Abstract
Aberrant expression of microRNAs (miRNAs) has been associated with clinical outcome in patients with chronic lymphocytic leukemia (CLL). To identify a powerful and easily assessable miRNA bio-marker of prognosis and survival, we performed quantitative reverse-transcription polymerase chain reaction (qRT-PCR) profiling in 104 CLL patients with a well-defined chromosome 17p status, and we validated our findings with miRNA microarray data from an independent cohort of 80 patients. We found that miR-15a, miR-21, miR-34a, miR-155, and miR-181b were differentially expressed between CLLs with chromosome 17p deletion and CLLs with normal 17p and normal karyotype, and that miR-181b was down-regulated in therapy-refractory cases. miR-21 expression levels were significantly higher in patients with poor prognosis and predicted overall survival (OS), and miR-181b expression levels significantly predicted treatment-free survival. We developed a 21FK score (miR-21 qRT-PCR, fluorescence in situ hybridization, Karyotype) to stratify patients according to OS and found that patients with a low score had a significantly longer OS time. When we evaluated the relative power of the 21FK score with the most used prognostic factors, the score was the most significant in both CLL cohorts. We conclude that the 21FK score represents a useful tool for distinguishing between good-prognosis and poor-prognosis CLL patients.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common type of leukemia in the Western world, accounting for approximately 30% of all leukemias in the United States.1,2 Recurrent chromosomal abnormalities without known protein-coding genes were observed in CLL for decades, suggesting the involvement of not yet deciphered common pathogenic pathways. The molecular basis of these correlations was largely unknown until recently, when it became clear that, in addition to being causal in the pathogenesis of CLL, the expression levels of microRNAs (miRNAs) were useful for predicting the clinical behavior of CLL.3 Chromosomal aberrations occur in approximately 80% of patients with CLL and include 13q deletion (13qDEL; containing miR-15a/miR-16-1 cluster; > 50%), 11q deletion (11qDEL; including miR-34b/miR-34c cluster; 18%), trisomy 12 (T12; 10%), and 17p deletion (17pDEL; containing TP53 suppressor; 7%).1–3 Moreover, genomic aberrations in CLL are important independent predictors of disease progression and survival. For example, patients with 17pDEL and 11qDEL experience more aggressive disease, whereas patients with 13qDEL as the sole abnormality and otherwise normal cytogenetics have an indolent clinical disease course.4,5 It was also shown that monoallelic TP53 inactivation is associated with poor prognosis, and survival was equally poor for patients with 17pDEL only, 17pDEL plus TP53 mutation, and TP53 mutation only.6 The lack of somatic mutations in the immunoglobulin heavy-chain variable-region gene (IgVH), high-level expression of the 70-kDa zeta-associated protein (ZAP-70), or high levels of β-2 microglobulin (B2M) are also associated with an aggressive clinical course.1,2,4
Structurally, miRNAs are short (19- to 25-nucleotide) RNAs, processed from hairpin loop structures (pre-miRNAs; 60-110 nucleotides in length) that regulate the expression of protein-coding genes as a result of imperfect complementarity with target messenger RNAs.7 A unique miRNA signature was found to be differentially expressed in patients with various IgVH and ZAP-70 kinase statuses8 and composed of the most frequently deregulated miRNAs in the different hematologic malignancies (such as miR-15/16, the miR-29 family, and miR-155). Other studies9–11 generated distinct miRNA signatures but consistently identified members of the miR-29b family that were found to be down-regulated in CLL and associated with selected predictors of aggressive disease.11
In the present study, in a large cohort of patient samples, we performed quantitative reverse-transcription polymerase chain reaction (qRT-PCR) to analyze the expression levels of miRNAs known to be important in the pathogenesis of CLL to identify a powerful marker of CLL prognosis and survival for easy, routine assessment. We hypothesized that by characterizing the group of patients with the worst prognosis (ie, patients with chromosome 17pDEL), we could identify markers for predicting patient outcome at the time of diagnosis and would be useful not only for these patients but also for patients with adverse outcomes in general. In fact, we found that miR-21 expression stratifies survival of patients with 17pDEL and that also, in independent cohorts of patients with CLL with various chromosomal aberrations, miR-21 expression predicts survival.
Methods
Patient sample selection
To study miRNA expression levels, we investigated the peripheral blood samples from 104 patients with CLL. For all patients receiving care at M. D. Anderson Cancer Center or at other CLL Research Consortium institutions, written informed consent was obtained, and the patient samples were de-identified for the molecular study in accordance with the Declaration of Helsinki. The investigation was approved by the institutional review board at M. D. Anderson Cancer Center. The median follow-up was approximately 20 months (range, 0-88 months). Briefly, blood was obtained from the patients, mononuclear cells were isolated through Ficoll-Hypaque gradient centrifugation (GE Healthcare), and the cells were processed for RNA extraction with the use of TRIzol reagent (Invitrogen) according to the manufacturer's protocols.8 The patients' clinical and biologic data are presented in Table 1 and supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Forty-three patients were previously treated. The TP53 mutation status was available in 18 patients. Cytogenetic data (fluorescence in situ hybridization [FISH] and/or karyotype) were available for all 104 patients in this study.
Table 1.
