Abstract
Listeria monocytogenes (Lm) is an important food-borne human pathogen that is able to strive under a wide range of environmental conditions. Its accessory gene regulator (agr) system was shown to impact on biofilm formation and virulence and has been proposed as one of the regulatory mechanisms involved in adaptation to these changing environments. The Lm agr operon is homologous to the Staphylococcus aureus system, which includes an agrD-encoded autoinducing peptide that stimulates expression of the agr genes via the AgrCA two-component system and is required for regulation of target genes. The aim of the present study was to identify the native autoinducing peptide (AIP) of Lm using a luciferase reporter system in wildtype and agrD deficient strains, rational design of synthetic peptides and mass spectrometry. Upon deletion of agrD, luciferase reporter activity driven by the PII promoter of the agr operon was completely abolished and this defect was restored by co-cultivation of the agrD-negative reporter strain with a producer strain. Based on the sequence and structures of known AIPs of other organisms, a set of potential Lm AIPs was designed and tested for PII-activation. This led to the identification of a cyclic pentapeptide that was able to induce PII-driven luciferase reporter activity and restore defective invasion of the agrD deletion mutant into Caco-2 cells. Analysis of supernatants of a recombinant Escherichia coli strain expressing AgrBD identified a peptide identical in mass and charge to the cyclic pentapeptide. The Lm agr system is specific for this pentapeptide since the AIP of Lactobacillus plantarum, which also is a pentapeptide yet with different amino acid sequence, did not induce PII activity. In summary, the presented results provide further evidence for the hypothesis that the agrD gene of Lm encodes a secreted AIP responsible for autoregulation of the agr system of Lm. Additionally, the structure of the native Lm AIP was identified.
Keywords: autoinducing peptide, accessory gene regulator, Listeria monocytogenes, peptide sensing
Introduction
The Gram-positive bacterium Listeria monocytogenes (Lm) is an opportunistic, intracellular pathogen that may cause severe, food-borne infections in high-risk groups such as immunocom promised persons, elderly people and pregnant women (Freitag et al., 2009). Lm is able to survive and replicate in a wide range of environments including soil, various food products, and different niches inside its human host (Freitag et al., 2009; Vivant et al., 2013; Ferreira et al., 2014; Gahan and Hill, 2014). In order to adapt to these changing conditions, L. monocytogenes possesses 15 complete two-component systems (Williams et al., 2005) and a number of regulatory circuits (Guariglia-Oropeza et al., 2014). The accessory gene regulator (agr) locus encodes one of these systems and has been shown to be involved in biofilm formation, virulence and survival in the environment (Autret et al., 2003; Rieu et al., 2007; Riedel et al., 2009; Vivant et al., 2015).
The prototype agr system was described for S. aureus and consists of the four gene operon agrBDCA (Novick and Geisinger, 2008). Of the four proteins encoded by the agr operon, AgrB is a membrane-bound peptidase that cleaves and processes the agrD-derived propeptide at the C-terminus, catalyzes formation of a thiolactone ring with a central cysteine, and, in combination with the signal peptidase SpsB, effects export and release of the active autoinducing peptide (AIP). Upon accumulation in the extracellular space, this AIP activates a two-component system consisting of AgrC (receptor-histidine kinase) and AgrA (response regulator). Expression of the operon is driven by the PII promoter upstream of agrB and is subject to autoregulation via AgrA. Target genes of the staphylococcal agr system are either directly regulated by AgrA or by a regulatory RNAIII transcribed in the opposite direction from the PIII promoter adjacent to PII (Thoendel et al., 2011).
Homologous agr systems have been identified in a number of Gram-positive microorganisms including streptococci, clostridia, lactobacilli, Bacillus sp., and Enterococcus faecalis (Wuster and Babu, 2008). The effects of agr regulation are pleiotropic. In S. aureus, the agr system regulates a wide range of genes involved in biofilm formation, virulence, and immune evasion (Queck et al., 2008; Thoendel et al., 2011). The agr system of Lactobacillus plantarum is involved in regulation of cell morphology and adhesion to glass surfaces (Sturme et al., 2005; Fujii et al., 2008). Similar to the staphylococcal system, the agr-like fsr system of E. faecalis and the agr system of Lm are involved in regulation of biofilm formation and virulence (Autret et al., 2003; Rieu et al., 2007; Riedel et al., 2009; Cook and Federle, 2014). Moreover, in Lm more than 650 genes are directly or indirectly regulated by the agr system as shown by transcriptional profiling of an agrD deletion mutant (Riedel et al., 2009). This suggests that agr systems represent rather global regulatory mechanisms.
Despite similarities on protein level, genetic organization, and phenotypic traits regulated, known agr systems differ regarding their mechanisms of target gene regulation. While in staphylococci, a significant number of agr-dependent genes are regulated by RNAIII (Thoendel et al., 2011), no information on RNAIII transcripts are available in other organisms. In E. faecalis and Lm, the genetic information upstream of the agr operon differs from that of staphylococci in that the preceding gene is transcribed in the same direction as the agr genes and no putative PIII promoters have been identified (Qin et al., 2001; Autret et al., 2003). Moreover, despite extensive bioinformatic approaches or transcriptional profiling a regulatory RNAIII has not been identified in Lm (Mandin et al., 2007; Toledo-Arana et al., 2009; Mellin and Cossart, 2012; Wurtzel et al., 2012). This suggests that in Lm (and E. faecalis) target genes are regulated by AgrA and/or other transcriptional regulators affected by AgrA-dependent regulation. However, it can not be excluded that the AIP signals through other two-component system besides AgrCA.
Structural information of AIPs is available only for a limited number of species. In S. aureus, four agr specificity groups with different AIPs varying in size from 7 to 9 amino acids (aa) are known (Novick and Geisinger, 2008). Similarly, three agr specificity groups exist in S. epidermidis with AIPs of 8–12 aa (Otto et al., 1998; Olson et al., 2014). The AIP of S. intermedius and S. lugdunensis are 9 and 7 aa in size, respectively (Ji et al., 1997; Kalkum et al., 2003). Outside the genus Staphylococcus, AIPs have been characterized for E. faecalis (11 aa), L. plantarum (5 aa), and C. acetobutylicum (6 aa) (Nakayama et al., 2001; Sturme et al., 2005; Steiner et al., 2012). Most of the known AIPs contain a thiolactone ring formed by the 5 C-terminal aa. Exceptions are the AIPs of C. acetobutylicum and E. faecalis, which have ring structures consisting of 6 and 9 aa, respectively (Nakayama et al., 2001; Steiner et al., 2012). Another common feature is a central cysteine, which is replaced by a serine in some cases, required for thiolactone ring formation.
