Abstract
Adverse events during the prenatal and early postnatal period of life are associated with development of cardiovascular disease in adulthood. Prenatal exposure to excess testosterone (T) in sheep induces adverse reproductive and metabolic programming leading to polycystic ovarian syndrome, insulin resistance and hypertension in the female offspring. We hypothesized that prenatal T excess disrupts insulin signaling in the cardiac left ventricle leading to adverse cardiac programming. Left ventricular tissues were obtained from 2-year-old female sheep treated prenatally with T or oil (control) from days 30–90 of gestation. Molecular markers of insulin signaling and cardiac hypertrophy were analyzed. Prenatal T excess increased the gene expression of molecular markers involved in insulin signaling and those associated with cardiac hypertrophy and stress including insulin receptor substrate-1 (IRS-1), phosphatidyl inositol-3 kinase (PI3K), Mammalian target of rapamycin complex 1 (mTORC1), nuclear factor of activated T cells –c3 (NFATc3), and brain natriuretic peptide (BNP) compared to controls. Furthermore, prenatal T excess increased the phosphorylation of PI3K, AKT and mTOR. Myocardial disarray (multifocal) and increase in cardiomyocyte diameter was evident on histological investigation in T-treated females. These findings support adverse left ventricular remodeling by prenatal T excess.
Organisms experiencing adverse intrauterine environment during a critical period of development have alterations at the molecular and gene expression level that start the process of increased susceptibility to adult disease and reduced ability to adapt to insult. It is well established that suboptimal intrauterine environments leads to increased incidence of cardiovascular disease1. One such suboptimal intrauterine environment is altered hormonal milieu that has been shown to lead to intrauterine programming of several organs of the growing fetus2. For instance, testosterone (T) excess in-utero, during critical windows of development, leads to adverse reproductive and metabolic programming in several animal models3,4. Many of these perturbations recapitulate the features seen in polycystic ovarian syndrome (PCOS), a common endocrine disorder in women of child-bearing age with a prevalence rate of 5–8%5. The exact etiology of PCOS is not clear but there is evidence that prenatal environment could play a significant role3. The clinical manifestations of the condition include hyperandrogenism, oligo-/anovulation, and multifollicular ovarian morphology5,6. Metabolic disturbances are highly prevalent in this population with insulin resistance in 50–70% of the patients7, impaired glucose tolerance in 40%, and type 2 diabetes in 10% of the patients8,9.
Women with PCOS are also at increased risk of cardiovascular disease and myocardial infarction10,11. They have higher incidences of hypertension, coronary artery disease and diastolic dysfunction12. Available evidence points to early onset of cardiac remodeling resulting in increased left ventricular mass in young women with PCOS13, and reduced left ventricular ejection fraction and diastolic dysfunction in older women with PCOS12. The underlying mechanisms leading to cardiovascular compromise in PCOS are unclear. Animal models of PCOS provide a unique opportunity to investigate underlying mechanisms. Prenatal T-treatment also causes hypertension in rats and sheep14,15, supportive of prenatal programming of vascular disease, and likely adverse cardiac programming. Because prenatal T excess also induces intrauterine growth restriction (IUGR) and metabolic perturbations such as insulin resistance in sheep4,16, attributes that are closely linked to adverse cardiovascular effects in humans17, any adverse cardiac programming in prenatally T-treated sheep may be the indirect consequence of IUGR and the insulin resistance induced by prenatal T excess16,18. Alternatively, effects of prenatal T excess may also be mediated by direct actions of androgen. Androgen receptors are present in cardiac tissue19 and T can induce cardiac hypertrophy both in-vitro and in-vivo20,21. Prenatally T-treated sheep are functionally hyperandrogenic4,22.
A number of molecular mediators involved in insulin signaling such as phosphatidyl inositol-3 kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin complex 1 (mTORC1) signaling pathway have been implicated in cardiac hypertrophy23,24. Evidence from in-vitro25 studies indicates that T triggers cardiac hypertrophy by activation of mTORC1 pathway. Considering that prenatal T induces functional hyperandrogenism and insulin resistance in sheep4,18, alters insulin signaling in liver and muscle26, and both T and insulin have the potential to alter cardiac function, we hypothesized that prenatal T excess also disrupts insulin signaling and upregulates molecular markers of cardiac hypertrophy leading to adverse cardiac remodeling.
Results
Body weights (kg; Mean ± SEM) were not different between control and T-treated sheep at the time of harvest (T-treated 69.4 ± 5.4 vs. control 63.09 ± 4.9 p = 0.40, cohen’s d = 0.44). Fasting insulin levels (IU/ml; Mean ± SEM) of prenatally T-treated females trended to be higher (T-treated 15.7 ± 4.7 vs. control 9.23 ± 2.12, p = 0.26), with Cohens analysis revealing a modest increase (d = 0.65). Fasting glucose (mg/dl; Mean ± SEM) levels averaged 58 ± 3.9 and 52 ± 1.7 for the T-treated females and controls, respectively (p = 0.28), with Cohens analyses revealing a modest effect size difference (d = 0.67). Insulin to glucose ratio also tended to be higher in the T-treated group (0.25 ± 0.06) compared to the control group (0.17 ± 0.04), with Cohen’s analyses showing a modest increase (d = 0.60). Left ventricular weights of the T-treated females also trended higher but did not differ statistically from control sheep (T-treated 164.9 ± 9.9 grams vs control 152 ± 7.2 grams, p = 0.30, cohen’s d = 0.54).
Mediators of insulin signaling in left ventricular tissue
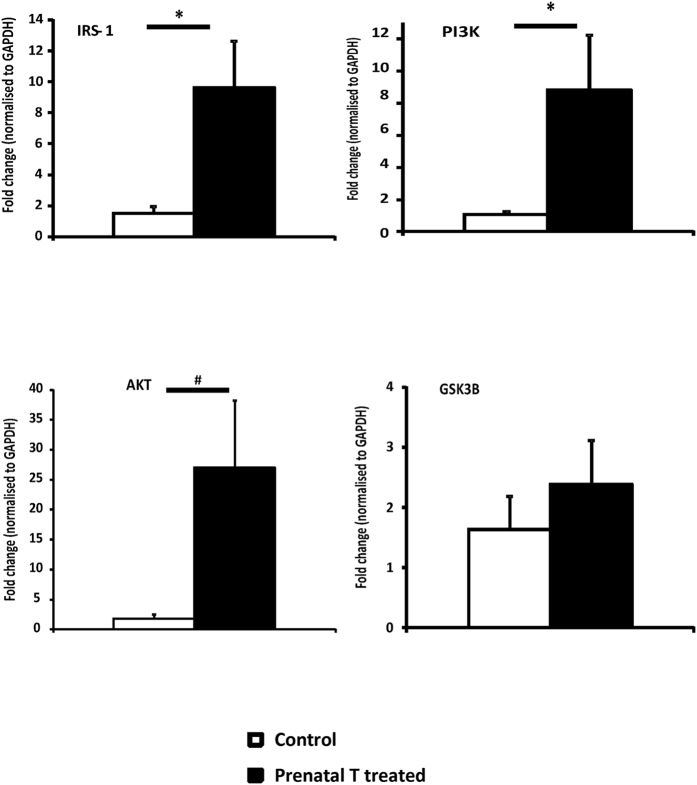
A significant increase in gene expression of proteins involved in the insulin signaling pathway was found in the left ventricle of prenatally T-treated females compared to controls. Prenatal T excess increased the expression level of IRS-1 by 6.4 fold (p = 0.027), PI3K by 8.5 fold (p = 0.04), and AKT by 27 fold (p = 0.06), compared to control (Fig. 1).
