Abstract
In biological systems, proteins catalyze the fundamental reactions that underlie all cellular functions, including metabolic processes and cell survival and death pathways. These biochemical reactions are rarely accomplished alone. Rather, they involve a concerted effect from many proteins that may operate in a directed signaling pathway and/or may physically associate in a complex to achieve a specific enzymatic activity. Therefore, defining the composition and regulation of protein complexes is critical for understanding cellular functions. In this chapter, we describe an approach that uses quantitative mass spectrometry (MS) to assess the specificity and the relative stability of protein interactions. Isolation of protein complexes from mammalian cells is performed by rapid immunoaffinity purification, and followed by in-solution digestion and high-resolution mass spectrometry analysis. We employ complementary quantitative MS workflows to assess the specificity of protein interactions using label-free MS and statistical analysis, and the relative stability of the interactions using a metabolic labeling technique. For each candidate protein interaction, scores from the two workflows can be correlated to minimize nonspecific background and profile protein complex composition and relative stability.
Keywords: Affinity isolation, Immunoprecipitation, Protein complexes, Protein interactions, Stable isotope labeling quantification, Label-free quantification, SAINT, I-DIRT
1 Introduction
Significant progress in the functional understanding of protein complexes has been made, due in large part to improvements in rapid biochemical affinity isolations using high-affinity epitope tags [1–5], such as FLAG, GFP and biotin, and to the increased sensitivity of detection by mass spectrometry (MS). Detection of low-abundance and/or transient interactions, along with posttranslational modifications within complexes, is now feasible [6–12]. Yet, increased sensitivity of detection also provides a greater number of identified proteins that co-isolate nonspecifically during the affinity capture of protein complexes. Therefore, it is critical that experimental designs employ appropriate negative controls. Such controls usually include isolation of the epitope tag alone from cell/tissue lysates and/or quantitative MS strategies to measure the specificity of interactions, i.e. enrichment of isolated proteins relative to contaminant datasets or to the background proteome [13, 14]. Since protein complexes are not static structures, MS-based proteomics has also been useful for studying the dynamics of protein complex regulation, which involve time-dependent changes in complex composition and subcellular localization in response to external and internal stimuli [8, 15–17].
A key technical advancement in the study of protein complex composition and dynamics was the development of quantitative MS for studying biological systems. Quantitative MS provides two technical approaches that are useful for studying proteins complexes. In one approach, quantification is achieved through the use of stable isotopes. The most prevalent strategies use 13C- and/or 15N-containing reagents to label whole-cell proteomes prior to sample processing [18], for example, using stable isotope labeling by amino acids in cell culture (SILAC) [19]. Alternatively, isotope-coded labeling reagents can be used, which are integrated after isolation of the protein complexes [20]. In a second approach, quantification is achieved using tandem MS fragmentation events (spectral counts) or high-resolution MS1 signals of intact peptide ions (peak area). Spectral counting- and peak area-based approaches are collectively termed “label-free” approaches [21, 22]. Both isotope labeling and label-free approaches can measure differences in relative protein abundance between different samples. These quantitative comparisons can inform on the changes in protein interactions under different biological conditions, such as different cell cycle stages, disease stages, or environmental stimuli. They also are invaluable for defining the composition of a protein complex by assessing the likely specificity of protein interactions. In this case, the comparison is performed between one sample representing the affinity isolated protein complexes and one representing a nonspecific control. Currently, label-free approaches are the most widely employed for determining interaction specificity as these methods (1) can be integrated into existing qualitative proteomic workflows with minimal modification, (2) have a broad range of applications, not being restricted to certain biological model systems, (3) are cost-effective when larger sample amounts are required, and (4) are well-suited for detection of large abundance differences, which are expected for the majority of specific interactions. Moreover, several computational tools for measuring and evaluating interaction specificities have been optimized for label-free metrics and are applicable to affinity isolation studies ranging from small-scale (a few target proteins) to large-scale (hundreds of targets) studies [21, 23, 24]. One such algorithm is SAINT (Significance Analysis of INTeractome) [25], which we have employed in our studies [8, 26] and is described in this chapter. Following immunoaffinity purification and proteomic analysis, SAINT employs a probabilistic approach using mixture models to analyze the spectral count or peak area distribution of a background (negative control) dataset compared to the experimental samples (Fig. 1a). For each identified protein, SAINT assigns a score representing its likelihood of being a specific interaction.
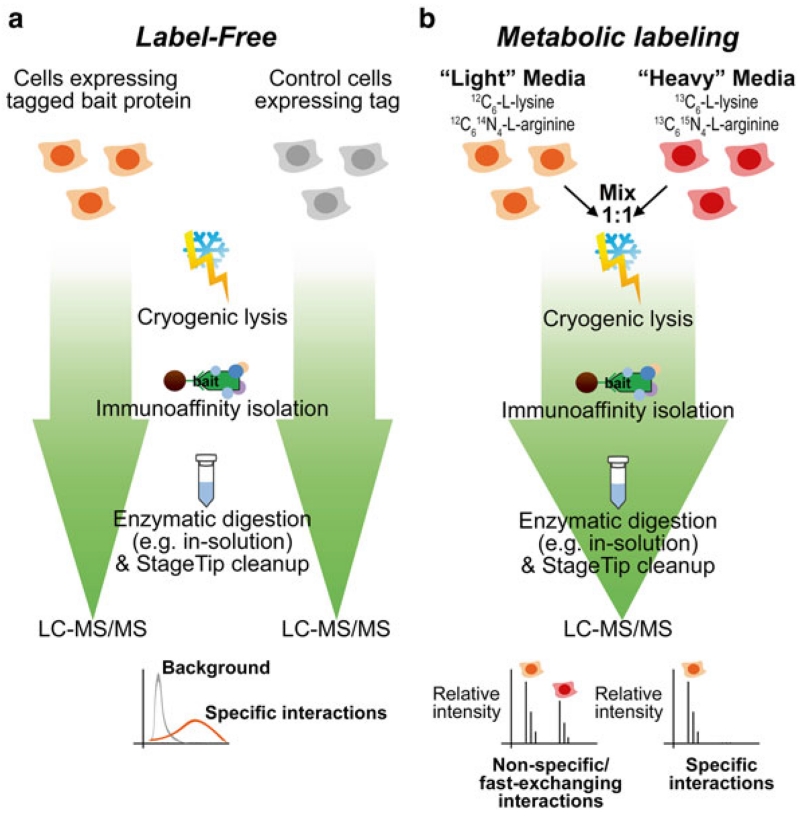
Fig. 1.