For staphylococci, E. faecalis and Lm, a contribution of the agr system to virulence gene regulation has been demonstrated and agr-deficient mutants are attenuated (Riedel et al., 2009; Thoendel et al., 2011; Cook and Federle, 2014). Consequently, interference with agr signaling was proposed as a therapeutic approach (Gray et al., 2013). Of note, the specificity of the interaction between the AIP and its cognate receptor AgrA has been used to device improved strategies by fusing the AIP to a bacteriocin to induce lysis of the targeted bacteria (Qiu et al., 2003). The structure of the native AIP of Lm has not been elucidated so far. With the present study, we aim closing this gap in order to further elucidate the components and mechanisms of the agr autoregulatory circuit of Lm and to facilitate future studies on strategies to interfere with cell–cell communication of this important human pathogen.
Materials and Methods
Bacterial Strains and Culture Conditions
All strains and plasmids used in this study are listed in Table 1. L. monocytogenes was generally incubated in Brain Heart Infusion broth (BHI, Oxoid Ltd) at 30°C. E. coli strains were grown in lysogeny broth (LB). For solid media, 15 g/l agar were added to the broth before autoclaving. Antibiotics were added if necessary. Where appropriate, kanamycin was used at a final concentration of 50 (for E. coli strains) and 15 μg/ml chloramphenicol were used for both species. For Lm strains carrying a chromosomal copy of pPL2 derivatives chloramphenicol was used at 7 μg/ml.
Table 1.
Bacterial strains and plasmids used in the present study.
| Strain/plasmid | Characteristics | Reference/source |
|---|---|---|
| Strains | ||
| Escherichia coli DH10B | Cloning host | Thermo Fisher Scientific |
| E. coli BL21 DE3 | Used for protein overexpression | New England Biolabs |
| E. coli BL21 DE3 pET29a_agrB | IPTG-inducible expression of agrB, Kanr | This study |
| E. coli BL21 DE3 pET29a_agrBD | IPTG-inducible expression of agrBD, Kanr | This study |
| Listeria monocytogenes EGD-e | Bécavin et al., 2014 | |
| L. monocytogenes ΔagrD | In-frame deletion of agrD in strain EGD-e | Riedel et al., 2009 |
| L. monocytogenesΔagrD::pIMK2agrD | pIMK2agrD integrated into the tRNAArg locus in the EGD-e chromosome, Kanr | Riedel et al., 2009 |
| L. monocytogenes EGD-e::pPL2luxPII | pPL2luxABCDEPII integrated into the tRNAArg locus in the EGD-e chromosome, Cmr | This study |
| L. monocytogenes EGD-e ΔagrD::pPL2luxPII | pPL2luxPII integrated into the tRNAArg locus in the EGD-eΔagrD chromosome, Cmr | This study |
| L. monocytogenes EGD-e ΔagrD pNZ44agrBD | Strain with constitutive, P44-driven expression of agrB and agrD, Cmr | This study |
| Plasmids | ||
| pPL2lux | Site-specific integrative vector to study promotor activity in L. monocytogenes, Cmr | Bron et al., 2006 |
| pPL2luxPII | Site-specific integrative vector for PII promoter activity analysis, Cmr | This study |
| pNZ44 | Plasmid for constitutive gene expression driven from the lactococcal promoter P44 | McGrath et al., 2001 |
| pNZ44agrBD | Plasmid for constitutive P44-driven expression of agrB and agrD in L. monocytogenes, Cmr | This study |
| pET29a(+) | Plasmid for strong IPTG inducible expression in E. coli, Kanr | Merck Millipore |
| pET29a_agrB | IPTG-inducible expression of agrB in E. coli, Kanr | This study |
| pET29a_agrBD | IPTG-inducible expression of agrBD in E. coli, Kanr | This study |
Generation of Recombinant Strains
Primers used for cloning or sequencing purposes are listed in Table 2. To study transcriptional activity of the agr operon, the PII promoter upstream of agrB (Rieu et al., 2007) was amplified with Phusion® polymerase (Thermo Fisher Scientific) using primers PII_fwd_SalI and PII_rev and chromosomal DNA of Lm EGD-e wildtype (WT) as template. The obtained PCR fragment was digested with SalI and cloned in frame in front of the luciferase reporter into SalI/SwaI-cut pPL2lux (Bron et al., 2006). The ligation mix was transformed into E. coli ElectroMaxTM DH10B (Thermo Fisher Scientific), and the resulting plasmid pPL2luxPII was verified by restriction analysis and amplification of the cloned PII promoter using primers PII_fwd_SalI and luxA_rev with subsequent Sanger sequencing of the PCR fragment by a commercial service provider (Eurofins, Germany). The plasmid was transformed into electrocompetent Lm EGD-e WT or ΔagrD (Riedel et al., 2009) as described previously (Monk et al., 2008) creating Lm EGD-e::pPL2luxPII and ΔagrD::pPL2luxPII. In both strains, successful chromosomal integration of pPL2luxPII at the correct site (tRNAArg) was verified using primers PL95 and PL102 (Lauer et al., 2002).
Table 2.
Primers used in this study.
| Name | Sequence | Reference/source |
|---|---|---|
| PII_fwd_SalI | CTGATGTCGACCTTCAAACAGAACAAGACG | This study |
| PII_rev | CAACTAATTCACCTCCACTAATATTTTACAACG | This study |
| luxA_rev | TACCTCTGTTTGAGAAAATTGGGGAGG | This study |
| PL95 | ACATAATCAGTCCAAAGTAGATGC | Lauer, 2002 |
| PL102 | TATCAGACCTAACCCAAACCTTCC | Lauer, 2002 |
| NZagrBD-fwd | AATTCCATGGGTAATTTTACTGCAAAAGTCCC | This study |
| NZagrBD-rev | GCATCGAGCTCTTATTTATTTTCGTTTTTTTC | This study |
| NZ-conf_fwd | CCATACAGGAGAAGGGACGATAGCAA | This study |
| NZ_colony_rev | CCTTGAGCCAGTTGGGATAGAGC | This study |
| agrBD_NdeI_fwd | GGAATTCCATATGAGTAATTTTACTGCAAAAGTCCC | This study |
| agrBD_BamHI_rev | CGCGGATCCATTAATCCTCCACTGTCTAAAATATCTAT | This study |
For homologous overexpression of agrBD, a PCR fragment containing both genes was amplified using primers NZagrBD_fwd and NZagrBD_rev and chromosomal DNA of Lm EGD-e as template. The PCR product was digested with NcoI and SacII and ligated as exact transcriptional fusion to the constitutive P44 promoter into NcoI/SacII digested pNZ44 (McGrath et al., 2001) to yield pNZ44agrBD. The product was transformed into E. coli DH10B. Clones were screened for plasmid containing the correct insert by PCR using primers NZ-confirm_fwd and NZ_colony_rev and sequencing of the PCR product. The correct plasmid as well as the empty vector (pNZ44) were transformed in electrocompetent Lm ΔagrD generated as described previously (Monk et al., 2008).
