Abstract
The RNA modification N6-methyladenosine (m6A) influences mRNA stability and cell-type-specific developmental programming, and is highly abundant in the adult brain. However, it has not been determined whether m6A is dynamically regulated by experience. Based on transcriptome-wide profiling of m6A, we report that the level of m6A increases in the medial prefrontal cortex (mPFC) of mice in response to behavioral experience. The modulation was enriched near the stop codon of mRNAs, including genes related to neuronal plasticity. In primary cortical neurons, in vitro, modulation of m6A by the RNA demethylase FTO influenced the degradation profiles of a subset of transcripts with modulated sites. In vivo, the expression of Fto and the m6A methyltransferase, Mettl3 correlated with the observed increase in m6A levels post-training. Furthermore, targeted knockdown of FTO in the mPFC led to enhanced consolidation of cued fear memory. Thus, together with its role in early development, the dynamic regulation of m6A in the adult brain serves as an important epitranscriptomic mechanism associated with behavioral adaptation.
SIGNIFICANCE STATEMENT N6-methyladenosine (m6A) is the most prevalent internal modification on RNA, however, its cellular dynamics in vivo remains elusive. Here we provide the first demonstration of m6A upregulation in the mouse medial prefrontal cortex (mPFC) following behavioral training. Knocking down the m6A demethylase FTO in the mPFC, which increases total m6A level, results in enhanced consolidation of fear memory. Our findings suggest that m6A is regulated in an activity-dependent manner in the adult brain, and may function to fine-tune mRNA turnover during memory-related processes.
Keywords: epigenetics, memory, mRNA stability, RNA methylation, synaptic plasticity, transcription
Introduction
The RNA modification N6-methyladenosine (m6A) has recently been shown to be highly abundant throughout the mammalian transcriptome, and appears to be involved in a variety of biological processes (Dominissini et al., 2012; Meyer et al., 2012; Fu et al., 2014). The discovery of RNA methyltransferases and demethylases highlights the dynamic and reversible nature of this epitranscriptomic mechanism. Despite this fact, little evidence exists to demonstrate whether m6A is influenced by experience. In the mouse brain, m6A is developmentally regulated and increases in adulthood (Meyer et al., 2012), suggesting a potential role in the post-transcriptional regulation associated with neural plasticity and behavioral adaptation. However, whether m6A can be dynamically regulated by experience and how it contributes to the regulation of activity-related gene expression remain equivocal.
It is widely accepted that the medial prefrontal cortex (mPFC) is actively engaged during the encoding of fear memory, and that synaptic plasticity-related transcriptional programming accompanies this process (Bero et al., 2014). To investigate the possible role of m6A as an epitranscriptomic mechanism within the mPFC, we performed transcriptome-wide mapping of m6A by methylated RNA immunoprecipitation (MeRIP-seq) in tissue derived from behaviorally trained mice. Moreover, we tested the functional significance of m6A in the adult brain by targeted knockdown of the RNA demethylase FTO in an associative fear-conditioning paradigm. We report an unprecedented increase in the locus-specific accumulation of m6A in response to behavioral training, and demonstrate that m6A signaling in the mPFC is involved in the consolidation of fear-related memory.
Materials and Methods
Mice
Adult male C57BL/6 mice (8–12 weeks old) were used for all experiments. Mice were housed on a 12 h light/dark schedule, and fed ad libitum. All testing was conducted during the light phase in red light-illuminated rooms following protocols approved by The University of Queensland Animal Ethics Committee.
DNA constructs and antibodies
The FTO shRNA construct was generated by inserting the Fto targeting sequence, 5′-AGATCGCCGCTGCATGTCA-3′ into the FG12 plasmid (Li et al., 2014). Fto was cloned from the mouse brain cDNA library into the pRK5-myc and FUW-myc plasmids (Anggono et al., 2013). FTO (5-2H10), tubulin (D65A4), GFP (JM-3999), and m6A (202-003) antibodies were purchased from Abcam, Cell Signaling Technology, MBL International, and Synaptic Systems, respectively.
