Abstract
Plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone), a natural naphthoquinone compound isolated from roots of Plumbago zeylanica L., has drawn a lot of attention for its plenty of pharmacological properties including antidiabetes and anti-cancer. The aim of this study was to investigate the effects of plumbagin on CYP1A2, CYP2B1/6, CYP2C9/11, CYP2D1/6, CYP2E1 and CYP3A2/4 activities in human and rat liver and evaluate the potential herb-drug interactions using the cocktail approach. All CYP substrates and their metabolites were analyzed using high-performance liquid chromatography–tandem mass spectrometry (LC-MS/MS). Plumbagin presented non-time-dependent inhibition of CYP activities in both human and rat liver. In humans, plumbagin was not only a mixed inhibitor of CYP2B6, CYP2C9, CYP2D6, CYP2E1 and CYP3A4, but also a non-competitive inhibitor of CYP1A2, with Ki values no more than 2.16 μM. In rats, the mixed inhibition of CYP1A2 and CYP2D1, and competitive inhibition for CYP2B1, CYP2C11 and CYP2E1 with Ki values less than 9.93 μM were observed. In general, the relatively low Ki values of plumbagin in humans would have a high potential to cause the toxicity and drug interactions involving CYP enzymes.
Plumbago zeylanica L. (family: Plumbaginaceae), also known as Chitrak, is a medicinal plant and mainly distributed around Africa and Asia including India and China1,2. It is originally documented in “Tang Ben Cao” in China and widely used in prescription for treating many diseases such as rheumatic joint pain, blood stasis amenorrhea and malignant sore in Traditional Chinese medicine3. Plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone, Fig. 1a) isolated from roots of Plumbago zeylanica L. is an active naphthoquinone and responsible for its therapeutic effects4. Previous studies have reported that plumbagin possesses lots of pharmacological properties including antidiabetic, anti-inflammatory, antibacterial and anti-immediate allergic reaction activities1,5. In particular, plumbagin presents significant anticancer activity6. For example, plumbagin can induce apoptosis and autophagy of human prostate cancer cells via sirtuin1- and PI3K/Akt/mTOR-mediated pathways7. And it also can kill human gastric cancer cells by induction of SH2-containing protein tyrosine phosphatase 18. Therefore, Plumbagin is considered to be a promising antitumor candidate for drug development.
Figure 1. Chemical structure of plumbagin (a) and the inhibitory effects of CYPs activities by plumbagin in pooled human liver microsomes (b) and pooled rat liver microsomes (c).
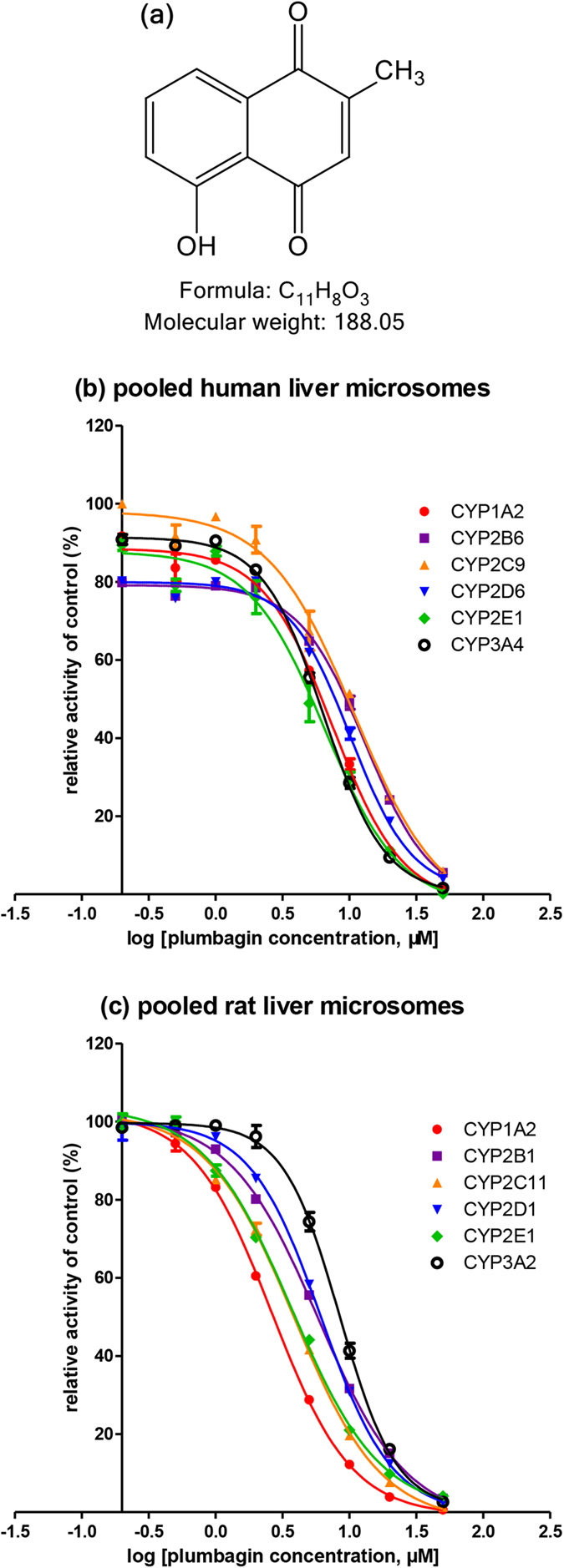
Chemical structure was produced using ChemDraw Professional 15.0. Results were mean ± SD of quadruplicate assays. The data was fit to log (plumbagin concentration) and normalized response equations using GraphPad Prism 5.0.
The cytochrome P450 (CYP) enzymes play an important role in metabolism of xenobiotics and endogenous substances. Almost 90% drugs are metabolized by CYP superfamily9,10. Among the CYP isoforms identified, the major human CYP isoforms (CYP1A2, CYP2B6, CYP2C9, CYP2D6, CYP2E1 and CYP3A4) are responsible for 80% of the CYP-mediated metabolism11,12,13. However, the inhibition and induction of CYP by herbs often occur, which is directly linked to herb-drug interactions (HDIs) and result in the toxicity and/or therapeutic failure14,15. In consequence, U.S. Food and Drug Administration (FDA) has recommended to use CYP-associated metabolic studies in vitro to predict the potential HDIs, which is the major attrition in drug development16. Recently, one study only found the inhibitory effects of plumbagin on CYP1A2, CYP2C19 and CYP3A417. However, the mechanisms of CYP inhibition remain unclear. Further studies should be carried out to illustrate the mechanisms of plumbagin on CYP enzymes.