Clinical data for patients with CLL with known 17p status*
| Characteristic | 17pDEL (N = 64) | NORM/NORM (N = 40) | P, χ2 test |
|---|---|---|---|
| No. of male/female | 46/18 | 26/14 | .131 |
| Median age at diagnosis, y (range) | 63 (37-89) | 61 (37-76) | .095 |
| Median time from diagnosis to sample collection, mo | 75.10 | 81.29 | |
| ZAP-70 expression | .215 | ||
| 20% or less | 21 | 17 | |
| More than 20% | 43 | 23 | |
| IgVH status* | .035 | ||
| Unmutated (≥ 98% homology) | 36 | 18 | |
| Mutated (< 98% homology) | 10 | 14 | |
| B2M expression* | .002 | ||
| No more than 4 mg/L | 27 | 29 | |
| More than 4 mg/L | 26 | 6 | |
| Therapy began after collection | |||
| No | 16 | 18 | .587 |
| Yes | 14 | 16 | |
| If therapy received, median time since collection, mo | 1.37 | 0.68 | |
| Treatment-refractory disease | 18 | 0 | < .001 |
ZAP-70 indicates 70-kDa ζ-associated protein; IgVH, immunoglobulin heavy-chain variable-region gene; and B2M, β-2 microglobulin.
Status of IgVH and B2M was known for 78 and 88 patients, respectively.
Assessment of prognostic factors and cytogenetic data
The assessment of prognostic factors was performed as previously described.12,13 ZAP-70 expression was assessed by immunoblotting and flow cytometric analysis, and IgVH mutation status was analyzed by direct sequencing. Cytogenetic data were available for all 104 patients in this study and performed in fewer than 30 days from sample collection. Conventional cytogenetic and FISH analyses were performed on peripheral blood samples that had been cultured for 24 hours without stimulation. For conventional analysis, up to 20 metaphases were analyzed, and the results were reported with the use of the International System for Human Cytogenetic Nomenclature. FISH analysis for abnormalities of ataxia-telangiectasia mutated (11q22.3), D13S319 (13q14.3), TP53 (17p13.1), and T12 (12p11.1-q11) were performed with the use of a commercial probe set (CLL panel; Vysis Inc). G-banding karyotype was performed according to standard procedures.
RNA isolation and miRNA expression by qRT-PCR detection
miRNA expression was measured with the TaqMan miRNA qRT-PCR method (Applied Biosystems) with the use of an IQ5 multicolor real-time PCR detection system (Bio-Rad). Briefly, 10 ng of total RNA was reverse transcribed with the use of the miRNA reverse transcription kit (Applied Biosystems) and a specific reverse-transcription stem-loop primer according to the manufacturer's protocol. All reactions were run in duplicate. The expression of each miRNA relative to the endogenous controls was determined with the 2−ΔCt method. miRNA levels were expressed in terms of the fold change of the target miRNA expression versus the small-nuclear RNAs U6 (Applied Biosystems). If expression values for the endogenous control used (U6) or for specific miRNAs were not obtained after 40 cycles of amplification in 2 successive experiments in duplicate wells each, then the specific values were considered to be not available. All the miRNA expression values were included in supplemental Table 2. The expression levels for the normalizer were similar for both groups of patients; for accuracy, for each amplified miRNA we performed U6 normalization on the same 96-well plate (supplemental Figure 1).
TP53 down-regulation by siRNA in MEG-01 leukemia cells
MEG-01 cells were reverse-transfected with 200nM On-target plus SMARTpool (Dharmacon) anti-TP53 or negative control small interfering RNA (siRNA) with the use of Lipofectamine 2000. Seventy-two hours after transfection, cells were collected and used for RNA and protein extraction. Total protein extracts were prepared in 0.5% NP40 lysis buffer. For immunoblotting, 40 μg of protein extracts were separated in 4% to 20% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (Criterion Precast Gel; Bio-Rad) and transferred to nitrocellulose membranes (Bio-Rad). Primary anti-p53 and anti-Vinculin antibodies were from BD PharMingen and were incubated overnight at 4°C. Secondary horseradish peroxidase–conjugated antibodies were from Amersham (anti–mouse) or Pierce (anti–goat) and were incubated for 2 hours at room temperature. Enhanced chemiluminescence kit for signal detection was from Amersham. Total RNA was isolated from transfected cells with the use of TRIzol reagent (Invitrogen). The quantification of transfected and/or endogenous mature miRNA levels was performed as described in “RNA isolation and miRNA expression by qRT-PCR detection.”
Statistical analysis
To determine the miRNA expression cutoff values (ie, to dichotomize the miRNA expression values as high or low), we calculated the median relative expression levels for each miRNA. The relationship between nondichotomized miRNA expression and treatment (refractory, treated and nonrefractory, and untreated patients at the time of collection) was assessed with the use of the Kruskal-Wallis test. Corresponding distribution plots were generated with the use of GraphPad Prism 5.0 (GraphPad Software). We identified miRNAs whose dichotomized expression was significantly related to the overall survival (OS) of the patient (defined as time from sample collection to time of death or last follow-up). We computed a significance level for each miRNA and clinical or biologic factor based on a univariate Cox proportional hazard regression model. For multivariate analysis, a full Cox proportional hazards model was initially fitted that included variables with a P value less than .1. A backward selection procedure was then used for model selection, with one variable removed at a time until all the variables retained in the model were significant at a P value less than .05. The OS and treatment-free survival (TFS; defined as time from sample collection to start of treatment) curves were then estimated with the use of the Kaplan-Meier method, and the resulting curves were compared with the use of the log-rank test. The joint effect of the covariates was examined with the use of the Cox proportional hazard regression model. For these models, we dichotomized patients according to their median age (≥ 62 years vs < 62 years); Rai stage (low Rai stage = 0, 1, or 2 vs high Rai stage = 3 or 4); ZAP-70 (≤ 20% vs > 20%); B2M (≤ 4 mg/L vs > 4 mg/L); absolute lymphocyte count (based on the median in 104 patients with CLL; 22.089 cells × 109/L [22 089 cells/μL]); and CD19+CD34+ expression (≤ 34% vs > 34%). Statistical analyses were performed with the use of SPSS 16.0 (SPSS Inc). All tests were 2-sided, and an effect was considered to be statistically significant at P less than .05. A description of the statistical tests used is included in supplemental Table 3.