Neuronal culture
Cortical neurons from embryonic day 17 mice were prepared and maintained in neurobasal medium containing 2% B27, 2 mm Glutamax, 50 U/ml penicillin, 50 μg/ml streptomycin, and 1% fetal bovine serum as described previously (Widagdo et al., 2015). Neurons were transduced with lentivirus at 8 days in vitro (DIV) and harvested for experiments at 13 DIV. Electroporation of plasmids was performed using the Amaxa Nucleofector system (Lonza) at 0 DIV (Widagdo et al., 2015).
Reverse transcription, real-time qPCR and mRNA decay analysis
RNA was reverse-transcribed using the QuantiTect kit (Qiagen) for gene-expression analysis or SuperScript II (Invitrogen) for MeRIP-quantitative PCR (qPCR) analysis. Enrichment of m6A targets was calculated relative to the input sample after normalizing to its Gapdh value. qPCR assay using Sybr green was performed on a RotorGeneQ (Qiagen). For messenger RNA (mRNA) decay analysis, 13 DIV neurons expressing either GFP alone (FG12 control), FTO shRNA or FTO cDNA (by electroporation or lentiviral transduction) were treated with 10 μg/ml of actinomycin-D for 15 min, followed by a 30 min incubation in 50 mm KCl-containing medium before being placed back in actinomycin D-containing medium for 0, 30, 60 or 180 min. Data were analyzed and fit into a one-phase exponential decay function (except for Rab33b, Arhgap39, Arhgef17 and Gria1 from the FTO overexpression datasets, which were fit with a sigmoid function) in GraphPad Prism, from which the time-constants (τ) were derived.
Lentiviral-mediated knockdown of FTO in the mPFC
Lentivirus was prepared as previously described (Anggono et al., 2013). The cannulation procedure has been described previously (Li et al., 2014). Double cannulae (PlasticsOne) were implanted into the mPFC (relative to bregma: 1.8 mm anterior–posterior, −1.75 mm dorsal–ventral). Virus was infused via two injections (total 2 μl) delivered at a rate of 0.1 μl/min within 48 h. Mice were returned to their home cage for at least 7 d before the start of the behavioral assays.
Behavioral assays
Cued-fear conditioning.
The training session started with a 2 min acclimation period, followed by pairings of a white noise conditioned stimulus (CS; 2 min, 80 dB) co-terminated with footshocks as the unconditioned stimulus (US; 1 s, 0.7 mA), with a 2 min intertrial interval as described by Li et al. (2014). Mice were returned to their home cage after 2 min. Training performed for sequencing experiments involved six pairings of the CS-US. The CS-only group received identical cues in the conditioning box without footshocks. For behavioral analysis, mice were given 3 CS-US pairings during the training session, followed by a memory recall test 24 h later in the same conditioning chambers with two 2 min CS presentations. The freezing scores during the first CS were taken as the measurement of fear memory recall. Animals with an off-target injection site or those that acquired <20% freezing score during the last CS (CS3) in the initial training were excluded from the analysis as they may be consequence of potential damage during the cannulation.
Open-field.
Individual mice were placed in a Plexiglas chamber (ENV-510, Med Associates) housed in a wooden box with a light intensity of 200 lux for 10 min. The amount of time spent in the center zone and the total distance traveled were computed automatically using activity monitoring software (Med Associates).
Elevated plus maze.
The day after the open-field test, mice were subjected to the elevated plus maze for a 10 min session. Data were analyzed with Ethovision XT 9 (SDR Scientific).
