ABSTRACT
Heritable endobacteria, which are transmitted from one host generation to the next, are subjected to evolutionary forces that are different from those experienced by free-living bacteria. In particular, they suffer consequences of Muller’s ratchet, a mechanism that leads to extinction of small asexual populations due to fixation of slightly deleterious mutations combined with the random loss of the most-fit genotypes, which cannot be recreated without recombination. Mycoplasma-related endobacteria (MRE) are heritable symbionts of fungi from two ancient lineages, Glomeromycota (arbuscular mycorrhizal fungi) and Mucoromycotina. Previous studies revealed that MRE maintain unusually diverse populations inside their hosts and may have been associated with fungi already in the early Paleozoic. Here we show that MRE are vulnerable to genomic degeneration and propose that they defy Muller’s ratchet thanks to retention of recombination and genome plasticity. We suggest that other endobacteria may be capable of raising similar defenses against Muller’s ratchet.
MOLECULAR EVOLUTION PATTERNS IN HERITABLE ENDOBACTERIA
Heritable endobacteria are an exceptional group of bacteria that reside within eukaryotic host cells and are transmitted vertically from one host generation to the next. They occupy the cytoplasmic niches of a variety of hosts, including arthropods, nematodes, and fungi (1, 2). These associations of bacteria and eukaryotic hosts often endow the partners with novel capabilities that could not be achieved in separation from each other, leading to evolutionary innovations. The extent of interdependence between the host and endobacteria can vary based on factors such as age of association and degree of coevolution: i.e., endosymbionts exhibit either facultative or obligate dependence on the host, and their impact is essential, nonessential, or antagonistic to the host’s survival. In addition, the antiquity and nature of the association between the partners determine whether endosymbiont transmission is exclusively vertical or punctuated by instances of horizontal transmission.
Vertically transmitted endobacteria undergo unique molecular evolution, resulting in genome structures not normally seen in their free-living relatives, with the exception of a select few marine bacteria (3). Endosymbiont genomes are reduced in size and exhibit accelerated sequence evolution (4). These features are consequences of living in the cytoplasmic niche of a host that is unlike a free-living environment; the intracellular compartment offers not only protection and absence of competition, but a metabolically rich milieu where many genes in the endobacterial genomes become redundant or unnecessary (5). Furthermore, exclusively vertical transmission has several consequences that affect endosymbiont population structure. In particular, population bottlenecks mark every transmission, since only a portion of the endosymbiont population is passed to the next generation of the host. Moreover, endosymbiont populations are subdivided by being restricted to individual host lineages. They do not engage in recombination and exhibit limited genetic diversity. These factors contribute to the reduction of endosymbiont effective population sizes relative to free-living bacteria. Based on the nearly neutral theory of molecular evolution, such population dynamics are predicted to limit the strength of purifying selection, resulting in the accumulation and fixation of slightly deleterious mutations in the endosymbiont genome (6), manifested by an overall increase in the ratio of nonsynonymous to synonymous substitutions in protein coding genes (7). Continual accumulation of slightly deleterious mutations over time leads to inactivation of the affected genes, including DNA repair machinery, which further accelerates mutation buildup in the genome. Due to deletional bias prevailing in bacterial genomes, mutation-compromised genes are eliminated, eventually resulting in the reduced and degenerate genomes typical for ancient heritable endobacteria (8).
The genes that are ultimately lost in reduced endobacterial genomes come from all functional categories—most notably metabolism, cell envelope biosynthesis, transcriptional regulation, and DNA repair and recombination (8). The genes that are retained, in addition to those important for symbiosis, are involved in essential cellular functions, such as DNA replication, RNA transcription, and protein translation. However, even within these essential categories, only minimal repertoires of genes required for basic functionality are maintained, with most endobacteria displaying the loss of accessory subunits otherwise present in these systems (5).
In heritable bacteria, changes in genome coding capacity and stability appear to complement gene content modifications associated with the transition to a life in the eukaryotic cell (8). Early stages of genome contraction are characterized by the abundance of pseudogenes, mobile genetic elements (MGEs), and genomic rearrangements. In contrast, the advanced stages found in ancient endobacteria with a reduced gene set are typified by the near absence of pseudogenes, the lack of MGEs, and relatively stable yet progressively deteriorating genomes.
MULLER’S RATCHET AND POPULATION EXTINCTION
Rapid fixation of slightly deleterious mutations and genome degeneration are not the only hazards that bedevil small asexual populations of heritable endobacteria. In the absence of recombination, they are vulnerable to extinction, as, by chance, the most-fit genotypes can be lost. This continual decrease in the mean fitness of a population was described by Hermann Joseph Muller (9) and is known as Muller’s ratchet. Each consecutive loss of the most-fit genotype advances the ratchet toward population collapse, as these genotypes cannot be recreated without recombination. Consequently, Muller’s ratchet is a powerful force that contributes to genome erosion in heritable endobacteria (7), ultimately leading to disappearance of endosymbiont lineages (10). While recombination is the principal mechanism capable of disabling the ratchet, evolutionary theory suggests that several other factors could decelerate its advance, including compensatory evolution, in which fitness losses caused by earlier mutations are restored by subsequent mutations (11), horizontal acquisition of foreign DNA (12), and host-level selection that maintains a network of endosymbiont populations associated with individual host lineages (13).
GENOME EVOLUTION IN THE MYCOPLASMA-RELATED ENDOBACTERIA OF FUNGI
Recently, genomic data (with GenBank accession numbers in brackets) were generated for a novel group of heritable endobacteria, the mycoplasma-related endobacteria (MRE), associated with four distinct Glomeromycota host species: MRE-CE (Claroideoglomus etunicatum [JPXH00000000]), MRE-DH (Dentiscutata heterogama [LN828718]), MRE-RC (Rhizophagus clarus [JPXG01000000]), and MRE-RV (Racocetra verrucosa [JQIB01000000]) (14, 15). MRE reside in the cytoplasm of fungi representing all major lineages of Glomeromycota (arbuscular mycorrhizal fungi [AMF]) and the Endogone lineage of Mucoromycotina (2, 16–18). AMF are asexual soil fungi that form obligate symbioses (mycorrhizae) with the majority of terrestrial plants (19). Fossil records date AMF to the Ordovician (~460 million years ago [MYA]) (20) and indicate that the mycorrhizal symbiosis existed as early as in the Devonian (~400 MYA) (21). In contrast, the genus Endogone encompasses a group of poorly understood fungi with diverse lifestyles (16). However, their ancestors are believed to have colonized the early Devonian plants simultaneously with AMF (22). The role of MRE in the biology of their fungal hosts is unknown.