Figure 1. Gene expression (Mean ± SEM) of proteins involved in insulin signaling pathway measured by qRT-PCR in left ventricle tissue obtained at 2 year of age (n = 6–8 per group).
Two tail student t-test was used to test differences between T-treated and control and * assigned to p < 0.05 and # for p value 0.06. The Cohen’s effect size analysis revealed large differences between T-treated and control group for genes involved in insulin signaling pathways (Cohen’s d: AKT = 1.11, IRS-1 = 1.33, PI3K = 1.29, GSK3b = 0.452). Abbreviations: IRS-1 insulin receptor substrate 1, PI3K Phosphoinositide 3-kinase, Akt protein kinase B, GSK3B Glycogen synthase kinase 3 beta.
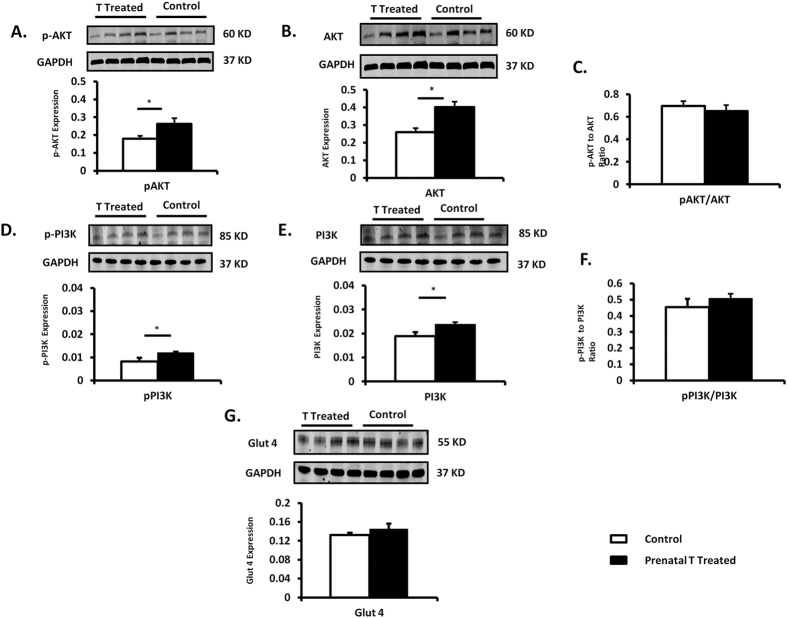
Under basal conditions, protein expression of total PI3K (p85 subunit) in the left ventricle was significantly different (p = 0.01) between prenatally T-treated (0.023 ± 0.0) and control females (0.019 ± 0.001); and pPI3K was also increased (p = 0.0008) in prenatally T-treated (0.01 ± 0.0005) compared to control (0.008 ± 0.0006). Ratio of pPI3K/PI3K was not statistically significant between groups (prenatal T-treated: 0.5 ± 0.03 vs control: 0.45 ± 0.05, p = 0.55). Total AKT (prenatal T: 0.40 ± 0.03 vs control 0.26 ± 0.02; p = 0.007) and pAKT (prenatal T: 0.26 ± 0.03 vs control: 0.18 ± 0.02; p = 0.028) were significantly increased in prenatally T-treated females compared to control. Ratio of pAKT/AKT (prenatal T-treated: 0.65 ± 0.05; vs control: 0.69 ± 0.04) between the two groups was not statistically different (p = 0.57). We did not see any difference in Glucose transporter 4 (GLUT 4) protein levels between prenatal T-treated vs. control (p = 0.33, Fig. 2).
Figure 2. Protein expression (Mean ± SEM) of total and phosphorylated proteins involved in insulin signaling pathway (n = 7–8 per group).
Two tail student t-test was used to test difference between T-treated and control and * assigned to p < 0.05. The Cohen’s effect size analysis revealed large differences between T-treated and control group for PI3K, AKT total and phosphorylated proteins involved in insulin signaling pathways (cohen’s d: PI3K = 1.68, pPI3K 2.53; AKT = 1.83, pAKT = 1.14) and small differences in GLUT 4 (cohen’s d: 0.57).
Mediators of left ventricular hypertrophy (LVH) and stress
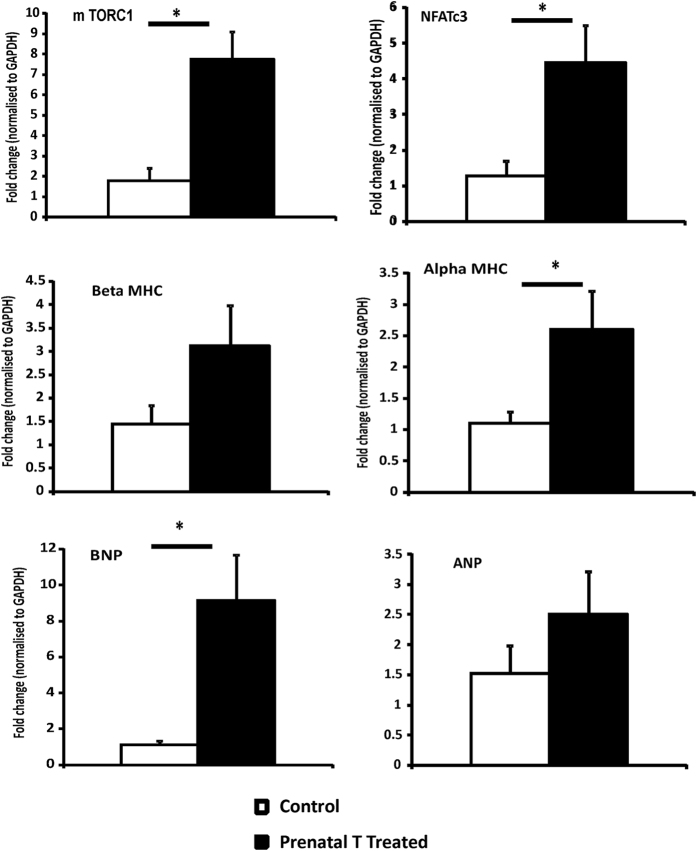
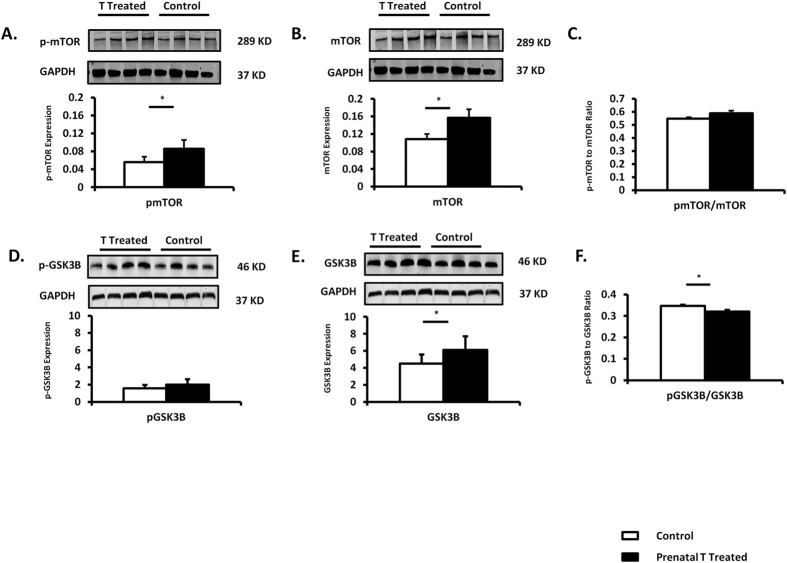
A 6.1 fold increase in mTORC1, marker of hypertrophic growth downstream from the insulin-signaling pathway that has shown to be activated by T in-vitro25,27 gene expression was found in the left ventricle of prenatally T-treated females compared to control (p = 0.002) (Fig. 3) and its protein expression was also increased in the left ventricle (total mTOR: prenatal T 0.16 ± 0.02 vs control 0.11 ± 0.01, p = 0.04; p mTOR: prenatal T: 0.09 ± 0.008 vs. control 0.056 ± 0.004; p = 0.01) (Fig. 4). GSK3ß, an important downstream target of AKT in the heart, which contributes to suppression of hypertrophic growth28, did differ between the two groups. Total GSK3 beta was significantly increased (p = 0.04) in prenatally T-treated females (6.1 ± 0.57) compared to control (4.5 ± 0.4); whereas p-GSK 3 beta was not different (p = 0.16) between prenatally T-treated (1.99 ± 0.22) and control (1.56 ± 0.15) groups. Ratio of p-GSK 3 beta/GSK3beta was significantly lower (p = 0.002) in prenatally T-treated females (0.32 ± 0.008) compared to control (0.35 ± 0.05) (Fig. 4).
