Abstract
C9orf72 is the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) in Caucasian populations. However, the relationship between C9orf72 repeats and Alzheimer's disease (AD) was not clear. Additionally, there were few articles assessing C9orf72 in other ethnicities with ALS. In this meta-analysis, we aimed to investigate the relationship between C9orf72 repeat expansions (≥30 repeats) and intermediate repeat copies (20–29 repeats) and AD or ALS. The results suggested positive correlations between C9orf72 repeat expansions and the risk of Alzheimer's disease (OR = 6.36, 95% CI = 3.13–12.92, and p < 0.00001), while intermediate repeat copies of C9orf72 gene were not associated with the risk of the disease. C9orf72 repeat expansions were positively correlated with the risk of familial and sporadic ALS (OR = 293.25, 95% CI = 148.17–580.38, and p < 0.00001; OR = 35.57, 95% CI = 19.61–64.51, and p < 0.00001). There was a positive correlation between the gene variations and ALS risk among Caucasians and Asians (OR = 57.56, 95% CI = 36.73–90.22, and p < 0.00001; OR = 6.35, 95% CI = 1.39–29.02, and p = 0.02).
1. Introduction
In 2006, two researchers reported a locus on the short arm of chromosome 9 which could be a causative for FTD, ALS, and ALS/FTD [1]. In 2011, hexanucleotide (GGGGCC) repeat expansions in the noncoding region of C9orf72 were confirmed to be the most common mutation of FTD, ALS, and ALS/FTD [2]. The frequencies of the mutation account for 29%, 50%, and 88% of the patients, respectively [3].
In human genome, C9orf72 gene is located on chromosome 9p21, spanning 27,546,543–27,573,864 base pairs. It encodes 11 exons and GGGGCC (G4C2) exists between noncoding exons 1a and 1b [4]. Some researchers suggested that G4C2 ≥ 30 repeats were pathological repeat expansions, while <20 repeat units were normal [5]. In neurodegenerative diseases such as ALS-FTD, the G4C2 repeat copies of C9orf72 could reach 700–1600 units [4]. In addition, some researchers demonstrated that G4C2 intermediate copies (20–29 repeats) could also contribute to the risk of neurodegenerative diseases [6]. The disease mechanisms of how C9orf72 expanded repeats lead to these diseases are still unknown. The possible hypotheses may be the loss of function of C9orf72 protein, the accumulation of toxic RNA foci, and the Repeat Associated Non-ATG Initiated Translation (RAN-Translation) [7–10].
Substantial clinical and pathological characteristics overlap among the common neurodegenerative diseases, FTD (frontotemporal dementia), ALS (amyotrophic lateral sclerosis), and AD (Alzheimer's disease). For example, ALS patients with C9orf72 repeat expansions can present with dementia which were common in FTD and AD patients [11, 12]. Tau positive pathology which is typical for AD can be found in FTD patients [13]. Patients presenting with behavior symptoms may have pathological features of AD. However, the relationship between C9orf72 and the risk of AD remains controversial. Kohli et al. and Beck et al. demonstrated that C9orf72 repeat expansions were risk factors for AD in the same year based on studies on large samples [13, 14]. However, Renton et al., Majounie et al., and some researchers stated that there were no associations between the gene mutation and the disease [5, 15].
C9orf72 repeat expansions vary strongly between different geographic regions. In countries such as Italy, United States, and Germany, the prevalence can be as high as 47% for familial ALS and 21% for sporadic ALS [2, 5, 16]. However, in some Asian countries such as China and Korea, the mutation cannot be found in ALS patients [17]. Although C9orf72 repeat expansions were considered pathological mutations of European ALS patients recently, the mutations were rare in Asians.
For better understanding the widening disease spectrum of C9orf72 repeat copies, we performed a meta-analysis to clarify the association between C9orf72 variations and AD or ALS.