Integrated label-free and metabolic label-based approach for profiling protein interactions. (a) Label-free workflow. Cells expressing the tagged protein of interest (bait protein) are cultured in parallel to cells expressing the tag alone. Following cryogenic lysis, immunoaffinity isolation of the tagged protein (with its interactions) is performed using antibody-conjugated magnetic beads. Captured proteins are subjected to enzymatic digestion, sample clean-up and mass spectrometry analysis. Mass spectrometry signals (e.g., spectrum counts or peak intensities) from the bait protein isolation versus control isolations are analyzed to set cut-offs for high-confidence specific interactions. (b) Metabolic labeling workflow. Cells expressing tagged bait protein are cultured in media containing “light” amino acids, while control wild type cells (not expressing a tag) are cultured in “heavy” amino acid media. Cells are mixed in a 1:1 ratio and subjected to cryogenic lysis, immunoaffinity purification, and proteomic analysis as in (a), except the MS data analysis calculates the relative ion intensities of the “heavy” and “light” peptide signals to determine the specificity of interactions
Whole-cell metabolic stable isotope labeling paired with affinity purification-MS has several key advantages relative to label-free approaches. First, the quantitative precision and accuracy of labeled approaches are often superior, allowing for detection of lower abundance interactions. Second, metabolic labeling can control for variability introduced during protein extraction and affinity isolation since the differentially labeled (light and heavy) samples are mixed prior to subsequent sample processing. This feature of metabolic labeling was leveraged in the I-DIRT (Isotopic Differentiation of Interactions as Random or Targeted) technique [27], which has been used to determine interaction specificity for affinity isolated epitope-tagged protein complexes in diverse biological contexts [7, 28–32]. In this approach, an unlabeled (light) whole-cell proteome expressing epitope-tagged protein complexes is mixed with an isotope-labeled (heavy) whole-cell proteome devoid of the epitope tag (wild-type control). After mixing, cell lysis and affinity purification and enzymatic digestion of epitope-tagged protein complexes are performed followed by nLC-MS/MS analysis (Fig. 1b). I-DIRT can also be used for analysis of nontagged protein complexes, but requires that the isotope-labeling be performed in a background control cell line with targeted knockdown of the protein of interest. By using I-DIRT affinity isolation, identified proteins that are nonspecific would derive equally from the light and heavy samples, while specific interactions would be detected only with light isotope signals. Overall, label-free and isotope-labeled approaches each have specific advantages in characterizing protein complexes.
In this chapter, we describe an approach that integrates label-free analysis using the SAINT algorithm and metabolic labeling using I-DIRT to provide increased confidence in the specificity of interactions as well as to provide a profile of the relative stability of interactions within isolated protein complexes. Although the I-DIRT approach provides a powerful tool for defining a core set of stable interactions, one caveat is that bona fide interactions that exchange on-and-off the complex during cell lysis and affinity isolation are excluded as nonspecific associations. In contrast, label-free affinity isolation approaches do not preclude fast-exchanging proteins from being detected as specific interactions. Therefore, when performed in parallel, these approaches can identify candidate interactions that are specific but may be less stable. Together, with functional studies or with prior knowledge about the function of the complex of interest, this complementary method can inform on the potential impact that an interaction’s relative stability has on its functional roles within the complex. Here, we illustrate this for the case of chromatin remodeling complexes containing human histone deacetylases in T cells, as we have reported in [8]. However, this integrated label-free and metabolic labeling approach is broadly applicable to studies of diverse protein complexes in a variety of cell types.
2 Materials and Equipment
2.1 Metabolic Labeling of CEM T Cells for I-DIRT Analysis
Custom “Heavy” isotope culture medium: l-arginine/l-lysine deficient RPMI-1640 media (Life Technologies) supplemented 10 % with dialyzed fetal bovine serum (Gibco, Life Technologies), 100 mg/L 13C6-l-lysine (Cambridge Isotopes), 100 mg/L 13C615N4-l-arginine (Cambridge Isotopes), and 1 % penicillin-streptomycin (Life Technologies).
Custom “Light” isotope culture medium: l-arginine/l-lysine deficient RPMI-1640 media (Life Technologies) supplemented 10 % with dialyzed fetal bovine serum (Life Technologies), 80 mg/L 12C6-l-lysine (Sigma), 80 mg/L 12C614N4-l-arginine (Sigma), and 1 % penicillin-streptomycin (Life Technologies).
Cell line: Human peripheral blood derived T lymphoblasts (CCRF-CEM, ATCC).
T75 flasks.
T300 flasks.
50 mL conical tubes.
Swinging bucket rotor (prechilled).
Dulbecco’s Phosphate Buffered Saline (D-PBS) (ice cold).
Protease inhibitor cocktail, 100× (Sigma).
Cell freezing buffer: 10 mM HEPES-NaOH, pH 7.4, containing 1.2 % polyvinylpyrrolidine. Supplement with protease inhibitor cocktail to 10× immediately before use.
Liquid nitrogen.
Styrofoam container with 50 mL conical tube rack insert.
2.2 CEM T Cell Culture for Label-Free Proteomic Analysis
Same reagents as above, except cells are passaged in the standard culture medium: RPMI-1640 media (Life Technologies) supplemented with 10 % fetal bovine serum (Life Technologies) and 1 % penicillin-streptomycin (Life Technologies).
2.3 Cell Lysis
Retsch MM 301 Mixer Mill with 2 × 10 mL jars and 2 × 20 mm (tungsten carbide or stainless steel) grinding balls (Retsch, Newtown, PA).
Liquid nitrogen.
Foam ice bucket.
Long forceps.
Windex.
Methanol.
10 % bleach solution
Ultrapure water.
Spatula (chilled by liquid nitrogen).
Dry ice.
50 mL conical tubes.
2.4 Affinity Isolation of Protein Complexes
2.4.1 Conjugation of Magnetic Beads
Dynabeads M-270 Epoxy (Invitrogen). Store at 4 °C.
Affinity purified antibodies against an epitope tag or protein of interest (e.g., anti-GFP antibodies described below for the isolation of GFP-tagged proteins) or Immunoglobulin G (for isolation of Protein A-tagged proteins). Store at −80 °C.
0.1 M Sodium Phosphate buffer, pH 7.4 (4 °C, filter sterilized). Prepare as 19 mM NaH2PO4, 81 mM Na2HPO4. Adjust pH to 7.4, if necessary.
3 M Ammonium Sulfate (filter sterilized). Prepare in 0.1 M Sodium Phosphate buffer, pH 7.4.
100 mM Glycine–HCl, pH 2.5 (4 °C, filter sterilized). Prepare in water and adjust to pH 2.5 with HCl.
10 mM Tris, pH 8.8 (4 °C, filter sterilized). Prepare in water and adjust to pH 8.8 with HCl.
100 mM Triethylamine: Prepare fresh in water. CAUTION: Triethylamine is toxic and extremely flammable, and must be handled in a chemical hood and disposed appropriately.
DPBS, pH 7.4 (Dulbecco’s Phosphate-Buffered Saline (1×), liquid) (Invitrogen).
DPBS containing 0.5 % Triton X-100. Prepare fresh in DPBS.
DPBS containing 0.02 % sodium azide. Prepare fresh in DPBS. CAUTION: Sodium azide is a toxic solid compound and must be handled in a chemical hood and disposed appropriately.
Rotator (at 30 °C).
Magnetic separation tube rack (Invitrogen).
Tube shaker e.g., TOMY micro tube mixer.
Safe-Lock tubes, 2 mL round bottom (Eppendorf).
Ultrapure water (e.g., from a Milli-Q Integral Water Purification System).
2.4.2 Immunoaffinity Isolation
Frozen cell powder (see Subheading 3.3.1). Store at −80 °C.
Optimized lysis buffer (see Subheading 3.3.2) prepared fresh prior to each experiment. Store on ice.
Magnetic beads conjugated with antibodies (see Subheading 3.4.1). Store at 4 °C.
50 mL conical tubes.
Polytron for tissue homogenization (e.g., PT 10–35 Polytron from Kinematica).
Centrifuge and rotor, compatible with 50 mL conical tubes and capable of 8000 × g at 4 °C.
Tube rotator at 4 °C.
Ultrapure H2O.
Safe-Lock tubes, 2 mL round bottom (Eppendorf).
Safe-Lock tubes, 1.5 mL (Eppendorf).
Bar magnets (for conical tubes) and magnetic separation rack (for micro tubes) (Invitrogen).
