Abstract
Background
The pink bollworm Pectinophora gossypiella (Saunders) (Lepidoptera: Gelechiidae) is one of the world’s most important pests of cotton. Insecticide sprays and transgenic cotton producing toxins of the bacterium Bacillus thuringiensis (Bt) are currently used to manage this pest. Bt toxins kill susceptible insects by specifically binding to and destroying midgut cells, but they are not toxic to most other organisms. Pink bollworm is useful as a model for understanding insect responses to Bt toxins, yet advances in understanding at the molecular level have been limited because basic genomic information is lacking for this cosmopolitan pest. Here, we have sequenced, de novo assembled and annotated a comprehensive larval midgut transcriptome from a susceptible strain of pink bollworm.
Findings
A de novo transcriptome assembly for the midgut of P. gossypiella was generated containing 46,458 transcripts (average length of 770 bp) derived from 39,874 unigenes. The size of the transcriptome is similar to published midgut transcriptomes of other Lepidoptera and includes up to 91 % annotated contigs. The dataset is publicly available in NCBI and GigaDB as a resource for researchers.
Conclusions
Foundational knowledge of protein-coding genes from the pink bollworm midgut is critical for understanding how this important insect pest functions. The transcriptome data presented here represent the first large-scale molecular resource for this species, and may be used for deciphering relevant midgut proteins critical for xenobiotic detoxification, nutrient digestion and allocation, as well as for the discovery of protein receptors important for Bt intoxication.
Electronic supplementary material
The online version of this article (doi:10.1186/s13742-016-0130-9) contains supplementary material, which is available to authorized users.
Keywords: Pectinophora gossypiella, Pink bollworm, RNA-Seq, Transcriptome, Midgut, Bacillus thuringiensis
Data description
Background
The pink bollworm Pectinophora gossypiella (Saunders) (Lepidoptera: Gelechiidae) is an important global pest of cotton. In many countries, transgenic cotton producing Bacillus thuringiensis (Bt) crystalline (Cry) proteins kills pests including the pink bollworm, thereby providing economic and environmental benefits. However, the evolution of pest resistance threatens the continued success of such Bt crops. While field populations of the pink bollworm in the USA have remained susceptible to two different Cry toxins produced simultaneously in Bt cotton, field-evolved practical resistance to Bt cotton has occurred widely in India [1–3].
Cry toxins kill susceptible pests like the pink bollworm by binding to protein receptors on the surface of midgut epithelial cells, eventually causing cell lysis [4]. In this study, we used Illumina sequencing of cDNA from the larval midgut of a Bt-susceptible strain to provide the first comprehensive view of the genes transcribed in this species. We generated over 18 million high-quality DNA sequence reads and >35 million bases that assembled into 21,715 unique transcripts. This transcriptome sequencing effort has dramatically increased the number of known genes for this insect, and provides an invaluable resource for the discovery of potential roles of proteins involved in various physiological and toxicological processes in the pink bollworm larval midgut.
Samples
Samples were derived from the APHIS-S strain of pink bollworm maintained at the US Department of Agriculture (USDA) Agricultural Research Service US Arid Land Agricultural Research Center in Maricopa, Arizona. APHIS-S is a Bt-susceptible strain that has been reared in the laboratory for more than 30 years without exposure to Bt toxins [5]. To generate samples, 120 newly emerged neonates were placed individually on ~5 g of wheat germ pink bollworm diet in 30 ml plastic cups and reared at 26 °C with ~30 % relative humidity and a photoperiod of 14 light:10 dark. After 9 days, midguts were dissected from three sets of ten female and ten male 4th instar larvae. Salivary glands, foregut, Malpighian tubules and hindguts were removed from each midgut in phosphate buffered saline buffer. Three biological replicates of 20 midguts each were pooled in 0.5 ml RNAlater (Sigma, St. Louis, USA), held overnight at 4 °C and stored at −80 °C. Frozen midguts in RNAlater were shipped to DuPont Pioneer in Johnston, Iowa, USA for RNA extraction, library preparation and DNA sequencing.