2. Materials and Methods
2.1. Search Strategy
EMBASE, PubMed, and Cochrane databases were searched for the comprehensive literature (the search was conducted on May 20, 2015). We used search terms as follows: (“AD,” “Alzheime∗/$,” or “Alzheimer's disease”)/(“ALS,” “amyotrophic lateral sclerosis,” “motor neuron disease”) and (“C9orf72”). The languages of the articles were limited to Chinese and English. The strategies were made by two researchers (L. Shu and QY Sun). If there were any disputes, we consulted another researcher (JF Guo).
2.2. Selection of Studies
Studies were selected when they met the following criteria: (1) original articles (when there were secondary articles, we added the useful original articles from them); (2) association studies between C9orf72 and AD or ALS; (3) sufficient data to calculate odds ratio (OR) and 95% confidence interval (95% CI); (4) study types being observational studies such as case-control studies and cohort studies; and (5) languages being limited to Chinese and English.
Studies were excluded when the following existed: (1) replicated data (if there were overlapped samples, we chose the largest samples); (2) incomplete data (no control group); (3) study types being reviews, case series, letters, editorials, and so forth; and (4) not human subjects (animal experiments, cell experiments, etc.). The PRISMA checklist is shown in Supplementary Table 1 (see Supplementary Table 1 in Supplementary Material available online at http://dx.doi.org/10.1155/2016/5731734).
2.3. Quality Assessment
The Newcastle-Ottawa Scale (NOS) was used to assess the quality of the selected original articles. The scale included three aspects: (1) subjects selection (0–4 points); (2) comparability between groups (0–2 points); and (3) exposure (0–3 points). The total points range from 0 to 9 [18]. The studies scored higher than 5 were considered of “high quality.” The assessments were conducted by two reviewers (L. Shu and QY Sun). The third author was consulted when agreements cannot be reached.
2.4. Data Extraction
Basic data were extracted for the statistical analysis including first author, publication year, ethnicity, country, age at onset, ages of cases and controls, genotyping methods, number of cases and controls, number of cases and controls who carried C9orf72 repeat expansions or intermediate repeat copies, diagnostic criteria, and NOS scores. Data were selected by two researchers (L. Shu and QY Sun). The third author was involved to solve the disagreements.
2.5. Statistical Analysis
The correlation between C9orf72 and neurodegenerative diseases was analyzed by pooled OR with 95% CI. The heterogeneity among the included studies was estimated by Q-test or I 2 statistic. If the Q-test showed p value ≤ 0.1, the heterogeneity was considered significant. We chose fixed-effects model for statistical analysis with low heterogeneity while we chose random-effects model with moderate to high heterogeneity. Z test was conducted to measure the association between C9orf72 and neurodegenerative diseases. p value < 0.05 indicated statistically significant difference. Funnel plot was visually inspected to assess the possible publication bias [19]. Sensitivity analysis was conducted by sequentially removing one publication to evaluate the influence of single publication on the whole results. RevMan 5.2 software was used for all the statistical analyses and graphics.
3. Results
3.1. Association between C9orf72 and AD
3.1.1. Eligible Studies
After searching of PubMed, EMBASE, and Cochrane databases, 299 articles were identified. 89 duplicated datasets were removed and a total of 210 articles were reviewed by title and abstract. 190 articles did not meet the inclusion criteria and only 20 of them were reviewed for full-test assessment. Ten articles were excluded: 5 conference abstracts, 1 not peripheral blood test, and 4 without controls. Thus, there were 10 articles left for the final statistical analysis [13–16, 20–26] (the detailed flowchart was shown in Supplementary Figure 1).
3.1.2. Characteristics of Studies
The detailed characteristics of the included studies are listed in Supplementary Table 2. From the table, there were 10 studies which had complete data about C9orf72 expansions in both case and control groups and 3 studies about C9orf72 intermediate repeat copies. The publication years of the included studies ranged from 2012 to 2014. The diagnostic criteria of AD were according to NINCDS-ADRDA criteria (Supplementary Table 2).