MeRIP-seq
The MeRIP procedure was performed according to Dominissini et al. (2012) with slight modifications. Briefly, the mPFC was dissected from the mice 2 h after behavioral training. Total RNA with DNase-treatment was extracted using the Direct-zol RNA mini-prep kit (Zymo). Eighteen micrograms of RNA were fragmented to ∼100 nt using ZnCl2, followed by ethanol precipitation. Before immunoprecipitation (IP), fragmented RNA was denatured at 75°C for 5 min, chilled on ice for 5 min, and then resuspended in 500 μl IP buffer containing the following (in mm): 150 NaCl, 10 Tris-HCl pH 7.4, 100 ribonuclease-vanadyl complex, 0.1% Igepal CA-630, and 200U RNAseOUT). All steps were performed at 4°C unless otherwise stated. Ten percent of the sample was saved as the input RNA fraction and the remainder was subjected to two rounds of IP, each with 3 μg of rabbit anti-m6A antibody pre-coupled to 30 μl of sheep anti-rabbit Dynabeads (Invitrogen) in IP buffer for 1 h. IP beads were combined and washed four times with IP buffer. Bound RNA was eluted in 300 μl elution buffer (5 mm Tris-HCl pH 7.5, 1 mm EDTA, 0.05% SDS) and 4.2 μl proteinase K (20 mg/ml), followed by Trizol (Invitrogen) extraction.
Libraries were generated using the Scriptseq v2 RNA-Seq kit (Epicenter) according to the manufacturer's ultra-low input protocol. Ribosomal RNA (rRNA) was depleted from input RNA with Ribo-Zero (Epicenter). Fifteen cycles of PCR were performed on the cDNA, each with unique index primers. Equal amounts of each library were pooled and subjected to AMPure XP (Beckman Coulter) purification to remove primer-dimers. The final libraries (size distribution from 150 to 400 bp, with the peak at ∼220 bp) were run on a single-flow cell on HiSeq 2000 (Illumina) for paired-end 100 bp sequencing. Standard Illumina Genome Analyzer software (Casava v1.8.2) and pipelines developed in-house were used for image processing and sequence extraction.
MeRIP-seq data analyses
Data access.
Sequencing data are accessible through the NCBI Sequence Read Archive with accession ID SRX982224.
Quality control of sequencing reads.
Cutadapt (https://pypi.python.org/pypi/cutadapt/) was used to trim off low-quality nucleotides (Phred quality <20) and Illuminaadaptor sequences at the 3′ end. The total coverage for all libraries ranges between 30 and 40 million reads. All reads were then aligned against a custom contaminant list to further filter reads mapped to rRNA or Phi X genomes using Bowtie2 (2.1.0; Langmead and Salzberg, 2012) and custom PERL scripts.
Alignment of quality filtered reads.
Tophat2 (2.0.10; Kim et al., 2013) was used to align the filtered paired-end reads to the mouse genome (mm10). Samtools (0.1.17; Li and Durbin, 2009) was used to keep the reads that were concordantly and uniquely aligned for downstream analyses. Duplicate paired-end reads were marked using Picard tools (1.72) and removed using Samtools, resulting in distinct paired-end reads, each of which was mapped to a unique location in the mm10 reference genome.
Detection of m6A sites and differential analysis.
Similar to a method described previously (Dominissini et al., 2012), analyses were limited to genes with at least 40 reads per 1000 nt in both replicates per group. Sliding windows of 100 nt with 50 nt overlap were scanned for each gene to search for putative peaks enriched in the IP sample compared with the input control using the winscore calculation. The winscore threshold of 2 was set for both biological replicates, and the corresponding false discovery rate was <2% (1.62% for the naive group, 1.99% for the CS-only group, and 0.46% for the fear-conditioned group). For the differential analysis, all putative m6A sites identified in each group were combined. If two or more peaks were within 150 bp (the length of 2 overlapping windows), only the peak with the largest number of covered fragments was retained in all samples. The number of covered fragments for each putative m6A site was calculated and normalized according to the number of final processed aligned reads. A Student's t test was performed to identify the differentially methylated sites in pairwise comparisons, naive versus CS-only and naive versus fear-conditioned with an unadjusted p value <0.05 (Fig. 1C).
