Abstract
The expression of BRDT, a member of the BET sub-family of double bromodomain-containing proteins, is restricted to the male germ line, specifically to pachytenediplotene spermatocytes and early spermatids. We previously showed that loss of the first bromodomain of BRDT by targeted mutagenesis (BrdtΔBD1) resulted in sterility and abnormalities in spermiogenesis, but little is known about BRDT's function at the molecular level. As part of studies designed to identify BRDT-interacting proteins we stably introduced a FLAG-tagged BRDT cDNA into 293T cells, which do not normally express BRDT. Affinity-purification of FLAG-tagged BRDT complexes indicated that BRDT has novel interactions with the histone deacetylase HDAC1, the arginine-specific histone methyltransferase 5 PRMT5, and the Tripartite motif-containing 28 protein TRIM28. Immunofluorescent microscopy revealed that BRDT co-localized with each of these proteins in round spermatids and co-immunoprecipitation of testicular extracts showed that these proteins interact with BRDT. Furthermore, they bind the promoter ofH1t, a putative target of BRDT-containing complexes. This binding of H1t was lost in mice expressing the BrdtΔBD1 mutant protein and concomitantly, H1t expression was elevated in round spermatids. Our study reveals a role for BRDT-containing complexes in the repression of gene expression in vivo that correlates with dramatic effects on chromatin remodeling and the progression of spermiogenesis.
Introduction
The highly conserved bromodomain motif is present in many nuclear transcriptional regulatory factors and has been implicated in chromatin remodeling through binding to acetylated lysines in histones and other proteins (Marmorstein and Berger 2001, Loyola and Almouzni 2004, Sanchez and Zhou 2009, Maze, Noh et al. 2014). There are over 30 bromodomain-containing proteins in mammals among which the BET (bromodomain and extra-terminal) family consisting of BRD2, BRD3, BRD4 and BRDT is characterized by the presence of two bromodomains (BD1 and BD2) and an additional extraterminal (ET) domain at the carboxyterminal end.
In both humans and mouse, the expression of the BET family protein BRDT is normally restricted to the male germ line, and targeted mutagenesis in the mouse model revealed that mice lacking the first bromodomain (BD1) of BRDT (BrdtΔBD1) are sterile, with severely disrupted spermiogenesis (Shang, Nickerson et al. 2007). Complete loss of BRDT function (Brdt−/−) results in an earlier and more severe phenotype, with spermatogenesis arresting in late meiotic prophase (Gaucher, Boussouar et al. 2012). Aberrant expression of BET family proteins, including BRDT, has been correlated with oncogenesis in humans (Muller, Filippakopoulos et al. 2011). For example, human BRDT has been implicated in non-small cell lung cancer (Scanlan, Altorki et al. 2000, Grunwald, Koslowski et al. 2006) and several other cancers (Scanlan, Altorki et al. 2000). However, very little is known about BRDT's normal function in cellular processes during spermatogenesis or its abnormal function in cancer.
As bromodomain-containing proteins function in transcriptional regulation not by binding to DNA but rather by their interaction with other proteins, it is important to identify their interacting proteins. The bromodomains have been clearly shown to bind acetylated lysines in histones and in other proteins (Strahl and Allis 2000, Marmorstein and Berger 2001, Loyola and Almouzni 2004, Sanchez and Zhou 2009), but very few of these proteins have been identified for BET proteins in general and BRDT in particular. Several bromodomain-containing proteins, including BET proteins, exhibit dual properties with regard to transcriptional regulation. That is, they have been shown to recruit either transcriptional co-activators or co-repressors, depending on the requirements of the signal transduction machinery and the promoter (Denis 2001, Denis, Nikolajczyk et al. 2010). For example, acetylation of H2A.Z and H4 results in the recruitment of BRD2 to chromatin during transcriptional activation (Draker, Ng et al. 2012). BRD2 is also a TBP-associated protein and a 26 amino acid peptide in the BD1 of BRD2 is essential for BRD2-TBP interaction (Peng, Dong et al. 2007). BRD3 is recruited by GATA1 to both active and repressed target genes and is required for GATA1 chromatin occupancy and normal erythroid maturation (Lamonica, Deng et al. 2011). BRD4 binding has been shown to reconstitute the active form of P-TEFB, which phosphorylates serine 2 of the C-terminal domain (CTD) of RNA polymerase II (RNAP II) along the chromatin template, and stimulates transcriptional elongation (Jang, Mochizuki et al. 2005, Yang, Yik et al. 2005). BRD4 has also been shown to recruit CDK9 to phosphorylate the C-terminal domain of RNAP II and facilitate transcription of NF-κB-dependent inflammatory genes (Huang, Yang et al. 2009). Conversely, BRD4 is also found in an HPV transcriptional silencing complex (Wu, Lee et al. 2006).
BRDT has been shown to recognize a diacetylated histone H4 peptide through its BD1 and has been implicated in the striking chromatin remodeling that follows histone hyperacetylation during spermiogenesis (Pivot-Pajot, Caron et al. 2003, Moriniere, Rousseaux et al. 2009). However, as it is known that bromodomain-containing proteins can interact with proteins other than histones, we wished to identify other physiologically relevant, in vivo interacting proteins for BRDT. As an initial approach to this goal, we generated cell lines stably expressing FLAG-tagged BRDT and identified BRDT-associated proteins by affinity purification of BRDT-containing complexes from these cells followed by mass spectrometry. Putative BRDT-interacting proteins HDAC1, PRMT5, and TRIM28 were identified and the physiological relevance of the interaction with BRDT confirmed by co-immunoprecipitation from testicular extracts and co-localization at the cellular level. Finally, we demonstrated a role for these proteins as part of BRDT-containing complexes that repress the expression of the putative target gene H1t in round spermatids.