For heterologous AIP production, agrBD or agrB alone were amplified using primer pairs agrBD_NdeI_fwd/agrBD_BamHI_rev and chromosomal DNA of Lm EGD-e WT or ΔagrD. Following restriction with NdeI and BamHI both PCR products were ligated into NdeI/BamHI digested pET29a(+) (Merck Millipore). This fuses the PCR products to the T7 promoter creating pET29a_agrB and pET29a_agrBD, respectively. Both plasmids were verified for correct cloning by restriction analysis and Sanger sequencing of inserts.
Luciferase Reporter Assays
For luciferase reporter assays, growth experiments were performed in white 96-well microtiter plates with transparent bottom (BRANDplates® pureGradeTM S). A single colony was inoculated into BHI and grown over night (o/N; i.e., approx. 16 h). Following o/N growth, cultures were diluted to an optical density at 600 nm (OD600) of 0.01 in fresh, sterile BHI. For co-cultivation of AIP producer and reporter strains, o/N cultures of both strains were used to inoculate BHI medium to a OD600 of 0.01 and then mixed at a 1:1 ratio. 200 μl aliquots of this mix were transferred into individual wells of the microtiter plates (each condition in triplicates). Plates were incubated at 30°C in a Tecan Infinite M200 plate reader and OD600 and luminescence intensity were measured every hour.
Synthetic Peptides
Synthetic peptides were purchased from Peptide Protein Research Ltd (UK) in lyophilized form with >70% purity. Peptides were reconstituted in dimethyl sulfoxide (DMSO) at 2 mM and stored at -20°C until further use. For experiments, these stocks were diluted as appropriate in 25% (v/v) DMSO in phosphate-buffered saline (PBS) to give the final concentrations as indicated. To test the effect of peptides on PII activity, reporter strains (Lm EGD-e::pPL2luxPII or ΔagrD::pPL2luxPII) were grown o/N and diluted to an OD600 of 0.01 in fresh BHI. 180 μl aliquots were distributed in 96 well microtiter plates (each condition in triplicate) and incubated at 30°C for 2 h. At this stage, 20 μl of diluted peptides were added to obtain the indicated final concentrations (5 nM–50 μM) and plates were incubated at 30°C in a Tecan Infinite M200 plate reader with hourly OD600 and luminescence intensity measurements.
AIP Production in E. coli
For heterologous AIP production, pET29a_agrB or pET29a_agrBD were transformed into E. coli BL21(DE3) (New England Biolabs) and transformants were selected on LB agar containing kanamycin. Four single colonies were streaked onto two LB agar plates containing kanamycin with or without 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). A clone showing good growth in the absence of IPTG but reduced growth in its presence was selected and a single colony was inoculated into 5 ml of LB medium and grown o/N on a rotary shaker at 37°C. Using the o/N culture, 500 ml LeMaster and Richards minimal medium (Paliy and Gunasekera, 2007) containing 50 mM glucose were inoculated to a final OD600 of 0.1 and incubated on a rotary shaker at 37°C to an OD600 of 0.8. At this stage, expression was induced by addition of 1 mM ITPG. Following incubation under the same conditions for an additional 2 h, bacterial cells were pelleted via centrifugation (3000 × g, 30 min and 4°C) and supernatants were collected, filter sterilized, frozen in liquid nitrogen and lyophilized. Lyophilized samples were stored at -20°C until further analysis by LC–MS/MS.
LC–MS/MS Analysis
The lyophilized supernatants of recombinant E. coli strains were reconstituted in a 25:35:35:5 H2O:Isopropanol:CH3CN:HCOOH mixture and diluted 1:10 in H2O. 5 μl were injected into a reverse-phase column with corresponding guard column (AerisTM PEPTIDE 3.6u XB-C18 150 × 2.1 mm, Security GuardTM ULTRA 2 × 2.1 mm guard column, Phenomenex). A constant flow rate of 0.4 ml/min was applied. Mobile phase A consisted of water with 0.2% (v/v) formic acid and mobile phase B was acetonitrile with 0.2% (v/v) formic acid. Elution program was: isocratic hold at 5% B for 5 min followed by a linear gradient from 5 to 45% B over 80 min. After each sample, the column was washed with 90% B for 10 min and equilibrated at starting conditions. Data was obtained in positive auto MS/MS mode on an Agilent 6540 Accurate-Mass Quadrupole (LC-Q-TOF/MS) with ESI Jet Stream Technology using the following conditions: drying gas flow rate of 10 l/min with a gas temperature of 250°C, nebulizer with 40 lb per square inch gauge, sheath gas flow rate of 10 l/min, sheath gas temperature of 300°C, capillary voltage of 4000 V, and fragmentor voltage of 170 V. The collision energy was set by formula with 4.5 slope and 10 offset. Data analysis was performed using Mass Hunter Workstation Software (Ver.B.05.519.0, Agilent Technologies) and the “Find compounds by formula” algorithms. Synthetic peptides were analyzed using the same conditions as the recombinant peptides expressed in E. coli to compare retention time, accurate mass and fragmentation patterns.
Invasion Assay
Invasion of Lm into Caco-2 cells was tested using a standard gentamycin protection assay essentially as described previously (Riedel et al., 2009). Briefly, Caco-2 cells were cultured in DMEM supplemented with 10% (v/v) fetal calf serum (FCS), 10 mM L-glutamine, 1% (v/v) penicillin/streptomycin and 1% (v/v) non-essential amino acids (NEAA) at 37°C and a 5% CO2 atmosphere. Cells were seeded to a density of 2 × 105 cells per well in a 24 well plate and cultivated to a monolayer for 4 days. One day prior to the experiment, culture media without antibiotics was added. A fresh o/N culture of the indicated bacterial strains was diluted 1:10 in 10 ml fresh BHI and grown to mid-exponential phase (OD600 = 0.8). Where appropriate, peptide R5T0 was added (5 μM final concentration). Bacteria were pelleted and diluted in DMEM containing 10 mM L-glutamine and 1% NEAA to 108 colony forming units per ml (cfu/ml) (OD600 = 0.5). 1 ml of this suspension was added to Caco-2 cells in quadruplicates (MOI = 100). Cells were incubated for 1 h to allow invasion of bacteria. To kill remaining extracellular bacteria, cells were washed once with PBS and 1 ml DMEM containing 10 μg/ml gentamicin (Gibco®) was added to the cells. After 1 h of incubation, cells were washed twice with PBS, lysed with ice-cold water and cfu/ml were determined by plating serial dilutions on BHI agar.
Statistical Analysis
All experiments were conducted in at least three biological replicates. Results were analyzed by Student’s t-test or ANOVA with Bonferroni post-test analysis to correct for multiple comparisons using GraphPad Prism (version 6) as indicated in figure legends and Supplementary Data Sheet 1. Differences between different strains or conditions were considered statistically significant at p < 0.05.