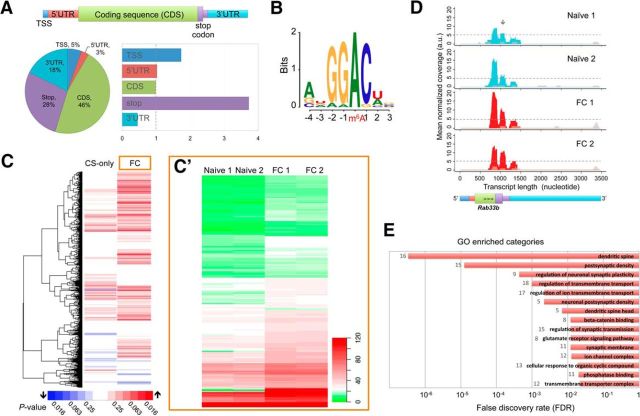
Figure 1.
Transcriptome-wide profiling reveals the dynamic landscape of m6A in the mouse mPFC following behavioral training. A, Distribution analysis of mRNA-associated m6A peaks identified in the mPFC shown as a percentage (pie chart) or as enrichment index after normalization to the relative length of each segment in the transcriptome (column graphs). B, Overall consensus motif associated with m6A peaks. C, Clustering and heat-map diagrams show the modulation of m6A peaks in the CS-only or fear-conditioned (FC) group relative to the naive group as the baseline control. Red and blue bars represent an increase and decrease, respectively (p values determined by Student's t test). C′, Heat-map diagram based on normalized count for 188 peaks identified to be differentially methylated in the FC versus naive comparison (p < 0.05). D, Representative MeRIP-seq traces of Rab33b showing consistent increase of m6A in a region near the stop codon in the FC mice (arrow). Cyan and red traces represent immunoprecipitated coverages, gray traces for the input coverages. E, Top 15 gene ontology categories associated with genes with modulated m6A peaks in the FC group (p < 0.05 or fold-change ≥1.5).
Analysis of m6A location.
Putative m6A sites were assigned to five non-overlapping transcript segments as described previously (Dominissini et al., 2012), according to the gene coordinates in the GTF file for mm10 from the Illumina iGenomes database.
Identification and clustering of enriched motifs.
The occurrences of each 4–6-nucleotide k-mer in the IPs and their corresponding input controls were counted based on a set of 51 bp windows centered on the putative m6A peaks or negative control peaks. For each k-mer length, the total number of k-mers of the same length were counted within each of the groups.
m6A immunoblotting
DNase-treated total RNA samples were run on a 1% agarose gel and transferred to a nylon membrane using NorthernMax-Gly kit (Invitrogen) as described previously (Meyer et al., 2012).
Results
m6A is dynamically regulated in the mPFC in an experience-dependent manner
MeRIP-seq analysis of RNA derived from the mPFC of naive, fear-conditioned, and conditioned stimulus-exposed (CS-only) mice revealed a total of 3936 putative m6A sites in the mouse transcriptome. These peaks were detected in 2650 genes, with a small proportion (∼1%) located in long noncoding RNAs, including the previously identified m6A targets Malat1, Neat1, and Miat (Meyer et al., 2012; Liu et al., 2015). The majority of the remaining peaks were detected in mRNAs (Fig. 1A), particularly within their coding sequence region (46%). However, following normalization to the average lengths of each mRNA segment in the transcriptome, the highest accumulation of m6A was found in the vicinity of the stop codon. This distribution pattern was relatively similar in all three groups. Sequence motif analyses also revealed the enrichment of the m6A consensus motif RRACH (R = G/A, H = A/C/U) in all samples, either individually or as a whole (Fig. 1B). Using naive mice as the baseline control, pairwise comparisons revealed increased intensities in a large proportion of m6A peaks post-behavioral training (Fig. 1C), particularly in fear-conditioned mice (Fig. 1C′). Using fold-change as a measurement, 256 peaks (248 genes) were consistently upregulated by at least 1.5-fold in both biological replicates. These modulated m6A sites were also highly enriched around the stop codon as exemplified by Rab33b, a member of the Rab family of GTPases, which has been implicated in neuronal trafficking (Cosker and Segal, 2014; Fig. 1D). Pathway analysis of these genes further revealed functional enrichment for dendritic and postsynaptic regulation, synaptic transmission, and transmembrane transport regulation (Fig. 1E).