Confirmation in independent cohort of patients with CLL
We downloaded the raw data previously published in Visone et al14 (E-TABM-762) and in Calin et al15 (E-TABM-184) from Array Express (http://www.ebi.ac.uk/microarray-as/ae/). After discarding the patients in common with our patient set and patients with less than 20% abnormal cells by FISH, we obtained a dataset of 80 patients that was normalized with the use of the quantile method (www.bioconductor.org) and imported into the Biometric Research Branch array tool, Version 3.7.2 (http://linus.nci.nih.gov/BRB-ArrayTools.html) for further analysis with the use of 3.3 (log2) intensity threshold as performed in the study of Visone et al.14 We retrieved and updated the OS, clinical, and cytogenetics data for all 80 patients (supplemental Table 4). miRNA expression values were extracted and dichotomized according to the median expression levels. Data were then imported into SPSS Version 16.0 (SPSS Inc) for survival analysis evaluation.
Results
Patients and microRNAs selection
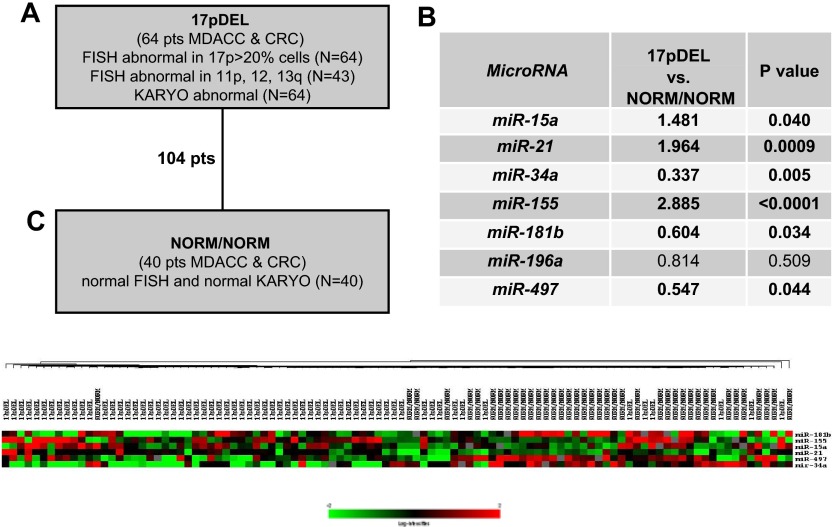
The patients with CLL were classified as 17pDEL (n = 64; defined by the deletion of 17p on FISH with or without abnormal 13q or 11q or trisomy 12 and with abnormal karyotype [KARYO]) or as NORM/NORM (n = 40; with normal FISH and normal KARYO) (Figure 1). The 2 categories of patients were significantly different in terms of their IgVH and B2M statuses and treatment-refractory disease but not in terms of their ZAP-70 levels (Table 1). Two criteria for collecting samples were strictly used. First, to avoid clone changes, the period from cytogenetic analysis to sample collection was fewer than 30 days. Second, we selected only patients who had 17pDEL in greater than 20% of nucleated blood cells, because the patients with 17pDEL in less than 20% of cells have survival and chemotherapy response rates similar to those of patients without 17pDEL.13,16
Figure 1.
microRNA signatures associated with 17pDEL in patients with CLL. (A) The definitions of patient categories. (B) The ratios and P values were obtained by applying the t test in the 7 studied miRNAs among the patients with CLL, classified according to the patients' properties on FISH and KARYO. (C) Clusters of differentially expressed miRNAs. Unsupervised clustering was performed on 104 patients with qRT-PCR expression of miR-15a, miR-21, miR-34a, miR-155, miR-181b, and miR-497. Single linkage and the Euclidean distance metric were used.
We did not aim at extensive testing of all previously published signatures; therefore, we selected a group of 6 miRNAs (miR-15a, miR-21, miR-34a, miR-155, miR-181b, and miR-196a) that were found to be important for CLL pathogenesis in at least 3 independent publications. MiR-16/miR-15 are tumor suppressor genes important for CLL predisposition that are also members of a signature linked to CLL prognosis.8,17,18 The miR-181 family was found to be related to aggressive CLL and to interact with the TCL1 oncogene,11,19,20 whereas miR-21, miR-155, and miR-196a were identified to be overexpressed in CLL samples.8,9,21 miR-34 family transcription was found to be regulated by TP53, and miR-34a was found differentially expressed in CLL cells versus normal CD5 B cells19 and recently linked to response to therapy in CLL.22,23 We also included miR-497, located on the short arm of chromosome 17, in our study, therefore analyzing a total of 7 genes.