Results
PII-Activity in Lm EGD-e
PII promoter activity was analyzed in Lm EGD-e::pPL2luxPII and ΔagrD::pPL2luxPII during growth in BHI medium at 30°C (Figure 1A). No differences in growth or final OD600 were observed between the two strains ruling out an effect of growth on luciferase activity. In the WT background, a significant increase in PII-dependent luciferase activity was observed during exponential growth with a peak in late exponential phase. By contrast, no luminescence above background could be detected for the agrD-deficient strain throughout the experiment. This suggests that the AIP is required for transcriptional activity of PII.
FIGURE 1.
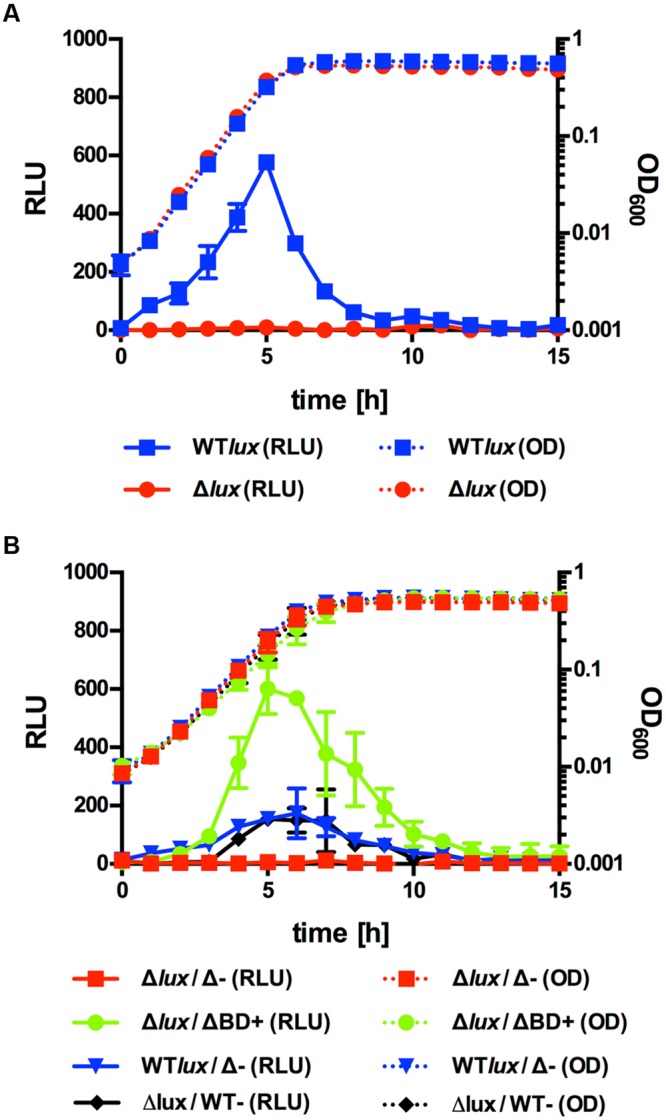
Growth (OD600) and luminescence (relative luminescence units; RLU) of Lm EGD-e::pPL2luxPII (WTlux) or ΔagrD::pPL2luxPII (Δlux) grown alone (A) or in co-culture (B) with ΔagrD pNZ44agrBD (ΔBD+), ΔagrD pNZ44 (Δ-), or EGD-e pNZ44 (WT-). Values are mean ± standard deviation of three independent experiments. Statistical analysis of RLU values was performed by Student’s t-test and results are provided in Supplementary Data Sheet 1.
AIPs are usually secreted into the extracellular environment. In order to confirm that the AIP of Lm is acting as an extracellular peptide, similar growth experiments were conducted using co-incubation of AIP producer and reporter strains in different combinations (Figure 1B). As expected, the agrD-deficient reporter strain showed no PII activity when incubated with Lm EGD-e ΔagrD. However, high levels of luminescence were observed using the same reporter strain in combination with Lm ΔagrD pNZ44agrBD, a ΔagrD derivative expressing agrBD from the P44 promoter on pNZ44. Luminescence in this setup was significantly higher compared to co-cultures of the WT reporter with the agrD deletion mutant or the agrD-deficient reporter strain with Lm EGD-e pNZ44 (i.e., the empty vector control) suggesting that AIP levels produced by Lm ΔagrD pNZ44agrBD are higher than that of the WT.
PII Activation by Synthetic AIP Candidates
Upon several attempts we were unable to identify the active AIP in supernatants of Lm EGD-e WT or the AIP overproducing strain ΔagrD pNZ44agrBD grown in either BHI or modified Welshimer’s broth. Sequence alignment of AIPs with a resolved structure, revealed that most AIPs consist of a 5 aa thiolactone ring with N-terminal tail varying from 0 to 7 aa (Figure 2A). Using this information, a range of peptides based on the AgrD sequence of Lm EGD-e were synthesized consisting of a thiolactone ring of 4–6 aa and an N-terminal tail of 0–5 aa (Figure 2B). The effect of these peptides on PII-driven luciferase activity was tested using the reporter strains Lm EGD-e::pPL2luxPII and ΔagrD::pPL2luxPII. At 5 μM, none of the peptides had a measurable effect on growth of the reporter strains (Supplementary Figures S1A,B). The peptide R5T0 consisting of a 5 aa thiolactone ring with no N-terminal tail slightly increased PII-driven luminescence in the WT reporter strain during the first 4 h of the experiment (Figure 3A). However, at later stages luminescence was comparable to the control, i.e., reporter without peptide. Interestingly, some of the tested peptides (R5T1, R5T2, R5T4, and R5T5) significantly inhibited luminescence of the WT reporter strain. More importantly, some of the peptides (R5T0, R5T1, R5T4, and R5T5) induced luminescence by the ΔagrD reporter strain (Figure 3B). The most potent inducer of PII activity was the peptide R5T0, i.e., a cyclic pentapeptide with the amino acid sequence Cys-Phe-Met-Phe-Val (CFMFV). At concentration of 5 and 50 μM, R5T0 also induced luminescence above control levels during the first 4 h in the WT reporter (Figure 3C) and for up to 7 h in the agrD-deficient reporter (Figure 3D). This suggests that the most likely candidate for the native AIP of Lm EGD-e is the peptide R5T0.
FIGURE 2.
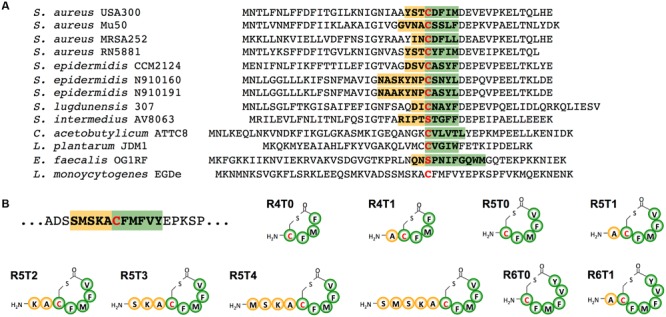
(A) Amino acid sequences of AgrD propetides with known structure compared with the AgrD sequence of Listeria monocytogenes. (B) Structure of synthetic cyclic peptides tested for autoinducing activity in L. monocytogenes. Amino acid residues of the native (A) or synthetic (B) peptides involved in thiolactone ring formation are labeled in green, those found in the N-terminal tails in yellow. The central cysteine or serine is marked by a red letter.