Activity-induced m6A is associated with the regulation of mRNA stability
To elucidate the functional relevance of activity-induced m6A in neurons, we adopted a primary cortical neuronal culture system under KCl-induced depolarizing conditions. KCl treatment at 50 mm did not modulate the overall m6A level in cortical neurons (data not shown). Locus-specific m6A modulation identified in the MeRIP-Seq analysis was then investigated using specific primers by MeRIP-qPCR assay. Stimulation of cultured neurons recapitulated the observed increase in the deposition of m6A on several plasticity-related genes identified by in vivo MeRIP-seq (Fig. 2A). The locus specificity was confirmed using primers against the negative non-m6A target region, which did not respond to KCl stimulation. Considering that the majority of the modulated m6A peaks reside in the vicinity of the stop codon, we next used these gene candidates to interrogate the regulatory effect of m6A on the mRNA stability in activated cortical neurons. To modulate the level of m6A, we knocked down or overexpressed the RNA demethylating enzyme FTO in neurons (Fig. 2B). Time course analyses of individual transcripts following transcriptional inhibition by actinomycin-D revealed differences in their degradation profiles in control, FTO knockdown, and/or FTO-overexpressing neurons (Fig. 2C). The decay rates of most of the examined transcripts appeared to be higher in overexpressing neurons and lower in knockdown cells (Rab33b, Ahrgap39, Arhgef17) as measured by tau (τ). However, the opposite increased τ was found for Crtc1 in knockdown neurons (Fig. 2D) suggesting that perturbation of FTO may involve a complex mechanism that leads to changes in the mRNA degradation rate depending on the status of the mRNA being targeted. It is also apparent that m6A mediates other RNA regulatory functions, as no changes in mRNA stability were detected in other genes, including members of the immediate early gene family, Egr1 and Arc (Fig. 2E).
Figure 2.
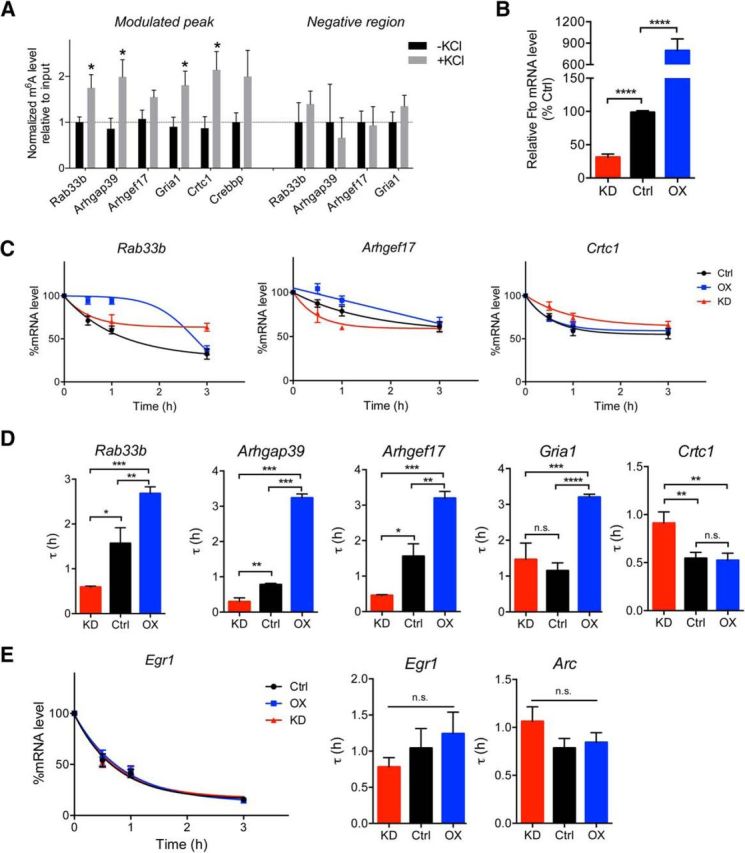
Activity-dependent regulation of locus-specific m6A in cultured neurons is associated with mRNA stability. A, MeRIP-qPCR analyses of KCl-depolarized neurons (50 mm, 30 min) showing modulation of locus-specific m6A (n = 3–10; Mann–Whitney U test; *p < 0.05 vs −KCl control). B, qPCR analysis of Fto in neurons transduced with FG12-FTO shRNA [knockdown (KD)], FG12 control, and FUW-FTO [overexpression (OX); n = 8–14; Mann–Whitney U test; ****p < 0.0001]. C, Representative plots showing mRNA decay profiles of Rab33b, Arhgef17, and Crtc1 in actinomycin-D pulse-chase experiments. D, mRNA decay time constant, τ for each transcript (n = 4–14, one-way ANOVA; *p < 0.05, **p < 0.01, ***p < 0.001). E, Decay plots for Egr1 and the calculated τ for Egr1 and Arc (n.s., not significant). Data represent mean ± SEM.