Sequencing
Total RNAs were isolated from frozen midgut pools using the Qiagen RNeasy kit (Hilden, Germany). Sequencing libraries from the resulting total RNAs were prepared using the TruSeq mRNA-Seq kit and protocol from Illumina, Inc. (San Diego, USA). Briefly, mRNAs were isolated via attachment to oligo(dT) beads, chemically fragmented and then reverse transcribed into cDNA via random hexamer priming. Resulting double stranded cDNA fragments were end-repaired to create blunt end fragments, 3′ A-tailed, ligated with Illumina indexed TruSeq adapters and PCR amplified using Illumina TruSeq primers. Purified PCR amplified libraries were assessed for quality and quantity on the Agilent Bioanalyzer DNA 7500 chip before normalization and sample pooling.
Sample pools were clustered and sequenced on the Illumina HiSeq 2500 system with Illumina TruSeq SBS Rapid v1 chemistry as per vendor protocols. Samples selected for transcriptome assembly were paired-end sequenced, with 76 cycles per read to a target depth of 40 million read pairs per sample. Raw quality was assessed and filtered with a custom pipeline that uses both the program FastQC and Trimmomatic (V 0.32), using the parameters ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:10 TRAILING:20 SLIDINGWINDOW:4:25 MINLEN:36 to remove adaptor sequence and filter by quality score. After filtering, approximately 18 million reads were obtained, totaling over 5 Gb or 2 × 72 bp paired-end data. The short read archive (SRA) accessions for data used in the assembly are as follows: SRX1164974, SRX1164977 and SRX1164978.
Transcriptome assembly
Before assembly, the three datasets were concatenated and read abundance was normalized to 50× coverage, using the in silico normalization tool in Trinity to improve assembly time and minimize memory requirements. Filtering and normalization reduced the dataset to 3 Gb, comprised of approximately 9 million read pairs that were then assembled using default parameters in Trinity (v.2.0.6) with the addition of the ‘-- jaccard clip’ flag to reduce the generation of transcript fusions from non-strand-specific data. Transcript expression levels were estimated with RSEM [6] and open reading frames were predicted using Transdecoder [7]. To remove bacterial contamination from the assembly, a BLASTx analysis of the newly assembled transcriptome was performed against a custom bacterial database containing all bacterial sequences deposited in NCBI (created 18 August 2015). After contamination filtration, the transcriptome was again filtered, sorted and prepared for NCBI transcriptome shotgun assembly (TSA) submission as previously described [8]. The resulting transcriptome was analyzed using TransRate (v.1.0.1), obtaining a TransRate score of 0.21, which indicates that the assembly is better than ~50 % of 155 published de novo transcriptomes available in the NCBI TSA [9].
Annotation
Functional annotation was performed at the peptide level using a custom pipeline [8] that defines protein products and assigns transcript names. Predicted proteins/peptides were analyzed using InterProScan5, which searched all available databases including Gene Ontology (GO) [10]. BLASTp analysis of the resulting proteins was performed with the UniProt Swiss-Prot database (downloaded 11 February 2015). Annie [11], a program that cross-references Swiss-Prot BLAST and InterProScan5 results to extract qualified gene names and products, was used to generate the transcript annotation file. The resulting .gff3 and .tbl files were further annotated with functional descriptors in Transvestigator [12].
Transcriptome comparisons
The assembled pink bollworm transcriptome was compared with midgut transcriptomes from three other lepidopterans, Plutella xylostella [13], Chilo suppressalis [14] and Heliothis virescens [15], and was found to have comparable metrics (Table 1). Specifically, the number of assembled contigs per 1000 reads was 2.5 for pink bollworm, 0.94 for C. suppressalis, 0.30 for H. virescens and 5.4 for P. xylostella. The number of BLASTx sequence hits (cutoff e-value of 10−5) in the non-redundant (nr) NCBI protein database per 1000 assembled reads was 1.2 for pink bollworm, 0.39 for C. suppressalis, 0.14 for H. virescens and 0.72 for P. xylostella (Table 1).
Table 1.