Origin of the MRE-fungus symbiosis.
MRE are members of the Mollicutes, a group characterized by minimal genomes, highly reduced metabolic capabilities, and the lack of a peptidoglycan cell wall. Phylogenetic reconstructions place MRE in the Mycoplasma pneumoniae group of the family Mycoplasmataceae (14) (Fig. 1), suggesting that MRE arose as a consequence of a host switch from animals to fungi. Such origin implies that vertically transmitted MRE associated with fungi share some genomic features with horizontally transmitted animal-associated mycoplasmas. Detection of MRE in both Glomeromycota and the Endogone lineage of Mucoromycotina, as well as molecular phylogeny data offering marginally significant support for codivergence between MRE and these two groups of fungi, gave rise to the hypothesis that the origin of the MRE association with fungi predates the divergence between Glomeromycota and Mucoromycotina during the early Paleozoic (16, 18). Alternatively, ancestral MRE could have invaded and spread concomitantly into representatives of these two groups of fungi when they cocolonized Devonian plants during early terrestrialization (22). Regardless of which scenario is accurate, the association between MRE and fungi may be one of the oldest heritable symbioses on the planet.
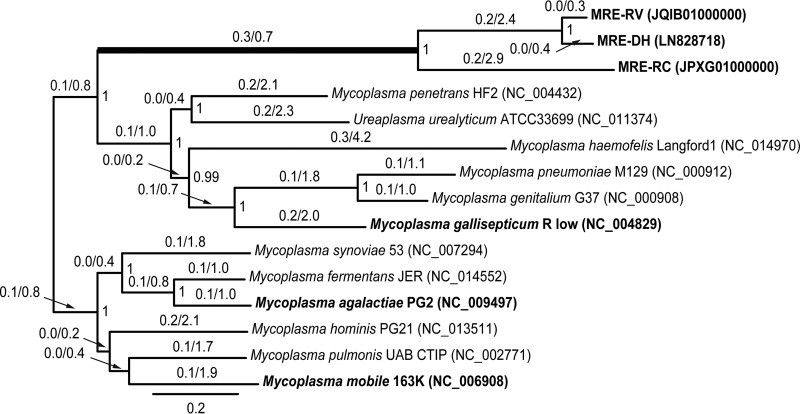
FIG 1 .
Bayesian phylogeny of MRE reconstructed based on the concatenated nucleotide sequences of the following genes: dnaG, infC, nusA, rplA, rplB, rplC, rplE, rplF, rplM, rplN, rplP, rplT, rpmA, rpsB, rpsC, rpsE, rpsJ, rpsS, and smpB. MRE were sampled from Dentiscutata heterogama (MRE-DH), Racocetra verrucosa (MRE-RV), and Rhizophagus clarus (MRE-RC). Bayesian posterior probabilities greater than 0.90 are shown at nodes; Bayesian analyses were conducted using MrBayes v.3.2 (64) under the nucleotide substitution model GTR + Γ + I with 1,000,000 generations and a 250,000 burn-in. The numbers above branches are the estimates of dN/dS ratios obtained using the codeml module of PAML v.4.8 (65), assuming a two-ratio model. The thickened branch leading to MRE indicates that, according to the likelihood ratio model testing conducted in codeml, the dN/dS ratio along it is significantly different from the background dN/dS ratio for all other branches (χ2 = 36.12, P < 0.001). Taxa in boldface were subjected to Tajima 1D relative rate tests (26) (Table 1).
MRE intrahost diversity.
Unexpectedly for heritable endobacteria, which tend to be genetically uniform within their hosts (8), MRE populations display unusually high diversity levels (2, 17, 18). There appear to be two sources of MRE intrahost diversity: (i) rapid accumulation of mutations and (ii) horizontal transmission between hosts. (i) Like in other mycoplasmas, mutation accumulation in MRE genomes can be attributed to the losses of DNA polymerase II (polB), the proofreading subunit of DNA polymerase III (dnaQ), and the methyl-directed mismatch repair system (MMRS) (mut genes) (Fig. 2) (14). The losses of dnaQ, polB, and MMRS genes have been shown to cause hypermutator phenotypes in other bacteria due to the inability to fix errors in their DNA sequences (23–25). To assess the extent of mutation accumulation in MRE, we compared the rates of evolution between MRE and Mycoplasma gallisepticum, which also represents the M. pneumoniae clade of the Mycoplasmataceae (Fig. 1), by conducting Tajima 1D relative rate test (26), implemented in MEGA7 (27), on DNA sequences at 19 protein coding loci (Table 1). With a mutation rate of 1.02 × 10−5 substitutions per site per year, M. gallisepticum is considered to be one of the fastest-evolving bacteria (28). We found that MRE genomes evolve more rapidly than that of M. gallisepticum (Table 1), which places MRE among the ultrafast-evolving microbes. (ii) The second source of MRE intrahost diversity appears to be horizontal transmission, inferred from analyses of codivergence patterns between MRE and AMF (18). The exact mechanisms of horizontal transmission are unknown. MRE dispersal across AMF of one species could be facilitated by fusions between hyphae of different fungal strains (29). MRE transmission across different AMF species could occur when hyphae are damaged due to grazing by soil fauna (30). In either case, horizontal transmission could potentially facilitate genetic exchanges among distinct MRE genotypes.