Figure 3. Gene expression (Mean ± SEM) of proteins involved in cardiac hypertrophy and stress measured by qRT-PCR in left ventricle tissue obtained at 2 year of age (n = 6–8 per group).
Two tail student t-test was used to test difference between T-treated and control and * assigned to p < 0.05 and # for p value 0.06. The Cohen’s effect size analysis revealed medium to large differences between T-treated and control group for genes involved in insulin signaling pathways (Cohen’s d: mTORC1 = 1.89, αMHC = 1.20, ßMHC = 0.89, mTORC1 = 1.89, BNP = 1.34, ANP = 0.61, NFATc3 = 1.46). Abbreviations: mTORC1 Mammalian target of rapamycin complex 1, MHC myosin heavy chain, BNP brain naturetic peptide, ANP atrial naturetic peptide, NFATc3 nuclear factor of activated T cells.
Figure 4. Protein expression (Mean ± SEM) of total and phosphorylated proteins for mTOR and GSK 3 beta (n = 7–8).
Two tail student t-test was used to test difference between T-treated and control and * assigned to p < 0.05. The Cohen’s effect size found large differences between T-treated and control groups for proteins involved in insulin signaling pathways (mTOR = 1.21, pmTOR = 1.82; GSK 3 beta = 1.12, pGSK3beta = 0.8).
Prenatal T treatment significantly upregulated other markers of cardiac hypertrophy including nuclear factor of activated T cells (NFATc3) a well-known marker of cardiac hypertrophy29. Prenatal T increased gene expression of NFATc3 by 3.5 fold above control (p = 0.02, Fig. 3). Measures of BNP and ANP, markers of cardiac hypertrophy that are released in response to cardiac wall strain, showed that BNP was elevated by 8.2 fold in the prenatally T-treated group relative to controls (p = 0.01, Fig. 3), while ANP levels did not differ (fold change 1.64: p = 0.28) between groups.
αMHC, the predominant myofilament protein in rodents, shown to be altered in rodent models of cardiac hypertrophy30,31,32, was elevated by 2.3 fold (p = 0.046) in the prenatally T-treated group compared to controls (Fig. 3). In contrast ßMHC the predominant myofilament protein in adult human left ventricle33 did not differ significantly (fold change 2.2: p = 0.10) between the 2 groups.
Steroid receptor expression
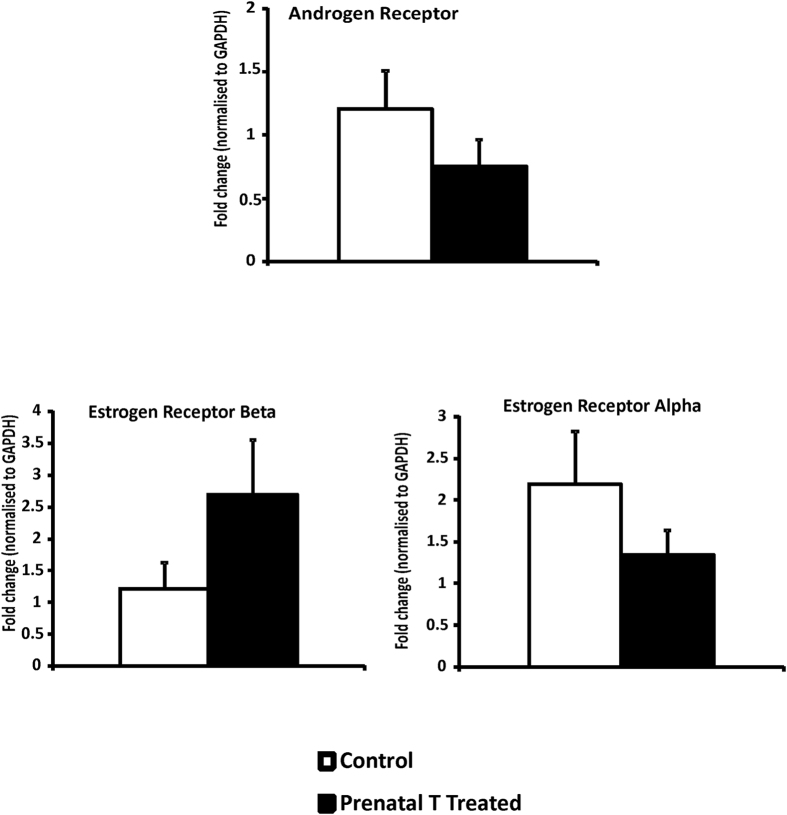
Although androgen and estrogen receptors were expressed in left ventricular tissue of sheep, prenatal T treatment did not significantly alter the gene expression of androgen receptor or estrogen receptor beta or alpha (AR fold change 0.88 p = 0.40, ER beta fold change 2.2: p = 0.17, ER alpha fold change 1.5: p = 0.64, Fig. 5).