Comparison of assembled lepidopteran transcriptomes
| Chilo suppressalis | Heliothis virescens | Plutella xylostella | Pectinophora gossypiella | |
|---|---|---|---|---|
| Platform | Illumina | Illumina, Roche, 454, Sanger | Illumina | Illumina |
| Assembled reads | 39,400,002 | 212,987,028 | 39,764,230 | 18,623,508 |
| Average read size (bp) | 90 | Variable | 90 | 72 |
| Number of contigs | 37,040 | 63,648 | 213,674 | 46,458 |
| Contigs per 1000 reads | 0.94 | 0.30 | 5.4 | 2.5 |
| Mean contig size (bp), range | 497, 201–9744 | 383, 80–2000 | 189, nr | 770, 224–14,619 |
| Sequences with e-value <10−5 | 15,446 | 29,978 | 28,768 | 21,715 |
| Number of e-value <10−5 hits Per 1000 reads | 0.39 | 0.14 | 0.72 | 1.2 |
| GC (%) | 42 | nra | nr | 39 |
| N50 transcript length (bp) | nr | 1031 | 262 | 1153 |
| Pipeline | Trinity | SeqMan Ngen v2.1 | Trinity | Trinity |
| Reference | [14] | [15] | [13] | This study |
a nr not reported
The quality of the pink bollworm assembly was further assessed by direct comparison of core statistics with the P. xylostella midgut transcriptome [13] (Table 2). We evaluated the completeness of both the P. xylostella and P. gossypiella transcripts using the program BUSCO (benchmarking universal single-copy orthologs) using the arthropod gene set [16]. The percentages of conserved genes from the P. gossypiella and P. xylostella transcriptomes recovered by the BUSCO analysis are ~34 and ~37 %, respectively. The overall BUSCO percentages are lower than previously reported for a reference Spodoptera frugiperda transcriptome [17], but are not surprising given these are single-organ (i.e., midgut) assemblies compared with assembled RNA sequences from whole larvae and tissue samples from multiple time points. Lastly, a tBLASTx analysis of the P. gossypiella transcriptome against the P. xylostella midgut transcriptome (representing the nearest phylogenetic relative lepidopteran relative with a currently available midgut transcriptome) revealed that 29 % (12,475 out of 46,458), 37 % (17,032 out of 46,458) and 91 % (42,089 out of 46,458) had matching hits at e-values of 10−5, 10−2 and 101 [18]. These results are not unexpected for tBLASTx at the more stringent e-values given the considerable phylogenetic distance between the two species of Lepidoptera [19].
Table 2.
Comparison of Pectinophora gossypiella and Plutella xylostella midgut transcriptomes by BUSCO analysisa
| Species | Complete (%) | Duplicated (%) | Fragment (%) | Missing (%) |
|---|---|---|---|---|
| P. gossypiella | 34 | 8.8 | 30 | 35 |
| P. xylostella | 37 | 12 | 24 | 38 |
aA total of 2675 total BUSCO groups were searched from the assembled P. gossypiella midgut transcriptome and the assembled P. xylostella midgut short read archive transcriptome
Gene ontology
Blast2GO [20, 21] was used to assign P. gossypiella transcripts with a minimum BLASTx e-value of 10−3 into putative functional groups or GO terms. A total of 12,762 transcript sequences were assigned GO terms (Additional file 1, [18]), including 7073 with hits at the Biological Process level, 6402 at the Cellular Component level and 7747 sequences at the Molecular Function level. Within the Biological Process GO category, the most abundant transcripts were assigned to ‘single-organism metabolic process’, ‘signal transduction’ and ‘cellular protein modification’ (Fig. 1a). ‘Integral component of membrane’, ‘nucleus’ and ‘intracellular organelle part’ were the most abundant GO terms for Cellular Component (Fig. 1b). For Molecular Function, ‘zinc ion binding’, ‘ATP binding’ and ‘DNA binding’ were the most prevalent, with several different types of hydrolases also highly represented (Fig. 1c). Overall, typical gut-specific functions, such as digestion and storage, energy metabolism, ion transport and gene regulation were indicated by GO terms (Additional file 1, [18]).
Fig. 1.