4× LDS elution buffer: Dissolve 0.666 g of Tris–HCl, 0.682 g of Tris–Base, 0.8 g of LDS, and 0.006 g of EDTA (free acid) in ultrapure H2O to a final volume of 10 mL. Aliquot and store at −20 °C.
10× reducing agent: 0.5 M TCEP, pH neutral (Pierce).
10× alkylating agent: 0.5 M chloroacetamide in water. Aliquot and store at −20 °C.
Heat block at 70 °C.
2.5 In-Solution Digestion of Immunoisolated Proteins
Store stock solutions in glass containers that have been thoroughly rinsed with ultrapure water. Avoid using glassware that has been washed with detergents.
Primary eluate from label-free affinity isolation (see Subheading 3.4.2).
Primary eluate from I-DIRT affinity isolation (see Subheading 3.4.2).
Refrigerated microcentrifuge capable of 14,000 × g (maintain at 20 °C).
LoBind pipet tips, 200 μL (Eppendorf).
Amicon Ultra-0.5 centrifugal filters, 30 kDa NMWL (Millipore).
MS grade water (Fisher).
10 % sodium deoxycholate (DOC): Prepare in MS grade water and protect from light.
Tris–HCl buffer: 0.2 M Tris–HCl, pH 8.0, in MS grade water. Store at 4 °C.
TUD wash buffer: Prepare buffer fresh before use by mixing 1.5 mL of 0.2 M Tris–HCl, 0.6 mL of 10 % DOC, and 1.44 g urea. Yields ~3 mL of buffer, sufficient for two samples.
ABC-DOC wash buffer: 0.05 M ABC, 2 % DOC.
Trypsin, lyophilized MS-grade (Pierce). After suspension, store at −80 °C.
Digestion buffer: Prepare 100 μL per sample by mixing 1 μL of 0.5 μg/μL trypsin stock and 99 μL of 0.05 M ABC. Prepare fresh immediately before use.
10 % trifluoroacetic acid (TFA). Prepare in MS grade water and store at 4 °C.
Ethyl acetate. CAUTION: Ethyl acetate is flammable and toxic. Handle in a chemical hood and dispose appropriately.
SpeedVac Concentrator.
2.6 Peptide Clean-Up and Fractionation Using SDB-RPS StageTips
Microcentrifuge.
LoBind pipet tips, 200 μL (Eppendorf).
14 gauge needle (Hamilton #90514).
Syringe plunger, 100 μL (Hamilton #1162-02).
Empore SDB-RPS disks (3 M #2241).
50 % ethyl acetate/0.5 % TFA in MS grade water.
0.5 % TFA in MS grade water.
Buffer 1: 0.10 M ammonium formate, 0.5 % formic acid, 40 % acetonitrile in water.
Buffer 2: 0.15 M ammonium formate, 0.5 % formic acid, 60 % acetonitrile in water.
Buffer 3: 5 % ammonium hydroxide and 80 % acetonitrile in water.
FA solution: 1 % formic acid and 4 % acetonitrile in water.
Autosampler vials.
2.7 Nanoliquid Chromatography Tandem Mass Spectrometry Analysis
Nanoflow HPLC system, e.g., Dionex Ultimate, Waters nano Acuity, or Agilent 1200 series.
Mobile phase A (MPA): 0.1 % FA/99.9 % water. Store in amber bottle for up to 6 months.
Mobile phase B (MPB): 0.1 % FA/97 % ACN/2.9 % water. Store in amber bottle for up to 6 months.
Analytical column, e.g., Acclaim PepMap RSLC 75 μm ID × 25 cm (Dionex).
LTQ-Orbitrap Velos hybrid mass spectrometer (Thermo Fisher Scientific).
Nanospray ESI source (Thermo Fisher Scientific).
SilicaTip Emitter, Tubing (OD × ID) 360 μm × 20 μm; Tip (ID) 10 μm (New Objective).
2.8 Data Analysis
Multi-core/multi-CPU 64-bit PC workstation with at least 12 GB of RAM and 2 TB of storage.
Software for generating peaklists and scoring PSMs, with support for precursor ion quantification e.g., Proteome Discoverer 1.4 (Thermo Fisher Scientific), Mascot 2.3 (Matrix Science), Scaffold 4.0 (Proteome Software).
SAINT (http://www.crapome.org/).
Spreadsheet software (e.g., Microsoft Excel).
3 Methods
This protocol involves two different quantitative MS workflows (see Fig. 1). The most significant differences are in the steps for cell culture; so, the respective steps for label-free and isotope-labeling workflows are described separately (Subheadings 3.1 and 3.2). The cell lysis, immunoaffinity isolation, and in-solution digestion are performed identically, independent of the quantitative workflow.
3.1 Metabolic Labeling of CEM T Cells for I-DIRT Analysis
Aliquot 1.2 × 107 wild-type CEM T cells into conical tube and pellet at 200 × g (see Note 1).
Resuspend the pellet in 10 mL of heavy isotope media and aliquot equally into 2× T75 flasks.
Add 25 mL of heavy media to each flask and culture in incubator (37 °C/5 % CO2) until cell concentration is 2 × 106/mL (~4 days).
Transfer each cell suspension (~30 mL) into a T300 flask and add 150 mL of heavy isotope media. Culture in incubator as above.
Divide total cell suspension (360 mL) equally into 50 mL conical tubes (8 × 45 mL).
Pellet cells at 200 × g at 4 °C.
Aspirate media and resuspend cell pellets in ice-cold D-PBS (10 mL). Pool cell suspensions in 2 × 50 mL conical tubes.
Pellet cells as above and aspirate media.
Wash cell pellets with D-PBS (20 mL), pool into a single pre-weighed 50 mL conical tube.
Pellet cells as above and aspirate media.
Repeat steps 9 and 10.
Weigh conical tube to determine wet cell pellet weight.
Keep cells on ice while preparing liquid nitrogen freezing bath.
Place a fresh 50 mL conical tube in Styrofoam container/rack. Fill conical tube with liquid nitrogen about halfway and leave uncovered. Fill bottom of rack with liquid nitrogen to slow evaporation in tube.
Add 100 μL of cell freezing buffer per gram of cells and pipet drop wise into the conical tube containing liquid nitrogen (see Note 2).
CRITICAL: Use a needle to create holes in the conical tube cap before re-capping the tube containing the frozen cell material. Secure the cap and gently agitate in a fume hood to allow liquid nitrogen evaporation. Heavy-labeled frozen cell pellets can be stored at −80 °C for up to several years.
Cell material for the complementary “light” I-DIRT sample (see Fig. 1) is generated as above, except the CEM T cells that stably express the affinity-tagged protein of interest are cultured in the custom “light” isotope medium (see Note 3).
3.2 CEM T Cell Culture for Label-Free Proteomic Analysis
-
Cell material for the label-free affinity isolation experiment is generated as described above in Subheading 3.1, except two separate CEM T cell lines, both grown in standard culture media, are required:
-
(a)
One cell line should stably express the affinity-tagged protein of interest.
-
(b)
The other cell line is the control, which stably expresses the affinity tag alone or an empty vector.
-
(a)
3.3 Cell Lysis
The procedures described below for immunoaffinity purification of protein complexes utilize mammalian cells as the starting material, which is cryogenically disrupted using a Mixer Mill. However, cell lysis can also be carried out using several alternative approaches, including direct homogenization in a detergent-containing lysis buffer or passage of the lysate through a needle. We prefer the method of cryogenic disruption described below, as we have observed that it leads to an increased efficiency of extraction and decreased level of nonspecific associations. This method has provided us with a reliable and effective means of cell lysis for isolating varied protein complexes [2, 5, 6, 8, 12, 15, 17, 26, 33–39] and has been described in detail elsewhere [40].