3.1.3. Cumulative Analysis
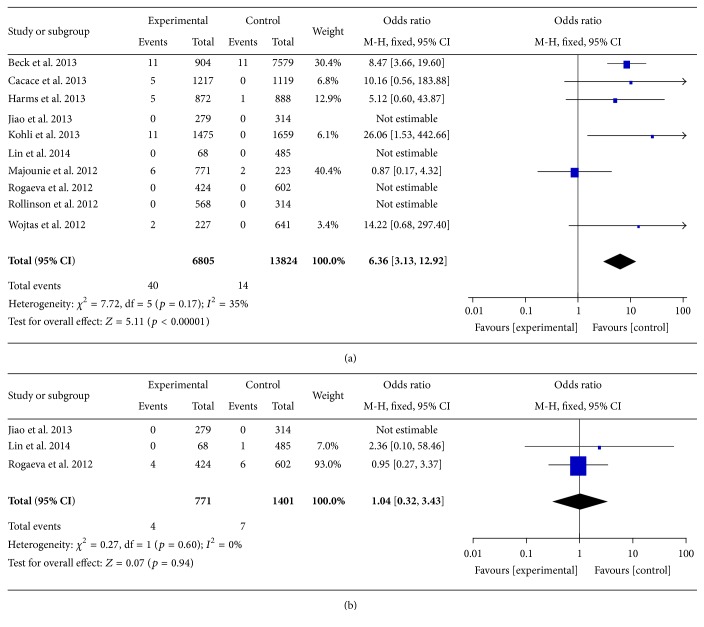
The results of the meta-analysis were present in Figures 1(a) and 1(b). There was no significant heterogeneity; thus a fixed-effects model was chosen for the meta-analysis. The results indicated that C9orf72 repeat expansions were related to the risk of AD (OR = 6.36, 95% CI = 3.13–12.92, and p < 0.00001, Figure 1(a)) while C9orf72 intermediate repeat copies were not correlated with the risk of AD (OR = 1.04, 95% CI = 0.32–3.43, and p = 0.94, Figure 1(b)).
Figure 1.
(a) Forest plot of the association between C9orf72 repeat expansions and AD. (b) Forest plot of the association between C9orf72 intermediate repeat copies and AD.
3.2. Association between C9orf72 and ALS
3.2.1. Eligible Studies
We conducted a comprehensive search on PubMed, EMBASE, and Cochrane databases. After the initial search, we got 1224 articles. 413 repeated datasets and 738 articles were excluded by abstracts and titles. 73 articles were left for full-test review. Articles were further excluded for the following reasons: incomplete date (6 studies), no controls (16 studies), other diseases (7 studies), and so forth. Finally, 24 articles were left for the cumulative analysis [5, 14, 16, 17, 20, 26–45] (details were shown in Supplementary Figure 2).
3.2.2. Characteristics of Studies
The details of the characteristics of the selected studies were present in Supplementary Table 3. There were 25 original articles included. Among them, 19 studies were conducted on Caucasians and 5 studies were performed on Asians. NOS scores of all the included studies are high except for three studies [5, 10, 33]. The publication years ranged from 2011 to 2014. The diagnostic criteria were according to El Escorial criteria or El Escorial revised criteria.