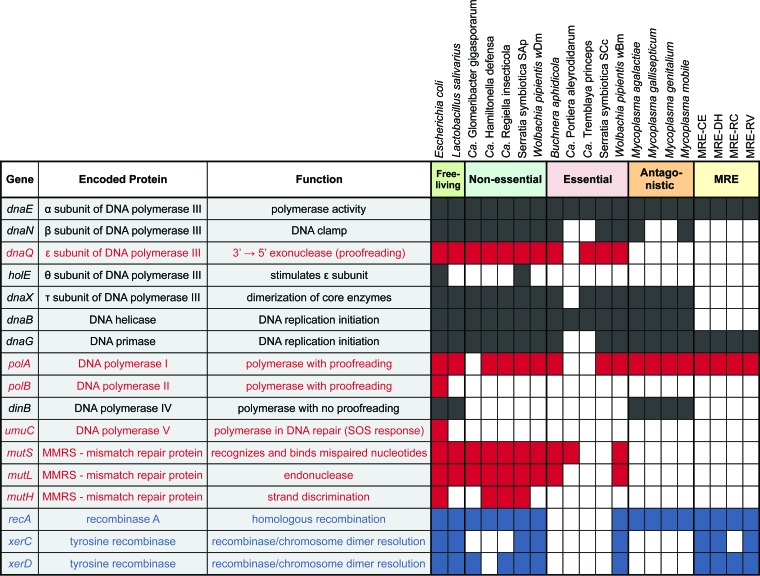
FIG 2 .
Loss and retention of select genes involved in DNA replication (black), repair (red), and recombination (blue). The four populations of the obligate endobacteria MRE are compared with free-living bacteria and other endobacteria, categorized as nonessential, essential, and antagonistic symbionts. Colored squares indicate the presence of the gene, and white squares indicate loss of the gene. The non-MRE bacterial strains used (with GenBank accession numbers in parentheses) are Escherichia coli O157:H7 strain Sakai (BA000007.2), Lactobacillus salivarius UCC118 (CP000233.1), “Candidatus Glomeribacter gigasporarum” BEG34 (CAFB00000000.1), “Candidatus Hamiltonella defensa” 5AT (CP001277.1), “Candidatus Regiella insecticola” LSR1 (ACYF00000000.1), Serratia symbiotica SAp (AENX00000000.1), Wolbachia pipientis endosymbiont of Drosophila melanogaster wDm (AE017196.1), Buchnera aphidicola APS (BA000003.2), “Candidatus Portiera aleyrodidarum” BT-QVLC (CP003867.1), “Candidatus Tremblaya princeps” PCVAL (CP002918.1), Serratia symbiotica SCc (CP002295.1), Wolbachia pipientis endosymbiont of Brugia malayi WBM (AE017321.1), Mycoplasma agalactiae PG2 (CU179680.1), Mycoplasma gallisepticum S6 (CP006916.2), Mycoplasma genitalium 6282 (CP003771.1), and Mycoplasma mobile 163K (AE017308.1).
TABLE 1 .
MRE exhibit molecular evolution rate acceleration relative to other mycoplasmasa
| Ingroup (GenBank accession no.) | Outgroup (GenBank accession no.) | Relative rate statisticb |
|---|---|---|
| MRE-DH (LN828718), M. gallisepticum R low (NC_004829) |
M. mobile 163K (NC_006908) | 123.65* |
| MRE-RC (JPXG01000000), M. gallisepticum R low (NC_004829) |
M. mobile 163K (NC_006908) | 126.42* |
| MRE-RV (JQIB01000000), M. gallisepticum R low (NC_004829) |
M. mobile 163K (NC_006908) | 128.82* |
| MRE-DH (LN828718), M. gallisepticum R low (NC_004829) |
M. agalactiae PG2 (NC_009497) | 140.48* |
| MRE-RC (JPXG01000000), M. gallisepticum R low (NC_004829) |
M. agalactiae PG2 (NC_009497) | 137.44* |
| MRE-RV (JQIB01000000), M. gallisepticum R low (NC_004829) |
M. agalactiae PG2 (NC_009497) | 142.19* |
Results are as indicated by the results of Tajima’s 1D relative rate test (26) for DNA sequences at 19 protein coding loci listed in Fig. 1.
The 1D relative rate statistic distribution is the same as the distribution of χ2. *, significant at P < 0.0001. If the value is significant, the null hypothesis of equal rates of sequence evolution can be rejected.
Are MRE genomes degenerate?
In terms of the size and extent of metabolic dependence on the host, MRE genomes are comparable to the genomes of other closely related animal-associated mycoplasmas. However, unlike in other Mycoplasma species, MRE transmission is predominantly vertical, which exposes MRE intrahost populations to demographic bottlenecks of about 1,000 cells that populate individual AMF spores (17). Such recurrent bottlenecks may contribute to a decline in effective population size and magnification of genetic drift relative to natural selection, which, in turn, could make MRE genomes vulnerable to progressive degeneration and expose their populations to Muller’s ratchet. In fact, the significant increase in the molecular evolution rate experienced by MRE relative to horizontally transmitted M. gallisepticum (Table 1) suggests that vertical transmission impacts MRE evolution. To assess the specific consequences of vertical transmission on MRE, we focused on (i) the ratio of the rate of nonsynonymous nucleotide substitutions to the rate of synonymous substitutions (dN/dS) in protein coding genes and (ii) the abundance of putative pseudogenes in the MRE genomes. (i) The genome-wide increases of dN/dS ratios in heritable endobacteria compared to those in free-living relatives are commonly used as an indicator of accumulation of slightly deleterious mutations (7). Our analysis of dN/dS ratios along the branches of the Mycoplasmataceae phylogeny reconstructed using sequences of 19 protein coding genes (Fig. 1) revealed that the dN/dS ratio along the branch leading to MRE is significantly higher than the background dN/dS ratio along all other branches of this phylogeny (χ2 = 36.12, P < 0.001). It is unlikely that this pattern represents adaptation to a new host, as the increased nonsynonymous substitution rate is apparent across several broadly conserved genes, including sequences encoding ribosomal proteins. Instead, this observation is consistent with the increased accumulation of slightly deleterious mutations after MRE had switched from horizontal to vertical transmission. Interestingly, dN/dS ratios along terminal branches leading to individual MRE genomes do not appear to be elevated, which suggests that, over time, MRE might have experienced refinement of the mechanisms that contribute to purging of slightly deleterious mutations. (ii) Proliferation of pseudogenes is a hallmark of rapid genome erosion due to accumulation of deleterious mutations (8). In the absence of proteomic data, putative pseudogenes can be identified by comparing open reading frames (ORFs) with their homologues in closely related taxa to detect changes that abolish their original function (31). We followed this approach in MRE and considered ORFs that were reduced in length by 50% relative to their homologues to harbor premature stop codons and ORFs that were over 150% longer than their homologues to have lost start/stop codons. This approach revealed that in MRE, putative pseudogenes constitute between 21 and 30% of all non-orphan ORFs (Table 2). These numbers are comparable to those observed in heritable nonessential mutualists of insects, such as Serratia symbiotica SAp in pea aphids, which originated 90 MYA, and 26% of its 2.8-Mb genome is made up by pseudogenes (32). The presence of pseudogenes in MRE suggests that their genomes are experiencing active degeneration. However, the evolutionary context of pseudogene formation in MRE appears to be different from that of other heritable endobacteria. In particular, the Mycoplasma parentage of the MRE lineage suggests that the genomes of MRE ancestors were already reduced in size at the time of their transition from animal to fungal hosts, an event that was also likely associated with a switch from horizontal to vertical transmission. Consequently, in contrast to bacteria in which pseudogenization represents early stages of progressive genome contraction, pseudogenization in MRE appears to be a steady-state process responsible for deactivation of genes continually acquired through horizontal transfer, not unlike in other mycoplasmas in which horizontal gene acquisition is one of the mechanisms that counter genome erosion (33). Collectively, the dN/dS values in protein coding genes and the abundance of putative pseudogenes in the MRE genomes suggest that the MRE lineage experienced a period of accelerated genome degradation followed by a refinement of mechanisms that allow for purging of slightly deleterious mutations, leading to present-day apparently steady-state genome erosion.