MATERIALS AND METHODS
Expression constructs
To generate plasmid pBABE-puromycin/BRDT-FLAG, a cDNA encoding full-length BRDT was PCR amplified from a testis cDNA library, using a forward primer (5′CCGGAATTCGAATTTGTAGACTTTTCCTGC-3′) that introduces an EcoRI restriction site, and a reverse primer (5′-CCGGAATTCTCATTTGTCATCGTCGTCCTTGTAGTCATCAAAGTTATTTTCAAACAT-3′) that introduces a FLAG epitope tag before a stop codon and EcoRI restriction site. The EcoRI-digested PCR fragment was next inserted into the corresponding site of the pBABE-puromycin vector.
Cell culture and establishment of stable cell lines
To establish the cell line 293T/BRDT-FLAG, 293T cells were transfected for 5 hr with 2 μg of pBabe/BRDT-FLAG using Lipofectamine (Invitrogen, Inc.). After 2 days, drug-resistant BRDT-FLAG cells were selected in the presence of 2.5 μg/ml puromycin. Several puromycin-resistant colonies were grown and positive clones were identified by immunoblotting using anti-FLAG (Santa Cruz) and BRDT antibodies. 293T/BRDT-FLAG cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) in the presence of puromycin at a final concentration of 2.5 μg/ml.
Isolation of populations of round spermatids and primary cell culture
Enriched populations of round spermatids were obtained using our previously described procedures with minor modifications (Wolgemuth, Gizang-Ginsberg et al. 1985). Briefly, testes were decapsulated into Dulbecco's modified Eagle's medium and the seminiferous tubules were dispersed and incubated in 0.5 mg/ml collagenase at 37°C for 10 min with agitation. The buffer was passed through a 70 μm filter and the seminiferous tubules were incubated in 0.5 mg/ml collagenase and 0.25 mg/ml trypsin at 37°C for 5 min with agitation, and the buffer was then passed through a 70 μm filter (BD Biosciences). The filtrate was centrifuged at 1500 rpm for 3 min. The supernatant was removed, and testicular cells in the pellet were washed with Dulbecco's modified Eagle's medium. The single cell suspension was separated on a 2-4% Bovine Serum Albumin (BSA) in DPBS gradient using gravity cell sedimentation. Populations of round spermatids of ≥90% purity were pooled. The final purity of the pooled populations was assessed by flow cytometric analysis using propidium iodide (Sigma cat#P4170) staining and analysis on a BD FACS Calibur Cell Analyzer. All steps of cell separation, fractionation, and pooling were performed at 4° C in order to maintain cell viability and maximize nucleic acid integrity. The isolated round spermatids were cultured using Dulbecco's modified Eagle's medium supplemented with 10% FBS serum and with or without 20 mM valproic acid (VPA, Sigma) at 37°C with 5% CO2 for 20 hr (Choi, Han et al. 2008). After 20 hr, cell viability was ascertained by staining with 0.4% trypan blue.
Protein purification and identification by mass spectrometry
FLAG-tagged BRDT-containing complexes were purified from nuclear extracts (Sif, Saurin et al. 2001, Pal, Vishwanath et al. 2004). Briefly, cells were lysed in buffer [10 mM Tris (pH 8.0), 10 mM KCl, 2 mM MgCl2, 50 mM β-glycerol phosphate, 0.1% NP40 and a protease inhibitor cocktail (Roche)] and then centrifuged for 5 min at 2500g at 4°C. Pelleted nuclei were resuspended and lysed in nuclei buffer [20 mM Tris (pH 8.0), 420 mM NaCl, 1 mM EDTA, 50 mM β-glycerol phosphate, 25% glycerol and a protease inhibitor cocktail (Roche)] on ice for 30 min. Nuclear extracts were cleared by centrifugation at 18 000g for 10 min at 4°C. FLAG-tagged BRDT-containing complexes were purified by incubating nuclear extracts with EZview red anti-FLAG M2 affinity gel (Sigma cat#F-2426) overnight at 4°C. The beads were loaded onto a column and washed extensively with buffer BC-0 [20 mM HEPES (pH 7.9), 20% glycerol, 2 mM EDTA, 1 mM DTT and 0.5 mM PMSF] supplemented with 0.15 M KCl, followed by a wash with buffer BC-0.3 (BC-0 containing 300 mM KCl). Bound proteins were eluted in BC-0.1 with FLAG peptide (Sigma cat#F-3290). Proteins followed by sodium dodecyl sulfatepolyacrylamide gel electrophoresis (SDS-PAGE) separation. After staining with Coomassie blue, the bands were excised and analyzed by mass spectrometry.
LC-MS/MS analysis was done on a Waters Ultima Q-Tof hybrid quadrupole/time-of-flight mass spectrometer with a nanoelectrospray source. Capillary voltage was set at 1.8kV and cone voltage 32V; collision energy was set according to mass and charge of the ion, from 18eV to 50eV. Chromatography was performed on an LC Packings HPLC with a C18 PepMap column using a linear acetonitrile gradient with flow rate of 200 nl/ min. Raw data files were processed using the MassLynx ProteinLynx software and .pkl files were submitted for searching at www.matrixscience.com using the Mascot algorithm.
Immunoblot analysis and immunoprecipitation
To examine Flag-tagged BRDT expression, 20 μg of nuclear, cytosolic, or whole-cell lysates from each cell line were loaded on an 8% SDS-polyacrylamide gel, transferred onto nitrocellulose membrane, and detected by enhanced chemiluminescence reagents according to the manufacturer's recommendations (Amersham Pharmacia Biotech, Inc.). For immunoprecipitation, approximately 200 μg of nuclear extract from either 293T-BRDT cells or testis was pre-cleared with 30 μl of a 50% slurry of protein A agarose beads for 2 hr at 4°C. Pre-cleared nuclear extracts were incubated with 20 μl of either pre-immune or immune anti-BRDT antibodies for an additional 2 hr at 4°C before the addition of 40 μl of pre-blocked protein A agarose beads. The samples were then incubated overnight at 4°C and washed four times with 1 ml of buffer A (40 mM Tris-HCl [pH 8.0], 250 mM NaCl, 0.5% NP-40), and bound proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by immunoblotting (Wang, Baiocchi et al. 2005). Antibodies specific for BRDT have been previously described (Shang, Nickerson et al. 2007), anti-FLAG M2, was purchased from Santa Cruz; anti-PRMT5 and anti-HDAC1 were purchased from Millipore (cat# 07-405, #06-720 ).