3.2.3. Cumulative Analysis
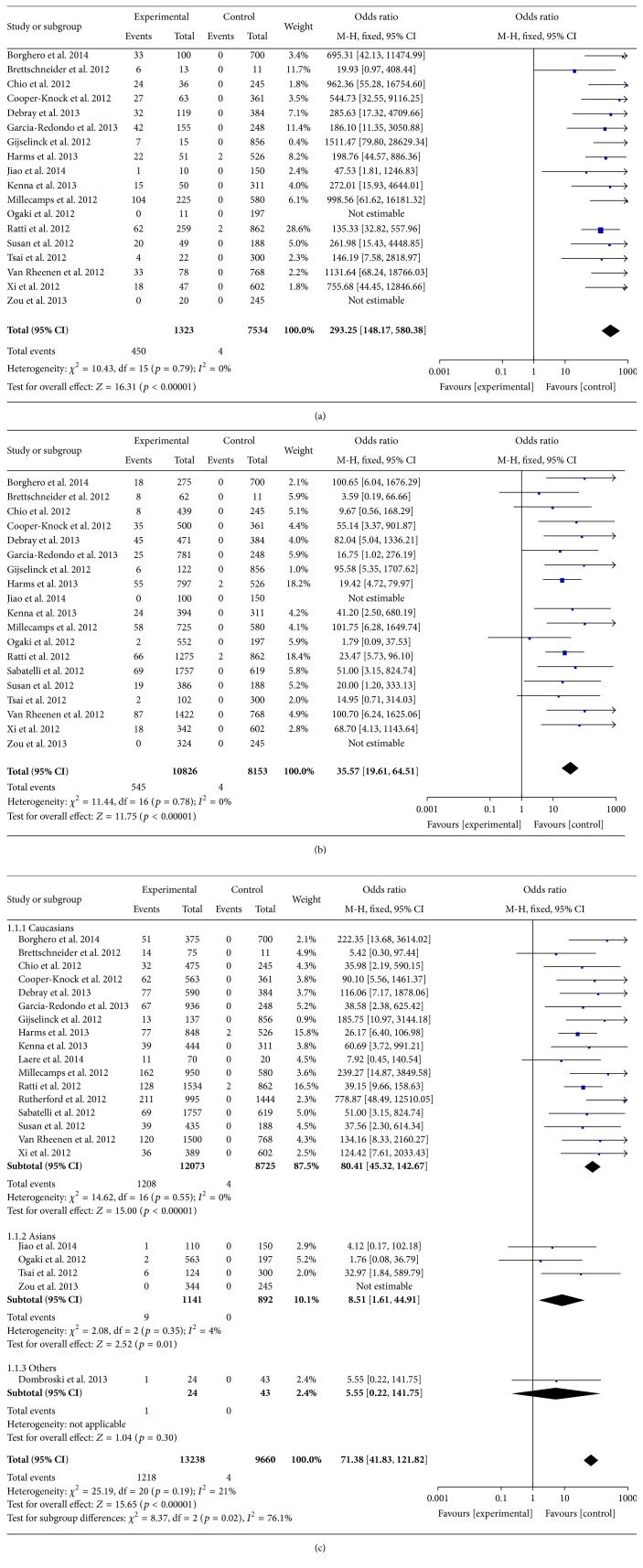
After excluding three studies with low NOS scores, we conducted the cumulative analyses with 22 articles. Of these 22 articles, 18 articles containing complete data about familial ALS and 19 articles containing full data about sporadic ALS were included for meta-analysis separately. Subgroup analyses including 17 articles about Caucasians and 4 articles about Asians were conducted.
The cumulative analyses suggested that C9orf72 repeat expansions were significantly correlated with the risk of ALS (OR = 71.38, 95% CI = 41.83–121.82, and p < 0.00001). C9orf72 repeat expansions were related to the risk of both familial and sporadic ALS (OR = 293.25, 95% CI = 148.17–580.38, and p < 0.00001; OR = 35.57, 95% CI = 19.61–64.51, and p < 0.00001). Subgroup analyses by ethnicity indicated that C9orf72 repeat expansions were risk factors for both Caucasian and Asian ALS patients (OR = 80.41, 95% CI = 45.32–142.67, and p < 0.00001; OR = 8.51, 95% CI = 1.61–44.91, and p = 0.01) (details were shown in Figure 2).
Figure 2.