Figure 5. Gene expression (Mean ± SEM) of androgen and estrogen receptor measured by qRT-PCR in left ventricle tissue obtained at 2 years of age (n = 7–8 per group).
The Cohen’s effect size found small differences between T-treated and control groups in gene expression of androgen and estrogen receptors (AR = 0.2, ER alpha 0.26 and ER beta 0.77) Abbreviations: AR Androgen receptor, ER estrogen receptor.
Histology results
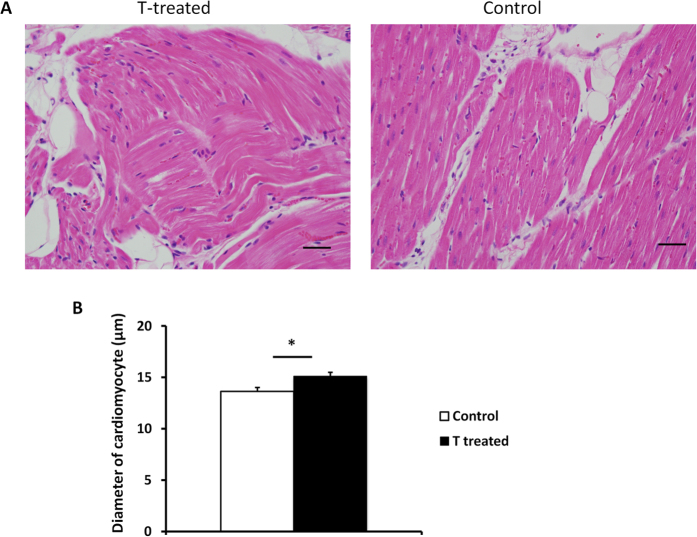
5 out of the 8 prenatally T-treated females had focal myocardial disarray in left ventricle on both H&E stain and Masson staining compared to controls that had no evidence of myocardial disarray (Fig. 6a). Prenatal T treatment increased cardiomyocyte size compared to control (T-treated 15.1 ± 0.4 μm vs 13.6 ± 0.4 μm, p = 0.02) (Fig. 6b).
Figure 6.
(A) Representative histological sections of H&E stained sections from left ventricle tissue from two-year-old prenatal T-treated sheep (A) and control (B). (B) Effect of prenatal T treatment on the mean diameter of cardiomyocytes. * Denotes p- value < 0.05. The Cohen’s effect size found large differences between T-treated and control groups with respect to cardiomyocyte width (Cohen’s d = 1.473).
Discussion
Findings from the current study, namely increased expression of genes and proteins involved in left ventricular hypertrophy and stress, together with histological evidence of focal myocardial disarray and an increase in cardiomyocyte size, suggest that prenatal exposure to excess T likely alters the trajectory of left ventricular tissue differentiation in the female sheep. This differentiation appears to involve upregulation of proteins involved in insulin signaling and downstream upregulation of mTOR, a known marker of hypertrophic growth. To our knowledge this is the first study to show perturbed molecular signaling in the left ventricle in a well-established sheep model of PCOS that also manifests IUGR during fetal development18,22. These perturbations in molecular signaling could lead to increased susceptibility of injury to the heart.
Increased left ventricular mass has been shown to predict increased cardiovascular morbidity and mortality34,35. Patients with hypertension and high plasma insulin are more likely to develop LVH36,37. Systemic insulin resistance has also been linked to LVH38,39,40. Consistent with this premise, Cohen’s effect size analysis revealed a modest increase in fasting insulin and insulin/glucose ratio in the prenatal T-treated females. The potential mechanism by which insulin resistance is linked to LVH is through metabolic dysregulation (hyperglycemia and hyperinsulinemia) that leads to altered myocardial protein degradation, altered matrix remodeling, increased cellular lipids and altered vascular compliance, eventually leading to LVH and remodeling40. Findings to date provide evidence that prenatal T-treatment leads to insulin resistance in insulin target tissues, hyperinsulinemia and hypertension15,18,26, which are all risk factors for development of LVH. Alternatively, some of the altered molecular signaling may occur in-utero secondary to the direct effect of T excess on the cardiac tissue and further accentuated by the perturbed systemic environment postnatally. Although prenatal T treatment did not lead to statistically significant increases in weight of the left ventricle the molecular changes and histological changes suggest that these early perturbation may eventually lead to such changes in LVH over time.
mTORC1 is one of the key regulators of protein synthesis in the cardiomyocyte and activation of this has been shown to lead to cardiac hypertrophy41,42. It can be activated by growth factors including insulin through its signaling pathway and also by T25,43. Activation of proteins downstream from the insulin receptor (PI3K/AKT/mTORC1) has been linked to “physiological hypertrophy” in the heart but there is also evidence for “pathological” hypertrophy with chronic activation of this pathway44,45,46,47,48 leading to disruption of coordinated tissue growth and angiogenesis48. Findings from this study showing increased expression of mTORC1, PI3K, and AKT suggests that prenatal T excess may underlie activation of cardiac insulin signaling pathway resulting in activation of mTORC1 culminating ultimately to cardiac hypertrophy. Increase in gene expression of BNP and NFATc3 both markers of pathological hypertrophy are suggestive of a maladaptive hypertrophy.
Because prenatal T excess leads to maternal hyperinsulinemia49 and postnatal insulin resistance18 in the offspring, it is likely that maternal hyperinsulinemia could also be a major player in programming prenatal T-induced cardiac hypertrophy in offspring. For instance, increase in PI3K/AKT/mTORC1 signaling is seen in newborns with congenital hyperinsulinemia50,51. Similarly, fetal hyperinsulinemia due to maternal obesity has been shown to lead to cardiac programming defects in the offspring with increase in markers of left ventricular hypertrophy and increase in AKT and also mTOR52.
While T can lead to cardiac hypertrophy indirectly via programming of hyperinsulinemia, the potential for direct effects of androgen on cardiac hypertrophy cannot be ignored. These animals are functionally hyperandrogenic and manifest increased expression of AR at the level of the hypothalamus53, pituitary (Cordoso and Padmanabhan, unpublished) and ovary54. Cardiomyopathy has been seen in newborns with congenital adrenal hyperplasia55, a condition where the fetus is exposed to excess endogenous androgen. Androgen and estrogen receptors are present in cardiac tissue both in the cytosol and the nucleus19. Sex hormone binding to their receptors have been found to lead to both genomic and non-genomic effects in the heart, and one mechanism is through activation of the PI3K/AKT pathway19 that leads to downstream activation of mTORC1 resulting in increased protein synthesis and eventually cardiac hypertrophy24. While androgen receptors are expressed in the left ventricle to facilitate direct effects of T, absence of changes in AR expression in prenatal T-treated females suggests that any direct effects of T may be mediated via the increase in ligand (T treatment) rather than the receptor.
