Abstract
Here we show that glioblastoma express high levels of branched-chain amino acid transaminase 1 (BCAT1), the enzyme that initiates the catabolism of branched-chain amino acids (BCAAs). Expression of BCAT1 was exclusive to tumors carrying wild-type isocitrate dehydrogenase 1 (IDH1) and IDH2 genes and was highly correlated with methylation patterns in the BCAT1 promoter region. BCAT1 expression was dependent on the concentration of α-ketoglutarate substrate in glioma cell lines and could be suppressed by ectopic overexpression of mutant IDH1 in immortalized human astrocytes, providing a link between IDH1 function and BCAT1 expression. Suppression of BCAT1 in glioma cell lines blocked the excretion of glutamate and led to reduced proliferation and invasiveness in vitro, as well as significant decreases in tumor growth in a glioblastoma xenograft model. These findings suggest a central role for BCAT1 in glioma pathogenesis, making BCAT1 and BCAA metabolism attractive targets for the development of targeted therapeutic approaches to treat patients with glioblastoma.
The essential BCAAs valine, leucine and isoleucine largely escape first-pass liver catabolism and remain available to peripheral organs. Metabolism of these BCAAs provides an important mechanism by which nitrogen moves throughout the body for the synthesis of nonessential amino acids1, whereas deregulation of BCAA catabolic pathways frequently results in neural dysfunction2. The first step in BCAA catabolism is a transfer of an α-amino group to α-ketoglutarate (α-KG) through the activity of cytosolic BCAT1 or mitochondrial BCAT2 isoenzymes, yielding glutamate and the respective branched-chain α-ketoacids3,4. Whereas BCAT2 expression is nearly ubiquitous, BCAT1 expression is restricted to a small number of tissues, including the brain5–7, where BCAAs serve as a major source of nitrogen for neurotransmitter glutamate synthesis8. After transamination, branched-chain α-ketoacids are further catabolized to acetyl coenzyme A (acetyl-CoA) and succinyl-CoA, which are subsequently oxidized in the tricarboxylic acid cycle (Fig. 1a) to provide macromolecule precursors and energy for mitochondrial ATP synthesis.
Figure 1.
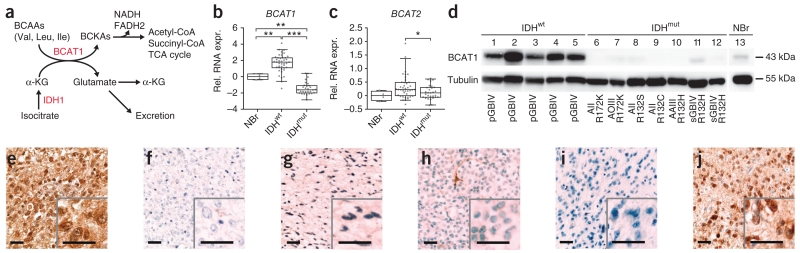
IDHwt astrocytic gliomas are characterized by high BCAT1 expression. (a) Schematic representation of BCAA catabolism. BCKAs, branched-chain ketoacids; TCA, tricarboxylic acid. (b,c) BCAT1 (b) and BCAT2 (c) RNA expression (relative RNA expression, rel. RNA exp.) in 70 astrocytic gliomas (41 IDHwt and 29 IDHmut) normalized to expression in normal brain (4 NBr). Whiskers mark the 5th and 95th percentiles. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed Student’s t test). (d) Western blot analysis showing BCAT1 protein expression in astrocytic gliomas with wild-type IDH1 and IDH2 (lanes 1–5), different mutations in IDH2 (lanes 6–7) or IDH1 (lanes 8–12) and normal brain tissue (lane 13). AII, diffuse astrocytoma WHO grade 2; AAIII, anaplastic astrocytoma WHO grade 3; sGBIV, secondary glioblastoma WHO grade 4; pGBIV, primary glioblastoma WHO grade 4; AOIII, anaplastic oligodendroglioma WHO grade 3. (e–h) Immunohistochemical stainings of BCAT1 in an IDHwt primary glioblastoma (e), a primary glioblastoma with the R132H IDH1 mutation (f), a diffuse astrocytoma with the R132C IDH1 mutation (g) and an anaplastic oligodendroglioma with the R172K IDH2 mutation (h). (i,j) Immunohistochemical staining of R132H IDH1 in the same tumors as those in e and f, respectively. Insets show representative areas at 2.5× relative magnification. Scale bars, 50 μm.