TABLE 2 .
Putative pseudogenes in the MRE genomesa
| ORF type | No. (%) of ORFs |
||
|---|---|---|---|
| MRE-CE | MRE-RV | MRE-RC | |
| Non-orphan ORFs | 323 (100) | 526 (100) | 390 (100) |
| ORFs with putative premature stop |
39 (12.1) | 61 (11.6) | 89 (22.8) |
| ORFs with putative loss of stop/start |
36 (11.1) | 49 (9.3) | 30 (7.7) |
Putative pseudogenes were identified by comparing MRE non-orphan open reading frames (ORFs) with their homologues. ORFs that were over 50% shorter (putative premature stop codon) and longer (putative loss of stop/start codon) are included.
Genetic recombination in MRE.
Unlike most ancient heritable endobacteria (8), MRE harbor various DNA recombination systems (14) (Fig. 2). The putative role of recombination in shaping the MRE population structure was indicated initially by the presence of recombination signatures in rRNA gene sequences sampled from MRE associated with diverse AMF hosts distributed globally (17, 18). To further explore the contribution of recombination to MRE evolution, we conducted coalescent population modeling using ClonalFrame (34). We estimated the per-site effect of recombination relative to mutation: i.e., the ratio of rates at which nucleotides become substituted as a result of recombination versus mutation (r/m) based on nucleotide sequences at 13 protein coding loci (rplA, rplB, rplC, rplM, rplN, rplP, rplT, rpmA, rpsB, rpsC, rpsE, rpsJ, and smpB) sampled from MRE-DH, MRE-RC, and MRE-RV. The MCMC analyses included 200,000 generations after an initial 50,000-generation burn-in; convergence was assessed using the Gelman-Rubin statistic with a cutoff of 1.1 (34). We found that in MRE, the r/m was 1.3 with a 95% confidence interval of 0.68 to 2.23. Given that the mutation rate is exceptionally high in MRE, and the recombination rate appears to exceed it, recombination is likely to play an important role in MRE evolution.
Mobile genetic elements and genome plasticity in MRE.
Similar to other mycoplasmas (35), MRE harbor a substantial number of MGEs, such as phages, insertion sequence elements (14), and integrative and conjugative elements (36). For example, between 1 and 8% of non-orphan genes in the MRE genomes encode transposases (14). Moreover, some of the MRE genomes show evidence of historical invasion by plectrovirus (14), a phage known to infect Spiroplasma species (35). The preponderance of MGEs in MRE presents a contrast with most other ancient heritable endobacteria with highly reduced genomes from which such elements appear to be absent (8) and suggests that MRE genomes may experience MGE-mediated genomic rearrangements (36).
One of the most striking features of MRE is the extreme plasticity of their genomes suggested by extensive chromosomal rearrangements apparent in MRE genomic assemblies (14). There appear to be two major sources of chromosomal rearrangements in MRE: recombination facilitated by simple sequence repeats (SSRs) and MGE activity (14). Both SSRs and MGEs often mark disruptions in gene synteny. The degree of plasticity in MRE genomes testifies to the activities of MGEs and recombination machineries. As these two mechanisms underlying MRE chromosomal rearrangements are common in other mycoplasmas (35, 37), MRE must have retained them even after the host switch from animals to fungi and the transition from horizontal to predominantly vertical transmission.
MRE SIMILARITIES TO OTHER HERITABLE ENDOBACTERIA
The trajectory of degenerative genome contraction in heritable endobacteria—with early stages characterized by a rapid loss of gene content, proliferation of pseudogenes, MGEs, and genomic rearrangements and advanced stages typified by minimal, relatively stable but gradually decaying genomes—was reconstructed from genomic comparisons across endobacteria of arthropods with different genome sizes and antiquities (8). For example, Buchnera aphidicola, which originated 160 to 280 MYA, is one of the oldest lineages with reduced and decayed genomes (38). Buchnera is an essential nutritional mutualist of aphids, and its transmission is exclusively vertical. The absence of MGEs from the 641-kb Buchnera genomes is believed to be responsible for the lack of genomic rearrangements between two strains that diverged over 50 MYA (39). Such lack of MGEs represents a stark contrast from the substantial mobilomes of nonessential mutualists with mixed transmission in which vertical transfers are occasionally punctuated by horizontal transfers. For example, the genomes of two sister species, “Candidatus Hamiltonella defensa” and “Candidatus Regiella insecticola,” are 2.1 and 2.0 Mb in size, with MGEs making up 34 and 20% of all coding sequences, respectively (40). While the antiquity of endosymbionts with mixed transmission is notoriously difficult to establish (41), the minimal age of these two nonessential defensive mutualists was estimated to be 100 million years (42). A similar pattern is apparent across different strains of Serratia symbiotica that form distinct types of mutualisms with their aphid hosts. For example, S. symbiotica SCc is an essential nutritional mutualist of unknown antiquity (43). Its 1.8-Mb genome lacks MGEs and does not show evidence of being able to support recombination (43). In contrast, S. symbiotica SAp, the previously mentioned nonessential defensive mutualist of aphids, which dates back to ~90 MYA, is considered to represent early stages of genome contraction (32). Its 2.8-Mb genome is capable of recombination and contains 4% MGEs (32).