(a) Forest plot of the association between C9orf72 repeat expansions and familial ALS. (b) Forest plot of the association between C9orf72 repeat expansions and sporadic ALS. (c) Forest plot of the association between C9orf72 repeat expansions and ALS (subgroup analysis).
4. Sensitivity Analysis
We performed sensitivity analyses by omitting single articles to test the stability of the results. After sequentially omitting single studies about ALS and AD, the total results were similar.
5. Publication Bias
There were no asymmetries of the funnel plot below and no significant publication biases of the meta-analysis about C9orf72 repeat expansions and AD or ALS (Supplementary Figure 3). We failed to do publication bias analysis on the association between C9orf72 intermediate copies and the risk of AD because of the scarcities of the included articles.
6. Discussion
There were clinical, pathological, and hereditary overlap between FTD and AD; therefore some researchers regarded them as the diseases of the same disease spectrum [46]. After discovering the most common genetic cause of FTD-C9orf72 repeat expansions, many researchers began to focus on the association between C9orf72 and AD. However, the disputes have not been solved yet. The results of our meta-analyses demonstrated that C9orf72 repeat expansions were positively correlated with the risk of AD while C9orf72 intermediate repeat copies were not related to the risk of AD. However, the limited number of the original articles about the association between C9orf72 intermediate repeat copies and AD influenced the validity of the analysis. Additionally, the diagnostic standards of the patients in the original articles were based on the clinical diagnosis.
C9orf72 has been discovered as the most common causative gene for ALS in white populations, which accounts for 40% of familial cases and 20% of sporadic cases in Finland [47]. While there were a large number of studies focusing on the identification of the gene in European countries, there were few studies reporting C9orf72 mutation in Asian ALS patients. Our meta-analysis selected high-quality studies and demonstrated that C9orf72 repeat expansions were related to the risk of ALS in Asians and Caucasians. We also proved that C9orf72 repeat expansions were not only correlated with the risk of familial ALS but also related to the risk of sporadic ALS. However, there were evidences that anticipation played a role in C9orf72 families. Gijselinck et al. demonstrated that there was a decreasing onset of age in younger generation in ALS families carrying longer G4C2 expansion. The original researches on the association between C9orf72 repeat expansions and ALS can be invalid because of the genetic anticipation and the wide range of age of onset of C9orf72 diseases [48]. Therefore, the different age of onset can cause bias in the meta-analysis of familial ALS. Additionally, there were few case-control studies about the relationship between C9orf72 repeat expansions or intermediate repeat copies and ALS in other populations. Further large sample studies are essential to clarify the association.
Despite the limitations stated above, there were some other limitations. First, the C9orf72 repeat expansions were analyzed by conventional PCR-based methods. When C9orf72 repeat copies were more than 60 copies, the accurate size cannot be gotten by conventional PCR-based methods. Other measures such as Southern blot were needed to detect the true repeat size [3]. Second, although our meta-analysis incorporated all the published case-control cohorts, some negative unpublished results were possibly neglected.
In conclusion, C9orf72 repeat expansions were risk factors for AD while C9orf72 intermediate repeat copies were not associated with the risk of AD. C9orf72 repeat expansions were correlated with the risk of ALS. However, inevitable limitations existed in our meta-analysis such as the limited number of original articles and possible publication biases. Further assessments were needed with enough case-control samples.
Supplementary Material
The flowchart of the selection of studies, the funnel plot and the detailed characteristics of the included studies are listed in the supplementary materials.
Acknowledgments
This work was supported by the State Key Program of National Natural Science Foundation of China (Grant no. 81430023), the Funds for International Cooperation and Exchange of the National Natural Science Foundation of China (Grant no. 81361120404), and the Major Research plan of the National Natural Science Foundation of China (Grant no. 91132000).