A miRNA fingerprint for CLLs with 17pDEL
By qRT-PCR we identified that the selected miRNAs were expressed at various levels in CLL samples (supplemental Figure 2) and that most of them were differentially expressed at statistically significant levels between samples with 17pDEL and NORM/NORM: miR-15a, miR-21, and miR-155 were high, whereas miR-34a and miR-181b were low in the 17pDEL samples (Figure 1), meaning that the expression variations of these miRNAs may be correlated to the presence of 17pDEL. In support of this, the “bystander” miR-497 located in the 17pDEL region, approximately 200 kilobase centromeric of TP53, was found to be significantly down-regulated in the 17pDEL group (Figure 1). When this group was divided according to abnormalities on FISH (17pDEL only, n = 21 vs 17pDEL associated with other FISH abnormalities, n = 43), no statistically significant differences in the miRNA levels were identified (data not shown).
To further understand whether the 5-miRNA signature is linked to loss of TP53 at 17pDEL, we treated the MEG-01 leukemia cells (wild-type TP53) with siRNA against TP53 and found that miR-34a was significantly down-regulated, whereas miR-155 was significantly up-regulated with respect to cells treated with scrambled siRNA control; when the complementary experiment was done in the same cells, ie, forced overexpression of TP53, the opposite variations of miR-34a and miR-155 were found (supplemental Figure 3). In support of this, in the 14 patients with 17DEL and TP53 mutations, we found that miR-155 was overexpressed approximately 5 times more than for the 40 NORM/NORM patients (data not shown), which is twice as high as the global 17pDEL group overexpression.
Because the main criterion for patient selection was the time from FISH and KARYO to sample collection, to obtain an adequate number of patients for statistical analyses, we collected samples from both treated and untreated patients (Table 1). To exclude the possibility that the miRNA signature was a result of treatment and not specifically to the presence or absence of 17pDEL, we used Student t test to compare the samples from patients who had already been treated when cells were collected (n = 40) with the samples from patients who were either treated after collection time or who were untreated (n = 64). We found that only miR-181b was significantly differentially expressed at a lower level in the treated CLL samples (P = .01; supplemental Figure 4), suggesting that miR-181b could be involved in response to therapy (for miR-21 the P = .44). Taken together, these data suggest that variations of miR-34a and miR-155 could be linked to loss of the TP53 gene, variations of miR-15a and miR-21 could be linked to the presence of 17pDEL, and variations of miR-181b could be linked to previous therapy.
miR-21 and miR-181b as survival prognostic factors
We hypothesized that the identified miRNA expression variations are not bystanders during leukemogenesis but are instead associated with the patients' clinical characteristics. Cox regression analysis was performed to evaluate the effect of miRNA-dichotomized data on OS and TFS (Table 2; supplemental tables 5-6). miR-21 was a univariate predictor of OS: patients with high levels of miR-21 had a higher risk of death than patients with low expression levels. The OS varied from 24.10 months to 12.86 months in patients with low versus high miR-21 expression (P = .033; hazard ratio [HR] = 2.283; 95% CI = 1.049-4.967). Also miR-181b was a univariate predictor of OS, with a mean OS from 12.21 to 21.3 in patients with low versus high miR-181b expression (P = .027; HR = 2.348; 95% CI = 1.075-5.102). Subsequently, on multivariate analysis, we found that miR-21, miR-181b, and Rai stage were significant predictors of OS (Table 2). miR-181b was also a univariate predictor of TFS: patients with low levels of miR-181b had higher probability of requiring treatment, with a mean TFS of 5.35 months versus 13.80 months in patients with high miR-181b expression (P = .006; HR = 2.770; 95% CI = 1.295-5.917) (Table 2). To reinforce our results, the survival analysis was performed among the 68 patients with CLL with low Rai stage (Rai = 0, 1, or 2). We found that miR-21 was still associated with OS and patient outcome, and miR-181b was still associated with TFS (supplemental Figure 5). All together these findings indicate that low miR-181b expression and high miR-21 expression were significantly unfavorable prognostic factors independent of other clinical-pathologic factors.
Table 2.
Overall survival and treatment-free survival results in 104 patients with CLL
| Variable | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| OS* | ||||
| FISH and KARYO (17pDEL vs NN)† | 8.944 (2.668-29.985) | < .001 | N/A | N/A |
| Rai [high (3,4) vs low (0,1,2)] | 4.144 (1.850-9.284) | .001 | 2.731 (1.136-6.565) | .025 |
| ABSLYM (> median vs ≤ median) | 1.724 (0.786-3.774) | .169 | NS | NS |
| IgVH (nonmutated vs mutated) | 10.000 (1.332-76.923) | .006 | NS | NS |
| ZAP-70 (> 20% vs ≤ 20%) | 1.326 (0.599-2.993) | .486 | NS | NS |
| B2M (> 4 mg/L vs ≤ 4 mg/L) | 4.484 (1.795-11.236) | .001 | NS | NS |
| miR-21 (high vs low) | 2.283 (1.049-4.967) | .033 | 3.474 (1.351-8.938) | .010 |
| miR-181b (low vs high) | 2.348 (1.075-5.102) | .027 | 2.717 (1.106-6.667) | .029 |
| 21FK score (2/2 vs 1/2 vs 0/2)† | 2.869 (1.626-5.062) | < .001 | N/A | N/A |
| TFS‡ | ||||
| FISH and KARYO (17pDEL vs NN)† | 1.182 (0.567-2.464) | .655 | N/A | N/A |
| Rai (high vs low) | 4.797 (2.071-11.113) | < .001 | 4.797 (2.071-11.113) | < .001 |
| ABSLYM (> median vs ≤ median) | 2.049 (0.970-4.329) | .055 | NS | NS |
| IgVH (nonmutated vs mutated) | 2.212 (0.806-6.061) | .114 | NS | NS |
| ZAP-70 (> 20% vs ≤ 20%) | 1.805 (0.809-4.184) | .140 | NS | NS |
| B2M (> 4 mg/L vs ≤ 4 mg/L) | 2.725 (1.175-6.329) | .015 | NS | NS |
| miR-21 (high vs low) | 1.291 (0.628-2.656) | .487 | NS | NS |
| miR-181b (low vs high) | 2.770 (1.295-5.917) | .006 | NS | NS |
HR indicates hazard ratio; CI, confidence interval; FISH, fluorescence in situ hybridization; NN, NORMAL/NORMAL; N/A, not available; ABSLYM, absolute lymphocyte count; NS, not statistically significant; IgVH, immunoglobulin heavy-chain variable-region gene; ZAP-70, 70-kDa ζ-associated protein; B2M, β-2 microglobulin; 21FK, miR-21, FISH, and Karyotype score; and 181bFK, miR-181b, FISH, and Karyotype score.