FTO knockdown in the mPFC promotes cued fear memory retention
Expression analysis of members of the m6A biogenesis pathway revealed a positive correlation between the levels of m6A and changes in the expression of the methyltransferase Mettl3 and Fto in the mPFC following novel context-cue exposure and fear conditioning, respectively (Fig. 3A). We then investigated whether m6A plays a role in mPFC-dependent cued fear learning by knocking down FTO. In cultured neurons, the expression of FTO was reduced, accompanied by an increase in the total m6A level, when transfected with a specific shRNA against FTO (Fig. 3B,C). Injection of FTO shRNA lentivirus into the mPFC (cingulate and prelimbic cortices) led to decreased FTO levels and a concomitant increase in global m6A levels (Fig. 3D–F). To investigate the effect of FTO knockdown in the mPFC on memory processes, cued fear conditioning was performed. The acquisition of fear acquisition was comparable in both groups (Fig. 3G); however, FTO knockdown led to a significant increase in fear memory when tested 24 h after training (Fig. 3H). This effect was not due to any detectable changes in locomotor activity or anxiety (Fig. 3I,J). Together, these data demonstrate the functional role of FTO, and by extension the accumulation of m6A, in modulating the consolidation of associative fear memory in mice.
Figure 3.
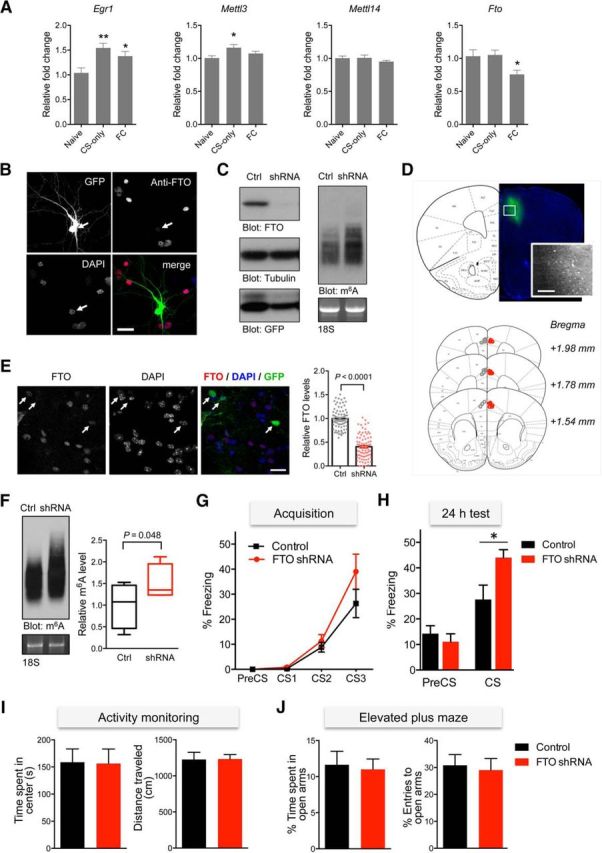
Targeted knockdown of FTO in the mPFC leads to enhanced fear retention. A, qPCR analyses of Egr1, Mettl3, Mettl14, and Fto in the mPFC following context-exposure (CS-only) or a fear conditioning (FC) paradigm (n = 6–8, one-way ANOVA with Dunnett's test relative to the naive controls, *p < 0.05, **p < 0.01). B, Cortical neurons were transfected with FG12-FTO shRNA, fixed and stained with anti-FTO antibody and DAPI. GFP expression indicates transfected neurons. Scale bar, 25 μm. C, Western blot (left) and m6A immunoblot (right) analyses of cortical neurons transduced with lentivirus particles expressing GFP control (Ctrl) or FTO shRNA. D, Representative image of mouse brain following lentiviral injections into the cingulate (Cgl) and prelimbic (PrL) cortical regions (placement for the control lentivirus indicated in gray and FTO shRNA in red). Scale bar, 200 μm. E, Immunohistochemical analysis of the mPFC region using anti-FTO antibody showing levels of FTO immunoreactivity in shRNA-expressing (GFP-positive) and GFP-negative control cells (n = 80 cells per group from 8 independent