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Vance C., Al-Chalabi A., Ruddy D., et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2-21.3. Brain. 2006;129(4):868–876. doi: 10.1093/brain/awl030. [DOI] [PubMed] [Google Scholar]
- 2.DeJesus-Hernandez M., Mackenzie I. R., Boeve B. F., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iguchi Y., Katsuno M., Ikenaka K., Ishigaki S., Sobue G. Amyotrophic lateral sclerosis: an update on recent genetic insights. Journal of Neurology. 2013;260(11):2917–2927. doi: 10.1007/s00415-013-7112-y. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y., Yu J.-T., Zong Y., Zhou J., Tan L. C9ORF72 mutations in neurodegenerative diseases. Molecular Neurobiology. 2014;49(1):386–398. doi: 10.1007/s12035-013-8528-1. [DOI] [PubMed] [Google Scholar]
- 5.Renton A. E., Majounie E., Waite A., et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nuytemans K., Bademci G., Kohli M. M., et al. C9orf72 intermediate repeat copies are a significant risk factor for Parkinson disease. Annals of Human Genetics. 2013;77(5):351–363. doi: 10.1111/ahg.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ash P. E. A., Bieniek K. F., Gendron T. F., et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freibaum B. D., Lu Y., Lopez-Gonzalez R., et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525(7567):129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang K., Donnelly C. J., Haeusler A. R., et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525(7567):56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zu T., Liu Y., Bañez-Coronel M., et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(51):E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith B. N., Newhouse S., Shatunov A., et al. The C9ORF72 expansion mutation is a common cause of ALS+/-FTD in Europe and has a single founder. European Journal of Human Genetics. 2013;21(1):102–108. doi: 10.1038/ejhg.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart H., Rutherford N. J., Briemberg H., et al. Clinical and pathological features of amyotrophic lateral sclerosis caused by mutation in the C9ORF72 gene on chromosome 9p. Acta Neuropathologica. 2012;123(3):409–417. doi: 10.1007/s00401-011-0937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohli M. A., John-Williams K., Rajbhandary R., et al. Repeat expansions in the C9ORF72 gene contribute to Alzheimer's disease in Caucasians. Neurobiology of Aging. 2013;34(5):1519.e5–1519.e12. doi: 10.1016/j.neurobiolaging.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beck J., Poulter M., Hensman D., et al. Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. The American Journal of Human Genetics. 2013;92(3):345–353. doi: 10.1016/j.ajhg.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majounie E., Abramzon Y., Renton A. E., et al. Repeat expansion in C9ORF72 in Alzheimer's disease. The New England Journal of Medicine. 2012;366(3):283–284. doi: 10.1056/nejmc1113592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gijselinck I., Van Langenhove T., van der Zee J., et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. The Lancet Neurology. 2012;11(1):54–65. doi: 10.1016/s1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 17.Zou Z.-Y., Li X.-G., Liu M.-S., Cui L.-Y. Screening for C9orf72 repeat expansions in Chinese amyotrophic lateral sclerosis patients. Neurobiology of Aging. 2013;34(6):1710.e5–1710.e6. doi: 10.1016/j.neurobiolaging.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 18.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European Journal of Epidemiology. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 19.Yang J.-P., Hyun M.-H., Yoon J.-M., Park M.-J., Kim D., Park S. Association between TNF-α-308 G/A gene polymorphism and gastric cancer risk: a systematic review and meta-analysis. Cytokine. 2014;70(2):104–114. doi: 10.1016/j.cyto.