The aim of this study was to investigate the effects of plumbagin on several major CYP activities both in human and rat liver, including CYP1A2, CYP2B1/6, CYP2C9/11, CYP2D1/6, CYP2E1 and CYP3A2/4, and further explore the mechanisms of plumbagin on CYP inhibitory properties. In this report, the cocktail approach using classical substrates including phenacetin (CYP1A2), bupropion (CYP2B1/6), tolbutamide (CYP2C9/11), dextromethorphan (CYP2D1/6), chlorzoxazone (CYP2E1) and midazolam (CYP3A2/4)16, alternatively named the n-in-one assay, was adopted to monitor several CYP activities in a single experiment, which was much comprehensive, time and resource efficient10,18. In this approach, a liquid chromatography tandem mass spectrometry (LC-MS/MS) was usually used for the simultaneous detection of the formation of metabolites of these probe substrates to calculate the enzymatic reaction rates, which could be used to evaluate the corresponding CYP enzyme activities19. To our knowledge, this is the first study to report the potential effects of plumbagin on CYP2B1/6, CYP2C9/11, CYP2E1 and CYP2D1/6, and demonstrate the inhibitory mechanisms of CYPs. Therefore, the present study could be helpful to evaluate the potential HDIs involved with plumbagin comprehensively.
Results
Inhibition effects of plumbagin on CYP1A2, CYP2B6, CYP2C9, CYP2D6, CYP2E1 and CYP3A4 activities in human liver
To investigate the effects of plumbagin on human CYP activities, various concentrations of plumbagin ranging from 0.05 μM to 50 μM were used to determine IC50 (Fig. 1b). Plumbagin inhibited CYP1A2-catalyzed phenacetin O-deethylation, CYP2B6-catalyzed bupropion hydroxylation, CYP2C9-catalyzed tolbutamide methyl-hydroxylation, CYP2D6-catalyzed dextromethorphan O-demethylation, CYP2E1-catalyzed chlorzoxazone 6-hydroxylation and CYP3A4-catalyzed midazolam 1-hydroxylation with IC50 values at 7.49 μM, 13.10 μM, 10.94 μM, 10.21 μM, 6.54 μM and 6.45 μM in HLMs, respectively (Table 1). These data certificated that plumbagin exhibited potently inhibitory effects on the major human six CYPs, including CYP1A2, CYP2B6, CYP2C9, CYP2D6, CYP2E1 and CYP3A4.
Table 1. Inhibitory effects of plumbagin on six CYP isoforms in HLMs and RLMs.
| CYP isoform | IC50 (μM) | Ki (μM) | αKi (μM) | α | Type | |
|---|---|---|---|---|---|---|
| Humans | 1A2 | 7.49 | 0.15 | 0.16 | 0.94 | non-competitive |
| 2B6 | 13.10 | 1.62 | 2.92 | 0.55 | mixed-type | |
| 2C9 | 10.94 | 2.16 | 1.46 | 1.48 | mixed-type | |
| 2D6 | 10.21 | 1.46 | 2.22 | 0.66 | mixed-type | |
| 2E1 | 6.54 | 0.65 | 2.64 | 0.25 | mixed-type | |
| 3A4 | 6.45 | 0.88 | 0.57 | 1.54 | mixed-type | |
| Rats | 1A2 | 2.65 | 2.64 | 8.51 | 0.31 | mixed-type |
| 2B1 | 5.86 | 9.93 | ‒ | ‒ | competitive | |
| 2C11 | 3.85 | 7.85 | ‒ | ‒ | competitive | |
| 2D1 | 6.17 | 9.49 | 13.79 | 0.69 | mixed-type | |
| 2E1 | 3.72 | 6.28 | ‒ | ‒ | competitive | |
| 3A2 | 8.47 | ‒ | ‒ | ‒ | atypical kinetics |
Inhibition effects of plumbagin on CYP1A2, CYP2B1, CYP2C11, CYP2D1, CYP2E1 and CYP3A2 activities in rat liver
In rat liver, plumbagin also inhibited CYP1A2-catalyzed phenacetin O-deethylation, CYP2B1-catalyzed bupropion hydroxylation, CYP2C11-catalyzed tolbutamide methyl-hydroxylation, CYP2D1-catalyzed dextromethorphan O-demethylation, CYP2E1-catalyzed chlorzoxazone 6-hydroxylation and CYP3A2-catalyzed midazolam 1-hydroxylation with IC50 values at 2.65 μM, 5.86 μM, 3.85 μM, 6.17 μM, 3.72 μM and 8.47 μM, respectively (Table 1). In brief, plumbagin strongly inhibited all selected CYP-catalyzed probe reactions in a concentration-dependent manner (Fig. 1c), which agreed with the data of humans.
Enzyme kinetic analysis for inhibition of CYPs by plumbagin in human liver
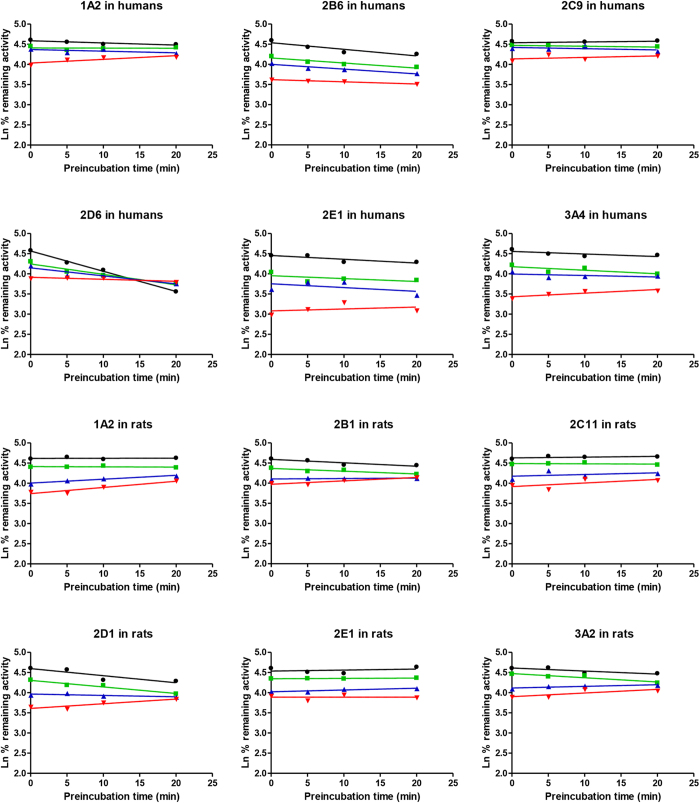
To investigate whether the inhibition of CYPs by plumbagin was time-dependent, CYP activities were determined by pre-incubating microsomal incubation mixtures with NADPH for 0, 5, 10 and 20 min at 37 °C. Plumbagin showed no apparent time-dependent inhibition on all CYPs (CYP1A2, CYP2B6, CYP2C9, CYP2D6, CYP2E1 and CYP3A4) (Fig. 2).