A dependence on glutamine catabolism for cell growth is frequently observed in malignant cells9 and has been intensively investigated in glioblastoma10–14. Under hypoxic conditions, glutamine can be metabolized to citrate and fatty acids through an isocitrate dehydrogenase–dependent pathway15–17. Mutations in cytoplasmic IDH1 and, less frequently, mitochondrial IDH2 are present in a majority of gliomas ranked as grade 2 or 3 by the World Health Organization (WHO) and secondary glioblastomas but are rare in the more common primary glioblastomas18–20. IDH1 and IDH2 mutations have a central role in glioma pathogenesis20,21 and constitute a key classifier distinguishing two major glioma subgroups identified on the basis of RNA expression and DNA methylation patterns22–27. Although IDH1 mutation status is a powerful prognostic factor that refines conventional diagnostic procedures for high-grade astrocytomas28, several studies have shown that tissue-specific knock in of mutant IDH (IDHmut) is insufficient to induce tumors in mice29,30, leaving the specific mechanisms by which IDH mutations contribute to tumor development poorly understood. Mutant IDH enzymes produce 2-hydroxyglutarate (2-HG) from α-KG, and depletion of α-KG, its competitive inhibition by 2-HG or both are believed to reduce the function of enzymes using α-KG31,32. Inhibition of histone demethylases and the TET family of 5-methylcytosine hydroxylases33–35 by 2-HG probably accounts for the widespread hypermethylation of gene promoters in IDHmut gliomas and leukemias24,33.
A better understanding of how differences in tumor metabolism are connected to growth patterns and the clinical behaviors of IDHmut gliomas and gliomas with wild-type IDH1 and IDH2 (IDHwt) is essential. In particular, it is important to know how IDHwt tumors fuel the rapid proliferation that accounts for the aggressive clinical phenotype of IDHwt glioblastomas. To address this issue, we analyzed transcriptional profiles generated during a previous study on astrocytic gliomas26 with a special emphasis on detecting molecular signatures characteristic of IDHwt tumors.
RESULTS
BCAT1 overexpression in IDHwt glioblastomas
Prediction analysis of gene expression data26 identified BCAT1 as the best classifier to distinguish primary glioblastoma from secondary glioblastoma, diffuse astrocytoma and anaplastic astrocytoma (Supplementary Tables 1 and 2). When classifying tumors on the basis of IDH mutation status, BCAT1 RNA expression levels were significantly higher in IDHwt gliomas relative to normal brain (P = 0.0051, two-tailed Student’s t test) and IDHmut gliomas (P < 0.0001; Fig. 1b). Compared to normal brain, BCAT1 RNA expression in IDHmut gliomas was downregulated (P = 0.0012). We did not observe comparable patterns of differential expression for BCAT2 (Fig. 1c). By mapping gene expression data26 onto the BCAA catabolic pathway using PathWave software36, we found that RNA expression levels for several other enzymes downstream of BCAT1 are also upregulated in IDHwt tumors relative to normal brain, indicating transcriptional activation of the entire pathway (Supplementary Fig. 1). Consistent with these RNA expression results, BCAT1 protein expression was high in IDHwt tumors but was essentially absent in IDHmut tumors regardless of the specific mutation in either IDH1 or IDH2 (Fig. 1d).
We further confirmed a tight correlation in IDH1 or IDH2 mutation status and BCAT1 expression through immunohistochemical staining of sections from 81 primary human gliomas (Fig. 1e–j and Supplementary Fig. 2a). We obtained independent confirmation through analysis of two published RNA expression data sets37,38 and a protein-level immunohistochemical analysis in an independent cohort of 210 gliomas (Supplementary Fig. 2b–d). These data clearly establish that high BCAT1 expression is a characteristic feature of IDHwt gliomas, which can be used to positively identify this tumor group in a diagnostic setting.