3.3.1 Cryogenic Cell Disruption
Clean one spatula, the Retsch Mixer Mill jars, and the grinding balls sequentially with Windex, ultrapure H2O, 10 % bleach solution, ultrapure dH2O, and 100 % methanol. Allow all parts to dry completely.
Cool the jars and balls in liquid nitrogen (e.g., using a foam ice bucket filled with liquid nitrogen). Once the liquid nitrogen no longer appears to be bubbling, the jars are sufficiently cool and can be removed from the liquid nitrogen using a pair of long forceps.
Quickly place the frozen cell pellets into the jar. For the label-free affinity isolation workflow, use cell pellets collected from a standard CEM T cell culture expressing the desired affinity-tagged protein (see Subheading 3.2). For the I-DIRT affinity isolation workflow, combine equal amounts of cell pellets collected from “light” and “heavy” CEM T cell cultures (see Subheading 3.1). Cell pellets can fill up to a maximum of one-third of the total volume of the jar for optimal cryogenic grinding (e.g., ~2.5 g frozen tissue pellets per 10 mL jar). Place a single chilled ball on top of the cell pellets, close the jar, and cool in the liquid nitrogen container.
Place the jars in the Retsch Mixer Mill holders. If only processing one sample, use an empty jar (without a ball) as a balance. Cryogenically lyse cells using ten cycles of 2 min 30 s each at a frequency of 30 Hz. In between each cycle, re-cool jars in liquid nitrogen and check that the jars remain securely closed.
Open the jar and use a chilled spatula to transfer the frozen cell powder to a 50 mL conical tube chilled on dry ice. Proceed rapidly to avoid thawing of the ground sample. Store the powder at −80 °C until the affinity isolation is to be performed.
3.3.2 Optimization of Lysis Buffer and Isolation Conditions
During cell lysis and protein isolation, the efficient extraction of the targeted protein in a soluble fraction, while maintaining its interactions, is the primary goal. As a result, the lysis buffer conditions should be optimized for each protein of interest before performing larger scale immunoaffinity isolations for proteomics studies. It is therefore recommended that small-scale experiments be performed to assess the efficiency of protein solubilization and efficiency of isolation by western blotting using at least three lysis buffer conditions with varied levels of stringency, as described in detail previously [13, 40].
3.4 Affinity Isolation of Protein Complexes
3.4.1 Conjugation of Magnetic Beads
This protocol has been optimized for the conjugation of M-270 Epoxy Dynabeads, but can also be applied for conjugation of additional types of magnetic beads with larger or smaller diameters (e.g., M-450 or MyOne Dynabeads). In such cases, the amount of antibody used for conjugation should be adjusted based on the binding capacity of the bead. This protocol can be used for conjugating beads with either high-affinity purified antibodies or commercially available antibodies. It is important to note that the storage of antibodies in buffers containing free amines (e.g., Tris) will limit the amount of antibody that will be covalently conjugated to the surface epoxy groups; so it is best to avoid such buffers.
It is optimal to begin this protocol in the afternoon and perform all washing steps (step 9) in the morning of the following day. All steps should be performed at room temperature, unless otherwise indicated. During the washing steps, the beads must not be allowed to dry out (i.e., proceed immediately from one wash step to the next and do not allow the beads to sit without a washing solution between individual steps).
Weigh out the necessary amount of magnetic Dynabeads in a round-bottom tube (see Note 4).
Add 1 mL Sodium Phosphate buffer (pH 7.4) over the top of the beads. Mix by vortexing for 30 s, followed by 15 min on a tube shaker (vigorous setting).
Place the tube on a magnetic rack or against a magnet. Remove and discard the buffer once the beads have settled towards the magnetic side.
Remove the tube from the rack. Add 1 mL Sodium Phosphate buffer (pH 7.4) and mix by vortexing for 30 s and remove the buffer as above.
-
Remove the tube from the rack. In the following order, add the necessary amount of antibodies, Sodium Phosphate buffer (pH 7.4), and Ammonium Sulfate solution (3 M).
-
(a)
The optimal total volume of the bead conjugation solution, which includes the antibodies, Sodium Phosphate buffer, and Ammonium Sulfate solution, is ~20 μL/mg beads.
-
(b)
The amount of antibody conjugated is 3–5 μg Ab/mg M-270 epoxy beads. If another type (or size) of bead is used, the amount of Ab should be optimized, as the binding capacity may be different.
-
(c)
The 3 M Ammonium Sulfate solution is added last to give a final concentration of 1 M (i.e., added at one-third of total final volume).
-
(d)
For example, a total volume of 360 μL is used to conjugate 18 mg beads. For an antibody:bead ratio of 3:1000, first add 54 μg antibody to the beads. Second, add 0.1 M Sodium Phosphate Buffer (such that the volume of 0.1 M Sodium Phosphate Buffer is equal to 360 μL minus the volumes of antibody and 3 M Ammonium Sulfate used). Finally, add 120 μL of 3 M Ammonium Sulfate.
-
(a)
Secure the tube with parafilm and incubate the bead slurry overnight on a rotator at 30 °C.
The next morning, place the tube with bead slurry against the magnetic rack.
OPTIONAL: Retain the supernatant to assess the efficiency of bead conjugation by SDS-PAGE.
-
Wash the beads sequentially with the following buffers:
-
(a)
1 mL of Sodium Phosphate buffer.
-
(b)
1 mL 100 mM Glycine–HCl, pH 2.5 (FAST).
-
(c)
1 mL 10 mM Tris–HCl pH 8.8.
-
(d)
1 mL 100 mM Triethylamine solution (FAST).
-
(e)
4 × 1 mL DPBS.
-
(f)
1 mL DPBS containing 0.5 % Triton X-100. Mix on a Tomy shaker with gentle agitation for 15 min.
-
(g)
1 mL DPBS.
-
(a)
Resuspend washed beads in 12.5 μL DPBS containing 0.02 % NaN3 per mg of beads. Measure the final volume of the bead slurry to determine the bead concentration (mg of beads/μL DPBS).
Beads can be used immediately or stored for up to 2 weeks at 4 °C. After 1 month of storage, their efficiency for isolation decreases by approximately 40 %.
3.4.2 Immunoaffinity Isolation
Prepare an appropriate volume of optimized lysis buffer as determined in Subheading 3.3.2. Pre-cool the buffer to 4 °C. Add protease inhibitors immediately prior to use. Prepare 10 mL of wash buffer per sample (used in steps 8 and 14–17), which is typically identical in composition to the optimized lysis buffer but lacks protease and phosphatase inhibitor cocktails.
Incubate the frozen cell/tissue powder on ice for 1 min. Proceed immediately to step 3.
Resuspend the frozen cell/tissue powder in the lysis buffer by first adding a small amount of lysis buffer and swirling the homogenate to solubilize pellet. Continue to add lysis buffer and gently mix by swirling or inversion until the powder is completely solubilized (see Note 5).
Run the Polytron homogenizer for 10 s in ultrapure dH2O to wash.
To avoid the spilling of the sample, ensure that the homogenate occupies a maximum of a 1/3 of the conical tube volume. Subject lysates to Polytron homogenization for 2 × 15 s (speed = 22.5 k), briefly incubating the sample on ice between homogenizations.