Complete data are included in supplemental Tables 5-6.
Univariate analysis, n = 99; 5 cases censored before the earliest event in a stratum. Multivariate analysis, n = 86; 4 cases censored before the earliest event in a stratum and 14 cases with missing values.
FISH and KARYO have not been included in multivariate analysis because they were used as criteria to select the patients. In addition, 21FK score has not been included in multivariate analyses because depends from miR-21 expression that was already included. These are indicated as N/A.
Univariate analysis, n = 63; 1 case censored before the earliest event in a stratum and 40 cases with missing values. Multivariate analysis, n = 62; 42 cases with missing values.
miR-21 improves risk stratification when combined with FISH and KARYO
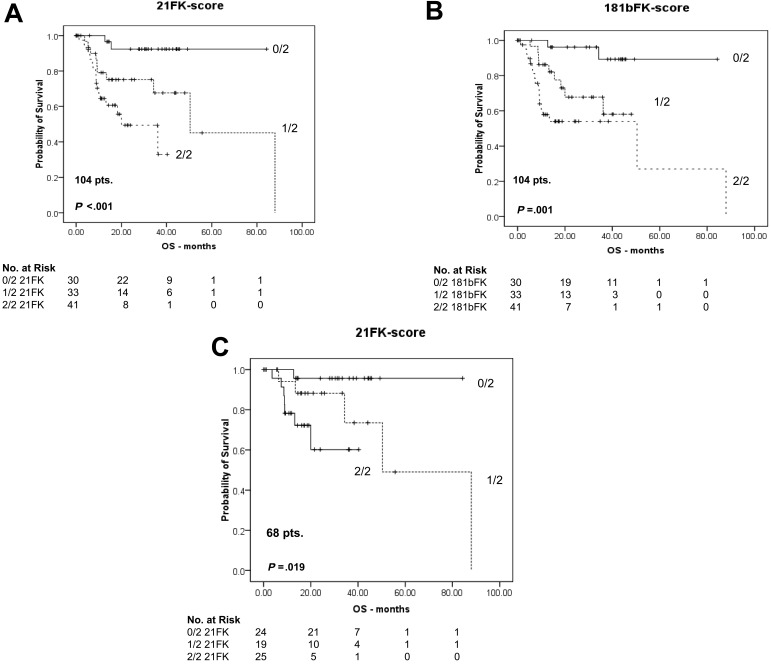
We then analyzed whether the studied miRNAs were good OS predictors when combined with FISH and KARYO, the selection criteria for our 104 patients with CLL. We found that miR-21 (1 point if high, or 0 points if low) on FISH and KARYO (1 point if 17pDEL or 0 points if NORM/NORM) when combined in a 21FK (miR-21 qRT-PCR, FISH, Karyotype) score of 0/2, 1/2, or 2/2 had an OS HR of 2.869 (P < .001; 95% CI = 1.626-5.062), meaning that when the score increased one unit the risk of death increased 2.87 times (Table 2; Figure 2). When we used 0/2 as the baseline in a univariate Cox model based on 104 patients, the HR for 21FK score of 1/2 versus 0/2 was 5.18 (95% CI = 1.11-24.2; P = .036), and the HR for 2/2 versus 0/2 was 11.1 (95% CI = 2.54 to 48.4; P = .001).
Figure 2.
Kaplan-Meier survival curves for 104 patients with CLL, including 68 CLLs with low Rai stage. (A) OS for patients with CLL was classified into 3 levels according to the 21FK score (0/2, 1/2, and 2/2). (B) OS for patients with CLL was classified into 3 levels according to the 181bFK score (0/2, 1/2, and 2/2). (C) Among the 104 patients, the 21FK score was validated in 68 patients with low Rai stage CLL. The 21FK score predicted patients' survival, even if they had a Rai disease stage of 0, 1, or 2. The survival data were compared, and the reported P values were calculated according to the log-rank test. In the main text, the P values were reported according to Cox regression analyses associated with the hazard ratio computation.
Further confirming this, in 68 patients with low Rai stage, the 21FK score was also significant in terms of OS: HR of 3.072 (95% CI = 1.286-7.341; P = .005) (Figure 2), meaning that the 21FK score can predict OS for early-stage disease. Using the same steps, we also tested the 181bFK score, which was statistically significant, with an OS HR of 2.733 (95% CI = 1.565-4.773; P < .001), but it was not significant for samples with low Rai stage. We then tested the 181bFK score on TFS without significant results, meaning that miR-181b is a good TFS prognostic factor but is not independent of FISH and KARYO properties.