Immunofluorescence
Immunofluorescence staining of single cell suspensions was carried out according to our laboratory protocols as previously published (Berkovits, Wang et al. 2012). Briefly, the tunica albuginea was removed from adult wild-type testes and the tubules were placed in cold PBS. The tubules were manually sheared with scissors and by pipetting, and then passed through a 40 μm filter. The resulting suspension was dropped on slides, air dried, and then fixed in 4:1 methanol: acetone for 10 min. BRDT antibodies generated by our lab (Shang, Nickerson et al. 2007) were used at a dilution of 1:300. The rabbit polyclonal anti-PRMT5 (Millipore, cat# 07-405), anti-HDAC1 (Millipore, cat#06-720), anti-TRIM28 (Gene Tex, cat#GTX102226) and H4R3me2s (Active motif, cat# 61187) primary antibodies were used at a 1:200 dilution. The following secondary antibodies were used: DyLight 594 Fab fragment donkey anti-rabbit IgG (Jackson ImmunoResearch cat# 711-517-003 04) at 1:300 dilution and Alexa Fluor-488 goat anti-rabbit IgG (Molecular Probes, cat#11008) at 1:300.
Reverse transcription (RT) real-time PCR
To perform quantitative real-time PCR, samples of purified control and BrdtΔBD1 (Shang, Nickerson et al. 2007) mutant round spermatids were prepared and RNA was extracted with Trizol reagent (Invitrogen, Inc.) according to the manufacturer's protocol. Real-time PCR was carried out according to our laboratory's standard protocol (Shang, Nickerson et al. 2007). The values were normalized to the expression of β-actin (Abcam) as an internal control. The primers were designed using the Primer3 program (http://frodo.wi.mit.edu/primer3) and synthesized by Integrated DNA Technologies. The following primers were used: H1t coding forward: GAGGAGAAACCTTCATCTAAAAGG; H1t coding reverse: ACCAGGACTCCTTTATTCACAAGT;
Chromatin immunoprecipitation
For chromatin immunoprecipitation (ChIP), a protocol from Upstate Biotechnology was followed. Briefly, seven testes were decapsulated and resuspended in 25 ml RPMI 1640 medium (Invitrogen, Inc.) containing 1.0 mg/ml collagenase and incubated at 33°C for 10 min. The cells were centrifuged and resuspended in 25 ml DMEM medium containing 0.25 mg/ml trypsin and incubated at 37°C for 5 min. The cells were cross-linked with 1% formaldehyde in RPMI 1640 for 10 min and the DNA was sheared to 300-500 bp with sonication. The debris was removed by centrifugation at 16,000 g for 10 min at 4°C and the supernatant was incubated with a C-terminal anti-BRDT antibody generated by our laboratory (Shang, Nickerson et al. 2007) or anti-IgG as a control and Dynabeads protein A (Invitrogen, Inc.) at 4°C overnight. The beads were washed successively with RIPA buffer (10 mM Tris, pH 7,6, 1 mM EDTA, 0.1% SDS, 0.1% Na-Deoxycholate, 1% Triton X-100), RIPA buffer containing 0.3 M NaCl, LiCl buffer (0.25 M LiCl, 0.5% NP40, 0.5% NaDOC), 1xTE contain 0.2% Triton X-100 and the chromatin was eluted with 1xTE buffer and reverse cross-linked by incubation at 65°C overnight. The DNA was purified and used as template for PCR detection using the primers as follows:
H1t promoter forward: TGTTATCATTTCCTAGCCCAAGGTGAC
H1t promoter reverse: AAAGCTGGAATCACAGTTTCCAATCTC
RESULTS
Establishment of a stable BRDT-expressing 293T cell line
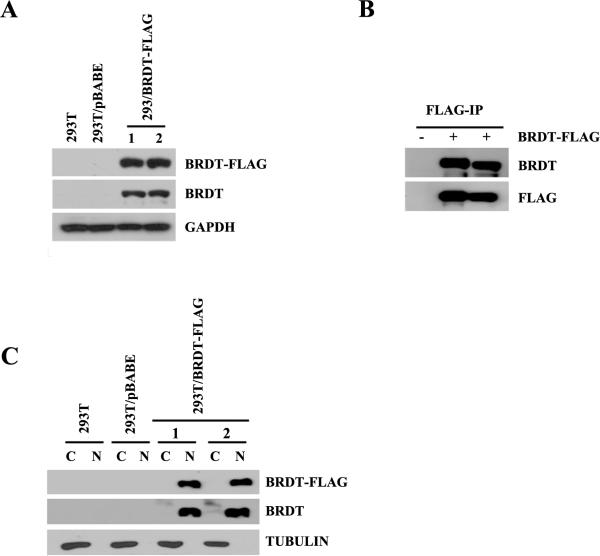
The construct pBABE-puromycin/BRDT-FLAG was used to establish cell lines that stably express BRDT as assessed by Western blot analysis using anti-FLAG and anti-BRDT antibodies. We were able to express BRDT in several 293T/BRDT-FLAG clones but not in the more physiologically relevant mouse germ cell line GC-4spc (Tascou, Nayernia et al. 2000) nor in NIH3T3, Hela, or mouse embryonic fibroblast cells. Figure 1A shows the detection of BRDT expression 293T/BRDT-FLAG but not in 293T cells alone or in 293T cells transfected with empty vector. We detected BRDT in FLAG-immunoprecipitates from the 293T-BRDT (or 293T/BRDT-FLAG) cell lines but not in untransfected cells or cells transfected with empty vector (Fig. 1B). The sub-cellular distribution of BRDT in the cell lines was determined by immunoblotting of nuclear and cytoplasmic extracts, and shown to be detected only in the nuclear compartment (Fig.1C). Collectively, these results demonstrate that we have successfully ectopically expressed BRDT in 293T cells.