If processing additional samples, rinse the homogenizer with ultrapure dH2O and run the Polytron in ultrapure dH2O to wash out any excess lysate residue. When finished with homogenization steps, perform a final methanol rinse.
Centrifuge the lysate at 8000 × g at 4 °C for 10 min.
While the lysates are centrifuging, place the tube containing antibody-conjugated magnetic beads against a magnetic rack for 30–60 s. Discard the storage buffer and wash with 3 × 1 mL wash buffer by gently pipetting up and down to resuspend the beads. Do not vortex the beads. Resuspend the beads in 100–200 μL of wash buffer.
Carefully pour the clarified lysates (supernatant) into new 50 mL conical tubes (see Note 6). RETAIN (1) the insoluble cell/tissue pellet and (2) 40 μL of the supernatant to serve as the input fraction for further analysis.
Mix the beads in solution by gently flicking the tube of antibody-conjugated beads. Pipette the appropriate amount of beads into the tube containing the clarified lysates.
Rotate the lysate–bead solution on a rotator at 4 °C for 1 h (see Note 7).
During the incubation step, prepare 1× LDS elution buffer.
Use a rubber band to attach a bar magnet to the tube holding the lysate-bead suspension. Incubate on ice for 5 min. RETAIN the flow-through (unbound) fraction by pouring the supernatant into a clean conical tube for further analysis.
Resuspend the beads in 1 mL of wash buffer and transfer the bead slurry to a clean round-bottom tube.
Place the tube against the magnetic rack to separate the beads from the buffer. Discard wash buffer. Perform this step between all subsequent wash steps.
Wash the beads 3 × 1 mL wash buffer. On the third wash, transfer the bead slurry to a clean round-bottom tube.
Wash the beads 2 × 1 mL with wash buffer.
Add 1 mL DPBS to beads and transfer slurry to a third clean round-bottom tube.
Wash once more with 1 mL of DPBS to remove residual detergent. Completely remove DPBS wash.
Add 40 μL of 1× LDS elution buffer to beads.
Incubate for 10 min at 70 °C, then 10 min at RT with agitation (see Note 8).
Isolate beads on the magnetic rack and transfer the primary eluate to a microcentrifuge tube. RETAIN the bead fraction.
Resuspend the bead fraction in 40 μL sample buffer and repeat step 21, except incubating the beads at 95 °C for 5 min.
Add 5 μL of 10× TCEP and 5 μL of 10× CAM to primary and secondary eluates. Heat at 95 °C for 5 min. RETAIN 10 % of primary and secondary eluates for analysis of isolation efficiency.
If immediately performing an in-solution digestion, proceed directly to Subheading 3.5 with the remaining 90 % of the primary eluate. Otherwise, samples can be stored at ≤−20 °C.
-
To assess the efficiency of immunoisolation, analyze equal percentages of the following fractions by Western blotting.
-
(a)
Cell pellet (step 9).
-
(b)
Input supernatant (step 9).
-
(c)
Flow-through (step 13).
-
(d)
Primary eluate (step 22).
-
(e)
Secondary eluate (step 23).
-
(a)
3.5 In-Solution Digestion of Immunoisolated Proteins
The in-solution digestion protocol described below uses a filter-aided sample preparation method [41] incorporating urea [42] and sodium deoxycholate [43] wash buffers to remove the LDS detergent and limit protein/peptide losses, respectively. Sodium deoxycholate is removed by organic phase extraction post-digestion [44]. Other digestion protocols may be used, but they must be capable of removing the LDS detergent prior to MS analysis.
Day 1
Set temperature of microcentrifuge to 20 °C (all subsequent spins are performed at this temperature).
Add 400 μL of TUD buffer to each unlabeled and isotope-labeled primary eluate (from Subheading 3.4.2).
Transfer each sample to a separate Amicon-0.5 filter and centrifuge at 14,000 × g for 10 min, or until volume is reduced to the minimum (~25 μL).
Discard flow-through and add 400 μL of TUD buffer to filter. Centrifuge as above.
Discard flow-through and add 300 μL of TUD buffer to filter. Centrifuge as above.
Discard flow-through and add 300 μL of TUD buffer to filter. Centrifuge as above.
Add 200 μL of ABC-DOC buffer to filter. Centrifuge at 14,000 × g for 10 min. Ensure that the retained volume is at the minimum.
Transfer filter units to fresh collection tubes and add 100 μL of Digestion buffer to the filter. Mix on TOMY shaker for 1 min.
Wrap the top of each tube in parafilm. Incubate in water bath overnight at 37 °C.
Day 2
Centrifuge filters at 14,000 × g for 5 min to recover digested peptides.
Add 25 μL of MS-grade water and centrifuge as above.
Add 25 μL of MS-grade water and centrifuge as above.
Discard filter unit and retain flow-through containing peptides.
Add an equal volume of ethyl acetate to the sample.
Adjust each sample to 0.5 % TFA.
Vortex, then mix on TOMY shaker for 2 min.
Centrifuge at 14,000 × g for 5 min.
Recover the denser aqueous phase, while avoiding the top organic (ethyl acetate) phase and interphase.
Proceed to “Sample Clean-up and Peptide Fractionation using StageTips” (Subheading 3.6) with the recovered aqueous phase.
3.6 Sample Clean-Up and Fractionation Using StageTips
For each sample, prepare one StageTip by depositing a single Empore SDB-RPS disk (cut using a 14 gauge needle) into the bottom of a 200 μL pipette tip using a syringe plunger (see Note 9).
Add half of the sample to the StageTip and centrifuge at 2000 × g until all solution has passed through the Empore disk (see Note 10).
Add the remaining sample to the StageTip and repeat centrifugation.
Wash disk with 100 μL of 50 % ethyl acetate/0.5 % TFA.
Wash disk with 100 μL of 0.5 % TFA.
Pass 50 μL of Elution buffer 1 over the disk and collect the eluate in an autosampler vial.
Repeat step 6 using Elution buffer 2 and again using Elution buffer 3. Collect each eluate in a separate autosampler vial.
Concentrate samples by vacuum centrifugation to near-dryness.
Add FA solution to achieve a final volume of 9 μL. Vortex briefly to mix.
Proceed immediately to nLC-MS/MS analysis (Subheading 3.7) or store at −80 °C for future analysis.
3.7 Nanoliquid Chromatography Tandem Mass Spectrometry Analysis
Many HPLC and MS system configurations are suitable for analyzing label-free and isotope-labeled peptides. However, to achieve optimal depth of analysis and quantitative precision, an LC system capable of low flow rates (<0.5 μL/min) and high pressure support (>400 bar) is highly preferable, as these capabilities allow the highest sensitivity of peptide detection using analytical columns with inner diameters ≤75 μm and lengths ≥25 cm. Additionally, a high-resolution and high-mass accuracy MS system with tandem MS fragmentation capability is required.
Ensure that the system is properly calibrated according to the manufacturer’s specifications.
Using MS instrument software, create an appropriate data-dependent acquisition method (see Note 11). For isotope-labeled samples it is critical that the precursor (MS1) scan be performed with high resolution (e.g., 60,000 at m/z 400).
Using the LC instrument software, create a reverse-phase method for label-free analysis. Program the method to separate peptides over 3 h using a linear gradient of 4–40 % mobile phase B.
For the analysis of isotope-labeled samples, the method should separate peptides over 6 h using the same mobile phase gradient parameters listed in step 3. The increase in LC run time is intended to compensate for increased spectral complexity due to isotopic labeling.
Create a shorter length (e.g., 60 min) gradient method to use for analysis of standard/quality control samples.