To evaluate the usefulness of the newly developed 21FK score, we evaluated the relative power of the classical prognostic factors, including B2M, ZAP-70, IgVH, and CD19+CD38+, and the 21FK score with the use of a multivariate Cox regression model. We found that the 21FK score was associated with the lowest P value (P = .007; HR = 3.514; 95% CI = 1.409-8.764; Table 3). The 21FK score was also the best performer by univariate analysis (supplemental Figure 6). Therefore, in a large multi-institutional set of patients, the 21FK score was a predictor of OS, and independent studies needs to be performed to exactly asses the performance with other previously described clinical or biologic prognostic factors.
Table 3.
Multivariate analyses comparing 21FK score with other survival prognostic factors in the CLL cohort and CLL validation cohort
| P | HR | 95% CI |
||
|---|---|---|---|---|
| Lower | Upper | |||
| CLL cohort (n = 104) | ||||
| B2M (≤ 4 mg/L vs > 4 mg/L) | .029 | 3.861 | 1.152 | 12.987 |
| CD19+CD38+ (≤ 34% vs > 34%) | .209 | 2.365 | 0.618 | 9.050 |
| IgVH (nonmutated vs mutated) | .026 | 10.638 | 1.330 | 83.333 |
| ZAP-70 (> 20% vs < 20%) | .120 | 2.642 | 0.778 | 8.976 |
| 21FK score (2/2 vs 1/2 vs 0/2) | .007 | 3.514 | 1.409 | 8.76 |
| CLL validation cohort (n = 80) | ||||
| IgVH (nonmutated vs mutated) | .640 | 7.082 | 0.002 | NA |
| ZAP-70 (≤ 20% vs > 20%) | .690 | 5.311 | 0.001 | NA |
| 21FK score (high vs low) | .013 | 5.217 | 1.408 | 19.327 |
CLL indicates chronic lymphocytic leukemia; HR, hazard ratio; CI, confidence interval; B2M, β-2 microglobulin; IgVH, immunoglobulin heavy-chain variable-region gene; ZAP-70, 70-kDa ζ-associated protein; 21FK, miR-21, FISH, and Karyotype score; and NA, not available.
Validation of 21FK score and miR-181b in an independent cohort of CLL patients
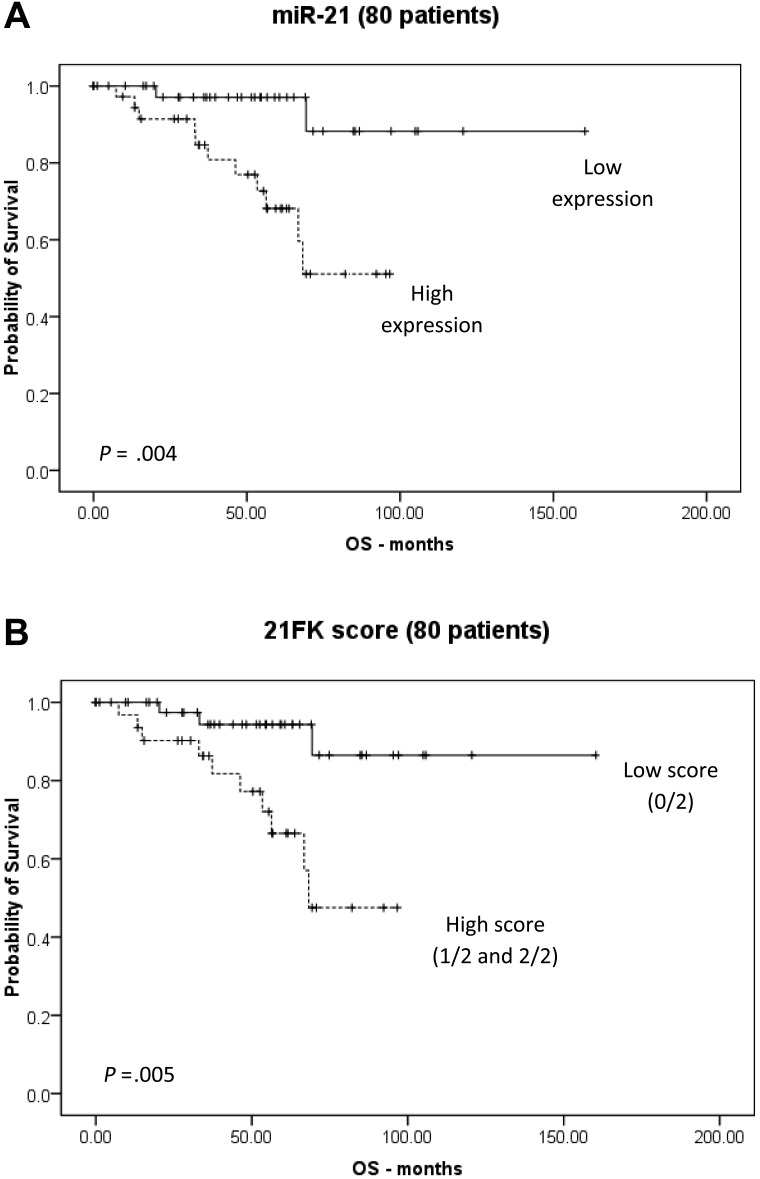
Because we identified the 21FK score by analyzing homogeneous populations of patients with CLL, we investigated whether the 21FK score has a wider significance and could be applied to nonhomogeneous sets of patients with CLL. To this aim, we used the sample set of 80 patients with CLL previously investigated by miRNA arrays (E-TABM-184 and E-TABM-762). We did not include the E-TABM-762 study in the selection of the presently analyzed miRNA (because it was published after we finished the data generation for our study); therefore, this represents a completely independent assessment of the validity of our data. The patients had either one chromosomal abnormality (13qDEL, 11qDEL, T12, 17pDEL) or normal karyotype (supplemental Table 4). Cox regression analysis showed that miR-21 was a significant predictor of OS, with an HR of 6.720 (95% CI = 1.484-30.443; P = .004; Figure 3). We then evaluated the 21FK score, dichotomized as low (0/2 and 1/2) and high (2/2) and found that it was able to stratify patients based on OS, with an HR of 5.321 (95% CI = 1.408-19.327; P = .013; Figure 3). ZAP-70 and IgVH status were also available (supplemental Table 4); when we performed a multivariate analysis with the use of the 21FK score, IgVH, and ZAP-70, the 21FK score was the only significant variable (HR = 5.217; 95% CI = 1.408-19.327; P = .013; Table 3). In conclusion, when independently tested in 2 sets of patients with the use of 2 different expression methods and with distinct, but overlapping, FISH characteristics, the 21FK score was the only rating among all of the analyzed variables able to stratify patients according to survival.