Figure 1. Expression of mouse BRDT in 293T cell lines.
(A) Analysis of ectopically expressed BRDT in selected 293T clones. Approximately 20 μg of whole extract isolated from either 293T, 293T/pBABE, or 293T/BRDT-FLAG cells was analyzed by immunoblotting using either anti-FLAG, BRDT, or GAPDH rabbit polyclonal antibodies. (B) Nuclear extracts from 293T/pBABE and 293T/BRDT-FLAG cells were FLAG-immunoprecipitated, and after extensive washing the retained proteins were analyzed by immunoblotting using anti-BRDT and anti-FLAG antibodies. (C) Immunoblot analysis of BRDT-FLAG nuclear and cytoplasmic expression in one of the stable 293T/BRDT-FLAG cell lines. Detection of tubulin served as a cytoplasmic extract control.
Identification of proteins associated with BRDT
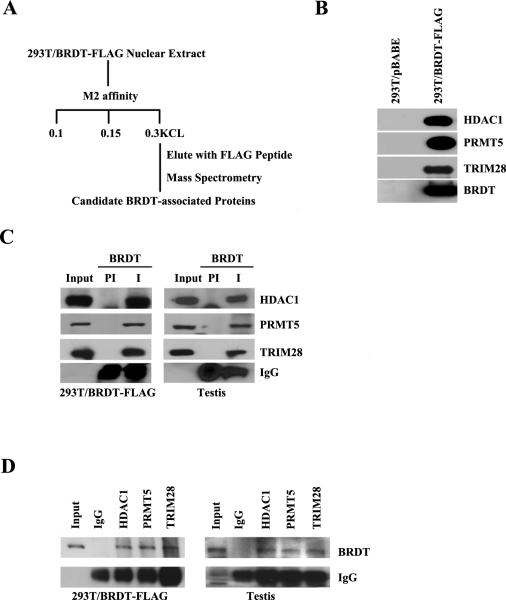
Our purpose for generating these 293T/BRDT-FLAG cell lines was to provide a system with which to identify proteins that interact with BRDT in vivo. We therefore affinity-purified FLAG-tagged BRDT from the 293T/BRDT-FLAG cells followed by electrophoretic separation and mass spectrometry (see cartoon in Fig. 2A). Nuclear extracts from 293T cells that express BRDT-FLAG were incubated with anti-FLAG M2 affinity gel and, after extensive washing, the retained proteins were eluted with a 20-fold molar excess of FLAG peptide (Fig. 2A). The presence of BRDT in the FLAG affinity purified fractions from 293T/BRDT-FLAG cells but not from control 293T fractions was confirmed using a highly specific anti-BRDT antibody (Fig. 2B). Protein mixtures purified by anti-FLAG affinity chromatography were separated by electrophoresis in a 10% SDS polyacrylamide gel. After staining with Coomassie blue, the bands were excised and analyzed by mass spectrometry. This analysis revealed that in addition to spliceosome complex components and other splicing proteins (Berkovits, Wang et al. 2012), the BRDT-containing complexes included components of repressive chromatin complexes, specifically HDAC1, PRMT5 and TRIM28.
Figure 2. BRDT associates with HDAC1, PRMT5 and TRIM28.
(A) Scheme for identifying interacting proteins in BRDT-containing complexes. Peptide microsequencing by mass spectrometry identified HDAC1, PRMT5 and TRIM28, among other proteins, as candidate BRDT-associated proteins. (B) The affinity-purified complexes from 293T/BRDT-FLAG cells were analyzed by immunoblotting and shown to co-immunoprecipitate endogenous HDAC1, PRMT5 and TRIM28 proteins. (C) Co-immunoprecipitation analysis with anti-BRDT followed by immunoblot analysis showing that HDAC1, PRMT5 and TRIM28 proteins complex in vivo with BRDT in testicular extracts as well as in 293T/BRDT-FLAG cells. (D) Reciprocal co-immunoprecipitation with antibodies against HDAC1, PRMT5 and TRIM28 followed by immunoblot analysis with anti-BRDT showing that their association with BRDT in vivo in testicular extracts as well as 293T/BRDT-FLAG cells.
BRDT is associated with HDAC1, PRMT5 and TRIM28 in lysates of 293T/ BRDT-FLAG cells and mouse testis
We next immunoprecipitated BRDT-FLAG complexes in 293T/BRDT-FLAG cell lines and confirmed the presence of HDAC1, PRMT5, and TRIM28 in anti-BRDT immunoprecipitated complexes but not in control pre-immune fractions (Fig. 2C). To begin to determine the physiological relevance of these interactions in vivo, we immunoprecipitated endogenous BRDT complexes from lysates of wild type round spermatids and checked for the presence of HDAC1, PRMT5 and TRIM28 (Fig. 2C). All three proteins were detected in anti-BRDT immunoprecipitates but not in control pre-immune fractions, suggesting that endogenous BRDT interacts with HDAC1, PRMT5 and TRIM28 in testicular cells. To further confirm that BRDT is associated with HDAC1, PRMT5 and TRIM28, the reciprocal co-immunoprecipitation was performed using HDAC1, PRMT5 and TRIM28 antibodies and extracts from 293T/ BRDT-FLAG cells and round spermatids of wild type mice. In each case, BRDT protein was co-immunoprecipitated (Fig. 2D).