FIGURE 3.
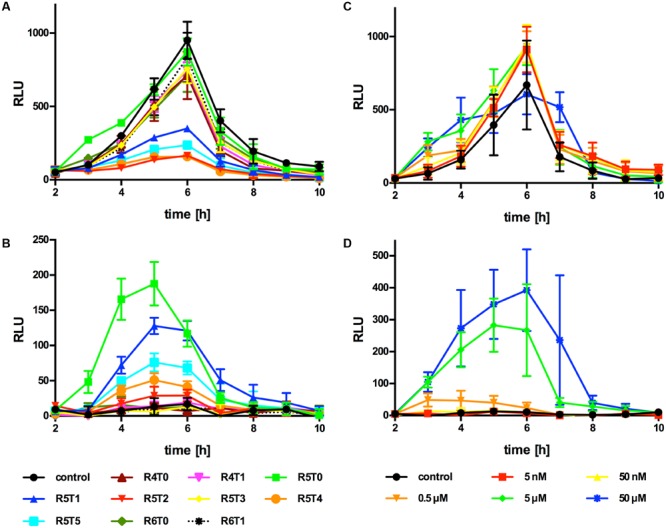
Luminescence (RLU) of Lm EGD-e::pPL2luxPII(A,C) or ΔagrD::pPL2luxPII(B,D) grown in the presence of different synthetic peptides (A,B; peptide concentration: 5 μM) or peptide R5T0 at the indicated concentrations (C,D). Values are mean ± standard deviation of three independent experiments. Statistical analysis of RLU values was performed by ANOVA and Bonferroni test to correct for multiple comparisons and results are provided in Supplementary Data Sheet 1.
The Synthetic AIP Restores the Invasion Defect of Lm ΔagrD
Deletion of ΔagrD and thus lack of a functional AIP results in reduced promoter activity of virulence factors and attenuated virulence (Riedel et al., 2009). In order to check if R5T0 is not only able to induce PII activity but also functionally complement the ΔagrD mutant, invasion assays were performed with Lm EGD-e ΔagrD grown in the presence and absence of R5T0 (Figure 4). As observed previously, deletion of agrD results in reduced invasion into Caco-2 intestinal epithelial cells and this defect was genetically complemented by integration of pIMK2agrD, i.e., a plasmid for constitutive expression of agrD (Riedel et al., 2009). More importantly, growth in the presence of 5 μM R5T0 completely restored invasion of Lm EGD-e ΔagrD to WT levels.
FIGURE 4.
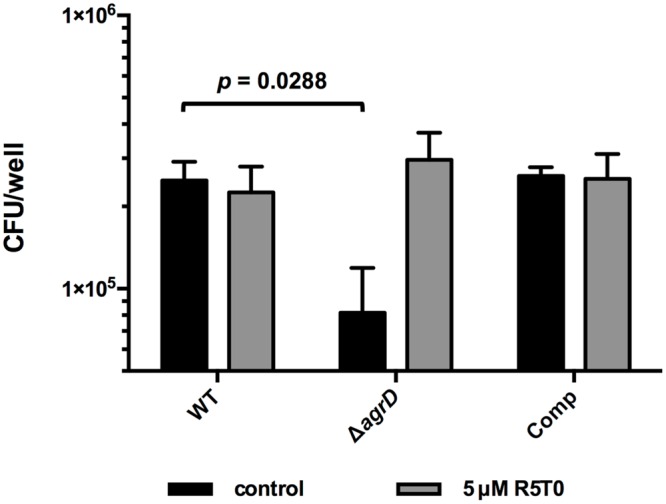
Invasion of Lm EGD-e (WT), EGD-e ΔagrD (ΔagrD), or EGD-e ΔagrD::pIMK2agrD (Comp) into Caco-2 cells. Bacteria were grown either in the absence (black bars) or presence of 5 μM peptide R5T0. Values are colony forming units per well (cfu/well) and are mean ± standard deviation of three independent experiments. Statistical analysis was performed by comparing all strains for a given condition (with or without peptide) by ANOVA. Bonferroni post tests were used to adjust p values for multiple comparisons.
Heterologous Production of the Lm AIP in E. coli
In a further approach to identify the AIP of Lm, the agrBD genes were expressed in E. coli using the IPTG-inducible pET29a system. Using LC–MS, a prominent signal was identified in supernatants of an induced culture of E. coli BL21 pET29a_agrBD (Figure 5A) with a mass of 627.2549 (Figure 5B). This signal was absent in the non-induced culture or supernatant of a control strain only expressing agrB (Supplementary Figure S2). In order to confirm the identity of the overexpressed peptide, analysis of the PII-activating synthetic peptide R5T0 was performed. Interestingly, the chromatogram of R5T0 yielded two peaks in close vicinity (Figure 5A). Both peaks correspond to peptides with identical mass and fragmentation pattern (Figure 5B). However, the different retention times and peak areas indicate that the two peaks represent stereoisomers or conformational isomers at different concentrations.
FIGURE 5.
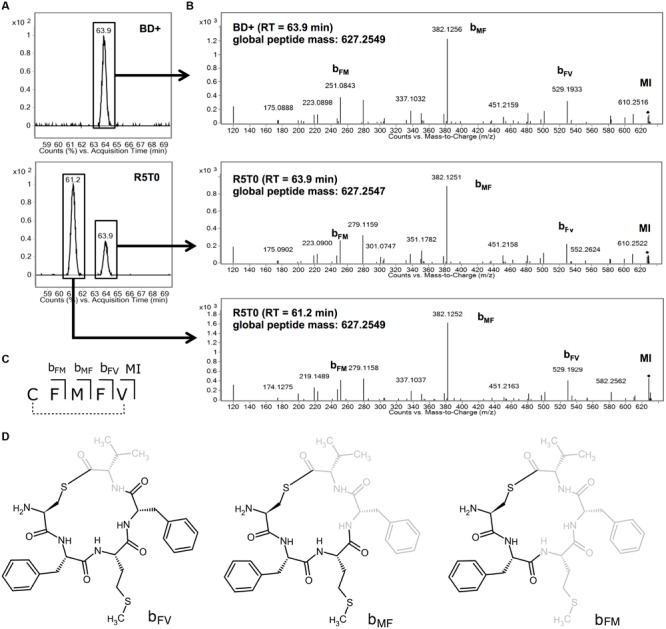
(A) Extracted ion chromatograms performed on culture supernatant of Escherichia coli BL21 DE3 pET29a_agrBD after induction with IPTG (BD+; upper panel) and the synthetic peptide R5T0 (lower panel; R5T0 molecular formula: C31H41N5O5S2). (B) Mass spectrometry fragmentation spectra for chromatographic peaks with retention times of 61.2 and 63.9 min (marked with a box in A). (C,D) Structure and assignment of fragments detected in MS/MS spectra to fragments of R5T0.