slices; Mann–Whitney U test; ****p < 0.0001). Scale bar, 20 μm. F, Representative m6A immunoblot analysis of RNA extracted from the mPFC tissues of mice injected with FG12 control or FTO shRNA lentivirus (n = 4; t test, p < 0.05). G, Cued fear conditioning revealed similar acquisition of fear between groups (control, n = 7; FTO shRNA, n = 8; two-way ANOVA; p = 0.1711). H, The freezing level measured in the same context 24 h after training was significantly higher in the knockdown group (one-way ANOVA with Tukey's post hoc analysis; in CS: control vs FTO shRNA; *p < 0.05). I, An open-field test revealed a comparable duration spent in the center area (left) and total distance traveled (right) by both groups (control, n = 14; FTO shRNA, n = 15). J, An elevated plus maze analysis revealed no differences in the percentage of time spent in (left) or the number of entries (right) to the open arms. Data represent mean ± SEM.
Discussion
Our findings highlight the dynamic, experience-dependent nature of m6A in the mPFC. An increase in the deposition of m6A on mRNA was evident following behavioral training. Observations made by others have suggested that the m6A landscape in mammalian cells may be relatively static upon exposure to various stimuli (Dominissini et al., 2012). However, its widespread upregulation in response to behavioral experience is strongly suggestive of an important role for m6A in regulating experience-dependent plasticity in the mPFC. In agreement with this idea, we demonstrated that FTO knockdown led to enhanced memory consolidation.
The activity-dependent nature of m6A in mammalian cells has only recently been reported by Zhou et al. (2015) in response to heat shock response in fibroblast and epithelial cells. In contrast to our findings, preferential upregulation of m6A was identified within the 5′ UTR in the majority of transcripts post-heat shock, suggesting that differential signaling pathways may regulate the functional influence of m6A in response to distinct stimuli. Consistent with the general observation of an effect of m6A on mRNA stability (Wang et al., 2014a,b; Liu et al., 2015), our data also reveal slower degradation rates for a subset of transcripts following FTO overexpression, although this does not apply to all the transcripts that we analyzed. The differential effects of m6A may depend on distinct cis-acting elements that are present on the RNA molecule which may interact with m6A, including micro-RNAs (Alarcón et al., 2015; Chen et al., 2015).