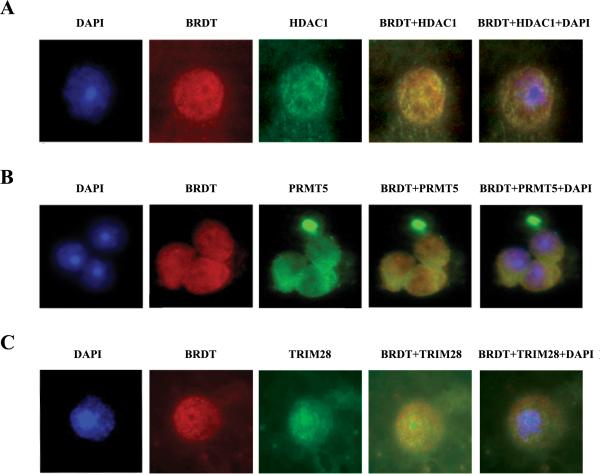
To further confirm that BRDT forms complexes with HDAC1, PRMT5 and TRIM28, we next sought to determine if they co-localize, at the cellular level, in round spermatids. To this end, we used immunofluorescent microscopy of cellular preparations of round spermatids simultaneously stained with anti-BRDT antibody and either anti-HDAC1, PRMT5 or TRIM28 antibodies and DAPI (Fig. 3). The data are presented as representative individual images for each staining and also as merged images in which yellow indicates the co-localization of BRDT/HDAC1, BRDT/PRMT5, and BRDT/TRIM28.
Figure 3. Co-localization of BRDT with HDAC1, PRMT5 and TRIM28 in round spermatids.
(A) Immunofluorescent detection of round spermatids stained for BRDT (red) and HDAC1 (green). Panels (B) and (C) depict the staining for BRDT and PRMT5 and TRIM28, respectively. Nuclei were counterstained with DAPI (blue). Photomicrographs were taken at 100× magnification.
BRDT, HDAC1, PRMT5 and TRIM28 are recruited to the H1t promoter in a BD1-dependent manner
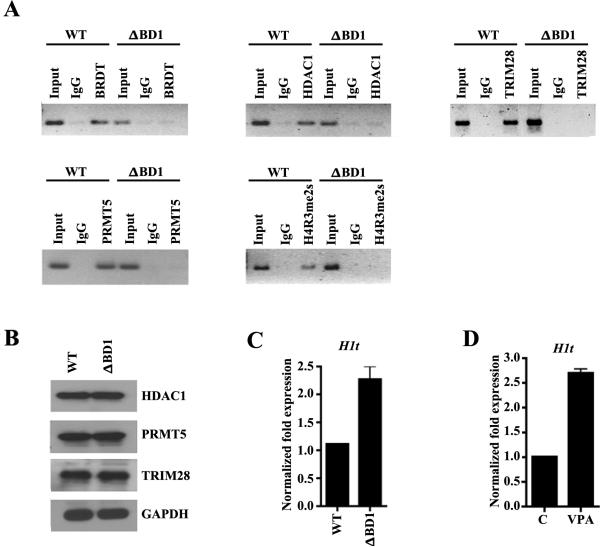
We have previously shown that BRDT-containing complexes bind to the promoter region of H1t (Shang, Nickerson et al. 2007). We therefore asked whether HDAC1, PRMT5 and TRIM28 are also recruited to the promoter region of BRDT target genes, such as H1t, using a chromatin immunoprecipitation (ChIP) approach. Cross-linked chromatin from round spermatids of WT mice was immunoprecipitated with either IgG or anti-BRDT or anti-HDAC1, PRMT5, or TRIM28 antibodies. We indeed detected binding of BRDT, HDAC1, PRMT5, and TRIM28 at the H1t promoter in wild type round spermatids (Fig. 4A). However, ChIP analysis using round spermatids from BrdtΔBD1 mutant testes revealed that loss of the BD1 of BRDT abrogated recruitment of BRDT and the interacting partners to the H1t promoter (Fig. 4A). The failure of HDAC1, PRMT5 and TRIM28 to be recruited to the H1t promoter is independent of their levels in the mutant spermatids (Fig. 4B). This suggested that the BD1 of BRDT is required for binding of the BRDT-interacting proteins HDAC1, PRMT5 and TRIM28. To determine if BRDT, HDAC1, PRMT5 and TRIM28 are directly involved in transcriptional repression of H1t in round spermatids, we analyzed the transcript levels of H1t in wild type and BrdtΔBD1 mutant round spermatids. Indeed, qRT-PCR analysis revealed that H1t was up-regulated 2.5 fold in the mutant round spermatids. (Fig. 4C).
Figure 4. BRDT, HDAC1, PRMT5 and TRIM28 are recruited to the H1t promoter and modulate its transcription and epigenetic modification of chromatin.
(A) ChIP assays were performed on cross-linked chromatin from round spermatids isolated from either WT or BrdtΔBD1 mutant (ΔBD1) mice using BRDT pre-immune (PI) or anti-BRDT antibody or using anti-HDAC1, anti-PRMT5, anti-H4R3me2s or anti-TRIM28 antibodies. Immunopurified DNA was amplified by PCR using H1t-specific primers. (B) Analysis of HDAC1, PRMT5 and TRIM28 expression in WT and BrdtΔBD1 round spermatids. Approximately 20 μg of extracts isolated from round spermatids were analyzed by immunoblotting using either anti-HDAC1, PRMT5 or TRIM28 antibodies; anti-GAPDH served as control. (C) Total RNA from WT and BrdtΔBD1 round spermatids was PCR-amplified using H1t gene-specific primers. (D) Total RNA from control and VPA-treated WT round spermatids (VPA) was used in RT-PCR using H1t gene-specific primers.
The arginine methyltransferase PRMT5 has been reported to mediate histone H4R3 methylation (Pal, Baiocchi et al. 2007, Zhao, Rank et al. 2009) and the repressive mark H4R3me2 is present in round spermatids (Eckert, Biermann et al. 2008). To begin to explore the mechanisms by which BRDT complexes might mediate repression of H1t, we next asked whether symmetrical dimethylation of arginine 3 on H4 could be detected in the promoter region of H1t in round spermatids. Our ChIP analysis showed the presence of H4R3me2 at the promoter of the H1t in round spermatids of WT but not BrdtΔBD1 mutant testes (Fig. 4A), similar to the failure of PRMT5 to be recruited to the H1t promoter in the absence of BD1.