T has been found to increase cardiomyocyte glucose uptake by augmenting GLUT4 translocation56 and although an increase in GLUT 4 protein expression was not seen in the left ventricle under basal condition, the increase in expression of insulin signaling proteins including IRS-1, PI3K and AKT is suggestive of increase in glucose metabolism within the left ventricle. Free fatty acids are the primary fuel for energy production in cardiac tissue but during fetal life and during times of stress, the heart does revert to glucose as its primary source of fuel57. As such, an increase in the gene expression of proteins involved in glucose uptake in prenatal T females may signify an adaptive response to increased metabolic demand on the heart since we also see an increase in the expression of BNP, a cardiac stress marker. The elevated mTORC1 expression suggests that T could lead to cardiac hypertrophy through increases in mTORC1 as suggested previously25.
Offspring that have experienced an adverse intrauterine environment during a critical period of development culminating in IUGR, such as that evident in prenatal T-treated sheep16,58 have alterations at the molecular level that builds a platform for increased susceptibility to cardiovascular disease later in life59,60. It is not clear whether the molecular changes seen in prenatally T-treated sheep involve similar mechanisms that lead to adverse cardiac programming in other models of IUGR. For instance, 50% nutrient restriction in sheep (a model of IUGR) leads to upregulation of a variety of genes in the fetal heart, some of which are involved with cardiac hypertrophy and metabolism61,62 and altered fatty acid content of the cardiomyocytes62. Functional changes in the heart have also been observed in offspring of rats treated with low protein diet63. Glucocorticoid excess prenatally has also been shown to lead to IUGR accompanied with hypertension and insulin resistance as seen with prenatal T excess64,65. Direct molecular effects on the myocardium have been seen with in-utero exposure to excess glucocorticoids including increased expression of cardiac glucose transporter 1 (one of the main glucose transporter in the heart)66 and upregulation in AKT/PKB pathway66, similar to what we see in our model.
From a translational standpoint, these findings may be of relevance to female offspring of women with PCOS. It has been shown that androgen and insulin levels are elevated in pregnant women with PCOS67,68 and there is some evidence with Spanish cohorts that they give birth to babies with reduced birth weight69. The female offspring of PCOS mothers have early metabolic derangement70 similar to that seen in our prenatal T model18, suggesting that as in the prenatally T-treated sheep model, presence of hyperandrogenism in PCOS women has the potential to adversely program several organ systems including the cardiovascular system in their offspring.
In summary, findings from this study provide evidence for cardiac reprogramming in a sheep model of PCOS. Further studies delineating the functional consequences of such changes are vital and will further our understanding the effect of prenatal T excess on cardiac health and assist in developing new treatment strategies towards preventing cardiac susceptibility to prenatal insults.
Material and Methods
Suffolk female lambs born to dams that received twice weekly intramuscular injections of 100 mg of T propionate (1.2 mg/kg; Sigma-Aldrich Corp., St. Louis, MO) in cottonseed oil from days 30 to 90 of pregnancy were used in the study. Control breeders were injected with the vehicle for the same duration. The concentrations of T achieved in maternal circulation with this mode of T delivery are in the range of adult males71. The concentrations of T achieved in female fetuses with this mode of T delivery are also within range seen in the control fetal males71. Mothers were maintained on pasture and supplemented with hay and concentrated pelleted food to meet the increased needs of the pregnant sheep. Breeder sheep diet ranged from 2.6–2.9 (Mcal/kg) of digestible energy based on NRC requirements throughout pregnancy as detailed in a previous publication27. All lambs were born by normal delivery and after full length gestation. After weaning (8 weeks of age), lambs were placed on an adjusted diet of commercial feed pellets (Shur-Gain, Elma, NY). A reduction in crude protein was implemented when they reached 40 kg of body weight as previously described72. Lambs were maintained with their mothers under natural photoperiod and with free access to water. All procedures for management and experimental methodologies were approved by the Institutional Animal Care and Use Committee of the University of Michigan, which are consistent with the National Research Council’s Guide for the Care and Use of Laboratory Animals.
At approximately 2 years of age, to avoid influence from differing levels of endogenous steroids, all animals were ovariectomized and implanted with a 1-cm-long SILASTIC capsule (inner diameter, 3.35 mm and outer diameter, 4.65 mm; Dow Corning Corp., Midland, MI) filled with 17β estradiol (estradiol; Sigma-Aldrich, St. Louis, MO, USA). Approximately 1 month later, two controlled internal drug release progesterone implants (InterAG, Hamilton, Waikato, New Zealand) were inserted subcutaneously into each animal. Progesterone implants were removed 14 days later and all animals received four additional 3-cm-long estradiol implants to achieve ovarian steroid levels found during the late follicular phase. Animals were euthanized ~20 hours after insertion of the estradiol implants during the presumptive late follicular phase of the cycle by administration of barbiturate overdose (Fatal Plus, Vortech Pharmaceuticals, Dearborn, MI). Hearts were harvested and portions of right and left ventricle were both quickly frozen and stored at −80 °C until processing or formalin fixed for histological processing. Plasma insulin and glucose were measured prior to euthanasia after 48 hours fast. Concentrations of insulin were determined by radioimmunoassay (insulin RIA Kit, MP Biomedicals). Assay sensitivity of insulin assay was 3.0 μU/L. Glucose concentrations were assessed by the glucose oxidase method (Pointe Scientific Inc.). Inter- and intra-assay CV for both the insulin and glucose assays were <10%.
Isolation of RNA and Real Time Polymerase Chain Reaction
RNA from left ventricular tissue (n = 8 prenatal T-treated and n = 7 control) was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA). Purification of RNA was done with deoxyribonuclease treatment. Purity of the RNA for each sample was validated by determining its RNA Integrity Number (RIN). Concentration was determined using a nanodrop spectrophotometer. First strand cDNA was synthesized from RNA using Superscript III first strand synthesis system (Invitrogen Cat. No: 18080-051). cDNA was prepared from 1 μg total RNA. Concentrations of cDNA over 1000 fold range was amplified using each target gene and GAPDH specific primers to determine the final concentration at which each primer sequence for both housekeeping gene and gene of interest were equally and >90% efficient. Gene specific primers were designed using primer select software and are detailed in supplemental material Table 1.
The SYBR Green real time PCR assay was performed using the cDNA generated. Negative reverse transcriptase control was run for each sample to rule out genomic DNA contamination of isolated RNA. Quantitative RT PCR reactions were run on Applied Biosystem’s 7200 Real Time PCR instrument, and were performed in triplicate. Results of the assay were quantified using the cycle threshold (CT) values of each sample. The CT values of each gene of interest were compared against those of the housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Only values less than 35 were considered positive. The results of each assay were verified by replicating the outcome in a second test.