The peptide present in the supernatant of E. coli BL21 pET29a_agrBD and both peaks of R5T0 had almost identical global masses (Figure 5B). Moreover, all three peptides showed highly similar fragmentation patterns (Figure 5B) and several signals of the MS/MS spectra correspond to fragments of R5T0 at a mass accuracy better than 2 ppm (Table 3; for corresponding structures see Figures 5C,D). These results clearly indicate that the listerial AIP is a cyclic pentapeptide with the amino acid sequence CFMFV forming a thiolactone ring, i.e., the structure of the synthetic peptide R5T0.
Table 3.
Mass-charge-ratios (m/z) of peptide fragments detected by MS/MS and difference to the m/z calculated according to the formula of the corresponding R5T0 fragment.
| Fragment | Formula | m/z (calculated) | m/z (measured)a | Difference (ppm) | Difference (mDa) |
|---|---|---|---|---|---|
| bFV | C26H33N4O4S2 | 529.19377 | 529.1933 | -0.9 | -0.47 |
| bFV-CO | C25H33N4O3S2 | 501.19886 | 501.1994 | 1.1 | 0.54 |
| bMF | C17H24N2S2 | 382.12536 | 382.1256 | 0.6 | 0.24 |
| bFM | C12H15N2O2S | 251.08487 | 251.0843 | -2.3 | -0.57 |
| bFM-CO | C11H15N2OS | 223.08996 | 223.0898 | -0.7 | -0.16 |
aMeasured m/z are derived from the MS/MS analysis shown in Figure 5B.
Specificity of the Lm AIP
Known AIPs differ greatly in sequence, length and structure among species and even strains (Figure 2A) and different AIPs of S. aureus display cross-inhibition (Ji et al., 1997). Similar to the AIP of Lm, the AIP of L. plantarum is a cyclic pentapeptide yet with a different sequence (Sturme et al., 2005). Further experiments were performed to test if PII activation is specific for the Lm AIP or if the L. plantarum AIP is also able to activate PII (Figure 6). As observed in the previous experiments, R5T0 slightly enhanced PII-driven luciferase activity in Lm EGD-e::pPL2luxPII (Figure 6A) and was a potent inducer of PII activity in the AIP-negative reporter strain Lm EGD-e ΔagrD::pPL2luxPII. By contrast, in both reporter strains the L. plantarum AIP had no effect on PII activity.
FIGURE 6.
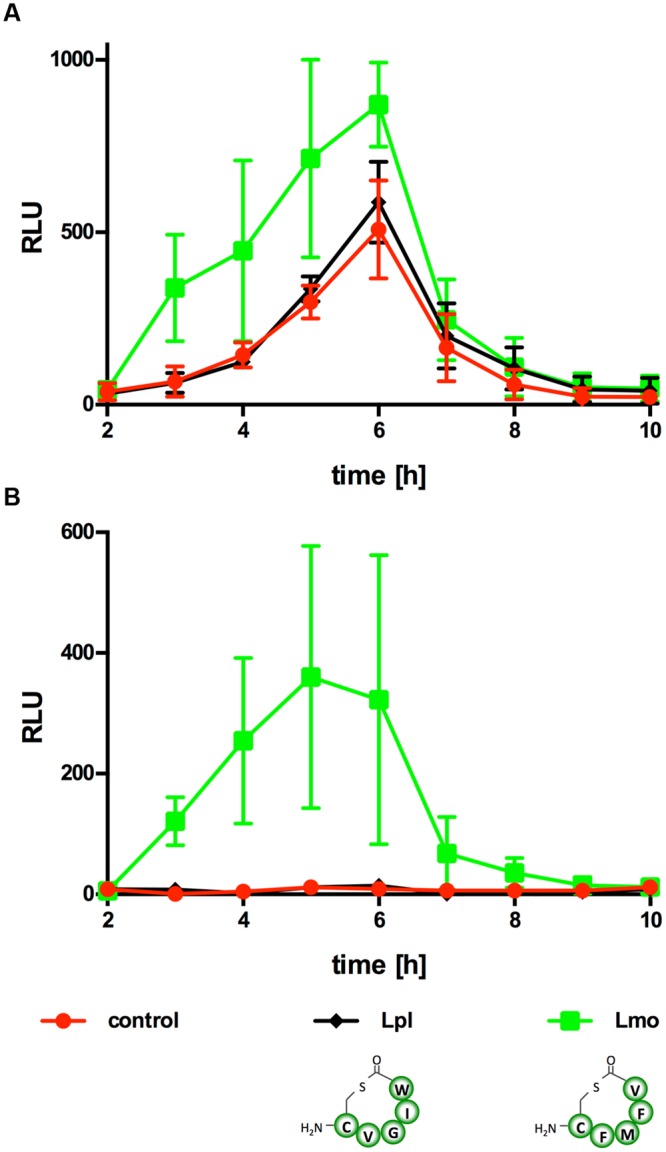
Luminescence (RLU) of Lm EGD-e::pPL2luxPII(A) or ΔagrD::pPL2luxPII(B) grown in the presence of the AIP of Lm (Lm; i.e., synthetic peptide R5T0) or L. plantarum (Lpl). As controls, bacteria were grown without peptides (control). Values are mean ± standard deviation of three independent experiments. Statistical analysis of RLU values was performed by ANOVA and Bonferroni test to correct for multiple comparisons and results are provided in Supplementary Data Sheet 1.
Discussion
Signaling peptides, also referred to as AIPs, are produced by a wide range of Gram-positive microorganisms (Wuster and Babu, 2008) and serve various purposes (Thoendel and Horswill, 2010). The best studied AIP system is the agr locus of S. aureus and homologous systems have been identified in a variety of Gram-positives (Wuster and Babu, 2008). In S. aureus, the agr system is a rather global regulatory circuit affecting a large number of genes and different phenotypic traits (Thoendel et al., 2011). Similarly, deletion of agrD in Lm affects more than 600 genes and phenotypically affects biofilm formation and virulence in vitro and in vivo (Rieu et al., 2007; Riedel et al., 2009). However, while absence of agr signaling is linked with enhanced biofilm formation of S. aureus (Vuong et al., 2000), agr mutants of Lm display reduced biofilm formation under the conditions monitored (Rieu et al., 2007; Riedel et al., 2009).