Figure 2. Time- and concentration-dependent inhibition of human and rat liver microsomal CYPs.
The figures were prepared by a semilogarithmic plot of percent remaining activity vs incubation time. Microsomal reaction mixtures were pre-incubated for 20 min without (⚫) or with plumbagin at 0.5 μM ( ), 1 μM (
), 1 μM ( ), 2 μM (
), 2 μM ( ) in humans or plumbagin at 0.2 μM (
) in humans or plumbagin at 0.2 μM ( ), 0.5 μM (
), 0.5 μM ( ), 1 μM (
), 1 μM ( ) in rats. Each data point represents the mean of quadruplicate determinations.
) in rats. Each data point represents the mean of quadruplicate determinations.
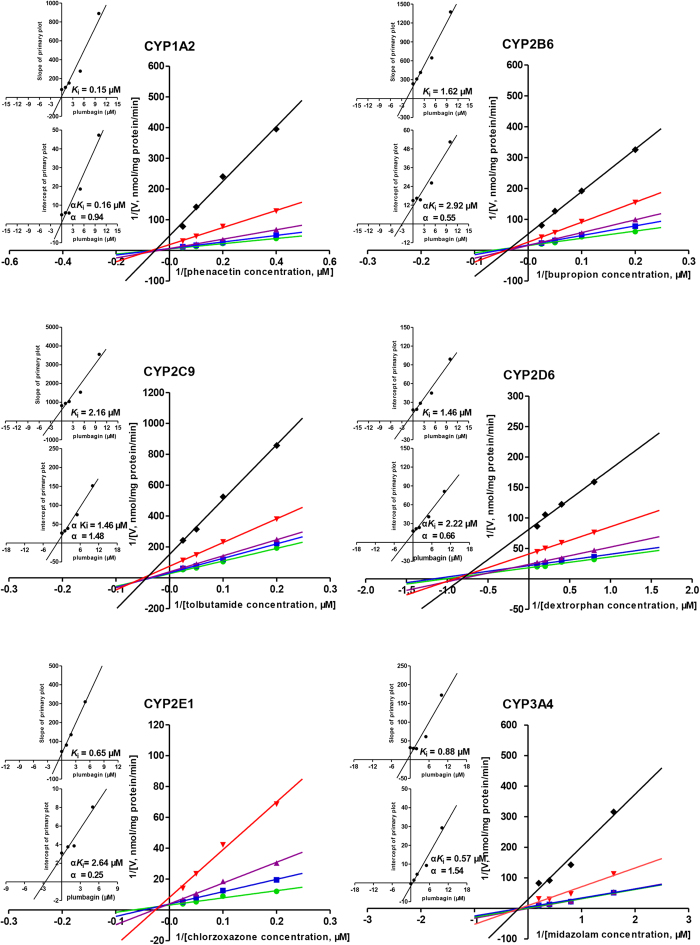
To further determine the types of CYP inhibition by plumbagin in HLMs, enzyme kinetics studies were carried out with various substrates concentrations in the presence or absence of plumbagin. The Lineweaver-Burk transformation of the enzyme velocities versus substrates concentrations showed that the types of inhibition by plumbagin for these selected CYPs were mixed inhibition on CYP2B6, CYP2C9, CYP2D6, CYP2E1 and CYP3A4, but non-competitive for CYP1A2. The Ki values were determined using the secondary Lineweaver–Burk plot, and then calculated at 0.15 μM, 1.62 μM, 2.16 μM, 1.46 μM, 0.65 μM and 0.88 μM for CYP1A2, CYP2B6, CYP2C9, CYP2D6, CYP2E1 and CYP3A4, respectively. From the secondary plot of Lineweaver–Burk plot for αKi, αKi values were 0.16 μM, 2.92 μM, 1.46 μM, 2.22 μM, 2.64 μM and 0.57 μM for CYP1A2, CYP2B6, CYP2C9, CYP2D6, CYP2E1 and CYP3A4, respectively. Since α values of CYP2B6, CYP2C9, CYP2D6, CYP2E1 and CYP3A4 were not equal to 1, the types of inhibition by plumbagin for these five CYPs were mixed inhibition. However, the type of inhibition by plumbagin for CYP1A2 was non-competitive inhibition due to α value of CYP1A2 approximately equal to 1 (Table 1, Fig. 3).
Figure 3. Primary Lineweaver–Burk plot, the secondary plot for Ki and the secondary plot for αKi in the inhibition of CYP-mediated probe substrate metabolism by various concentrations of plumbagin (0 , 1
, 1 , 2
, 2 , 5
, 5  , 10♦ μM) in pooled human liver microsomes.
, 10♦ μM) in pooled human liver microsomes.
Phenacetin was used at concentrations of 2.5, 5, 10 and 20 μM; Bupropion was used at 5, 10, 20 and 40 μM; Tolbutamide was used at 5, 10, 20 and 40 μM; Dextromethorphan was used at 1.25, 2.5, 5 and 10 μM; Chlorzoxazone was used at 5, 10, 20 and 40 μM; Midazolam was used at 0.625, 1.25, 2.5 and 5 μM. Each data point represents the mean of quadruplicate determinations.
Enzyme kinetic analysis for inhibition of CYPs by plumbagin in rat liver
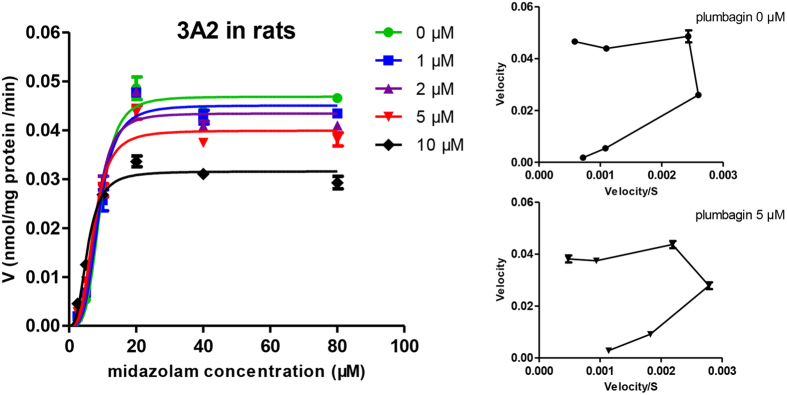
The time-dependent inhibition experiments were also performed and no time-dependent inhibition was observed in RLMs (Fig. 2). The Lineweaver-Burk transformation of the enzyme velocities versus substrates concentrations showed that the types of inhibition by plumbagin for these selected CYPs were mixed inhibition on CYP1A2 and CYP2D1, but competitive for CYP2B1, CYP2C11 and CYP2E1. The Ki values were calculated to be 2.64 μM, 9.93 μM, 7.85 μM, 9.49 μM and 6.28 μM for CYP1A2, CYP2B1, CYP2C11, CYP2D1 and CYP2E1, respectively. From the secondary plot of Lineweaver-Burk plot for αKi, αKi values were 8.51 μM and 13.79 μM for CYP1A2 and CYP2D1, respectively. The types of inhibition by plumbagin on CYP1A2 and CYP2D1 were mixed inhibition, because α values of CYP1A2 and CYP2D1 were not equal to 01 (Table 1, Fig. 4). Moreover, the enzyme inhibition kinetics of CYP3A2 did not conform to the classical Michaelis–Menten kinetics (Fig. 5), which would be discussed in Discussion section.