Western Blot
Sheep left ventricular tissue was crushed under liquid nitrogen and protein lysate was generated using lysis buffer (Triton 1%, sodium vanadate (1 nM), sodium fluoride (50 mM) and sodium pyrophosphate 10 mM and protease inhibitor cocktail 10 mM). Each sample was normalized to contain 2 μg/μl by adding lysis buffer, Laemelli buffer (Bio-rad, Cat. no: 161-0737) and Beta-mercaptoethanol to the lysate. 15 μl of the sample was loaded onto 4–12% Bis-Tris gels (Life Technologies Cat. No: NP0321BOX) and gel electrophoresis was performed using the XCell Sure Lock chamber (Invitrogen, Carlsbad, CA). The proteins were then transferred to Optitran Reinforced Nitrocellulose (Whatman, Cat. no: 10439396) at 100 Volts for one hour. The blots were blocked for one hour in Odyssey Blocking Buffer (Li-Cor, Lincoln, NE. Cat. No: 927-40000). Primary antibody (polyclonal rabbit) as detailed in supplemental material Table 2, for the AKT, p-AKT (serine 473), p85 subunit of PI3Kinase and p-PI3K and insulin-inducible, glycogen synthase kinase-3ß (GSK3ß), glucose transporter type 4 (GLUT 4) were obtained from Cell Signaling Technology (Danvers, MA) and the housekeeping gene GAPDH (mouse monoclonal) antibody was obtained from Sigma-Aldrich (St. Louis, MO). The primary antibodies were left to incubate overnight at 4 °C. Blots were then washed and incubated with Li-COR Secondary antibody (1:10000) of Donkey anti- Rabbit (680RD) and Donkey anti- Mouse (800 CW). Western blots were developed with the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE). Band intensities were quantified using the Odyssey software (LI-COR Biosciences) and normalized to GAPDH.
Histopathological analysis
Left ventricular tissues were fixed in formaldehyde in PBS pH 7.4. The tissue was then dehydrated through serial alcohols, cleared in xylenes, and embedded in paraffin. Paraffin sections with a thickness of 5 μm were cut and stained with hematoxylin and eosin (H&E) for gross appearance analysis. Periodic acid-Schiff counterstained with hematoxylin (PAS-H) was used to facilitate quantification of cardiomyocyte size by our Investigative Histopathology laboratory at Michigan State University. Histological sections were reviewed in random order by two independent veterinary pathologists that were blinded to treatment groups.
Histomorphometry
Following previously established protocols73,74 left ventricular wall cardiomyocyte mean diameter of each sample was determined by measuring the short axis of 100 cardiomyocytes in nucleated transverse section with nearly circular capillary profiles in 10 randomly selected microscopic fields at 400X magnification. Measurements were obtained with CellSens software.
Statistical analysis
The cardiomyocyte diameters between treatment groups were compared using two tailed student t-test. To determine changes in gene expression, ΔCT formula (Ct for target gene-Ct for GADH gene) was used to normalize each target gene Ct value for control and T-treated animals. Relative gene expressions were determined using formulae ΔΔCt = ΔCtT-treated − ΔCtcontrol and relative target mRNA level was analyzed following the 2−ΔΔCt method as previously described75. Results are presented as mean 2−ΔΔCt ± SEM. For each gene, the difference between control and prenatally T-treated females was determined using two-tailed Student’s t-test. Student two-tailed t-test was also used to compare differences between protein expressions of control vs. prenatally T-treated females after normalizing the protein of interest to GAPDH. P-values are reported for each test. In addition, to complement findings from the t-test, Cohen’s effect size analysis was performed to address the magnitude of changes. This analysis allows comparison of the means between two treatments with respect to the magnitude of difference between them. The computed statistic is Cohen’s d value, and values above 0.2, 0.5, and 0.8 were considered as small, medium, and large effect sizes, respectively76 Cohens values for data presented in the figures are provided in the legend.
Additional Information
How to cite this article: Vyas, A. K. et al. Prenatal programming: adverse cardiac programming by gestational testosterone excess. Sci. Rep. 6, 28335; doi: 10.1038/srep28335 (2016).
Supplementary Material
Acknowledgments
We thank Jeff Landgraff at the genomic core at Michigan State University for assistance with designing primers for this study and efficiency curve for each primer. We thank Douglas Doop and Gary McCalla for expert animal care, facility management and help with generation and maintenance of the experimental animals; Ms. Carol Herkimer, Mr. Evan M. Beckett, Mr. Jacob Moeller, Drs. Bachir Abi-Salloum, Teresa Steckler and Almudena Veiga-Lopez for assistance with prenatal treatments and/or help with tissue harvest; Drs. Fred Karsch and Robert Goodman for help with ovariectomies. RCMI Core grant number: G12MD007585 for use of Tuskegee core facilities equipment. This work was supported by NIH: P01 HD44232 (VP), BIRCWH K 12 HD065879 (AKV).
Footnotes
Author Contributions A.K.V. and V.P. conceived the study idea. A.K.V. and V.H. involved in conducting the molecular studies. E.G. and K.A.M. involved in interpretation of the histological data. A.K.V., E.G., K.A.M., V.P. and V.H. were involved in preparation of the manuscript. All authors read and approved the final manuscript.