Previous studies have already indicated that, like the staphylococcal system, the Lm agr locus is subject to positive autoregulation involving a diffusible factor, probably the agrD-encoded AIP involved in regulation. Transcription levels of the agr operon were greatly reduced in agr-deficient Lm mutants (Rieu et al., 2007; Riedel et al., 2009; Garmyn et al., 2012). Also, the biofilm defect of a ΔagrD mutant was complemented when bacteria were grown in the reconstituted culture supernatants of the WT or in the presence of small amounts of WT bacteria (Riedel et al., 2009). The presented results further strengthen the hypothesis that agrD encodes a secreted AIP that positively regulates the agr system of Lm. In the ΔagrD mutant, no activity of the agr promoter could be observed (Figure 1A) and promoter activity was restored when the agrD-deficient reporter strain was co-cultured with a strain carrying a plasmid for constitutive expression of agrBD (Figure 1B).
The presented results provide further evidence that, in Lm, agrD actually encodes the propeptide, which is processed released into the extracellular environment where it acts as an AIP. Moreover, our data suggests that the native AIP is a cyclic pentapeptide R5T0 consisting of the amino acids (from N- to C-terminus) Cys, Phe, Met, Phe, Val. A peptide with this structure was found in the culture supernatant of a recombinant E. coli strain expressing AgrBD (Figure 5) and a synthetic peptide with identical structure was able to potently induce activity of the PII promoter of the agr system (Figure 3) and to functionally complement the invasion defect in a ΔagrD mutant (Figure 4).
Induction of luciferase activity in the ΔagrD reporter upon co-cultivation with the AIP producing WT strain (Figure 1B) indicates that at least some of the AIP must be present in culture supernatants. However, we were unable to identify the native peptide in supernatants of Lm EGD-e grown in complex media (brain heart infusion) or modified Welshimer’s broth. This may be explained by the high levels of peptides in brain heart infusion, which makes identification impossible by LC–MS/MS. In modified Welshimer’s broth Lm only grows to low final optical densities and thus any secreted peptide will also be present at low concentrations especially when subject to positive autoregulation and fully induced only at high cell densities. Further studies will be needed to quantify actual AIP concentrations in culture supernatants and the threshold required to activate PII and target gene regulation.
Interestingly, four different synthetic peptides with a five-membered thiolactone ring and varying tail length had inhibitory activity on the agr promoter in the WT reporter strain, which itself is able to produce the native AIP. Since agr mutants of Lm display attenuated virulence (Autret et al., 2003; Riedel et al., 2009), this suggests that these peptides are antagonists of the native AIP and may represent a potential supplementary or alternative therapeutic approach as proposed for S. aureus and other pathogens (Gray et al., 2013). Interestingly, they also exhibited PII activation in the ΔagrD reporter to varying degrees. This may indicate that these peptides compete with R5T0 or the native AIP for binding to the receptor but their affinity and/or activity is lower. Thus, of the four candidate peptides, the best antagonist of the native AIP is probably R5T2, which efficiently blocks PII activity in the WT but activates luminescence inly marginally in the mutant reporter.
A striking difference between the agr systems of S. aureus and Lm is the structural diversity of the AIPs. Within the species S. aureus, four specificity groups of strains with different AIP are found and these groups show cross-inhibition (Novick and Geisinger, 2008). By contrast, the AgrD propeptides of the genus Listeria are rather conserved and the species Lm, L. innocua, L. ivanovii, L. welshimeri, L. seeligeri, and L. marthii have identical (predicted) AIP sequences (Supplementary Figure S3A) suggesting cross-reactivity. Moreover, phylogenetic analysis based on 16S rRNA gene sequences reveals that Listeria sp. that share identical AIP sequences form a cluster that separates from the other species indicating that they are more closely related (Supplementary Figure S3B).
With the exception of C. acetobutylicum, phylogenetic trees calculated using concatenate AgrA, AgrB, AgrC, and AgrD sequences are in line with trees inferred from 16S sequences (Wuster and Babu, 2008). This suggests that agr systems are generally inherited vertically. It has been proposed that C. acetobutylicum, whose AgrD sequence is almost identical to that of Listeriaceae, is the only known case of horizontal transfer of an agr system (Wuster and Babu, 2008). Further experimental data comparing the Lm AIP with the AIP of L. plantarum, which also consist of a five cyclic pentapeptide although with different aa composition, indicates that the Lm agr system is specific for the AIP of those Listeria sp. that share a conserved AgrD sequence but does not respond to the cyclic pentapeptide AIPs of other organisms. This also suggests that intervention strategies based on antagonistic peptides targeting the agr systems of Lm (and other organisms) are specific for organisms with identical AIPs.
In summary, the presented data shows that the agrD of Lm EGD-e encodes a secreted peptide consisting of a five-membered thiolactone ring, which has autoinducing activity. Moreover, the identification of several synthetic peptides with antagonistic activity proposes a potential option to treat Lm infections or inhibit biofilm formation as suggested by others previously.