Substrate-dependent expression of BCAT1
We found that BCAT1 expression in glioblastoma cell lines is upregulated by hypoxia (Supplementary Fig. 3) and the addition of increasing concentrations of cell-permeable dimethyl–α-KG substrate to the culture medium (Fig. 2a). Conversely, shRNA-mediated knockdown of IDH1, a major source of α-KG in the cytoplasm39, led to strong downregulation of BCAT1 expression (Fig. 2b). This downregulation through IDH1 knockdown was almost completely reversed when dimethyl–α-KG was supplied in the culture medium (Fig. 2c). The 2-HG produced by IDHmut in glioblastomas is a structural analog of α-KG that has been shown to inhibit α-KG–dependent enzymes33,35. To determine whether 2-HG affects BCAT1 activity, we compared its inhibitory effect to those of α-ketoisocaproate (KIC) and α-KG in brain detergent extracts from Bcat2 knockout mice40, in which only Bcat1 activity is retained (Fig. 2d). A 40-fold excess of α-KG to α-ketoisovalerate (KIV) and a tenfold molar excess of KIC to KIV inhibited Bcat1 activity by 65% and 85%, respectively. Using up to 10 mM 2-HG, which is equivalent to a 100-fold molar excess relative to KIV, we achieved a maximum inhibition of 13%, indicating that BCAT1 is largely insensitive to 2-HG.
Figure 2.
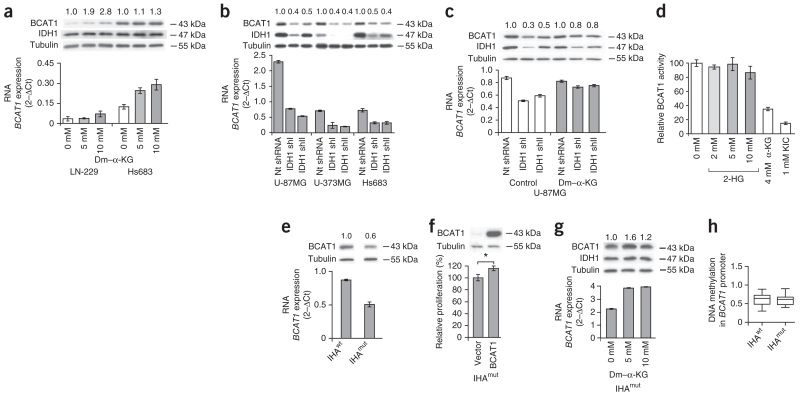
BCAT1 shows substrate-dependent expression in glioblastoma cell lines. (a–c) Effects of α-KG on BCAT1 expression in glioma cell lines. The numbers above the western blots indicate the fold ratios of protein expression relative to control cells after normalization to the tubulin loading control. The mRNA expression values represent the mean ± s.d. of triplicate samples. (a) Effect of cell-permeable dimethyl–α-KG (dm–α-KG) for 24 h on BCAT1 expression. (b) BCAT1 expression after lentiviral knockdown of α-KG–producing cytoplasmic IDH1 with two different shRNAs (shI and shII). nt, nontargeting. (c) BCAT1 expression after shRNA-mediated knockdown of IDH1 and additional supplementation of the culture medium with 1 mM cell-permeable dm–α-KG for 6 d. (d) Effect of branched-chain α-ketoacid substrates and 2-HG on brain BCAT1 activities. Values are the means ± s.d. of three independent experiments. (e) Western blot analysis showing BCAT1 protein expression in IHAwt and IHAmut cells. (f) Effect of lentiviral overexpression of BCAT1 in IHAmut cells on proliferation determined using the Click-iT EdU cell proliferation assay. Values in the graphs (e,f) represent the mean ± s.d. for n = 3 replicates. *P < 0.05 compared to empty vector control (Student’s t test). (g) Effect of dm–α-KG treatment for 24 h on BCAT1 expression in IHAmut cells. Values in the graph represent the mean ± s.d. for n = 3 replicates. (h) DNA methylation of CpGs (n = 19) in the BCAT1 promoter in IHAwt and IHAmut cells (Gene Expression Omnibus GSE30338). The average absolute methylation levels are shown. Box plots represent the median, the 25th and 75th percentiles (boxes) and the 5th and 95th percentiles (whiskers).