Figure 3.
Validation by microarray data evaluation: miR-21 and 21FK score stratify OS risk in an independent validation cohort. Kaplan-Meier survival curves have been reported. miR-21 expression levels were measured by microarray, and cutoffs were based on the median expression levels. (A) Patients with high miR-21 expression had a significantly poorer prognosis in terms of OS. (B) Patients with a high 21FK score (2/2 and 1/2) showed a significantly poorer prognosis in terms of OS than for patients with a low 21FK score (0/2). Statistical differences between curves were calculated with the use of the log-rank test. In the main text the P values were reported according to Cox regression analyses associated to hazard ratio computation.
Discussion
In the present study, we first identified an miRNA signature specific for CLL samples with 17pDEL. This signature comprised high miR-21, miR-155, and miR-15a expression and low miR-34a and miR-181b expression. We confirmed the findings from previous reports that miR-34a was transactivated by TP5324 and expanded this network by adding miR-155 to the list of miRNAs regulated by TP53. The identified signature is complex, because it contains miRNAs whose expressions are correlated with TP53 deletion (miR-34a and miR-155), 17pDEL (miR-21 and miR-15a), and both 17pDEL and response to therapy (miR-181b). The molecular basis of such complex interrelations needs to be further investigated. Furthermore, in the 2 analyzed groups of patients we found significant heterogeneity with respect to VH-mutation status, B2M levels, and proportion of treatment refractory cases; therefore, the exact effect of each of these factors on miRNA expression should be assessed.
The use of the prognostic factors in patients with CLL is essential because CLL therapy is started not at diagnosis but at signs of aggressive evolution.1,2 In the present study we identified a composite score that is based on miR-21 expression and 2 routine cytogenetics procedures (FISH and KARYO) that can predict the mortality risk for patients with CLL. Patients with a higher 21FK score had a significantly higher risk of death than patients with a lower 21FK score. The use of this score in clinical practice could have several advantages: the detection of miR-21 expression by qRT-PCR is technically easy to perform and uses a small amount of starting material (10 ng of total RNA), and, more important, the 21FK score has a better performance on multivariate analyses than the currently used survival and prognostic markers (such as ZAP-70 and IgVH status). High miR-21 expression has been previously associated with poor survival and poor therapeutic outcome in patients with colorectal cancers.25 Because the same miRNA has clinical significance in a very different type of tumor represents an indirect argument that miR-21 is involved in essential survival and/or proliferation pathways important for different types of cells. In fact, in the most comprehensive miRNA expression study published on solid cancers to date, miR-21 was found to be overexpressed in all 6 types of cancers analyzed.26 Furthermore, another study found that miR-21 is involved in down-regulation of PTEN in CLL, which could not be explained by loss of heterozygosity.27 Recently, 4 human genes were reported to be down-regulated in patients with CLL with 17pDEL: GPS2, DPH1/OVCA1, CCND2, and GABARAP.28 Using the RNA22 target prediction program, we found that CCND2 and DPH1/OVCA1 are possibly targeted by miR-21 (data not shown). Furthermore, CCND2 was reported to control cell cycle progression in CLL cells; DPH1 is a candidate tumor suppressor gene in ovarian and breast cancers and is localized telomerically to TP53 and lost in a large fraction of tumors that retain TP53.29 Experimental in vivo data have shown that DPH1 can modify TP53-induced tumorigenesis, suggesting that DPH1 acts as a regulator of cell cycle progression. In particular, the loss of one copy of DPH1 accelerated tumorigenesis in a Tp53+/− mouse model and increased tumor burden in a Tp53−/− background.30
Last but not least, we found that the expression levels of miR-181b can predict TFS. We previously found that the miR-181 family is down-regulated in patients with CLL who have a poor prognosis8 and that miR-181b targets the TCL1 oncogene, which is overexpressed in patients with aggressive CLL.20 Other targets could also be involved in this pathogenic link: eg, using RNA22, we predicted that miR-181b could target both PER2 and CRY1, circadian genes that have already been reported to predict the clinical outcome in patients with CLL, as measured by TFS.31
In conclusion, the 21FK score represents a useful marker to distinguishing between good-prognosis and poor-prognosis patients with CLL. In both the good- and poor-prognosis subgroups, miR-181b levels identified patients who need early therapy and who thus require a closer follow-up.