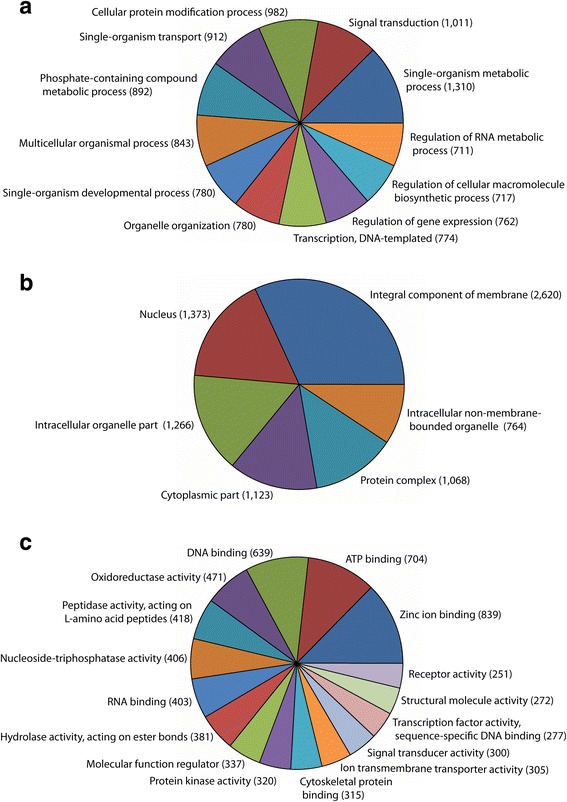
Classification of Pectinophora gossypiella midgut transcripts based on predicted Gene Ontology (GO) terms. a Biological Process, (b) Cellular Component and (c) Molecular Function GO terms were determined using Blast2Go [19, 20] with an e-value cutoff of 10−3, and minimum sequence filters set at 707 sequences for Biological Process, 640 for Cellular Component and 250 for Molecular Function for generating pie charts. Note that individual categories can have multiple mappings, resulting in a sum greater than the total number of transcript sequences assigned GO terms
Abbreviations
Bt, Bacillus thuringiensis; BUSCO, benchmarking universal single-copy orthologs; Cry protein, crystalline protein; GO, gene ontology; NCBI, National Center for Biotechnology Information; nr, non-redundant; SRA, short read archive; TSA, transcriptome shotgun assembly; USDA, US Department of Agriculture
Acknowledgements
Funding was provided by USDA-ARS and JAF, BET, and YC received partial support from DuPont-Pioneer (agreement #58-3K95-4-1666). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer.
Availability of supporting data
The filtered and annotated transcriptome was deposited at the NCBI TSA under the accession GDQN00000000, associated with BioProject PRJNA293973. Datasets further supporting the results of this article are available in the GigaScience repository, GigaDB [18].
Authors’ contributions
JAF conceived, designed and performed the experiments, analyzed the data, evaluated the conclusions and wrote the paper. YC conceived and designed the experiments and evaluated the conclusions. BET conceived and designed the experiments and evaluated the conclusions. JM performed the experiments and evaluated the conclusions. GZH performed the experiments and evaluated the conclusions. MEN performed the experiments and evaluated the conclusions. GW evaluated the conclusions. JLF evaluated the conclusions. EET analyzed the data, evaluated the conclusions and wrote the paper. All authors read and approved the final manuscript.
Competing interests
This is a cooperative investigation between the USDA Agricultural Research Service, the University of Arizona and DuPont-Pioneer, with JAF, BET and YC receiving partial funding from DuPont-Pioneer to support this work (agreement #58-3K95-4-1666). JAF is coauthor of a patent “Cadherin Receptor Peptide for Potentiating Bt Biopesticides” (patent numbers: US20090175974A1, US8354371, WO2009067487A2, WO2009067487A3). BET is a coauthor of a patent on modified Bt toxins, “Suppression of Resistance in Insects to Bacillus thuringiensis Cry Toxins, Using Toxins that do not Require the Cadherin Receptor” (patent numbers: CA2690188A1, CN101730712A, EP2184293A2, EP2184293A4, EP2184293B1, WO2008150150A2, WO2008150150A3). Bayer CropScience, Dow AgroSciences, Monsanto and Syngenta did not provide funding to support this work, but may be affected financially by publication of this paper and have funded other work by some of the authors.