Perform duplicate (at a minimum) injections of a peptide standard to ensure that the system is performing at an acceptable level prior to injecting experimental samples.
For experimental samples, inject 4 μL of each fraction using the appropriate LC-MS/MS method designed above. The injection order should be selected to analyze the label-free samples first, followed by the isotope-labeled samples (see Note 12).
After experimental sample injections are complete, inject the standard peptide mixture to confirm that instrument performance has been maintained throughout the analysis.
3.8 Data Analysis
3.8.1 Peptide Identification and Protein Assignment for Label-Free Datasets
Extract all MS/MS spectra from raw mass spectrometry data, removing MS/MS spectra that do not contain at least ten peaks.
-
Generate instrument and experiment-specific database search parameters.
-
(a)
Define static peptide modification for cysteine carbamidomethylation.
-
(b)
Define variable modification for methionine oxidation (see Note 13).
-
(a)
Submit spectra to an appropriate workflow to obtain pep-tide spectrum matches and protein group assignments (see Note 14).
Select peptide and protein scoring filters to achieve a desired false discovery rate (e.g., ≤1 %).
Export data tables containing, at minimum, protein group descriptions with respective accession numbers and total spectrum counts. This output will be used interaction specificity analysis using the SAINT algorithm.
3.8.2 I-DIRT Isotope Labeling Quantification
-
To analyze I-DIRT datasets, which include both light- and heavy-labeled lysine and arginine containing peptides, create a duplicate analysis workflow from the workflow created above, and modify as follows:
-
(a)
Define additional variable modifications for heavy 13C6 lysine and heavy 13C6-15N4 arginine.
-
(b)
Include additional necessary modules and associated parameters for the extraction of light and heavy peptide signals and their integration over the LC elution peak.
-
(a)
Calculate I-DIRT peptide ratios as (Heavy Peptide Signal)/(Light Peptide + Heavy Peptide Signal).
Calculate protein group level ratios as the median of peptide ratios.
If available in the software, protein ratios can be median normalized to correct for non-equal mixing of light and heavy-labeled cells. This option should only be used if the majority of identified proteins are nonspecific interactions.
Export data tables containing, at minimum, protein group descriptions, accession numbers, I-DIRT protein ratios, ratio variances, and number of quantified peptides.
3.8.3 SAINT Interaction Specificity Analysis Using Label-Free Spectral Counts
Access the website www.crapome.org and register for a free user account to enable the full SAINT analysis functionality (see Note 15).
Select “Workflow 3: Analyze Your Data”.
OPTIONAL: If desired (e.g., if control isolations have not been performed), select negative controls from the CRAPOME database (see Note 16).
Using the label-free analysis data tables exported above, generate a compatible SAINT matrix input file, as specified in the workflow step 2 (Upload Data).
Upload SAINT matrix file and proceed to step 3, Data Analysis.
Under the “Analysis Options”, enable “Probability Score”, choose the “SAINT” model, and increase the “n-iter” option to 10,000 (see Note 17). Run Analysis.
After the analysis has completed, save and open output file, which reports the individual and average SAINT scores (AvgP) for each identified protein. Scores range from 0 to 1 (least to most specific).
Evaluate the performance of SAINT in distinguishing between specific interactions and nonspecific background. If many interactions are already known for a particular protein of interest, the sensitivity and specificity of the analysis can be estimated by constructing ROC plots. If no prior interaction knowledge is available, then construct a histogram for the distribution of SAINT scores (see Note 18 and Data Interpretation section below). Use these analyses to select a SAINT score cut-off that eliminates the majority of nonspecific interactions (false positives), while retaining the highest scoring interactions.
3.8.4 Interpretation of Label-Free and Labeling Results
For each candidate protein interaction, the output of SAINT and I-DIRT provides a score ranging from 0-to-1 and 0.5-to-1, respectively. As illustrated in the graph in Fig. 2, higher SAINT scores represent increased probability of a specific interaction, while higher I-DIRT scores represent greater interaction stability and specificity. An important step that requires careful consideration is the selection of appropriate cutoff scores to both maximize true positive interactions and minimize false positives. Also, it should be noted that this workflow does measure exchange rates or binding affinities directly. If more rigorous determination of interaction stability is required, one could measure the isotope exchange in a time series after mixing a light-labeled immunoisolated complex with a heavy-labeled whole-cell lysates. Alternatively, a more targeted analysis of relative protein abundance under different isolation stringencies can be performed by Western blotting.
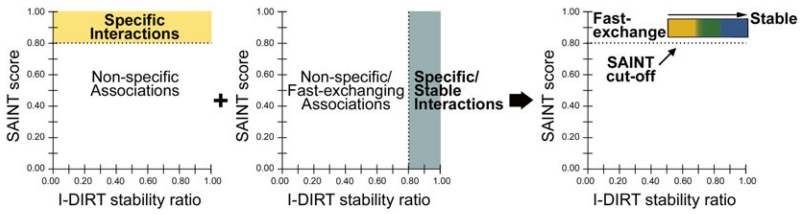
Fig. 2.
Integration of SAINT and I-DIRT methods allows the simultaneous investigation of specificity and relative stability of protein interactions. Specificity determination using the SAINT algorithm assigns interaction specificity scores to individual proteins (SAINT score), while I-DIRT metabolic labeling distinguishes between stable/specific interactions and nonspecific/background contaminants by calculation of isotope ratios (I-DIRT stability ratio). Integrating these two methods (right panel) provides insight into the relative stability and/or fast-exchanging nature of specific protein interactions of a given bait protein
For SAINT, the selected threshold defines nonspecific versus specific interactions. One of the most effective approaches for selecting SAINT scoring thresholds is to generate a Receiver Operating Characteristic (ROC) curve for each bait protein using previously known interactions (e.g. from the BioGRID repository [45]). This approach allows empirical selection of a SAINT score threshold to balance true versus false positives, as in [8]. However, this is not always feasible, especially in the case of proteins that lack known protein interactions. Alternatively, the distribution of SAINT scores can be examined to select an appropriate threshold [26]. Overall, for the majority of datasets, we have found as a general guide that a SAINT score threshold between 0.8 and 0.95 is appropriate.
For I-DIRT datasets, protein scores represent the fraction of the protein abundance from the tagged bait condition versus the total abundance (tagged bait + wild-type background). Therefore, values closer to 1.0 represent specific interactions that are very stable. Values less than 1.0 and decreasing progressively down to 0.5 reflect increasingly nonspecific interactions. However, a subset of these proteins may exhibit fast-exchange within their respective complexes and represent false negatives. Therefore, an important aspect of interpreting results from I-DIRT experiments is that they alone cannot distinguish nonspecific versus fast-exchanging interactions.