With their origin at over 400 MYA (18), the ultrarapid evolution rate, and the incessant interplay of genome erosion and restoration, MRE appear to defy predictions of the degenerative genome contraction model. Importantly, this departure from the degenerative evolution model is not a consequence of host factors, which could be expected to differentiate fungus-associated MRE from the arthropod-associated endobacteria, such as Buchnera or Serratia SCc. The absence of such host effects is evidenced by the intrahost genetic diversity that distinguishes MRE not only from most arthropod-associated endobacteria but also from another heritable endosymbiont of AMF, “Candidatus Glomeribacter gigasporarum,” which displays genetic homogeneity of intrahost populations (17, 41).
Recent studies driven by rapid accumulation of genomic data from diverse endobacteria suggest that departures from the degenerative model of endosymbiont evolution may be also found in other organisms. For example, genome rearrangements were detected in highly reduced and decayed genomes of essential mutualists of insects, “Candidatus Portiera aleyrodidarum” in whiteflies (44) and “Candidatus Tremblaya princeps” in mealybugs (45); both of these associations date back to 100 to 200 MYA (46). In the 358-kb genome of “Ca. Portiera aleyrodidarum” BT, large-scale structural polymorphisms exist even within individual insects, and this variation is likely mediated by recombination across identical repeats that are maintained by gene conversion (44). Similarly, the 139-kb genome of “Ca. Tremblaya princeps” exists within single insects in two forms, differentiated by a genomic inversion present in both orientations (45). Likewise, MGEs have been suggested to be important in the essential mutualist of nematodes, Wolbachia pipientis wBm, due to their ability to act as gene conversion sites or sites of homologous recombination (47). Wolbachia wBm dates back to 50 to 55 MYA (48). It is capable of recombination, and its 1.1-Mb genome contains 2.4% apparently inactive MGEs (47, 49). In addition to ancient mutualists with exclusively vertical transmission, MRE share several similarities with Wolbachia reproductive parasites of arthropods. Genomes of these arthropod-associated Wolbachia are also reduced and degenerate (50) while retaining the ability to recombine (51) and undergo extensive rearrangements (52), a process often mediated by MGEs (53). Like MRE, arthropod-associated Wolbachia strains engage occasionally in horizontal transmission, and distinct Wolbachia genotypes have been observed to coexist in host individuals (51). However, with the estimated age of 50 to 55 million years (48), the association of Wolbachia with their arthropod hosts is considerably younger than the symbiosis between MRE and fungi (18). Moreover, Wolbachia genomes retain a larger repertoire of DNA repair mechanisms than those present in MRE (Fig. 2). Collectively, while these examples suggest that other heritable endobacteria may share some evolutionary mechanisms with MRE, none of these lineages appears to be equally ancient.
MAINTENANCE OF RECOMBINATION AND GENOME PLASTICITY TO OVERCOME MULLER’S RATCHET
We equate the maintenance of recombination machinery in the MRE genome to the evolution of sex in eukaryotic systems. Despite its inherent costs, sex is maintained in the majority of eukaryotes, with models explaining its significance that range from the creation of novel genotypes to resist pathogen infection in the Red Queen hypothesis, through acceleration of adaptation by eliminating competition among beneficial mutations in the Fisher-Muller hypothesis, to the prevention of population extinction via Muller’s ratchet in finite populations (54). Though the exact mechanism is unclear, MRE are capable of genomic recombination (17, 18), perhaps through the uptake of DNA from dead MRE cells present in the same host cytoplasmic niche. Given that some highly divergent MRE populations appear to be products of horizontal transmission (18), such DNA acquisitions could generate novel high-fitness genotypes. Yet, being subjected to predominantly vertical transmission, the MRE genomes inevitably erode through Muller’s ratchet (Fig. 1; Table 2). Nevertheless, they appear to be able to purge the deleterious mutations and generate new genotypes through DNA recombination and genome plasticity (Fig. 3). Furthermore, theoretical modeling indicates that recombination-mediated horizontal gene transfer of foreign DNA in bacterial populations can stall the advance of Muller’s ratchet, even if the foreign genes contain more deleterious mutations than the recipient cells (12). If uptake and recombination of foreign DNA are frequent enough, a diverse bacterial population can resist Muller’s ratchet better than a genetically homogenous population of the same size.
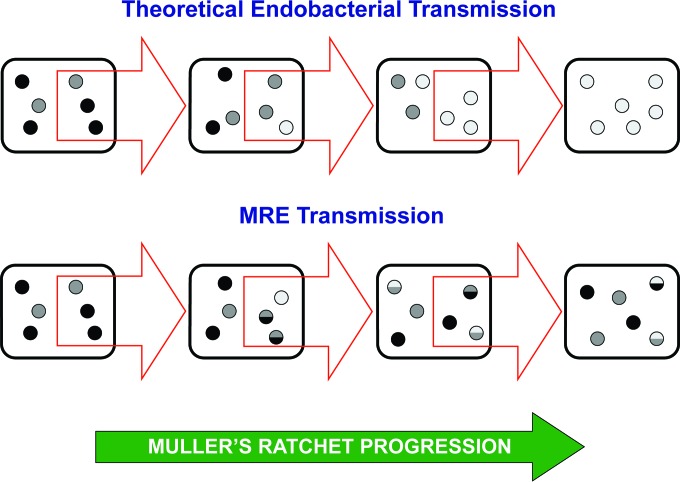
FIG 3 .