HDAC1 inhibitors enhance the expression of H1t in round spermatids
Based on our finding that BRDT is associated with HDAC1, we next explored the relationship between the BRDT complex target gene H1t and HDAC1 function during spermiogenesis. Specifically, we isolated round spermatids from mouse testes and treated them with a well-known HDAC inhibitor, VPA, known to be specific for class I HDACs and to interrupt co-repressor-associated proteins (Gottlicher, Minucci et al. 2001). qRT-PCR analysis of the levels of expression of H1t upon treatment with VPA revealed a 2-fold increased expression of H1t in VPA-treated round spermatids as compared to controls (Fig 4D).
Discussion
The BET family member BRDT is testis-specific in both mice and humans and has been shown to play an important role in spermatogenesis in the mouse model (Shang, Nickerson et al. 2007, Gaucher, Boussouar et al. 2012). We have shown that mice expressing a truncated BRDT protein, lacking only the N-terminal region and the BD1 are sterile, with severely disrupted spermiogenesis (Shang, Nickerson et al. 2007). Our microarray analysis revealed both up- and down-regulated genes in BrdtΔBD1 mutant testes compared with WT, demonstrating that BD1 has a role in both gene activation and repression (Berkovits, Wang et al. 2012). Our ongoing ChIP-Seq analysis has further identified thousands of BRDT-containing complex binding sites to promoter regions of genes in pachytene spermatocytes and round spermatids (L. Wang et al., in preparation).
In the present study, we used FLAG-tagged BRDT expressed in a cultured cell line to identify endogenous proteins that can interact with BRDT. We then confirmed the physiological relevance by showing that the identified proteins interact with BRDT in vivo, in testicular cells. The screen yielded several interesting candidates, including several splicing-related proteins (Berkovits, Wang et al. 2012), and presented here, three proteins known to be involved in transcriptional repression, HDAC1, PRMT5, and TRIM28, each of which is expressed in the testis. Importantly, the expression of BRDT, PRMT5, HDAC1, and TRIM28 overlap from pachytene spermatocytes through round spermatids (Weber, Cammas et al. 2002, Shang, Nickerson et al. 2007, Choi, Han et al. 2008, Eckert, Biermann et al. 2008).
We had previously shown that BRDT binds to the promoter region of the H1t gene to repress its expression and that this binding depends on its BD1 (Shang, Nickerson et al. 2007). However, the molecular nature of this putative repressive complex was not known. The present study provides evidence that suggests that BRDT forms functional complexes with the transcription repressors HDAC1, PRMT5 and TRIM28 to repress expression of the BRDT target gene H1t in round spermatids. Moreover, our ChIP results showed that the association of HDAC1, PRMT5, and TRIM28 with the H1t promoter in round spermatids from BrdtΔBD1 mutant testes is decreased, and concomitantly, H1t expression level is increased.
HDAC1 is a class I histone deacetylase found as a component of multiple transcriptional co-repressor complexes (Fischle, Kiermer et al. 2001, Liang, Wan et al. 2008, Ma and Schultz 2008, Hwang, Roe et al. 2012). HDAC1-mediated repression has been demonstrated in the testis as well (Hong, Suh et al. 2005, Lui, Wong et al. 2007) and HDAC1 mis-regulation has been implicated in impaired spermatogenesis (Omisanjo, Biermann et al. 2007, Pacheco, Houseman et al. 2011). HDAC1 is expressed from the spermatogonial stage until step 8 spermatids. Thereafter its absence is permissive to the hyperactylation of canonical histones that is required for the deposition of transition proteins and then protamines (Meistrich, Trostle-Weige et al. 1992, Hazzouri, Pivot-Pajot et al. 2000).
PRMTs regulate chromatin structure and expression of a wide spectrum of target genes (Karkhanis, Hu et al. 2011). PRMT5 in particular works in concert with a variety of cellular proteins including ATP-dependent chromatin remodelers and co-repressors to induce epigenetic silencing and control growth-promoting and pro-survival pathways (Karkhanis, Hu et al. 2011). PRMT5 is an arginine-specific histone methyltransferase that mediates symmetrical dimethylation of arginine 3 in histone H2A and/or H4 tails (H2A/H4R3me2) and other non-histone proteins (Pal, Baiocchi et al. 2007). Interestingly, it has been shown that H4R3 methylation is involved in the suppression of somatic differentiation programs in primordial germ cells through a complex containing BLIMP1 and PRMT5 (Ancelin, Lange et al. 2006). The germ cell lineage is maintained and propagated by the BLIMP1–PRMT5 complex through symmetrical methylation of arginine 3 on histone H2A and H4 (H2A/H4R3me2s). BLIMP1-PRMT5 co-localization in the nucleus results in high levels of H2A/H4R3 methylation in PGCs at E8.5. However, at E11.5, BLIMP1-PRMT5 translocates from the nucleus to the cytoplasm, resulting in the loss of H2A/H4 R3 methylation at the time of extensive epigenetic reprogramming of germ cells.
TRIM28 (TIF1β, KAP1) is a transcriptional co-repressor that can act as a scaffold for various heterochromatin-induced factors, such as heterochromatin binding protein (HP1 α, β, γ), the histone methyltransferase SETB1, the nucleosome remodeling and histone deacetylation (NuRD) complex, and the nuclear receptor co-repressor complex 1 (N-CoR1) (Schultz, Friedman et al. 2001, Schultz, Ayyanathan et al. 2002, Sripathy, Stevens et al. 2006). Through the formation of these complexes, TRIM28 mediates repression pathways that are involved in the local heterochromatinization of DNA. During spermatogenesis, TRIM28 was reported be expressed in pachytene and diplotene spermatocytes and round spermatids and its ablation produces testicular degeneration due to shedding of immature germ cells (Weber, Cammas et al. 2002).