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Harms M., Benitez B. A., Cairns N., et al. C9orf72 hexanucleotide repeat expansions in clinical alzheimer disease. JAMA Neurology. 2013;70(6):736–741. doi: 10.1001/2013.jamaneurol.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cacace R., Van Cauwenberghe C., Bettens K., et al. C9orf72 G4C2 repeat expansions in Alzheimer's disease and mild cognitive impairment. Neurobiology of Aging. 2013;34(6):1712–e7. doi: 10.1016/j.neurobiolaging.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 22.Jiao B., Guo J.-F., Wang Y.-Q., et al. C9orf72 mutation is rare in Alzheimer's disease, Parkinson's disease, and essential tremor in China. Frontiers in Cellular Neuroscience. 2013;7, article 164 doi: 10.3389/fncel.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin C.-H., Chen T.-F., Chiu M.-J., Lin H.-I., Wu R.-M. Lack of C9orf72 repeat expansion in Taiwanese patients with mixed neurodegenerative disorders. Frontiers in Neurology. 2014;5, article 59 doi: 10.3389/fneur.2014.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rollinson S., Halliwell N., Young K., et al. Analysis of the hexanucleotide repeat in C9ORF72 in Alzheimer's disease. Neurobiology of Aging. 2012;33(8):1846.e5–1846.e6. doi: 10.1016/j.neurobiolaging.2012.01.109. [DOI] [PubMed] [Google Scholar]
- 25.Wojtas A., Heggeli K. A., Finch N., et al. C9ORF72 repeat expansions and other FTD gene mutations in a clinical AD patient series from Mayo Clinic. American Journal of Neurodegenerative Disease. 2012;1(1):107–118. [PMC free article] [PubMed] [Google Scholar]
- 26.Xi Z., Zinman L., Grinberg Y., et al. Investigation of C9orf72 in 4 neurodegenerative disorders. Archives of Neurology. 2012;69(12):1583–1590. doi: 10.1001/archneurol.2012.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borghero G., Pugliatti M., Marrosu F., et al. Genetic architecture of ALS in Sardinia. Neurobiology of Aging. 2014;35(12):2882.e7–2882.e12. doi: 10.1016/j.neurobiolaging.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brettschneider J., Van Deerlin V. M., Robinson J. L., et al. Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathologica. 2012;123(6):825–839. doi: 10.1007/s00401-012-0970-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byrne S., Elamin M., Bede P., et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. The Lancet Neurology. 2012;11(3):232–240. doi: 10.1016/s1474-4422(12)70014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiò A., Borghero G., Restagno G., et al. Clinical characteristics of patients with familial amyotrophic lateral sclerosis carrying the pathogenic GGGGCC hexanucleotide repeat expansion of C9ORF72. Brain. 2012;135(3):784–793. doi: 10.1093/brain/awr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper-Knock J., Hewitt C., Highley J. R., et al. Clinico-pathological features in amyotrophic lateral sclerosis with expansions in C9ORF72. Brain. 2012;135(3):751–764. doi: 10.1093/brain/awr365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debray S., Race V., Crabbé V., et al. Frequency of C9orf72 repeat expansions in amyotrophic lateral sclerosis: a Belgian cohort study. Neurobiology of Aging. 2013;34(12):2890.e7–2890.e12. doi: 10.1016/j.neurobiolaging.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Dombroski B. A., Galasko D. R., Mata I. F., et al. C9orf72 hexanucleotide repeat expansion and Guam amyotrophic lateral sclerosis-Parkinsonism-dementia complex. JAMA Neurology. 2013;70(6):742–745. doi: 10.1001/jamaneurol.2013.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.García-Redondo A., Dols-Icardo O., Rojas-García R., et al. Analysis of the C9orf72 gene in patients with amyotrophic lateral sclerosis in Spain and different populations worldwide. Human Mutation. 2013;34(1):79–82. doi: 10.1002/humu.22211. [DOI] [PubMed] [Google Scholar]