MRE transmission and Muller’s ratchet progression compared to theoretical predictions for heritable endobacteria. Due to transmission bottlenecks and fixation of slightly deleterious mutations, heritable endobacteria are expected to degenerately progress to a relatively homogeneous population of unfit individuals. MRE escape Muller’s ratchet by retention of recombination and genome plasticity to purge some of the slightly deleterious mutations and maintain a genetically diverse population. Intrahost selection is expected to eliminate low-fitness genotypes generated by recombination. Black outlined rectangles represent host cells, with red arrows indicating transmission bottlenecks. Endobacteria are represented as small circles, with darker shading depicting the most-fit individuals to lighter shading depicting the least-fit individuals. Circles with two-tone color depict recombination events in MRE.
It is unclear how MRE are able to prevent their recombination machinery from being eroded, as seen in the majority of other heritable endobacteria. This may be a general feature of the genome structure in all mycoplasmas. Their limited gene set and rapid mutation accumulation may be sources of strong selective pressure to maintain recombination genes. Rapid genetic change afforded by active recombination machinery is also likely favored given a mycoplasma lifestyle of antagonistic dependence on eukaryotic hosts whose evolving defensive responses necessitate continuous evasion (55). Accordingly, despite their minimal genomes, Mycoplasma species appear to have retained their sexual competence and ability for horizontal gene transfer among cells sharing the same niche (56). For example, a recent study has shown that Mycoplasma agalactiae is able to exchange and transfer nearly every fragment of its chromosome through conjugation, including long stretches of DNA containing up to 80 genes (57). Whether a similar mechanism operates in MRE remains to be investigated.
While recombination is considered to be an unequivocally beneficial evolutionary force, understanding the role of MGEs in evolution is more nuanced (58). MGEs are selfish entities that impose on the host cell costs of their replication. More importantly, integration events into new sites of the host genome may disrupt important regulatory and metabolic functions (59). Despite these downsides, the presence of MGEs is viewed as advantageous in mycoplasmas (35). It allows for acquisition of new genetic information and contributes to genomic rearrangements associated with novel phenotypes (60). In MRE, facilitation of chromosomal rearrangements seems to be the defining role of MGEs (14). Furthermore, homologous MGEs scattered throughout the MRE genomes could act as sites of recombination or gene conversions (36). Such recombinogenic roles of MGEs have also been observed in other bacteria (47, 61, 62). Consequently, the potential of MGEs to create sites of homology, in conjunction with the retention of recombination genes, provides MRE with a mechanism for escaping Muller’s ratchet to maintain evolutionary longevity.
Mycoplasmas are now seen as a highly adaptable rather than a degenerate group of bacteria driven only by reductive evolution (56, 57, 63). Likewise, MRE appear to have maintained horizontal transmission and recombination abilities as well as MGE activity that allow for maintenance of a dynamic population structure. Consequently, we propose that MRE utilize bacterial sex and genome plasticity as a mechanism for decelerating Muller’s ratchet, a phenomenon likely linked to their mollicute ancestry and relatedness to the mycoplasmas. In particular, it cannot be excluded that, like other mycoplasmas, MRE are antagonists of their hosts, a lifestyle that favors rapid genetic change to either avoid or frustrate host defenses.
CONCLUSIONS
Our exploration of the novel lineage of ancient heritable endobacteria of fungi revealed evidence of genomic degeneration accompanied by active maintenance of recombination genes and genome plasticity, which is responsible for intrahost genetic diversity. We propose that these diversity-generating mechanisms hinder the impending advance of Muller’s ratchet (Fig. 3), a luxury not possible in other endobacteria that have lost, through genome erosion, their recombinant abilities and other mechanisms underlying genome plasticity.
It is clear that genome evolution in endobacteria is influenced by multiple factors, and thus the trajectory of endobacterial populations may not be as simple and predictable as once believed. The dynamics of endobacterial populations have profound effects not only on their evolutionary fate but also on the hosts that harbor them. MRE are exceptional endosymbionts that appear to have retained bacterial sex and genome plasticity and, as a consequence, are able to prevent the effects of Muller’s ratchet that imperil other ancient heritable endobacteria.
ACKNOWLEDGMENTS
We thank two anonymous reviewers for helpful comments.
This work was supported by National Science Foundation grant IOS-1261004.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Naito M, Pawlowska TE. 2016. Defying Muller’s ratchet: ancient heritable endobacteria escape extinction through retention of recombination and genome plasticity. mBio 7(3):e02057-15. doi:10.1128/mBio.02057-15.