Figure 4. Primary Lineweaver–Burk plot, the secondary plot for Ki and the secondary plot for αKi in the inhibition of CYP-mediated probe substrate metabolism by various concentrations of plumbagin (0 , 1
, 1 , 2
, 2 , 5
, 5  , 10 ♦ μM) in rat liver microsomes.
, 10 ♦ μM) in rat liver microsomes.
Phenacetin was used at concentrations of 2.5, 5, 10 and 20 μM; Bupropion was used at 5, 10, 20 and 40 μM; Tolbutamide was used at 5, 10, 20 and 40 μM; Dextromethorphan was used at 1.25, 2.5, 5 and 10 μM; Chlorzoxazone was used at 5, 10, 20 and 40 μM. Each data point represents the mean of six rats.
Figure 5. Sigmoidal auto-activation kinetics curve of CYP3A2 by various concentrations of plumbagin (0  , 1
, 1 , 2
, 2 , 5
, 5  , 10 ♦ μM) in rat liver microsomes.
, 10 ♦ μM) in rat liver microsomes.
Midazolam was used at concentrations of 2.5, 5, 10, 20, 40 and 80 μM. Insets show the corresponding Eadie-Hofstee plots for CYP3A2 kinetic profile.
Discussion
Plumbago zeylanica L. has been widely used as traditional herbal medicine among many Chinese minorities for many years. Meanwhile, the medicinal plant is the main ingredient of Qubai Babuqi Pian (Sinopharm XinJiang Pharmaceutical Co., Ltd.) which is used to treat leucoderma in Uighur medicine. Nowadays, herbal medicines, especially being consumed along with modern drugs, are increasingly popular when patients are in several diseases or conditions11,14. When they are co-administered over overlapping time periods, the possibility of HDIs mediated by CYPs might exist20,21. However, the inhibition effects of plumbagin on CYP activities were rarely concerned. Among all CYP isoforms in humans, CYP3A4, CYP2C9, CYP1A2, CYP2E1, CYP2D6 and CYP2B6 are relative abundant, which accounts about for 40%, 20%, 13%, 10%, 2% and 2% in total CYPs respectively, and responsible for metabolism of the majority of clinically prescribed drugs14,22,23,24. CYP1A2 is involved in the metabolism of a variety of clinically using drugs, such as clozapine, tacrine, tizanidine, theophylline, and several important endogenous compounds22,24,25,26. CYP2B6 is responsible for approximately 3–8% of widely used pharmaceuticals and also reported to have high expression in breast tumors23,27,28. CYP2C9 contributes to the metabolism of not only commonly prescribed drugs which accounts for 10–20%, but also fatty acids, prostanoids and steroid hormones22. CYP2D6 expressing in liver and extrahepatic organs mediates approximately 25% of marketed drugs, such as antidepressants and antipsychotics29. Meanwhile, CYP2D6 with decreased activity is implicated in Parkinson disease in Caucasian populations29. CYP2E1 as a natural ethanol-inducible enzyme metabolically activates various carcinogens. Increased CYP2E1 activity is linked to the carcinogenic process30. Furthermore, more than 50% of drugs were metabolized by CYP3A4 including antibiotics, antiviral drugs, benzodiazepines, calcium channel blockers, statins and immunosuppressive drugs31,32. However, to our knowledge, the inhibition effects of plumbagin on these important CYPs have not been reported except for CYP1A2 and CYP3A417. In particular, the mechanisms of inhibition of all six CYPs were completely investigated for the first time. The purpose of our work was to investigate the effects of plumbagin on the major CYP activities both in human and rat liver and assess the potential HDIs involved plumbagin or Plumbago zeylanica L.
To evaluate the potential HDIs, the cocktail approach, frequently used for CYP inhibition studies33,34,35, has been developed for monitoring CYP activities in a single run by LC-MS/MS with high sensitivity and selectivity. According to FDA guidance, classical probe substrates including phenacetin (CYP1A2), bupropion (CYP2B1/6), tolbutamide (CYP2C9/11), dextromethorphan (CYP2D1/6), chlorzoxazone (CYP2E1) and midazolam (CYP3A2/4) were selected. The concentrations of probe substrates were close to the Km values of each CYP-catalyzed reaction according to published papers16,33. Given that the linear initial rates of metabolites formation for all selected CYPs with minimal substrates depletion and the automatic inactivation of CYPs in 37 °C comprehensively, the incubation time was chosen to 20 min. Moreover, the microsomal protein concentration in HLMs was 0.5 mg/ml, which was the most frequently employed, and high protein concentrations ought to be avoided because of the possibility of protein binding18.
To investigate whether inhibition of CYPs by plumbagin was time-dependent, time-dependent inhibition assays were performed with various plumbagin concentrations close to IC25, which was the most sensitive point for detecting a time-dependent inhibitor36,37. Too low concentrations of plumbagin may miss inactivation, but too high concentrations of plumbagin may cause much reversible inhibition to make the effect of time-dependent inhibition not obvious36. Meanwhile, considering not only automatic inactivation of some CYPs but also weak inhibitors which could not be identified within a short inactivation time, 20 min was used for this time-dependent inhibition assays. From Fig. 2, the activity of CYP2D6 in HLMs decreased slightly in a time-dependent manner without any inhibitor, which could be explained for easily automatic inactivation of CYP2D6 in HLMs. On the other hand, the inhibition effects on several CYPs by plumbagin were reduced with pre-incubation with NADPH, which could be due to metabolic depletion of plumbagin leading to diminished inhibitory capability36,38. Therefore, no time-dependent inhibition was observed in both HLMs and RLMs, which showed reversible inhibition of CYP1A2, CYP2B1/6, CYP2C9/11, CYP2D1/6, CYP2E1 and CYP3A2/4 by plumbagin.
