubMed] [Google Scholar]
- Xue B, Robinson RC. Guardians of the actin monomer. European journal of cell biology. 2013;92:316–332. doi: 10.1016/j.ejcb.2013.10.012. [DOI] [PubMed] [Google Scholar]
- Yamashiro S, Speicher KD, Speicher DW, Fowler VM. Mammalian tropomodulins nucleate actin polymerization via their actin monomer binding and filament pointed end-capping activities. The Journal of biological chemistry. 2010;285:33265–33280. doi: 10.1074/jbc.M110.144873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JW, Czech T, Felizardo M, Baumgartner C, Lubec G. Aberrant expression of cytoskeleton proteins in hippocampus from patients with mesial temporal lobe epilepsy. Amino acids. 2006;30:477–493. doi: 10.1007/s00726-005-0281-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.