References
- Barker D. J. Fetal programming of coronary heart disease. Trends in endocrinology and metabolism: TEM 13, 364–368 (2002). [DOI] [PubMed] [Google Scholar]
- Fowden A. L. & Forhead A. J. Endocrine mechanisms of intrauterine programming. Reproduction 127, 515–526 (2004). [DOI] [PubMed] [Google Scholar]
- Dumesic D. A., Abbott D. H. & Padmanabhan V. Polycystic ovary syndrome and its developmental origins. Rev. Endocr Metab Disord 8, 127–141 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan V. & Veiga-Lopez A. Animal models of the polycystic ovary syndrome phenotype. Steroids 78, 734–740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azziz R. et al. The Androgen Excess and PCOS Society criteria for the polycystic ovary syndrome: the complete task force report. Fertil Steril 91, 456–488 (2009). [DOI] [PubMed] [Google Scholar]
- Pasquali R. et al. PCOS Forum: research in polycystic ovary syndrome today and tomorrow. Clinical endocrinology 74, 424–433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamanti-Kandarakis E. & Dunaif A. Insulin resistance and the polycystic ovary syndrome revisited: an update on mechanisms and implications. Endocr Rev. 33, 981–1030 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legro R. S., Kunselman A. R., Dodson W. C. & Dunaif A. Prevalence and predictors of risk for type 2 diabetes mellitus and impaired glucose tolerance in polycystic ovary syndrome: a prospective, controlled study in 254 affected women. J Clin Endocrinol Metab 84, 165–169 (1999). [DOI] [PubMed] [Google Scholar]
- Ehrmann D. A., Barnes R. B., Rosenfield R. L., Cavaghan M. K. & Imperial J. Prevalence of impaired glucose tolerance and diabetes in women with polycystic ovary syndrome. Diabetes Care 22, 141–146 (1999). [DOI] [PubMed] [Google Scholar]
- Dahlgren E., Janson P. O., Johansson S., Lapidus L. & Oden A. Polycystic ovary syndrome and risk for myocardial infarction. Evaluated from a risk factor model based on a prospective population study of women. Acta Obstet Gynecol Scand 71, 599–604 (1992). [DOI] [PubMed] [Google Scholar]
- Mani H. et al. Diabetes and cardiovascular events in women with polycystic ovary syndrome: a 20-year retrospective cohort study. Clinical endocrinology 78, 926–934 (2013). [DOI] [PubMed] [Google Scholar]
- Randeva H. S. et al. Cardiometabolic aspects of the polycystic ovary syndrome. Endocr Rev. 33, 812–841 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. T. et al. Polycystic ovary syndrome is associated with higher left ventricular mass index: the CARDIA women’s study. J. Clin. Endocrinol Metab 97, 4656–4662 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathishkumar K., Elkins R., Yallampalli U., Balakrishnan M. & Yallampalli C. Fetal programming of adult hypertension in female rat offspring exposed to androgens in utero. Early Hum Dev. 87, 407–414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. J. et al. Hypertension caused by prenatal testosterone excess in female sheep. Am J. Physiol Endocrinol Metab 292, E1837–1841 (2007). [DOI] [PubMed] [Google Scholar]
- Manikkam M. et al. Fetal programming: prenatal testosterone excess leads to fetal growth retardation and postnatal catch-up growth in sheep. Endocrinology 145, 790–798 (2004). [DOI] [PubMed] [Google Scholar]
- Barker D. J. Deprivation in infancy and risk of ischaemic heart disease. Lancet 337, 981 (1991). [DOI] [PubMed] [Google Scholar]
- Padmanabhan V., Veiga-Lopez A., Abbott D. H., Recabarren S. E. & Herkimer C. Developmental programming: impact of prenatal testosterone excess and postnatal weight gain on insulin sensitivity index and transfer of traits to offspring of overweight females. Endocrinology 151, 595–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J. R., Bernasochi G. B., Varma U., Raaijmakers A. J. & Delbridge L. M. Sex and sex hormones in cardiac stress–mechanistic insights. J Steroid Biochem Mol. Biol. 137, 124–135 (2013). [DOI] [PubMed] [Google Scholar]
- Marsh J. D. et al. Androgen receptors mediate hypertrophy in cardiac myocytes. Circulation 98, 256–261 (1998). [DOI] [PubMed] [Google Scholar]
- Nahrendorf M. et al. Effect of testosterone on post-myocardial infarction remodeling and function. Cardiovasc Res. 57, 370–378 (2003). [DOI] [PubMed] [Google Scholar]
- Padmanabhan V. & Veiga-Lopez A. Sheep models of polycystic ovary syndrome phenotype. Molecular and cellular endocrinology 373, 8–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyagi T. & Matsui T. Phosphoinositide-3 kinase signaling in cardiac hypertrophy and heart failure. Current pharmaceutical design 17, 1818–1824 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke J. & Molkentin J. D. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 7, 589–600 (2006). [DOI] [PubMed] [Google Scholar]
- Altamirano F. et al. Testosterone induces cardiomyocyte hypertrophy through mammalian target of rapamycin complex 1 pathway. J Endocrinol 202, 299–307 (2009). [DOI] [PubMed] [Google Scholar]
- Nada S. E., Thompson R. C. & Padmanabhan V. Developmental programming: differential effects of prenatal testosterone excess on insulin target tissues. Endocrinology 151, 5165–5173 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemi O. J. et al. Activation or inactivation of cardiac Akt/mTOR signaling diverges physiological from pathological hypertrophy. J. Cell Physiol. 214, 316–321 (2008). [DOI] [PubMed] [Google Scholar]
- Sugden P. H., Fuller S. J., Weiss S. C. & Clerk A. Glycogen synthase kinase 3 (GSK3) in the heart: a point of integration in hypertrophic signalling and a therapeutic target? A critical analysis. Br. J. Pharmacol 153 Suppl 1, S137–153 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J. D. et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93, 215–228 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J. E. et al. Beta-myosin heavy chain is induced by pressure overload in a minor subpopulation of smaller mouse cardiac myocytes. Circulation research 109, 629–638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenz M. & Robbins J. Impact of beta-myosin heavy chain expression on cardiac function during stress. Journal of the American College of Cardiology 44, 2390–2397 (2004). [DOI] [PubMed] [Google Scholar]
- Huang Y., Liu H. & Li Y. Alterations in myosin heavy chain isoform gene expression during the transition from compensatory hypertrophy to congestive heart failure in rats. Chin. Med. J. (Engl) 114, 183–185 (2001). [PubMed] [Google Scholar]
- Reiser P. J., Portman M. A., Ning X. H. & Schomisch Moravec C. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. American Journal of Physiology. Heart and Circulatory Physiology 280, H1814–1820 (2001). [DOI] [PubMed] [Google Scholar]
- Koren M. J., Devereux R. B., Casale P. N., Savage D. D. & Laragh J. H. Relation of left ventricular mass and geometry to morbidity and mortality in uncomplicated essential hypertension. Ann. Intern. Med. 114, 345–352 (1991). [DOI] [PubMed] [Google Scholar]
- Levy D., Garrison R. J., Savage D. D., Kannel W. B. & Castelli W. P. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N. Engl. J. Med. 322, 1561–1566 (1990). [DOI] [PubMed] [Google Scholar]
- Karason K., Sjostrom L., Wallentin I. & Peltonen M. Impact of blood pressure and insulin on the relationship between body fat and left ventricular structure. Eur. Heart J. 24, 1500–1505 (2003). [DOI] [PubMed] [Google Scholar]
- Shigematsu Y. et al. Relation of genetic predisposition and insulin resistance to left ventricular hypertrophy in hypertension. Am. J. Hypertens 18, 457–463 (2005). [DOI] [PubMed] [Google Scholar]
- Lind L., Andersson P. E., Andren B., Hanni A. & Lithell H. O. Left ventricular hypertrophy in hypertension is associated with the insulin resistance metabolic syndrome. J. Hypertens 13, 433–438 (1995). [PubMed] [Google Scholar]
- Sasson Z., Rasooly Y., Bhesania T. & Rasooly I. Insulin resistance is an important determinant of left ventricular mass in the obese. Circulation 88, 1431–1436 (1993). [DOI] [PubMed] [Google Scholar]
- Rutter M. K. et al. Impact of glucose intolerance and insulin resistance on cardiac structure and function: sex-related differences in the Framingham Heart Study. Circulation 107, 448–454 (2003). [DOI] [PubMed] [Google Scholar]
- McMullen J. R. et al. Inhibition of mTOR signaling with rapamycin regresses established cardiac hypertrophy induced by pressure overload. Circulation 109, 3050–3055 (2004). [DOI] [PubMed] [Google Scholar]
- Shioi T. et al. Rapamycin attenuates load-induced cardiac hypertrophy in mice. Circulation 107, 1664–1670 (2003). [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R. & Hall M. N. TOR signaling in growth and metabolism. Cell 124, 471–484 (2006). [DOI] [PubMed] [Google Scholar]
- Condorelli G. et al. Akt induces enhanced myocardial contractility and cell size in vivo in transgenic mice. Proc. Natl. Acad. Sci. USA 99, 12333–12338 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T. et al. Phenotypic spectrum caused by transgenic overexpression of activated Akt in the heart. J. Biol. Chem. 277, 22896–22901 (2002). [DOI] [PubMed] [Google Scholar]
- Shimizu I. et al. Excessive cardiac insulin signaling exacerbates systolic dysfunction induced by pressure overload in rodents. J. Clin. Invest. 120, 1506–1514 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioi T. et al. Akt/protein kinase B promotes organ growth in transgenic mice. Mol. Cell Biol. 22, 2799–2809 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiojima I. et al. Disruption of coordinated cardiac hypertrophy and angiogenesis contributes to the transition to heart failure. J. Clin. Invest. 115, 2108–2118 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi Salloum B., Veiga-Lopez A., Abbott D. H., Burant C. F. & Padmanabhan V. Developmental programming: exposure to testosterone excess disrupts steroidal and metabolic environment in pregnant sheep. Endocrinology (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmo S. et al. Pathologic ventricular hypertrophy in the offspring of diabetic mothers: a retrospective study. Eur. Heart J. 28, 1319–1325 (2007). [DOI] [PubMed] [Google Scholar]
- Huang T., Kelly A., Becker S. A., Cohen M. S. & Stanley C. A. Hypertrophic cardiomyopathy in neonates with congenital hyperinsulinism. Arch. Dis. Child Fetal Neonatal Ed 98, F351–354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Twinn D. S. et al. The programming of cardiac hypertrophy in the offspring by maternal obesity is associated with hyperinsulinemia, AKT, ERK, and mTOR activation. Endocrinology 153, 5961–5971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernea M., Lee T., Cheng G., Padmanabhan V., Lehman M. N. & Coolen L. M. Excess prenatal testosterone increases androgen receptor expression in hypothalamic areas of the female sheep brain. Program of the Annual Meeting of the Society for Neuroscience, Washington DC, USA, p 499.420 (2011).