We recognize that conducting this interaction screen in 293T cells likely missed information about testis-specific or testis-abundant interacting proteins that might be physiologically relevant to BRDT's function during spermatogenesis. As mentioned earlier, we were unable to express full-length BRDT in the more physiologically relevant mouse germ cell line GC-4spc but were successful using 293T cells. Curiously, we noted that the FLAG-293T cells used in the present study grew more slowly than the control parent 293T cells (LW and DJW, unpublished observations). Recently, SMARCE1, a member of the SWI/SNF family and a component of multimeric ATP-dependent chromatin remodeling complexes regulating subunit interactions and transcriptional activation, was identified as an interacting partner of BRDT in rat testes (Dhar, Thota et al. 2012). While BRDT was reported to be present in the elongating stages of spermiogenesis in rat, in mouse testis, we detected BRDT only in pachytene spermatocytes and round spermatids (Shang, Nickerson et al. 2007, Berkovits and Wolgemuth 2011). Whether this reflects pattern of expression differences between species or is an antibody-specific phenomenon remains to be determined.
Although the screen in the 293T/BRDT-FLAG cultured cell line we generated yielded several interesting candidates, including several splicing-related proteins (Berkovits, Wang et al. 2012) and three proteins known to be involved in transcriptional repression (this study), it is curious that transcriptional activators were not identified. Our recent ChIP-Seq data indicate that ~89% and ~70% of the BRDT promoter binding sites associated with the active transcription mark H3K4me3 in pachytene spermatocytes and round spermatids, respectively, and also associated with the active histone variants H2A.Lap1 and H2A.Z. (Li Wang et al., in preparation). Furthermore, this ChIP-Seq analysis and microarray data (Berkovits, Wang et al. 2012) also showed enrichment of BRDT binding in promoter regions of genes that are highly expressed in pachytene spermatocytes and round spermatids. Advances in the sensitivity of proteomic approaches should allow us to undertake ‘interactome’ analysis using IP followed by mass spectrometry of extracts from pachytene spermatocytes and round spermatids.
Acknowledgements
We thank Dr. Said Sif for providing PRMT5 antibody, Dr. Mary Ann Gawinowicz in Columbia University Medical Center's Protein Core facility for performing the mass spectrometry, and Dr. Enyuan Shang for critical comments on the manuscript.
Funding
This work was supported in part by a grant from the NIH, GM081767 to DJW.
REFERENCES
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8(6):623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Berkovits BD, Wang L, Guarnieri P, Wolgemuth DJ. The testisspecific double bromodomain-containing protein BRDT forms a complex with multiple spliceosome components and is required for mRNA splicing and 3'-UTR truncation in round spermatids. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits BD, Wolgemuth DJ. The first bromodomain of the testisspecific double bromodomain protein Brdt is required for chromocenter organization that is modulated by genetic background. Dev Biol. 2011;360(2):358–368. doi: 10.1016/j.ydbio.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Han C, Park I, Lee B, Jin S, Choi H, Kim do H, Park ZY, Eddy EM, Cho C. A novel germ cell-specific protein, SHIP1, forms a complex with chromatin remodeling activity during spermatogenesis. J Biol Chem. 2008;283(50):35283–35294. doi: 10.1074/jbc.M805590200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis GV. Duality in bromodomain-containing protein complexes. Front Biosci. 2001;6:D849–852. doi: 10.2741/denis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis GV, Nikolajczyk BS, Schnitzler GR. An emerging role for bromodomain-containing proteins in chromatin regulation and transcriptional control of adipogenesis. FEBS Lett. 2010;584(15):3260–3268. doi: 10.1016/j.febslet.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar S, Thota A, Rao MR. Insights into role of bromodomain, testisspecific (Brdt) in acetylated histone H4-dependent chromatin remodeling in mammalian spermiogenesis. J Biol Chem. 2012;287(9):6387–6405. doi: 10.1074/jbc.M111.288167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draker R, Ng MK, Sarcinella E, Ignatchenko V, Kislinger T, Cheung P. A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet. 2012;8(11):e1003047. doi: 10.1371/journal.pgen.1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D, Biermann K, Nettersheim D, Gillis AJ, Steger K, Jack HM, Muller AM, Looijenga LH, Schorle H. Expression of BLIMP1/PRMT5 and concurrent histone H2A/H4 arginine 3 dimethylation in fetal germ cells, CIS/IGCNU and germ cell tumors. BMC Dev Biol. 2008;8:106. doi: 10.1186/1471-213X-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Kiermer V, Dequiedt F, Verdin E. The emerging role of class II histone deacetylases. Biochem Cell Biol. 2001;79(3):337–348. [PubMed] [Google Scholar]
- Gaucher J, Boussouar F, Montellier E, Curtet S, Buchou T, Bertrand S, Hery P, Jounier S, Depaux A, Vitte AL, Guardiola P, Pernet K, Debernardi A, Lopez F, Holota H, Imbert J, Wolgemuth DJ, Gerard M, Rousseaux S, Khochbin S. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J. 2012;31(19):3809–3820. doi: 10.1038/emboj.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S, Sleeman JP, Lo Coco F, Nervi C, Pelicci PG, Heinzel T. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20(24):6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald C, Koslowski M, Arsiray T, Dhaene K, Praet M, Victor A, Morresi-Hauf A, Lindner M, Passlick B, Lehr HA, Schafer SC, Seitz G, Huber C, Sahin U, Tureci O. Expression of multiple epigenetically regulated cancer/germline genes in nonsmall cell lung cancer. Int J Cancer. 2006;118(10):2522–2528. doi: 10.1002/ijc.21669. [DOI] [PubMed] [Google Scholar]
- Hazzouri M, Pivot-Pajot C, Faure AK, Usson Y, Pelletier R, Sele B, Khochbin S, Rousseaux S. Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases. Eur J Cell Biol. 2000;79(12):950–960. doi: 10.1078/0171-9335-00123. [DOI] [PubMed] [Google Scholar]