The present study showed that the types of inhibition by plumbagin were mixed inhibition on CYP2B6, CYP2C9, CYP2D6, CYP2E1 and CYP3A4, but non-competitive for CYP1A2 in HLMs and the Ki values were 1.62 μM, 2.16 μM, 1.46 μM, 0.65 μM, 0.88 μM and 0.15 μM, respectively. Correspondingly, mixed inhibition on CYP1A2 and CYP2D1, but competitive for CYP2B1, CYP2C11 and CYP2E1 were observed in RLMs and the Ki values were 2.64 μM, 9.49 μM, 9.93 μM, 7.85 μM, and 6.28 μM, respectively. Thus, plumbagin was a potent inhibitor on the above CYPs in both humans and rats.
Based on the inhibition results of HLMs and RLMs, some differences should be noticed such as the types of CYP inhibition and inhibitory intensity. The Ki values of inhibition on CYPs by plumbagin in HLMs are generally lower than those in RLMs. It may be attributed to variations in CYP levels, inducibilities and the existence of different CYP isoforms of the same protein family or subfamily in different species39. For example, CYP2C11 accounts for about 50% in total rat CYPs while human CYP2C9 being a homologue of the rat CYP2C11 with 77% homology only accounts for 20% of total human CYPs40. In addition, the enzyme inhibition kinetic of CYP3A2 disagreed with the classical Michaelis–Menten kinetics in RLMs, but the enzyme inhibition kinetic of CYP3A4 in HLMs conformed to that when the concentrations of midazolam were no more than 5 μΜ. The differences between CYP3A2 and CYP3A4 could also be a reflection of species differences. Consequently, extrapolating results from animals to humans should be cautious.
As mentioned above, when investigating the enzyme inhibition kinetic of CYP3A2, we found it did not conform to the classical Michaelis–Menten kinetics in RLMs. In fact, metabolic activation was achieved by incubation of plumbagin in the presence of CYP3A2 when the concentrations of midazolam are less than 5 μΜ in RLMs41. In this report, however, the unusual effects of plumbagin on CYP3A2 could be explained using an atypical kinetics model. Atypical Michaelis–Menten kinetics including substrate inhibition, partial inhibition, activation, auto-activation and biphasic metabolism has been demonstrated for some CYP3A-mediated reactions42,43. In this study, CYP3A2 displayed auto-activation kinetics. Multisite hypothesis was a possible mechanistic model of atypical kinetics. CYP3A enzyme except for two or more midazolam-binding sites contains an activator binding site which is closely related to the substrate binding sites42. Plumbagin might lead to metabolic activation by binding to the activator binding sites. These speculations still need to be further proved by experiments.
Previous pharmacokinetic study of plumbagin by oral administration (100 mg/kg) in rats showed that the peak concentration (Cmax), half-life (t1/2) and area under the curve (AUC) of plumbagin in the plasma was 0.35 μg/ml (1.86 μM), 1028 min and 271.9 min*μg/ml, respectively3. Therefore, the rat plasma concentrations were close to the IC50 and Ki values determined in this study, thus suggesting plumbagin could quite probably decrease the activities of CYP1A2, CYP2B1, CYP2C11, CYP2D1, CYP2E1 and CYP3A2 in vivo. In fact, these CYP homologues in humans are responsible for 80% of the CYP-mediated metabolism including a number of drugs with narrow therapeutic ranges. Therefore the potential HDIs should be brought to attention and further studies in vivo are needed to confirm the interactions of plumbagin with CYP isoforms in humans.
In conclusion, the data in this study demonstrated that plumbagin exhibited potently broad-spectrum inhibitory effects toward CYP enzymes including CYP1A2, CYP2B1/6, CYP2C9/11, CYP2D1/6, CYP2E1 and CYP3A2/4 with lower Ki values in both human and rat liver. In particular, plumbagin was not only a mixed inhibitor of CYP2B6, CYP2C9, CYP2D6, CYP2E1 and CYP3A4, but also a non-competitive inhibition of CYP1A2 in humans. These results provided useful guidance for safe and effective usage of plumbaign in clinic, especially in combination with other drugs.
Methods
Materials and chemicals
Pooled human liver microsomes (n = 20) were obtained from Corning Gentest Corporation (Woburn, MA, USA) and stored at −150 °C until use. All the experimental procedures involving humans have been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) guidelines. Phenacetin, bupropion, tolbutamide, dextromethorphan, midazolam, 3-acetamidophenol (internal standard), chlorpropamide (internal standard), glucose 6-phosphate (G6P), glucose 6-phosphate dehydrogenase (G6PDH), β-nicotinamide adenine dinucleotide phosphate (NADP) and tris (hydroxymethyl) aminomethane hydrochloride (Tris-HCl) were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Chlorzoxazone was purchased from Alfa Aesar (Massachusetts, USA). 4-acetamidophenol, hydroxybupropion, 4-hydroxytolbutamide, dextrorphan, 6-hydroxychlorzoxazone and 1-hydroxymidazolam were obtained from Toronto Research Chemical (North York, Canada). Mebendazole (internal standard) was obtained from Aladdin Industrial Co. (California, USA). Plumbagin (purity > 98%) was purchased from Dalian Meilun Biotech Co., Ltd (Dalian, China). Acetonitrile and methanol (all HPLC grade) were purchased from Fisher Chemicals (Leicester, UK). Formic acid (HPLC grade) was purchased from TEDIA (Ohio, USA). Distilled water was purified in a Millipore system Milli Q (Millipore Corp., Bedford, MA, USA).
Animals
Male Sprague-Dawley rats (200–250 g) were supplied by National Rodent Laboratory Animal Resources, Shanghai Branch of China. The animals were kept in a specific pathogen-free facility and with access to rodent cubes and sterile water, with 12 h light-dark cycles. All the methods in animals were carried out in accordance with the National Institutes of Health standards established in the ‘Guidelines for the Care and Use of Experimental Animals’. All experimental protocols in animals were approved by the Ethics Committee on Animal Experimentation of East China Normal University (Shanghai, China).
Preparation of rat liver microsomes
In this study, the rats were fasted overnight and killed by cervical dislocation before removal of the liver. The liver was excised, rinsed with ice-cold saline (0.9% NaCl w/v), weighed and homogenized in a 0.05 mM Tris/KCl buffer (pH 7.4). Subsequently, the homogenate was centrifuged at 10,000 × g at 4 °C for 30 min. The supernatant was then centrifuged at 105,000 × g at 4 °C for 60 min. The pellet was reconstituted with 0.05 mM Tris/KCl buffer (pH 7.4) and stored at −150 °C until use. The protein concentration of the liver microsomes was determined by a protein quantitative assay using bicinchoninic acid22.