Additional file
Bollworm_GO_26APR16.csv. CSV file of gene ontology terms for pink bollworm transcripts. (CSV 2338 kb)
References
- 1.Dhurua S, Gujar GT. Field-evolved resistance to Bt toxin Cry1Ac in the pink bollworm, Pectinophora gossypiella (Saunders) (Lepidoptera: Gelechiidae), from India. Pest Manag Sci. 2011;67:898–903. doi: 10.1002/ps.2127. [DOI] [PubMed] [Google Scholar]
- 2.Fabrick JA, Ponnuraj J, Singh A, Tanwar RK, Unnithan GC, Yelich AJ, et al. Alternative splicing and highly variable cadherin transcripts associated with field-evolved resistance of pink bollworm to Bt cotton in India. PLoS One. 2014;9:e97900. doi: 10.1371/journal.pone.0097900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohan KS, Ravi KC, Suresh PJ, Sumerford D, Head GP. Field resistance to the Bacillus thuringiensis protein Cry1Ac expressed in Bollgard® hybrid cotton in pink bollworm, Pectinophora gossypiella (Saunders), populations in India. Pest Manag Sci. 2016;72:738–46. doi: 10.1002/ps.4047. [DOI] [PubMed] [Google Scholar]
- 4.Adang MJ, Crickmore N, Jurat-Fuentes JL. Diversity of Bacillus thuringiensis crystal toxins and mechanism of action. In: Dhadialla TS, Gill SS, editors. Advances in insect physiology vol. 47. Oxford: Academic; 2014. pp. 39–87. [Google Scholar]
- 5.Liu YB, Tabashnik BE, Dennehy TJ, Patin AL, Sims MA, Meyer SK, et al. Effects of Bt cotton and Cry1Ac toxin on survival and development of pink bollworm (Lepidoptera: Gelechiidae) J Econ Entomol. 2001;94:1237–42. doi: 10.1603/0022-0493-94.5.1237. [DOI] [PubMed] [Google Scholar]
- 6.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sim SB, Calla B, Hall B, DeRego T, Geib SM. Reconstructing a comprehensive transcriptome assembly of a white-pupal translocated strain of the pest fruit fly Bactrocera cucurbitae. Gigascience. 2015;4:14. doi: 10.1186/s13742-015-0053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith-Unna RD, Boursnell C, Patro R, Hibberd JM, Kelly S. TransRate: reference free quality assessment of de-novo transcriptome assemblies. bioRxiv. 2015. http://dx.doi.org/10.1101/021626. Accessed 17 July 2015. [DOI] [PMC free article] [PubMed]
- 10.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–40. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tate R, Hall B, DeRego T. Annie the functional annotator - initial release. ZENODO. 2014. http://doi.org/10.5281/zenodo.10470. Accessed 02 Sept 2015.
- 12.DeRego T, Hall B, Tate R, Geib S. Transvestigator early release. ZENODO. 2014. http://doi.org/10.5281/zenodo.10471. Accessed 02 Sept 2015.
- 13.Xie W, Meng QS, Wu QJ, Wang SL, Yang X, Yang NN, et al. Tissue-specific transcriptome profiling of Plutella xylostella third instar larval midgut. Int J Biol Sci. 2012;8:1142–55. doi: 10.7150/ijbs.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma W, Zhang Z, Peng C, Wang X, Li F, Lin Y. Exploring the midgut transcriptome and brush border membrane vesicle proteome of the rice stem borer, Chilo suppressalis (Walker) PLoS One. 2012;7:e38151. doi: 10.1371/journal.pone.0038151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perera OP, Shelby KS, Popham HJR, Gould F, Adang MJ, Jurat-Fuentes JL. Generation of a transcriptome in a model lepidopteran pest, Heliothis virescens, using multiple sequencing strategies for profiling midgut gene expression. PLoS One. 2015;10:e0128563. doi: 10.1371/journal.pone.0128563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;13:3210–2. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 17.Legeai F, Gimenez S, Duvic B, Escoubas JM, Gosselin Grenet AS, Blanc F, et al. Establishment and analysis of a reference transcriptome for Spodoptera frugiperda. BMC Genomics. 2014;15:704. doi: 10.1186/1471-2164-15-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tassone ET, Zastrow-Hayes G, Mathis J, Nelson ME, Wu G, Flexner JL, Carriere Y, Tabashnik BE, Fabrick JA. Supporting data for “Sequencing, de novo assembly, and annotation of a pink bollworm larval midgut transcriptome”. GigaScience Database. 2016. http://dx.doi.org/10.5524/100203. Accessed 15 May 2016. [DOI] [PMC free article] [PubMed]
- 19.Ramirez-Rios V, Franco-Sierra ND, Alvarez JC, Saldamando-Benjumea CI, Villanueva-Mejia DF. Mitochondrial genome characterization of Tecia solanivora (Lepidoptera: Gelechiidae) and its phylogenetic relationship with other lepidopteran insects. Gene. 2016;581:107–16. doi: 10.1016/j.gene.2016.01.031. [DOI] [PubMed] [Google Scholar]
- 20.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 21.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–35. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]