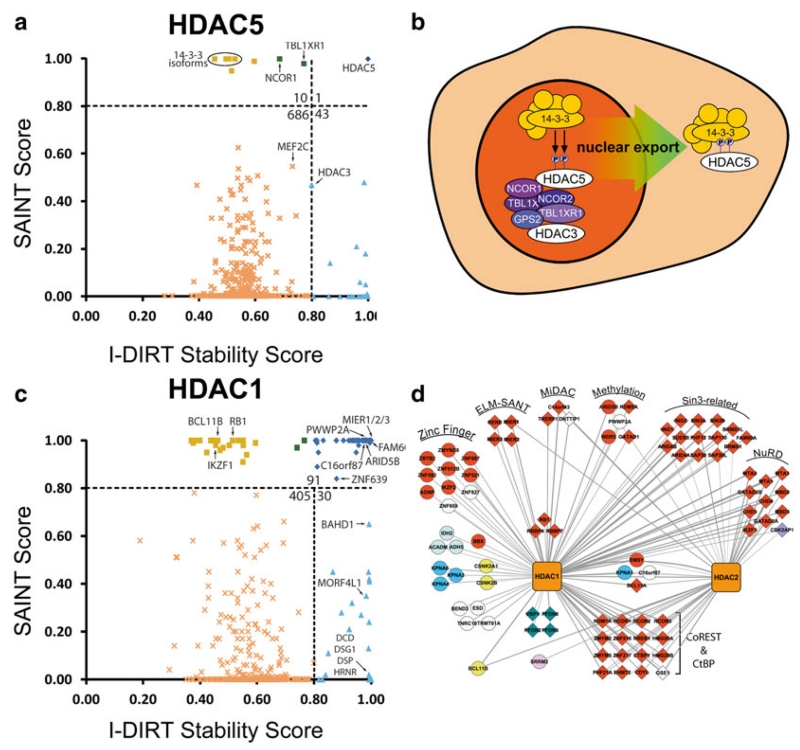
However, since the label-free SAINT workflow is performed separately for control and experimental samples, exchange does not influence SAINT scoring. Therefore, by cross-comparing proteins with low I-DIRT values but high SAINT scores, candidate interactions that are specific but fast-exchanging can be classified with higher confidence. A proof-of-concept example is illustrated for well-established proteins interactions of histone deacetylase 5 (HDAC5), such as 14-3-3 and the nuclear receptor corepressor 1 protein, which have low I-DIRT scores but high SAINT scores (Fig. 3a). These results are consistent with the known regulation and cellular roles of HDAC5, which involve shuttling of HDAC5 between the nucleus and the cytoplasm concomitant with dynamic changes in its interactions (Fig. 3b) [6, 17]. In addition, using the well-studied histone deacetylase 1 protein, we illustrate that, if prior functional knowledge of known and unknown protein interactions is available (Fig. 3d), then the specificity and the stability data can be used together to form hypotheses about the roles of these proteins within particular complexes (Fig. 3c). Finally, if these data are acquired for interactions shared between different affinity enriched proteins, one could formulate hypotheses whether a given protein is similarly or differentially regulated within distinct complexes.
Fig. 3.
SAINT/I-DIRT analysis of HDAC5 and HDAC1 reflect their subcellular localizations and functions in transcriptional regulation and chromatin remodeling. (a) SAINT/I-DIRT plot for immunoisolated HDAC5 highlights fast-exchanging interactions with 14-3-3 chaperone proteins and components of the nuclear corepressor complex. (b) Transient interactions predicted by SAINT/I-DIRT scoring are consistent with the nucleo-cytoplasmic shuttling of HDAC5. In the nucleus, HDAC5 associates with the nuclear NCoR proteins (purple). During nuclear export, HDAC5 can dissociate from the NCoR complex and increase its interaction with 14-3-3 chaperone proteins. (c) SAINT/I-DIRT plot for immunoisolated HDAC1 allows classification of known and novel protein associations of HDAC1, highlighting transient association with proteins associated with transcription and stable association with numerous chromatin remodeling complexes. (d) HDAC1 associates specifically with chromatin remodeling complexes (e.g. NuRD, Sin3a/b, CoREST), the transcriptional regulatory complex CtBP, and the mitotic deacetylase complex MiDAC. The integrated SAINT/I-DIRT method provides functional insight into the relative stabilities of individual proteins within known complexes reported to associate with HDAC1
Another advantage of this complementary interaction workflow is the identification of a subclass of candidate interactions with low specificity scores by SAINT but with high I-DIRT values (Fig. 2, lower-right quadrant). When using spectrum counts to assess the specificity of interaction, small proteins or low-abundance interactions may be detected as false negatives (low SAINT scores). Yet, these candidates are “rescued” by the I-DIRT analysis, which can provide reliable quantification with only a few sequenced peptides. Alternatively, this subset of candidates can also be environmental sample contaminants, as they would be present only with a light isotope signal. These environmental contaminants can be excluded by performing an additional I-DIRT experiment in which the isotope labels are swapped.
Overall, this integrative label-free and isotope-labeling approach generates interaction datasets that inform on both the specificity and the relative stability of protein interactions. Additionally, isotope-labeling experiments are now more cost-effective, making integrating both complementary approaches into a single experimental design feasible. This hybrid approach is broadly applicable to investigating the dynamic interactions within protein complexes in different cell types and across different biological conditions. We expect that the future development of improved quantitative mass spectrometry techniques will continue to shed light on the intricacies of protein complex regulation in space and time.
Acknowledgements
We are grateful for funding from NIH grants R01GM114141, R21AI102187, and R21 HD073044, an NJCCR postdoctoral fellowship to TMG, and a NSF graduate research fellowship to AJG.
Footnotes
This protocol describes the characterization of protein complexes from a specific cell type: mammalian CEM T cells, which are grown in suspension. However, this method can be applied in any cellular/tissue model system that achieves sufficient incorporation of metabolic labels.
For cell amounts less than 1 g, add 100 μL of cell freezing buffer.
To generate the “light” cell material in the I-DIRT workflow, standard culture medium with nondialyzed serum can be used as an alternative method. However, the growth rates and signaling pathways may vary significantly when compared to methods using dialyzed serum, depending on the cell type used.
Round-bottom tubes are the preferred tube shape, which minimizes bead trapping during the conjugation. The required amount of beads is dependent on both the experimental objective and the abundance of the protein to be immunoaffinity purified. As an approximate guide, 1–2 mg beads are appropriate for small-scale optimization experiments (as described in Subheading 3.2), 5–7 mg beads are usually sufficient for single immunoaffinity purifications, and 10–20 mg beads may be suitable for proteins of high abundance.
After suspending the cell/tissue powder in lysis buffer, the lysate solution may be slightly turbid; however, the solution should be devoid of cell/tissue clumps or aggregates. Do not proceed to Polytron (step 4) homogenization until a homogenous suspension is observed. If necessary, additional rotation for 10–20 min at 4 °C can be performed to promote solubilization.
If insoluble particles are present in supernatant after centrifugation, a pipette can be used to selectively transfer supernatant to a clean 50 mL conical tube.
Longer incubation times tend to promote the accumulation of nonspecific binders and the loss of weak interacting partners [2].
We have found that when using high-affinity antibodies (e.g. anti-GFP) stringent heat and detergent denaturing conditions are required for efficient recovery of the target proteins from the beads.
Ensure that the disk makes a seal with the walls of the pipette tip and is located a few mm above the tapered end of the tip. Each disk can bind ~25 μg of peptides. If greater capacity is needed, additional disks can be layered in the same StageTip and the number of washes increased to be equal to the number of total disks used.
Sample loading and washing of StageTips can be performed manually by applying pressure with a small plastic syringe or by centrifugation of the StageTip in a collection tube with an adapter.
Many considerations are required when designing an LC-MS/MS method, many of which are instrument-specific. However, in general the MS acquisition cycle should be designed based on the performance characteristics of the LC system. It is critical that the MS cycle time, determined largely by the number of full and tandem MS scans, permits acquisition of multiple full scans over the average LC elution peak. For example, given LC peak widths of 15–30 s, an optimal time for a single acquisition cycle would be in the range of 2–3 s for data-dependent methods.
It is recommended to perform several “blank” or “wash” runs between unlabeled and isotope-labeled samples and/or samples from different target protein isolations to minimize run-to-run carry-over.