Microsomal incubation
Microsomal incubation was carried out at 37 °C in a mixture of 200 μl in 0.05 M Tris/HCl buffer (pH 7.4) consisting of pooled HLMs (0.5 mg/ml) or RLMs (1 mg/ml) and an NADPH-regenerating system including MgCl2 (5 mM), G6P (10 mM), G6PDH (0.4 U/ml) and NADP (1 mM). Substrates were added into the incubation mixture and obtained the final concentrations (10 μM phenacetin for CYP1A2, 20 μM bupropion for CYP2B1/6, 20 μM tolbutamide for CYP2C9/11, 5 μM dextromethorphan for CYP2D1/6, 20 μM chlorzoxazone for CYP2E1 and 10 μM midazolam for CYP3A2/4), which were close to Km respectively. The proportion of organic solvent was not higher than 1% (v/v) in the incubation mixture18,44. After pre-incubation for 5 min, the reaction was initiated by addition of NADP. Incubations were performed for 20 min, stopped by adding 200 μl ice-cold acetonitrile. And then 20 μl internal standard solution was added (3-acetamidophenol 2 μg/ml, chlorpropamide 2 μg/ml and mebendazole 2 μg/ml). The solution was mixed thoroughly for 3 min and centrifuged for 10 min at 16,000 × g at 4 °C. The supernatant (60 μl) was transferred to autosampler vials, and 2 μl was injected into LC-MS/MS system.
Determination of IC50 values for inhibition
The IC50 (concentration of inhibitor to cause 50% inhibition of original enzyme activity) values were determined with the same incubation condition above. The specific concentrations of plumbagin were ranged from 0.1 μM to 50 μM. The IC50 values were calculated by plotting relative activities over the logarithm of plumbagin concentrations using GraphPad Prism 5.0 (GraphPad software Inc., CA, USA).
Mechanism of inhibition of CYPs by plumbagin
To explore the time-dependent inhibition of plumbagin on CYPs, inactivation incubation containing plumbagin whose final concentration close to IC25 (concentration of inhibitor to cause 25% inhibition of original enzyme activity)37, pooled HLMs (0.5 mg/ml) or RLMs (1 mg/ml), MgCl2 (5 mM), G6P (10 mM), G6PDH (0.4 U/ml), NADP (1 mM) and 0.05 M Tris/HCl buffer (pH 7.4) was performed for 0, 5, 10 and 20 min at 37 °C. Cocktail probe substrates were then added and incubated for 20 min. Reactions were stopped with 200 μl ice-cold acetonitrile. The samples were treated according to the microsomal incubation method and then injected into LC-MS/MS for analysis.
Kinetic studies were performed for determining the types of inhibition on CYP by plumbagin. Pooled HLMs (0.5 mg/ml) or RLMs (1 mg/ml) with plumbagin (1 μM, 2 μM, 5 μM and 10 μM) were incubated in 0.05 M Tris/HCl buffer (pH 7.4) for 20 min at 37 °C. A series of concentrations for cocktail probe substrates were used: 2.5–20 μM phenacetin for CYP1A2; 5–40 μM bupropion for CYP2B1/6; 5–40 μM tolbutamide for CYP2C9/11; 1.25–10 μM dextromethorphan for CYP2D1/6; 5–40 μM chlorzoxazone for CYP2E1; 2.5–80 μM midazolam for CYP3A2; and 0.625–5 μM midazolam for CYP3A4. Enzyme kinetics as well as statistical analysis were performed using GraphPad Prism 5.0.
LC-MS/MS analysis
LC-MS/MS is a powerful tool to detect the concentrations of several CYP substrates and metabolites in a short time, and to calculate the enzymatic reaction rate, which is used to evaluate the corresponding CYP enzyme activities. In this study, an Agilent 1290 LC system consisting of a degasser, a binary pump, an autosampler and a thermostatic column compartment was coupled with a 6460 triple-quadrupole mass spectrometer (Agilent Technologies, USA), which was equipped with an Agilent Jet Stream electrospray ionization (ESI) source and operated with Agilent MassHunter version 5.0.280.1 software (Agilent Technologies, USA). Chromatography separation was performed on a Phenomenex Kinetex XB-C18 column (100 × 3.00 mm, 2.6 μM) protected by a Phenomenex C18 guard column (Torrance, CA, USA). The mobile phase consisted of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile) using gradient elution at a flow rate of 0.3 mL/min. The optimum condition for elution was as follows: 0–2.2 min, 10–11% B; 2.2–8.5 min, 11–90% B; 8.5–9 min, 90–92% B; 9–9.3 min, 92% B; 9.3–9.6 min, 92–10% B; 9.6–11.5 min, 10% B. The temperature of column oven was maintained at 30 °C, and the injection volume was 2 μL. The mass spectrometer was operated in the positive/negative ion-switching ESI mode, and the detection of the ions was performed in the multiple reaction monitoring (MRM) mode. Each MRM transition and the compound dependent parameters are presented in Table 2. MS/MS conditions were optimized as follows: gas temperature, 350 °C; gas flow, 10 L/min; nebulizer, 35 psi; sheath gas temperature, 350 °C; sheath gas flow, 11 L/min; collision gas (N2), 1.6 MPa. A divert valve was used to divert the eluent to waste from 0 to 2 min, 10 to 11.5 min, and to MS from 2 to 10 min. Representative multiple reaction monitoring chromatograms of analytes were shown in Fig. 6.
Table 2. Analytical parameters for the individual substrate, metabolite and internal standard (IS).
| Analyte | Ionization mode | Precursor ion (m/z) | Product ion (m/z) | Fragmentor (V) | Collision energy (eV) |
|---|---|---|---|---|---|
| Phenacetin | ESI+ | 180.1 | 110.2 | 110 | 21 |
| 4-acetamidophenol | ESI+ | 152.1 | 110.1 | 90 | 15 |
| Bupropion | ESI+ | 240.1 | 184.1 | 90 | 8 |
| Hydroxybupropion | ESI+ | 256.1 | 238.1 | 70 | 8 |
| Tolbutamide | ESI− | 269.2 | 170.0 | 100 | 12 |
| 4-hydroxytolbutamide | ESI− | 285.2 | 186.0 | 100 | 12 |
| Dextromethorphan | ESI+ | 272.3 | 147.1 | 150 | 33 |
| Dextrorphan | ESI+ | 258.1 | 157.1 | 150 | 42 |
| Chlorzoxazone | ESI− | 168.0 | 132.0 | 110 | 20 |
| 6-hydroxychlorzoxazone | ESI− | 184.1 | 120.0 | 90 | 17 |
| Midazolam | ESI+ | 326.1 | 291.0 | 170 | 28 |
| 1-hydroxymidazolam | ESI+ | 342.1 | 324.0 | 130 | 20 |
| 3-acetamidophenol (IS) | ESI+ | 152.1 | 93.0 | 80 | 26 |
| Chlorpropamide (IS) | ESI− | 275.1 | 190 | 100 | 11 |
| Mebendazole (IS) | ESI+ | 296.1 | 264.1 | 150 | 22 |
Figure 6. Representative multiple reaction monitoring chromatograms of CYP probe substrates, metabolites and internal standards.