Because of the versatility of m6A in regulating a plethora of RNA functions, the phenotypic consequence of FTO perturbation in mice is likely to be a combination of multiple regulatory effects exerted by m6A in the mPFC. Although the functional roles of m6A in alternative splicing, translational dynamics, and mRNA transport in vivo remain to be investigated, we postulate that one of the functions for the increase in m6A in response to learning is to constrain the sorting efficiencies or turnover of nascent mRNAs. Modification by m6A has been shown to direct its target mRNAs for both degradation and translation in the cytoplasm (Wang et al., 2015; Zhou et al., 2015). With FTO knockdown, the increased targeting of plasticity-related genes by m6A may allow them to be efficiently translated, then rapidly degraded to bring the mRNA pool down to a minimum, thereby reducing “transcription noise” in neurons (Siciliano et al., 2013).
Recent explorations into the functional implications of m6A regulation have relied on perturbation of m6A-mediating enzymes in biological systems. It is clear that molecular understanding by which FTO regulates memory consolidation will benefit from better characterization of FTO. In the current study, we observed a predominant nuclear localization of FTO (Fig. 3E) as widely reported; however, our biochemical fractionation assay also detected significant levels of FTO in the cytosolic fraction of mouse brain lysate (data not shown). Thus, this opens possibilities for FTO function outside the nucleus, either to induce localized RNA demethylation upon neuronal activation or to perform secondary functions that are independent of m6A. The latter possibility cannot currently be ruled out. Using a more systematic and sensitive approach, future studies should determine whether the FTO-mediated effect on memory consolidation occurs through direct m6A modulation on target RNAs, in vivo. Furthermore, targeting the m6A writer and its relevant readers in a temporally and spatially specific manner in the brain will also contribute toward a better understanding of the role of m6A in behavior and cognition.
Footnotes
This work was supported by grants from the National Institutes of Mental Health (1R01MH109588-01 to T.W.B.), Australian National Health and Medical Research Council (APP1042051 and APP1062570 to T.W.B.), the Australian Research Council (DP1096148 to T.W.B.), and the John T. Reid Charitable Trusts (to V.A.); a fellowship from the Australian National Health and Medical Research Council (477108 to V.A.); and postgraduate scholarships from the Australian Government and Queensland Brain Institute, The University of Queensland to P.A.S. We thank Dr. Danay Baker-Andresen for assisting with behavioral experiments and Rowan Tweedale for helpful editing of this paper.
The authors declare no competing financial interests.
References
- Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggono V, Koç-Schmitz Y, Widagdo J, Kormann J, Quan A, Chen CM, Robinson PJ, Choi SY, Linden DJ, Plomann M, Huganir RL. PICK1 interacts with PACSIN to regulate AMPA receptor internalization and cerebellar long-term depression. Proc Natl Acad Sci U S A. 2013;110:13976–13981. doi: 10.1073/pnas.1312467110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bero AW, Meng J, Cho S, Shen AH, Canter RG, Ericsson M, Tsai LH. Early remodeling of the neocortex upon episodic memory encoding. Proc Natl Acad Sci U S A. 2014;111:11852–11857. doi: 10.1073/pnas.1408378111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, Li A, Yang Y, Jin KX, Zhao X, Li Y, Ping XL, Lai WY, Wu LG, Jiang G, Wang HL, et al. m6A RNA methylation is regulated by micro RNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- Cosker KE, Segal RA. Neuronal signaling through endocytosis. Cold Spring Harb Perspect Biol. 2014;6:a020669. doi: 10.1101/cshperspect.a020669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wei W, Zhao QY, Widagdo J, Baker-Andresen D, Flavell CR, D'Alessio A, Zhang Y, Bredy TW. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc Natl Acad Sci U S A. 2014;111:7120–7125. doi: 10.1073/pnas.1318906111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano V, Garzilli I, Fracassi C, Criscuolo S, Ventre S, di Bernardo D. MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat Commun. 2013;4:2364. doi: 10.1038/ncomms3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014a;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014b;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo J, Chai YJ, Ridder MC, Chau YQ, Johnson RC, Sah P, Huganir RL, Anggono V. Activity-dependent ubiquitination of GluA1 and GluA2 regulates AMPA receptor intracellular sorting and degradation. Cell Rep. 2015;10:783–795. doi: 10.1016/j.celrep.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]