Author Contributions
CR conceived the study. MZ, MW, and AS-K carried out experiments. MZ, AS-K, BB, and CR analyzed data. MZ, AS-K, BB, and CR drafted the manuscript and all the authors contributed to preparing the final version of the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The study was conducted by intramural funding of the Universities of Ulm and Stuttgart. The funders had no role in design of the study or analysis and interpretation of the data.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00989
References
- Autret N., Raynaud C., Dubail I., Berche P., Charbit A. (2003). Identification of the agr locus of Listeria monocytogenes: role in bacterial virulence. Infect. Immun. 71 4463–4471. 10.1128/IAI.71.8.4463-4471.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécavin C., Bouchier C., Lechat P., Archambaud C., Creno S., Gouin E., et al. (2014). Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. MBio 5 e969–14. 10.1128/mBio.00969-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron P. A., Monk I. R., Corr S. C., Hill C., Gahan C. G. M. (2006). Novel luciferase reporter system for in vitro and organ-specific monitoring of differential gene expression in Listeria monocytogenes. Appl. Environ. Microbiol. 72 2876–2884. 10.1128/AEM.72.4.2876-2884.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook L. C., Federle M. J. (2014). Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol. Rev. 38 473–492. 10.1111/1574-6976.12046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira V., Wiedmann M., Teixeira P., Stasiewicz M. J. (2014). Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J. Food Prot. 77 150–170. 10.4315/0362-028X.JFP-13-150 [DOI] [PubMed] [Google Scholar]
- Freitag N. E., Port G. C., Miner M. D. (2009). Listeria monocytogenes – from saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 7 623–628. 10.1038/nrmicro2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T., Ingham C., Nakayama J., Beerthuyzen M., Kunuki R., Molenaar D., et al. (2008). Two homologous Agr-like quorum-sensing systems cooperatively control adherence, cell morphology, and cell viability properties in Lactobacillus plantarum WCFS1. J. Bacteriol. 190 7655–7665. 10.1128/JB.01489-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahan C. G. M., Hill C. (2014). Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front. Cell. Infect. Microbiol. 4:9 10.3389/fcimb.2014.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmyn D., Augagneur Y., Gal L., Vivant A.-L., Piveteau P. (2012). Listeria monocytogenes differential transcriptome analysis reveals temperature-dependent Agr regulation and suggests overlaps with other regulons. PLoS ONE 7:e43154 10.1371/journal.pone.0043154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray B., Hall P., Gresham H. (2013). Targeting agr- and agr-Like quorum sensing systems for development of common therapeutics to treat multiple gram-positive bacterial infections. Sensors (Basel) 13 5130–5166. 10.3390/s130405130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guariglia-Oropeza V., Orsi R. H., Yu H., Boor K. J., Wiedmann M., Guldimann C. (2014). Regulatory network features in Listeria monocytogenes-changing the way we talk. Front. Cell. Infect. Microbiol. 4:14 10.3389/fcimb.2014.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G., Beavis R., Novick R. P. (1997). Bacterial interference caused by autoinducing peptide variants. Science 276 2027–2030. 10.1126/science.276.5321.2027 [DOI] [PubMed] [Google Scholar]
- Kalkum M., Lyon G. J., Chait B. T. (2003). Detection of secreted peptides by using hypothesis-driven multistage mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 100 2795–2800. 10.1073/pnas.0436605100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer P., Chow M. Y. N., Loessner M. J., Portnoy D. A., Calendar R. (2002). Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184 4177–4186. 10.1128/JB.184.15.4177-4186.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P., Repoila F., Vergassola M., Geissmann T., Cossart P. (2007). Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 35 962–974. 10.1093/nar/gkl1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath S., Fitzgerald G. F., van Sinderen D. (2001). Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl. Environ. Microbiol. 67 608–616. 10.1128/AEM.67.2.608-616.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin J. R., Cossart P. (2012). The non-coding RNA world of the bacterial pathogen Listeria monocytogenes. RNA Biol. 9 372–378. 10.4161/rna.19235 [DOI] [PubMed] [Google Scholar]
- Monk I. R., Gahan C. G. M., Hill C. (2008). Tools for functional postgenomic analysis of Listeria monocytogenes. Appl. Environ. Microbiol. 74 3921–3934. 10.1128/AEM.00314-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Cao Y., Horii T., Sakuda S., Akkermans A. D., de Vos W. M., et al. (2001). Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41 145–154. 10.1046/j.1365-2958.2001.02486.x [DOI] [PubMed] [Google Scholar]
- Novick R. P., Geisinger E. (2008). Quorum sensing in staphylococci. Annu. Rev. Genet. 42 541–564. 10.1146/annurev.genet.42.110807.091640 [DOI] [PubMed] [Google Scholar]
- Olson M. E., Todd D. A., Schaeffer C. R., Paharik A. E., Van Dyke M. J., Büttner H., et al. (2014). Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J. Bacteriol. 196 3482–3493. 10.1128/JB.01882-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M., Süssmuth R., Jung G., Götz F. (1998). Structure of the pheromone peptide of the Staphylococcus epidermidis agr system. FEBS Lett. 424 89–94. 10.1016/S0014-5793(98)00145-8 [DOI] [PubMed] [Google Scholar]
- Paliy O., Gunasekera T. S. (2007). Growth of E. coli BL21 in minimal media with different gluconeogenic carbon sources and salt contents. Appl. Microbiol. Biotechnol. 73 1169–1172. 10.1007/s00253-006-0554-8 [DOI] [PubMed] [Google Scholar]
- Qin X., Singh K. V., Weinstock G. M., Murray B. E. (2001). Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183 3372–3382. 10.1128/JB.183.11.3372-3382.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X.-Q., Wang H., Lu X.-F., Zhang J., Li S.-F., Cheng G., et al. (2003). An engineered multidomain bactericidal peptide as a model for targeted antibiotics against specific bacteria. Nat. Biotechnol. 21 1480–1485. 10.1038/nbt913 [DOI] [PubMed] [Google Scholar]
- Queck S. Y., Jameson-Lee M., Villaruz A. E., Bach T.-H. L., Khan B. A., Sturdevant D. E., et al. (2008). RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32 150–158. 10.1016/j.molcel.2008.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C. U., Monk I. R., Casey P. G., Waidmann M. S., Gahan C. G. M., Hill C. (2009). AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in Listeria monocytogenes. Mol. Microbiol. 71 1177–1189. 10.1111/j.1365-2958.2008.06589.x [DOI] [PubMed] [Google Scholar]
- Rieu A., Weidmann S., Garmyn D., Piveteau P., Guzzo J. (2007). Agr system of Listeria monocytogenes EGD-e: role in adherence and differential expression pattern. Appl. Environ. Microbiol. 73 6125–6133. 10.1128/AEM.00608-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E., Scott J., Minton N. P., Winzer K. (2012). An agr quorum sensing system that regulates granulose formation and sporulation in Clostridium acetobutylicum. Appl. Environ. Microbiol. 78 1113–1122. 10.1128/AEM.06376-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturme M. H. J., Nakayama J., Molenaar D., Murakami Y., Kunugi R., Fujii T., et al. (2005). An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J. Bacteriol. 187 5224–5235. 10.1128/JB.187.15.5224-5235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoendel M., Horswill A. R. (2010). Biosynthesis of peptide signals in gram-positive bacteria. Adv. Appl. Microbiol. 71 91–112. 10.1016/S0065-2164(10)71004-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoendel M., Kavanaugh J. S., Flack C. E., Horswill A. R. (2011). Peptide signaling in the staphylococci. Chem. Rev. 111 117–151. 10.1021/cr100370n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459 950–956. 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- Vivant A.-L., Garmyn D., Gal L., Hartmann A., Piveteau P. (2015). Survival of Listeria monocytogenes in soil requires agra-mediated regulation. Appl. Environ. Microbiol. 81 5073–5084. 10.1128/AEM.04134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivant A.-L., Garmyn D., Piveteau P. (2013). Listeria monocytogenes, a down-to-earth pathogen. Front. Cell. Infect. Microbiol. 3:87 10.3389/fcimb.2013.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C., Saenz H. L., Götz F., Otto M. (2000). Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182 1688–1693. 10.1086/317606 [DOI] [PubMed] [Google Scholar]
- Williams T., Bauer S., Beier D., Kuhn M. (2005). Construction and characterization of Listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infect. Immun. 73 3152–3159. 10.1128/IAI.73.5.3152-3159.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O., Sesto N., Mellin J. R., Karunker I., Edelheit S., Bécavin C., et al. (2012). Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol. 8 583 10.1038/msb.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuster A., Babu M. M. (2008). Conservation and evolutionary dynamics of the agr cell-to-cell communication system across firmicutes. J. Bacteriol. 190 743–746. 10.1128/JB.01135-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