REFERENCES
- 1.Hoffmeister M, Martin W. 2003. Interspecific evolution: microbial symbiosis, endosymbiosis and gene transfer. Environ Microbiol 5:641–649. doi: 10.1046/j.1462-2920.2003.00454.x. [DOI] [PubMed] [Google Scholar]
- 2.Naumann M, Schüssler A, Bonfante P. 2010. The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the Mollicutes. ISME J 4:862–871. doi: 10.1038/ismej.2010.21. [DOI] [PubMed] [Google Scholar]
- 3.Morris JJ, Lenski RE, Zinser ER. 2012. The Black Queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3:e00036-12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 5.Moya A, Peretó J, Gil R, Latorre A. 2008. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet 9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- 6.Ohta T. 1973. Slightly deleterious mutant substitutions in evolution. Nature 246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- 7.Moran NA. 1996. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A 93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 9.Muller HJ. 1964. The relation of recombination to mutational advance. Mutat Res 1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 10.Koga R, Bennett GM, Cryan JR, Moran NA. 2013. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol 15:2073–2081. doi: 10.1111/1462-2920.12121. [DOI] [PubMed] [Google Scholar]
- 11.Poon A, Otto SP. 2000. Compensating for our load of mutations: freezing the meltdown of small populations. Evolution 54:1467–1479. doi: 10.1111/j.0014-3820.2000.tb00693.x. [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi N, Kaneko K, Koonin EV. 2014. Horizontal gene transfer can rescue prokaryotes from Muller’s ratchet: benefit of DNA from dead cells and population subdivision. G3 4:325–339. doi: 10.1534/g3.113.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pettersson ME, Berg OG. 2007. Muller’s ratchet in symbiont populations. Genetica 130:199–211. doi: 10.1007/s10709-006-9007-7. [DOI] [PubMed] [Google Scholar]
- 14.Naito M, Morton JB, Pawlowska TE. 2015. Minimal genomes of mycoplasma-related endobacteria are plastic and contain host-derived genes for sustained life within Glomeromycota. Proc Natl Acad Sci U S A 112:7791–7796. doi: 10.1073/pnas.1501676112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres-Cortés G, Ghignone S, Bonfante P, Schüßler A. 2015. Mosaic genome of endobacteria in arbuscular mycorrhizal fungi: transkingdom gene transfer in an ancient mycoplasma-fungus association. Proc Natl Acad Sci U S A 112:7785–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desirò A, Faccio A, Kaech A, Bidartondo MI, Bonfante P. 2015. Endogone, one of the oldest plant-associated fungi, host unique Mollicutes-related endobacteria. New Phytol 205:1464–1472. doi: 10.1111/nph.13136. [DOI] [PubMed] [Google Scholar]
- 17.Desirò A, Salvioli A, Ngonkeu EL, Mondo SJ, Epis S, Faccio A, Kaech A, Pawlowska TE, Bonfante P. 2014. Detection of a novel intracellular microbiome hosted in arbuscular mycorrhizal fungi. ISME J 8:257–270. doi: 10.1038/ismej.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toomer KH, Chen X, Naito M, Mondo SJ, den Bakker HC, VanKuren NW, Lekberg Y, Morton JB, Pawlowska TE. 2015. Molecular evolution patterns reveal life history features of mycoplasma-related endobacteria associated with arbuscular mycorrhizal fungi. Mol Ecol 24:3485–3500. doi: 10.1111/mec.13250. [DOI] [PubMed] [Google Scholar]
- 19.Smith SE, Read DJ. 2008. Mycorrhizal symbiosis, 3rd ed. Academic Press, New York, NY. [Google Scholar]
- 20.Redecker D, Kodner R, Graham LE. 2000. Glomalean fungi from the Ordovician. Science 289:1920–1921. doi: 10.1126/science.289.5486.1920. [DOI] [PubMed] [Google Scholar]
- 21.Remy W, Taylor TN, Hass H, Kerp H. 1994. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci U S A 91:11841–11843. doi: 10.1073/pnas.91.25.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strullu-Derrien C, Kenrick P, Pressel S, Duckett JG, Rioult J-P, Strullu D-G. 2014. Fungal associations in Horneophyton ligneri from the Rhynie Chert (c. 407 million year old) closely resemble those in extant lower land plants: novel insights into ancestral plant-fungus symbioses. New Phytol 203:964–979. doi: 10.1111/nph.12805. [DOI] [PubMed] [Google Scholar]
- 23.Scheuermann R, Tam S, Burgers PM, Lu C, Echols H. 1983. Identification of the epsilon-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc Natl Acad Sci U S A 80:7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowosielska A, Janion C, Grzesiuk E. 2004. Effect of deletion of SOS-induced polymerases, Pol II, IV, and V, on spontaneous mutagenesis in Escherichia coli mutD5. Environ Mol Mutagen 43:226–234. doi: 10.1002/em.20019. [DOI] [PubMed] [Google Scholar]
- 25.Prunier AL, Leclercq R. 2005. Role of mutS and mutL genes in hypermutability and recombination in Staphylococcus aureus. J Bacteriol 187:3455–3464. doi: 10.1128/JB.187.10.3455-3464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tajima F. 1993. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S, Stecher G, Tamura K 22 March 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:863–869 doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delaney NF, Balenger S, Bonneaud C, Marx CJ, Hill GE, Ferguson-Noel N, Tsai P, Rodrigo A, Edwards SV. 2012. Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet 8:e1002511. doi: 10.1371/journal.pgen.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purin S, Morton JB. 2013. Anastomosis behavior differs between asymbiotic and symbiotic hyphae of Rhizophagus clarus. Mycologia 105:589–602. doi: 10.3852/12-135. [DOI] [PubMed] [Google Scholar]
- 30.McGonigle TP, Fitter AH. 1988. Ecological consequences of arthropod grazing on VA mycorrhizal fungi. Proc R Soc Edinb Sec B Biol Sci 94B:25–32. [Google Scholar]
- 31.Ochman H, Davalos LM. 2006. The nature and dynamics of bacterial genomes. Science 311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- 32.Burke GR, Moran NA. 2011. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol 3:195–208. doi: 10.1093/gbe/evr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Harrison PM, Kunin V, Gerstein M. 2004. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol 5:R64. doi: 10.1186/gb-2004-5-9-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marenda MS. 2014. Genomic mosaics, p 15–54. In Browning GF, Citti C (ed), Mollicutes: molecular biology and pathogenesis. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 36.Naito M, Pawlowska TE. 2016. The role of mobile genetic elements in evolutionary longevity of heritable endobacteria. Mobile Genet Elem 6:e1136375. doi: 10.1080/2159256X.2015.1136375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mrázek J. 2006. Analysis of distribution indicates diverse functions of simple sequence repeats in Mycoplasma genomes. Mol Biol Evol 23:1370–1385. doi: 10.1093/molbev/msk023. [DOI] [PubMed] [Google Scholar]