Acknowledgments
We thank Alyson Todd (Department of Scientific Publications, M. D. Anderson Cancer Center) for editing this manuscript.
G.A.C. is supported as a Fellow of The University of Texas M. D. Anderson Research Trust, as a Research Scholar of The University of Texas System Regents, and by the Ladjevardian Regents Research Scholar Fund. M.F. is supported by a 2009 Kimmel Scholar Award.
The funding organizations had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit the manuscript for publication
Footnotes
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: G.A.C. and M.J.K. designed the strategy of the study and experimental flow; S.R., M.S., E.B., M.S.N., F.D., D.S., and M.F. performed experiments and analyzed data; L.L.B., L.Z.R., S.L., L.J., L.X., J.H., P.S., G.Z., S.V., M.N., W.W., T.J.K., W.P., K.R.C., L.V.A., M.J.K., and G.A.C. analyzed data; and S.R., K.R.C., L.V.A., M.J.K., and G.A.C. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: George A. Calin, Department of Experimental Therapeutics, Unit 36, The University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: gcalin@mdanderson.org; and Michael J. Keating, Department of Leukemia, Unit 428, The University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: mkeating@mdanderson.org.
References
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Zenz T, Mertens D, Küppers R, Döhner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10(1):37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. Chronic lymphocytic leukemia: interplay between non-coding RNAs and protein-coding genes. Blood. 2009;114(23):4761–4770. doi: 10.1182/blood-2009-07-192740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dohner H, Stilgenbaue S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 6.Zenz T, Krober A, Scherer K, et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood. 2008;112(8):3322–3329. doi: 10.1182/blood-2008-04-154070. [DOI] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 9.Fulci V, Chiaretti S, Goldoni M, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109(11):4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 10.Marton S, Garcia MR, Robello C, et al. Small RNAs analysis in CLL reveals a deregulation of miRNA expression and novel miRNA candidates of putative relevance in CLL pathogenesis. Leukemia. 2008;22(2):330–338. doi: 10.1038/sj.leu.2405022. [DOI] [PubMed] [Google Scholar]
- 11.Stamatopoulos B, Meuleman N, Haibe-Kains B, et al. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113(21):5237–5245. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
- 12.Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351(9):893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 13.Tam CS, Shanafelt TD, Wierda WG, et al. De novo deletion 17p13.1 chronic lymphocytic leukemia shows significant clinical heterogeneity: the MD Anderson and Mayo Clinic experience. Blood. 2009;114(5):957–964. doi: 10.1182/blood-2009-03-210591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visone R, Rassenti LZ, Veronese A, et al. Karyotype specific microRNA signature in chronic lymphocytic leukemia. Blood. 2009;114(18):3872–3879. doi: 10.1182/blood-2009-06-229211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calin GA, Liu CG, Ferracin M, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 16.Catovsky D, Richards S, Matutes E, et al. Assessment of fludarabine plus cyclophosphamide for patients with chronic lymphocytic leukaemia (the LRF CLL4 Trial): a randomised controlled trial. Lancet. 2007;370(9583):230–239. doi: 10.1016/S0140-6736(07)61125-8. [DOI] [PubMed] [Google Scholar]
- 17.Calin GA, Cimmino A, Fabbri M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105(13):5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raveche ES, Salerno E, Scaglione BJ, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109(12):5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calin GA, Liu CG, Sevignani C, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101(32):11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pekarsky Y, Santanam U, Cimmino A, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66(24):11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 21.Wang M, Tan LP, Dijkstra MK, et al. miRNA analysis in B-cell chronic lymphocytic leukaemia: proliferation centres characterized by low miR-150 and high BIC/miR-155 expression. J Pathol. 2008;215(1):13–20. doi: 10.1002/path.2333. [DOI] [PubMed] [Google Scholar]
- 22.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network–another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7(11):819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zenz T, Mohr J, Eldering E, et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood. 2009;113(16):3801–3808. doi: 10.1182/blood-2008-08-172254. [DOI] [PubMed] [Google Scholar]
- 24.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leupin N, Cenni B, Novak U, et al. Disparate expression of the PTEN gene: a novel finding in B-cell chronic lymphocytic leukaemia (B-CLL). Br J Haematol. 2003;121(1):97–100. doi: 10.1046/j.1365-2141.2003.04227.x. [DOI] [PubMed] [Google Scholar]
- 28.Fabris S, Mosca L, Todoerti K, et al. Molecular and transcriptional characterization of 17p loss in B-cell chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2008;47(9):781–793. doi: 10.1002/gcc.20579. [DOI] [PubMed] [Google Scholar]
- 29.Schultz DC, Vanderveer L, Berman DB, Hamilton TC, Wong AJ, Godwin AK. Identification of two candidate tumor suppressor genes on chromosome 17p13.3. Cancer Res. 1996;56(9):1997–2002. [PubMed] [Google Scholar]
- 30.Chen CM, Behringer RR. Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev. 2004;18(3):320–332. doi: 10.1101/gad.1162204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abruzzo LV, Barron LL, Anderson K, et al. Identification and validation of biomarkers of IgV(H) mutation status in chronic lymphocytic leukemia using microfluidics quantitative real-time polymerase chain reaction technology. J Mol Diagn. 2007;9(4):546–555. doi: 10.2353/jmoldx.2007.070001. [DOI] [PMC free article] [PubMed] [Google Scholar]