Other variable modifications may be included in the primary database search, such as phosphorylation, acetylation, or deamidation. However, as addition of modifications increases both search time and space, it is recommended to retain only those modifications that are present in the sample.
When selecting an analysis workflow, ensure that it incorporates the ability to control for false positive sequence matches, e.g., by performing database searching against reversed protein sequences to estimate false discovery rates. If available, it is highly recommended to use a software platform that also controls false identification rates at the protein level.
An alternative strategy to using the online SAINT algorithm is to download the latest version of the SAINT source files (www.sourceforge.com) and compile it for your appropriate operating system. This strategy allows the SAINT algorithm to be run locally in the command-line, but requires additional computational knowledge. For a more detailed description of the underlying SAINT algorithm and its associated parameters see [25].
To compute meaningful SAINT specificity scores for the unlabeled/label-free datasets, at least two biological replicates of the experimental and control isolations are required. Ideally, control isolations are “user” controls performed in parallel to the experimental samples; however, user controls can be replaced and/or supplemented with negative control data from the CRAPOME database [24] to provide additional stringency. These datasets are easily added when using the online SAINT workflow #3.
Several user-defined options are available when running SAINT. A thorough discussion of their recommended usage can be found in [25].
References
- 1.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17(10):1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 2.Cristea IM, et al. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4(12):1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Sueda S, Tanaka H, Yamagishi M. A biotin-based protein tagging system. Anal Biochem. 2009;393(2):189–195. doi: 10.1016/j.ab.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 4.Lambert JP, et al. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J Proteomics. 2015;118:81–94. doi: 10.1016/j.jprot.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaltenbrun E, et al. A Gro/TLE-NuRD corepressor complex facilitates Tbx20-dependent transcriptional repression. J Proteome Res. 2013;12(12):5395–5409. doi: 10.1021/pr400818c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greco TM, et al. Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol Cell Proteomics. 2011;10(2) doi: 10.1074/mcp.M110.004317. M110.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domanski M, et al. Improved methodology for the affinity isolation of human protein complexes expressed at near endogenous levels. Biotechniques. 2012;0(0):1–6. doi: 10.2144/000113864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi P, et al. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol. 2013;9:672. doi: 10.1038/msb.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pflieger D, et al. Quantitative proteomic analysis of protein complexes: concurrent identification of interactors and their state of phosphorylation. Mol Cell Proteomics. 2008;7(2):326–346. doi: 10.1074/mcp.M700282-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Hubner NC, et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189(4):739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blagoev B, et al. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21(3):315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 12.Mathias RA, et al. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159(7):1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miteva YV, Budayeva HG, Cristea IM. Proteomics-based methods for discovery, quantification, and validation of protein-protein interactions. Anal Chem. 2013;85(2):749–768. doi: 10.1021/ac3033257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nesvizhskii AI. Computational and informatics strategies for identification of specific protein interaction partners in affinity purification mass spectrometry experiments. Proteomics. 2012;12(10):1639–1655. doi: 10.1002/pmic.201100537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cristea IM, et al. Tracking and elucidating alphavirus-host protein interactions. J Biol Chem. 2006;281(40):30269–30278. doi: 10.1074/jbc.M603980200. [DOI] [PubMed] [Google Scholar]
- 16.Collins BC, et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat Methods. 2013;10(12):1246–1253. doi: 10.1038/nmeth.2703. [DOI] [PubMed] [Google Scholar]
- 17.Guise AJ, et al. Aurora B-dependent regulation of class IIa histone deacetylases by mitotic nuclear localization signal phosphorylation. Mol Cell Proteomics. 2012;11(11):1220–1229. doi: 10.1074/mcp.M112.021030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oda Y, et al. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci U S A. 1999;96(12):6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong SE, Kratchmarova I, Mann M. Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC) J Proteome Res. 2003;2(2):173–181. doi: 10.1021/pr0255708. [DOI] [PubMed] [Google Scholar]
- 20.Ranish JA, et al. The study of macromolecular complexes by quantitative proteomics. Nat Genet. 2003;33(3):349–355. doi: 10.1038/ng1101. [DOI] [PubMed] [Google Scholar]
- 21.Sardiu ME, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105(5):1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tate S, et al. Label-free quantitative proteomics trends for protein-protein interactions. J Proteomics. 2013;81:91–101. doi: 10.1016/j.jprot.2012.10.027. [DOI] [PubMed] [Google Scholar]
- 23.Choi H, et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011;8(1):70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellacheruvu D, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10(8):730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi H, et al. Analyzing protein-protein interactions from affinity purification-mass spectrometry data with SAINT. Curr Protoc Bioinformatics. 2012 doi: 10.1002/0471250953.bi0815s39. Chapter 8, Unit8.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diner BA, et al. The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Mol Syst Biol. 2015;11(2):787. doi: 10.15252/msb.20145808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tackett AJ, et al. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J Proteome Res. 2005;4(5):1752–1756. doi: 10.1021/pr050225e. [DOI] [PubMed] [Google Scholar]
- 28.Byrum SD, et al. ChAP-MS: a method for identification of proteins and histone post-translational modifications at a single genomic locus. Cell Rep. 2012;2(1):198–205. doi: 10.1016/j.celrep.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bottermann K, et al. Systematic analysis reveals elongation factor 2 and alpha-enolase as novel interaction partners of AKT2. PLoS One. 2013;8(6):e66045. doi: 10.1371/journal.pone.0066045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Virgilio M, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339(6120):711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramanagoudr-Bhojappa R, et al. Physical and functional interaction between yeast Pif1 helicase and Rim1 single-stranded DNA binding protein. Nucleic Acids Res. 2013;41(2):1029–1046. doi: 10.1093/nar/gks1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai YC, et al. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics. 2012;11(5):60–76. doi: 10.1074/mcp.A111.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moorman NJ, et al. A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol Cell Proteomics. 2010;9(5):851–860. doi: 10.1074/mcp.M900485-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140(5):678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carabetta VJ, Silhavy TJ, Cristea IM. The response regulator SprE (RssB) is required for maintaining poly(A) polymerase I-degradosome association during stationary phase. J Bacteriol. 2010;192(14):3713–3721. doi: 10.1128/JB.00300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niepel M, et al. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol Biol Cell. 2013;24(24):3920–3938. doi: 10.1091/mbc.E13-07-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mann JM, et al. Complex formation and processing of the minor transformation pilins of Bacillus subtilis. Mol Microbiol. 2013;90(6):1201–1215. doi: 10.1111/mmi.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castellana M, et al. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol. 2014;32(10):1011–1018. doi: 10.1038/nbt.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miteva YV, Cristea IM. A proteomic perspective of Sirtuin 6 (SIRT6) phosphorylation and interactions and their dependence on its catalytic activity. Mol Cell Proteomics. 2014;13(1):168–183. doi: 10.1074/mcp.M113.032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conlon FL, et al. Immunoisolation of protein complexes from Xenopus. Methods Mol Biol. 2012;917:369–390. doi: 10.1007/978-1-61779-992-1_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manza LL, et al. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 2005;5(7):1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 42.Wisniewski JR, et al. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 43.Erde J, Loo RR, Loo JA. Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J Proteome Res. 2014;13(4):1885–1895. doi: 10.1021/pr4010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen EI, et al. Optimization of mass spectrometry-compatible surfactants for shot-gun proteomics. J Proteome Res. 2007;6(7):2529–2538. doi: 10.1021/pr060682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]