- 38.Moran NA, Munson MA, Baumann P, Ishikawa H. 1993. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc Lond Ser B Biol Sci 253:167–171. doi: 10.1098/rspb.1993.0098. [DOI] [Google Scholar]
- 39.Tamas I, Klasson L, Canbäck B, Näslund AK, Eriksson AS, Wernegreen JJ, Sandström JP, Moran NA, Andersson SG. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 40.Degnan PH, Leonardo TE, Cass BN, Hurwitz B, Stern D, Gibbs RA, Richards S, Moran NA. 2010. Dynamics of genome evolution in facultative symbionts of aphids. Environ Microbiol 12:2060–2069. doi: 10.1111/j.1462-2920.2009.02085.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mondo SJ, Toomer KH, Morton JB, Lekberg Y, Pawlowska TE. 2012. Evolutionary stability in a 400-million-year-old heritable facultative mutualism. Evolution 66:2564–2576. doi: 10.1111/j.1558-5646.2012.01611.x. [DOI] [PubMed] [Google Scholar]
- 42.Russell JA, Latorre A, Sabater-Muñoz B, Moya A, Moran NA. 2003. Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol Ecol 12:1061–1075. doi: 10.1046/j.1365-294X.2003.01780.x. [DOI] [PubMed] [Google Scholar]
- 43.Manzano-Marín A, Latorre A. 2014. Settling down: the genome of Serratia symbiotica from the aphid Cinara tujafilina zooms in on the process of accommodation to a cooperative intracellular life. Genome Biol Evol 6:1683–1698. doi: 10.1093/gbe/evu133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sloan DB, Moran NA. 2013. The evolution of genomic instability in the obligate endosymbionts of whiteflies. Genome Biol Evol 5:783–793. doi: 10.1093/gbe/evt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCutcheon JP, von Dohlen CD. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol 21:1366–1372. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baumann P. 2005. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- 47.Cordaux R. 2009. Gene conversion maintains nonfunctional transposable elements in an obligate mutualistic endosymbiont. Mol Biol Evol 26:1679–1682. doi: 10.1093/molbev/msp093. [DOI] [PubMed] [Google Scholar]
- 48.Clark MA, Moran NA, Baumann P. 1999. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol Biol Evol 16:1586–1598. doi: 10.1093/oxfordjournals.molbev.a026071. [DOI] [PubMed] [Google Scholar]
- 49.Foster J, Ganatra M, Kamal I, Ware J, Makarova K, Ivanova N, Bhattacharyya A, Kapatral V, Kumar S, Posfai J, Vincze T, Ingram J, Moran L, Lapidus A, Omelchenko M, Kyrpides N, Ghedin E, Wang S, Goltsman E, Joukov V, Ostrovskaya O, Tsukerman K, Mazur M, Comb D, Koonin E, Slatko B. 2005. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol 3:e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambert JD, Moran NA. 1998. Deleterious mutations destabilize ribosomal RNA in endosymbiotic bacteria. Proc Natl Acad Sci U S A 95:4458–4462. doi: 10.1073/pnas.95.8.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellegaard KM, Klasson L, Näslund K, Bourtzis K, Andersson SG. 2013. Comparative genomics of Wolbachia and the bacterial species concept. PLoS Genet 9:e1003381. doi: 10.1371/journal.pgen.1003381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brelsfoard C, Tsiamis G, Falchetto M, Gomulski LM, Telleria E, Alam U, Doudoumis V, Scolari F, Benoit JB, Swain M, Takac P, Malacrida AR, Bourtzis K, Aksoy S. 2014. Presence of extensive Wolbachia symbiont insertions discovered in the genome of its host Glossina morsitans morsitans. PLoS Negl Trop Dis 8:e2728. doi: 10.1371/journal.pntd.0002728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cerveau N, Leclercq S, Leroy E, Bouchon D, Cordaux R. 2011. Short- and long-term evolutionary dynamics of bacterial insertion sequences: insights from Wolbachia endosymbionts. Genome Biol Evol 3:1175–1186. doi: 10.1093/gbe/evr096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kondrashov AS. 1993. Classification of hypotheses on the advantage of amphimixis. J Hered 84:372–387. [DOI] [PubMed] [Google Scholar]
- 55.Jaenike J. 1978. A hypothesis to account for the maintenance of sex within populations. J Evol Theory 3:191–194. [Google Scholar]
- 56.Sirand-Pugnet P, Lartigue C, Marenda M, Jacob D, Barré A, Barbe V, Schenowitz C, Mangenot S, Couloux A, Segurens B, de Daruvar A, Blanchard A, Citti C. 2007. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet 3:e75. doi: 10.1371/journal.pgen.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dordet-Frisoni E, Sagné E, Baranowski E, Breton M, Nouvel LX, Blanchard A, Marenda MS, Tardy F, Sirand-Pugnet P, Citti C. 2014. Chromosomal transfers in mycoplasmas: when minimal genomes go mobile. mBio 5:e01958-14. doi: 10.1128/mBio.01958-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rankin DJ, Rocha EP, Brown SP. 2011. What traits are carried on mobile genetic elements, and why? Heredity 106:1–10. doi: 10.1038/hdy.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thiaucourt F, Manso-Silvan L, Salah W, Barbe V, Vacherie B, Jacob D, Breton M, Dupuy V, Lomenech AM, Blanchard A, Sirand-Pugnet P. 2011. Mycoplasma mycoides, from “mycoides Small Colony” to “capri”. A microevolutionary perspective. BMC Genomics 12:114. doi: 10.1186/1471-2164-12-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szczepanek SM, Tulman ER, Gorton TS, Liao X, Lu Z, Zinski J, Aziz F, Frasca S Jr, Kutish GF, Geary SJ. 2010. Comparative genomic analyses of attenuated strains of Mycoplasma gallisepticum. Infect Immun 78:1760–1771. doi: 10.1128/IAI.01172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marini E, Palmieri C, Magi G, Facinelli B. 2015. Recombination between Streptococcus suis ICESsu32457 and Streptococcus agalactiae ICESa2603 yields a hybrid ICE transferable to Streptococcus pyogenes. Vet Microbiol 178:99–104. doi: 10.1016/j.vetmic.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 62.Garriss G, Waldor MK, Burrus V. 2009. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet 5:e1000775. doi: 10.1371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tardy F, Mick V, Dordet-Frisoni E, Marenda MS, Sirand-Pugnet P, Blanchard A, Citti C. 2015. Integrative conjugative elements are widespread in field isolates of Mycoplasma species pathogenic for ruminants. Appl Environ Microbiol 81:1634–1643. doi: 10.1128/AEM.03723-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]