Data analysis
All data were presented as mean ± SD. The IC50 values were fitted by non-linear regression analysis using GraphPad Prism 5.0. The kinetic parameters were obtained by fitting velocity data to kinetic models (Michaelis-Menten) using GraphPad Prism 5.0. The Lineweaver-Burk Plot (the reciprocal of reaction velocities versus the reciprocal of substrates concentrations) was used to determine the quality of fit to a specific inhibition model. The inhibition constant (Ki) was obtained by a secondary plot using the slopes of the primary Lineweaver-Burk Plot (Km/Vmax versus inhibitor concentrations). αKi was obtained from a secondary plot using the y-intercepts of the Lineweaver-Burk plot (1/Vmax versus inhibitor concentrations). All results were analyzed in quadruplicate. One-way analysis of variance was used to estimate the significance of differences. There was statistical significance between control and test groups if p < 0.05.
Additional Information
How to cite this article: Chen, A. et al. Evaluation of the inhibition potential of plumbagin against cytochrome P450 using LC-MS/MS and cocktail approach. Sci. Rep. 6, 28482; doi: 10.1038/srep28482 (2016).
Acknowledgments
This work was partially supported by grants from the National Natural Science Foundation of China (No. 81301908), and the Science and Technology Commission of Shanghai Municipality (Nos 15140904700, 13ZR1412600 and 14DZ2270100).
Footnotes
The authors declare no competing financial interests.
Author Contributions M.L. and X.W. designed experiments. A.C., X.Z. and S.T. performed experiments. A.C., X.Z. and X.W. analyzed the data. A.C., X.Z. and X.W. wrote the manuscript and prepared figures. M.L. and X.W. revised and finalized this manuscript.
References
- Sunil C., Duraipandiyan V., Agastian P. & Ignacimuthu S. Antidiabetic effect of plumbagin isolated from Plumbago zeylanica L. root and its effect on GLUT4 translocation in streptozotocin-induced diabetic rats. Food and chemical toxicology: an international journal published for the British Industrial Biological Research Association 50, 4356–4363 (2012). [DOI] [PubMed] [Google Scholar]
- Olagunju J. A., Jobi A. A. & Oyedapo O. O. An Investigation into the Biochemical Basis of the Observed Hyperglycaemia in Rats Treated with Ethanol Root Extract of Plumbago zeylanica. Phytotherapy research 13, 346–348 (1999). [DOI] [PubMed] [Google Scholar]
- Hsieh Y. J., Lin L. C. & Tsai T. H. Measurement and pharmacokinetic study of plumbagin in a conscious freely moving rat using liquid chromatography/tandem mass spectrometry. J Chromatogr B-Analyt Technol Biomed Life Sci 844, 1–5 (2006). [DOI] [PubMed] [Google Scholar]
- Nayak P., Sharma M., Behera S. N., Thirunavoukkarasu M. & Chand P. K. High-performance liquid chromatographic quantification of plumbagin from transformed rhizoclones of Plumbago zeylanica L.: inter-clonal variation in biomass growth and plumbagin production. Applied biochemistry and biotechnology 175, 1745–1770 (2015). [DOI] [PubMed] [Google Scholar]
- Saritha K., Rajesh A., Manjulatha K., Setty O. H. & Yenugu S. Mechanism of antibacterial action of the alcoholic extracts of Hemidesmus indicus (L.) R. Br. ex Schult, Leucas aspera (Wild.), Plumbago zeylanica L., and Tridax procumbens (L.) R. Br. ex Schult. Frontiers in microbiology 6, 577 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand J. M. et al. Plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone), isolated from Plumbago zeylanica, inhibits ultraviolet radiation-induced development of squamous cell carcinomas. Carcinogenesis 33, 184–190 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. W. et al. Induction of apoptosis and autophagy via sirtuin1- and PI3K/Akt/mTOR-mediated pathways by plumbagin in human prostate cancer cells. Drug design, development and therapy 9, 1511–1554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Joo M. K. et al. Antitumorigenic effect of plumbagin by induction of SH2-containing protein tyrosine phosphatase 1 in human gastric cancer cells. International journal of oncology 46, 2380–2388 (2015). [DOI] [PubMed] [Google Scholar]
- Rendic S. Summary of information on human CYP enzymes: human P450 metabolism data. Drug metabolism reviews 34, 83–448 (2002). [DOI] [PubMed] [Google Scholar]
- Dahlinger D. et al. Development and validation of an in vitro, seven-in-one human cytochrome P450 assay for evaluation of both direct and time-dependent inhibition. Journal of pharmacological and toxicological methods 77, 66–75 (2015). [DOI] [PubMed] [Google Scholar]
- Arora S. et al. In vivo prediction of CYP-mediated metabolic interaction potential of formononetin and biochanin A using in vitro human and rat CYP450 inhibition data. Toxicology letters 239, 1–8 (2015). [DOI] [PubMed] [Google Scholar]
- Daly A. K. Pharmacogenetics of the cytochromes P450. Current topics in medicinal chemistry 4, 1733–1744 (2004). [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends in pharmacological sciences 25, 193–200 (2004). [DOI] [PubMed] [Google Scholar]
- Zheng L. et al. Evaluation of the impact of Polygonum capitatum, a traditional Chinese herbal medicine, on rat hepatic cytochrome P450 enzymes by using a cocktail of probe drugs. Journal of ethnopharmacology 158, 276–282 (2014). [DOI] [PubMed] [Google Scholar]
- Posadzki P., Watson L. & Ernst E. Herb-drug interactions: an overview of systematic reviews. British journal of clinical pharmacology 75, 603–618 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research. Drug Interaction Studies — Study Design, Data Analysis, Implications for Dosing, and Labeling Recommendations. Draft guidance (2012). Available at: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM292362.pdf (Accessed: 24th March 2016).