- Ortega H. H., Salvetti N. R. & Padmanabhan V. Developmental programming: prenatal androgen excess disrupts ovarian steroid receptor balance. Reproduction (Cambridge, England) 137, 865–877 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minette M. S. et al. Cardiac function in congenital adrenal hyperplasia: a pattern of reversible cardiomyopathy. J Pediatr 162, 1193–1198, 1198 e1191 (2013). [DOI] [PubMed] [Google Scholar]
- Wilson C. et al. Testosterone increases GLUT4-dependent glucose uptake in cardiomyocytes. J Cell Physiol 228, 2399–2407 (2013). [DOI] [PubMed] [Google Scholar]
- Abel E. D. Glucose transport in the heart. Front Biosci 9, 201–215 (2004). [DOI] [PubMed] [Google Scholar]
- Beckett E. M., Astapova O., Steckler T. L., Veiga-Lopez A. & Padmanabhan V. Developmental programing: impact of testosterone on placental differentiation. Reproduction 148, 199–209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan V., Lee J.S., Anthony R. V., Mohankumar S. & Mohankumar P. S. Fetal programming: prenatal testosterone excess leads to compromised cardiac development. In Proceedings of the 86th annual Meeting of the Endocrine Society page 322 (Endocrine Society, New Orleans, 2004).
- Barker D. J., Osmond C., Golding J., Kuh D. & Wadsworth M. E. Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. Bmj 298, 564–567 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. & Lubo Z. Fetal programming of cardiac function and disease. Reprod Sci 14, 209–216 (2007). [DOI] [PubMed] [Google Scholar]
- Tappia P. S., Nijjar M. S., Mahay A., Aroutiounova N. & Dhalla N. S. Phospholipid profile of developing heart of rats exposed to low-protein diet in pregnancy. Am J Physiol Regul Integr Comp Physiol 289, R1400–1406 (2005). [DOI] [PubMed] [Google Scholar]
- Cheema K. K., Dent M. R., Saini H. K., Aroutiounova N. & Tappia P. S. Prenatal exposure to maternal undernutrition induces adult cardiac dysfunction. Br J Nutr 93, 471–477 (2005). [DOI] [PubMed] [Google Scholar]
- Dodic M. et al. Programming effects of short prenatal exposure to cortisol. Faseb J 16, 1017–1026 (2002). [DOI] [PubMed] [Google Scholar]
- Moss T. J. et al. Programming effects in sheep of prenatal growth restriction and glucocorticoid exposure. Am J Physiol Regul Integr Comp Physiol 281, R960–970 (2001). [DOI] [PubMed] [Google Scholar]
- Langdown M. L., Holness M. J. & Sugden M. C. Early growth retardation induced by excessive exposure to glucocorticoids in utero selectively increases cardiac GLUT1 protein expression and Akt/protein kinase B activity in adulthood. J Endocrinol 169, 11–22 (2001). [DOI] [PubMed] [Google Scholar]
- Sir-Petermann T. et al. Maternal serum androgens in pregnant women with polycystic ovarian syndrome: possible implications in prenatal androgenization. Human reproduction 17, 2573–2579 (2002). [DOI] [PubMed] [Google Scholar]
- Crisosto N. et al. Improvement of hyperandrogenism and hyperinsulinemia during pregnancy in women with polycystic ovary syndrome: possible effect in the ovarian follicular mass of their daughters. Fertil Steril 97, 218–224 (2012). [DOI] [PubMed] [Google Scholar]
- Sir-Petermann T. et al. Birth weight in offspring of mothers with polycystic ovarian syndrome. Human reproduction 20, 2122–2126 (2005). [DOI] [PubMed] [Google Scholar]
- Sir-Petermann T. et al. Early metabolic derangements in daughters of women with polycystic ovary syndrome. J Clin Endocrinol Metab 92, 4637–4642 (2007). [DOI] [PubMed] [Google Scholar]
- Veiga-Lopez A. et al. Developmental programming: impact of excess prenatal testosterone on intrauterine fetal endocrine milieu and growth in sheep. Biol Reprod 84, 87–96 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikkam M., Steckler T. L., Welch K. B., Inskeep E. K. & Padmanabhan V. Fetal programming: prenatal testosterone treatment leads to follicular persistence/luteal defects; partial restoration of ovarian function by cyclic progesterone treatment. Endocrinology 147, 1997–2007 (2006). [DOI] [PubMed] [Google Scholar]
- Hayashi T. et al. Role of gp91phox-containing NADPH oxidase in left ventricular remodeling induced by intermittent hypoxic stress. American journal of physiology 294, H2197–2203 (2008). [DOI] [PubMed] [Google Scholar]
- Barrick C. J., Rojas M., Schoonhoven R., Smyth S. S. & Threadgill D. W. Cardiac response to pressure overload in 129S1/SvImJ and C57BL/6J mice: temporal- and background-dependent development of concentric left ventricular hypertrophy. American journal of physiology 292, H2119–2130 (2007). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Nakagawa S. & Cuthill I. C. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biol Rev Camb Philos Soc 82, 591–605 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.