- Hong CY, Suh JH, Kim K, Gong EY, Jeon SH, Ko M, Seong RH, Kwon HB, Lee K. Modulation of androgen receptor transactivation by the SWI3-related gene product (SRG3) in multiple ways. Mol Cell Biol. 2005;25(12):4841–4852. doi: 10.1128/MCB.25.12.4841-4852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29(5):1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang IY, Roe JS, Seol JH, Kim HR, Cho EJ, Youn HD. pVHLmediated transcriptional repression of c-Myc by recruitment of histone deacetylases. Mol Cells. 2012 doi: 10.1007/s10059-012-2268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem Sci. 2011;36(12):633–641. doi: 10.1016/j.tibs.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamonica JM, Deng W, Kadauke S, Campbell AE, Gamsjaeger R, Wang H, Cheng Y, Billin AN, Hardison RC, Mackay JP, Blobel GA. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci U S A. 2011;108(22):E159–168. doi: 10.1073/pnas.1102140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10(6):731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. Bromodomains in living cells participate in deciphering the histone code. Trends Cell Biol. 2004;14(6):279–281. doi: 10.1016/j.tcb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Lui WY, Wong EW, Guan Y, Lee WM. Dual transcriptional control of claudin-11 via an overlapping GATA/NF-Y motif: positive regulation through the interaction of GATA, NF-YA, and CREB and negative regulation through the interaction of Smad, HDAC1, and mSin3A. J Cell Physiol. 2007;211(3):638–648. doi: 10.1002/jcp.20970. [DOI] [PubMed] [Google Scholar]
- Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol. 2008;319(1):110–120. doi: 10.1016/j.ydbio.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, Berger SL. Structure and function of bromodomains in chromatin-regulating complexes. Gene. 2001;272(1-2):1–9. doi: 10.1016/s0378-1119(01)00519-4. [DOI] [PubMed] [Google Scholar]
- Maze I, Noh KM, Soshnev AA, Allis CD. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat Rev Genet. 2014;15(4):259–271. doi: 10.1038/nrg3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich ML, Trostle-Weige PK, Lin R, Bhatnagar YM, Allis CD. Highly acetylated H4 is associated with histone displacement in rat spermatids. Mol Reprod Dev. 1992;31(3):170–181. doi: 10.1002/mrd.1080310303. [DOI] [PubMed] [Google Scholar]
- Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, Krijgsveld J, Khochbin S, Muller CW, Petosa C. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461(7264):664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- Muller S, Filippakopoulos P, Knapp S. Bromodomains as therapeutic targets. Expert Rev Mol Med. 2011;13:e29. doi: 10.1017/S1462399411001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omisanjo OA, Biermann K, Hartmann S, Heukamp LC, Sonnack V, Hild A, Brehm R, Bergmann M, Weidner W, Steger K. DNMT1 and HDAC1 gene expression in impaired spermatogenesis and testicular cancer. Histochem Cell Biol. 2007;127(2):175–181. doi: 10.1007/s00418-006-0234-x. [DOI] [PubMed] [Google Scholar]
- Pacheco SE, Houseman EA, Christensen BC, Marsit CJ, Kelsey KT, Sigman M, Boekelheide K. Integrative DNA methylation and gene expression analyses identify DNA packaging and epigenetic regulatory genes associated with low motility sperm. PLoS One. 2011;6(6):e20280. doi: 10.1371/journal.pone.0020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26(15):3558–3569. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24(21):9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Dong W, Chen L, Zou T, Qi Y, Liu Y. Brd2 is a TBP-associated protein and recruits TBP into E2F-1 transcriptional complex in response to serum stimulation. Mol Cell Biochem. 2007;294(1-2):45–54. doi: 10.1007/s11010-006-9223-6. [DOI] [PubMed] [Google Scholar]
- Pivot-Pajot C, Caron C, Govin J, Vion A, Rousseaux S, Khochbin S. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol Cell Biol. 2003;23(15):5354–5365. doi: 10.1128/MCB.23.15.5354-5365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel. 2009;12(5):659–665. [PMC free article] [PubMed] [Google Scholar]
- Scanlan MJ, Altorki NK, Gure AO, Williamson B, Jungbluth A, Chen YT, Old LJ. Expression of cancer-testis antigens in lung cancer: definition of bromodomain testis-specific gene (BRDT) as a new CT gene, CT9. Cancer Lett. 2000;150(2):155–164. doi: 10.1016/s0304-3835(99)00385-7. [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zincfinger proteins. Genes Dev. 2002;16(8):919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ., 3rd Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15(4):428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang E, Nickerson HD, Wen D, Wang X, Wolgemuth DJ. The first bromodomain of Brdt, a testis-specific member of the BET sub-family of doublebromodomain- containing proteins, is essential for male germ cell differentiation. Development. 2007;134(19):3507–3515. doi: 10.1242/dev.004481. [DOI] [PubMed] [Google Scholar]
- Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15(5):603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26(22):8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tascou S, Nayernia K, Samani A, Schmidtke J, Vogel T, Engel W, Burfeind P. Immortalization of murine male germ cells at a discrete stage of differentiation by a novel directed promoter-based selection strategy. Biol Reprod. 2000;63(5):1555–1561. doi: 10.1095/biolreprod63.5.1555. [DOI] [PubMed] [Google Scholar]
- Wang L, Baiocchi RA, Pal S, Mosialos G, Caligiuri M, Sif S. The BRG1- and hBRM-associated factor BAF57 induces apoptosis by stimulating expression of the cylindromatosis tumor suppressor gene. Mol Cell Biol. 2005;25(18):7953–7965. doi: 10.1128/MCB.25.18.7953-7965.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P, Cammas F, Gerard C, Metzger D, Chambon P, Losson R, Mark M. Germ cell expression of the transcriptional co-repressor TIF1beta is required for the maintenance of spermatogenesis in the mouse. Development. 2002;129(10):2329–2337. doi: 10.1242/dev.129.10.2329. [DOI] [PubMed] [Google Scholar]
- Wolgemuth DJ, Gizang-Ginsberg E, Engelmyer E, Gavin BJ, Ponzetto C. Separation of mouse testis cells on a Celsep (TM) apparatus and their usefulness as a source of high molecular weight DNA or RNA. Gamete Res. 1985;12:1–10. doi: 10.1002/mrd.1120120102. [DOI] [PubMed] [Google Scholar]
- Wu SY, Lee AY, Hou SY, Kemper JK, Erdjument-Bromage H, Tempst P, Chiang CM. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20(17):2383–2396. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19(4):535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM, Jane SM. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16(3):304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]