- 35.Jiao B., Tang B., Liu X., et al. Identification of C9orf72 repeat expansions in patients with amyotrophic lateral sclerosis and frontotemporal dementia in mainland China. Neurobiology of Aging. 2014;35(4):936–e22. doi: 10.1016/j.neurobiolaging.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Kenna K. P., McLaughlin R. L., Byrne S., et al. Delineating the genetic heterogeneity of ALS using targeted high-throughput sequencing. Journal of Medical Genetics. 2013;50(11):776–783. doi: 10.1136/jmedgenet-2013-101795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konno T., Shiga A., Tsujino A., et al. Japanese amyotrophic lateral sclerosis patients with GGGGCC hexanucleotide repeat expansion in C9ORF72. Journal of Neurology, Neurosurgery and Psychiatry. 2013;84(4):398–401. doi: 10.1136/jnnp-2012-302272. [DOI] [PubMed] [Google Scholar]
- 38.Millecamps S., Boillée S., Le Ber I., et al. Phenotype difference between ALS patients with expanded repeats in C9ORF72 and patients with mutations in other ALS-related genes. Journal of Medical Genetics. 2012;49(4):258–263. doi: 10.1136/jmedgenet-2011-100699. [DOI] [PubMed] [Google Scholar]
- 39.Ogaki K., Li Y., Atsuta N., et al. Analysis of C9orf72 repeat expansion in 563 Japanese patients with amyotrophic lateral sclerosis. Neurobiology of Aging. 2012;33(10):2527.e11–2527.e6. doi: 10.1016/j.neurobiolaging.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 40.Ratti A., Corrado L., Castellotti B., et al. C9ORF72 repeat expansion in a large Italian ALS cohort: evidence of a founder effect. Neurobiology of Aging. 2012;33(10):e7–e14. doi: 10.1016/j.neurobiolaging.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 41.Rutherford N. J., Heckman M. G., DeJesus-Hernandez M., et al. Length of normal alleles of C9ORF72 GGGGCC repeat do not influence disease phenotype. Neurobiology of Aging. 2012;33(12):2950.e5–2950.e7. doi: 10.1016/j.neurobiolaging.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabatelli M., Conforti F. L., Zollino M., et al. C9ORF72 hexanucleotide repeat expansions in the Italian sporadic ALS population. Neurobiology of Aging. 2012;33(8):1848.e15–1848.e20. doi: 10.1016/j.neurobiolaging.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai C.-P., Soong B.-W., Tu P.-H., et al. A hexanucleotide repeat expansion in C9ORF72 causes familial and sporadic ALS in Taiwan. Neurobiology of Aging. 2012;33(9):2232.e11–2232.e18. doi: 10.1016/j.neurobiolaging.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Van Laere K., Vanhee A., Verschueren J., et al. Value of 18fluorodeoxyglucose-positron-emission tomography in amyotrophic lateral sclerosis: a prospective study. JAMA Neurology. 2014;71(5):553–561. doi: 10.1001/jamaneurol.2014.62. [DOI] [PubMed] [Google Scholar]
- 45.van Rheenen W., van Blitterswijk M., Huisman M. H. B., et al. Hexanucleotide repeat expansions in C9ORF72 in the spectrum of motor neuron diseases. Neurology. 2012;79(9):878–882. doi: 10.1212/wnl.0b013e3182661d14. [DOI] [PubMed] [Google Scholar]
- 46.Van Der Zee J., Sleegers K., Broeckhoven C. V. Invited article: the Alzheimer disease-frontotemporal lobar degeneration spectrum. Neurology. 2008;71(15):1191–1197. doi: 10.1212/01.wnl.0000327523.52537.86. [DOI] [PubMed] [Google Scholar]
- 47.Alavi A., Nafissi S., Rohani M., et al. Repeat expansion in C9ORF72 is not a major cause of amyotrophic lateral sclerosis among iranian patients. Neurobiology of Aging. 2014;35(1):267.e1–267.e7. doi: 10.1016/j.neurobiolaging.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 48.Gijselinck I., van Mossevelde S., van der Zee J., et al. The C9orf72 repeat size correlates with onset age of disease, DNA methylation and transcriptional downregulation of the promoter. Molecular Psychiatry. 2015 doi: 10.1038/mp.2015.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The flowchart of the selection of studies, the funnel plot and the detailed characteristics of the included studies are listed in the supplementary materials.