- Sumsakul W., Chaijaroenkul W. & Na-Bangchang K. In vitro inhibitory effects of plumbagin, the promising antimalarial candidate, on human cytochrome P450 enzymes. Asian Pacific journal of tropical medicine 8, 914–918 (2015). [DOI] [PubMed] [Google Scholar]
- Spaggiari D., Geiser L., Daali Y. & Rudaz S. A cocktail approach for assessing the in vitro activity of human cytochrome P450s: an overview of current methodologies. Journal of pharmaceutical and biomedical analysis 101, 221–237 (2014). [DOI] [PubMed] [Google Scholar]
- Frye R. F. Probing the world of cytochrome P450 enzymes. Molecular interventions 4, 157–162 (2004). [DOI] [PubMed] [Google Scholar]
- Wang X., Cheung C. M., Lee W. Y., Or P. M. & Yeung J. H. Major tanshinones of Danshen (Salvia miltiorrhiza) exhibit different modes of inhibition on human CYP1A2, CYP2C9, CYP2E1 and CYP3A4 activities in vitro. Phytomedicine: international journal of phytotherapy and phytopharmacology 17, 868–875 (2010). [DOI] [PubMed] [Google Scholar]
- Lin J. H. & Lu A. Y. H. Inhibition and induction of cytochrome P450 and the clinical implications. Clin Pharmacokinet 35, 361–390 (1998). [DOI] [PubMed] [Google Scholar]
- Sun M., Tang Y., Ding T., Liu M. & Wang X. Inhibitory effects of celastrol on rat liver cytochrome P450 1A2, 2C11, 2D6, 2E1 and 3A2 activity. Fitoterapia 92, 1–8 (2014). [DOI] [PubMed] [Google Scholar]
- Justenhoven C. et al. CYP2B6*6 is associated with increased breast cancer risk. International journal of cancer. Journal international du cancer 134, 426–430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragia G. et al. Frequency of CYP450 enzyme gene polymorphisms in the Greek population: review of the literature, original findings and clinical significance. Drug metabolism and drug interactions 29, 235–248 (2014). [DOI] [PubMed] [Google Scholar]
- Wang X., Lee W. Y., Or P. M. & Yeung J. H. Effects of major tanshinones isolated from Danshen (Salvia miltiorrhiza) on rat CYP1A2 expression and metabolism of model CYP1A2 probe substrates. Phytomedicine: international journal of phytotherapy and phytopharmacology 16, 712–725 (2009). [DOI] [PubMed] [Google Scholar]
- Wang X. & Yeung J. H. Investigation of cytochrome P450 1A2 and 3A inhibitory properties of Danshen tincture. Phytomedicine: international journal of phytotherapy and phytopharmacology 19, 348–354 (2012). [DOI] [PubMed] [Google Scholar]
- Zheng L. et al. Imperatorin is a mechanism-based inactivator of CYP2B6. Drug metabolism and disposition: the biological fate of chemicals 43, 82–88 (2015). [DOI] [PubMed] [Google Scholar]
- Yuce-Artun N., Kose G. & Suzen H. S. Allele and genotype frequencies of CYP2B6 in a Turkish population. Molecular biology reports 41, 3891–3896 (2014). [DOI] [PubMed] [Google Scholar]
- Koh K. H. et al. Kruppel-like factor 9 promotes hepatic cytochrome P450 2D6 expression during pregnancy in CYP2D6-humanized mice. Molecular pharmacology 86, 727–735 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X. H. et al. Association between the CYP2E1 polymorphisms and lung cancer risk: a meta-analysis. Molecular genetics and genomics: MGG 290, 545–558 (2015). [DOI] [PubMed] [Google Scholar]
- Loue C. & Tod M. Reliability and extension of quantitative prediction of CYP3A4-mediated drug interactions based on clinical data. The AAPS journal 16, 1309–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. & Yeung J. H. Effects of Salvia miltiorrhiza extract on the liver CYP3A activity in humans and rats. Phytotherapy research: PTR 25, 1653–1659 (2011). [DOI] [PubMed] [Google Scholar]
- Dinger J., Meyer M. R. & Maurer H. H. Development and validation of a liquid-chromatography high-resolution tandem mass spectrometry approach for quantification of nine cytochrome P450 (CYP) model substrate metabolites in an in vitro CYP inhibition cocktail. Analytical and bioanalytical chemistry 406, 4453–4464 (2014). [DOI] [PubMed] [Google Scholar]
- Liu L. Y. et al. A sensitive and high-throughput LC-MS/MS method for inhibition assay of seven major cytochrome P450s in human liver microsomes using an in vitro cocktail of probe substrates. Biomedical chromatography: BMC 29, 437–444 (2015). [DOI] [PubMed] [Google Scholar]
- Otten J. N., Hingorani G. P., Hartley D. P., Kragerud S. D. & Franklin R. B. An in vitro, High Throughput, Seven CYP Cocktail Inhibition Assay for the Evaluation of New Chemical Entities Using LC-MS/MS. Drug Metabolism Letters 5, 17–24 (2011). [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan K., Obach R. S. & Rostami-Hodjegan A. Mechanism-based inactivation of human cytochrome P450 enzymes: strategies for diagnosis and drug–drug interaction risk assessment. Xenobiotica; the fate of foreign compounds in biological systems 37, 1225–1256 (2007). [DOI] [PubMed] [Google Scholar]
- Obach R. S., Walsky R. L. & Venkatakrishnan K. Mechanism-based inactivation of human cytochrome p450 enzymes and the prediction of drug-drug interactions. Drug metabolism and disposition: the biological fate of chemicals 35, 246–255 (2007). [DOI] [PubMed] [Google Scholar]
- Stresser D. M., Mao J., Kenny J. R., Jones B. C. & Grime K. Exploring concepts of in vitro time-dependent CYP inhibition assays. Expert opinion on drug metabolism & toxicology 10, 157–174 (2014). [DOI] [PubMed] [Google Scholar]
- Lewis D. F. V., Loannides C. & Parke D. V. Cytochromes P450 and Species Differences in Xenobiotic Metabolism and Activation of Carcinogen. Environmental Health Perspectives 106, 633–641 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. & Yeung J. H. Inhibitory effect of tanshinones on rat CYP3A2 and CYP2C11 activity and its structure-activity relationship. Fitoterapia 82, 539–545 (2011). [DOI] [PubMed] [Google Scholar]
- Kolrep F. et al. Differences in metabolism of the marine biotoxin okadaic acid by human and rat cytochrome P450 monooxygenases. Archives of toxicology (2015). [DOI] [PubMed] [Google Scholar]
- He F. et al. Mechanism of Action of Panaxytriol on Midazolam 1 ′-Hydroxylation and 4-Hydroxylation Mediated by CYP3A in Liver Microsomes and Rat Primary Hepatocytes. Biol. Pharm. Bull. 38, 1470–1477 (2015). [DOI] [PubMed] [Google Scholar]
- Hutzler J. M. & Tracy T. S. Atypical kinetic profiles in drug metabolism reactions. Drug metabolism & disposition 30, 355–362 (2002). [DOI] [PubMed] [Google Scholar]
- Peng Y. et al. A comprehensive assay for nine major cytochrome P450 enzymes activities with 16 probe reactions on human liver microsomes by a single LC/MS/MS run to support reliable in vitro inhibitory drug-drug interaction evaluation. Xenobiotica; the fate of foreign compounds in biological systems 1–17 (2015). [DOI] [PubMed] [Google Scholar]