To more directly test the effects of mutant IDH1 on BCAT1 expression, we used immortalized human astrocytes (IHAs) stably overexpressing either wild-type IDH1 (IHAwt) or R132H mutant IDH1 (IHAmut)38. Analysis of published gene expression data38 revealed that BCAT1 is downregulated in IHAmut cells. We showed by western blot and quantitative RT-PCR (qRT-PCR) analyses that BCAT1 expression levels were 40% lower in IHAmut compared to IHAwt cells (Fig. 2e), indicating that IDH1mut expression suppresses BCAT1 expression in this cell line model. To determine whether BCAT1 expression can accelerate the growth rate of astrocytic cells, we ectopically overexpressed BCAT1 in IHAmut cells, resulting in a significant increase in proliferation rate (P = 0.0141, two-tailed Student’s t test; Fig. 2f). The substrate dependence of BCAT1 expression was maintained in IHAmut astrocytes (Fig. 2g). In analyzing published data38, we could not detect any differences in BCAT1 promoter methylation between IHAmut and IHAwt cells (Fig. 2h). This finding is consistent with our observation that IHAmut cells maintain an intermediate level of BCAT1 expression (Fig. 2e).
BCAT1 promoter methylation and RNA expression in gliomas
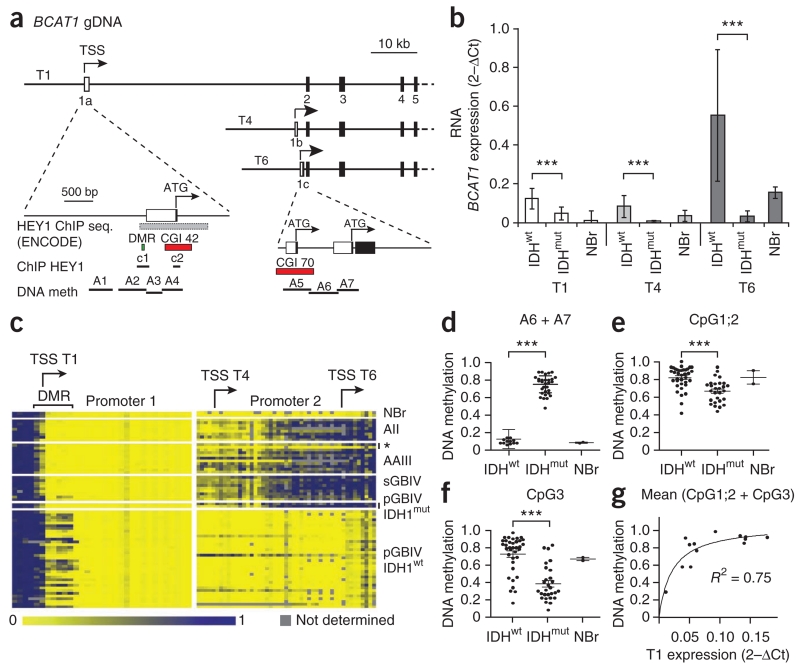
To better define BCAT1 expression patterns in IDHwt and IDHmut gliomas, we quantified transcript isoform expression in tumor samples from patients with glioma. Using RT-PCR and sequencing, we detected expression of three BCAT1 transcripts (T1, T4 and T6) coding for 386-, 398- and 385-residue proteins, respectively, in tumor and normal brain tissues (Supplementary Fig. 4). These transcripts, which differ only in their first exons, originate from two alternative promoters (Fig. 3a) and correspond to the single protein band of 43 kDa identified using western blot analysis. Transcript-specific qRT-PCR identified T6 as the predominant transcript in primary gliomas (Fig. 3b). Notably, expression of all BCAT1 transcripts (T1, T4 and T6) was significantly (P ≤ 0.001) higher in IDHwt compared to IDHmut tumors. Using sequence analysis of the BCAT1 promoter and coding regions from 20 gliomas, we found no nonsense or missense mutations that might affect BCAT1 expression levels in the patient tumors (Supplementary Table 3).
Figure 3.
Expression levels of the three BCAT1 transcripts are associated with differential methylation of two alternative promoters. (a) Schematic drawing of exons 1–5 of BCAT1 showing the exon structure of the three transcripts, T1, T4 and T6. The two alternative promoters, 1 and 2 with exons 1a and 1c, respectively, are shown in the enlarged sections on the lower left and lower right, respectively. gDNA, genomic DNA; TSS, transcription start site; seq., sequence; meth, methylation; CGI, CpG island; c1 and c2, amplicons for HEY1 chromatin immunoprecipiation assay. (b) qRT-PCR quantification of the RNA expression of transcripts T1, T4 and T6 in astrocytic gliomas (13 IDHwt and 18 IDHmut) and a pool of RNAs from normal brain tissues (23 NBr). Values represent the mean ± s.d. of triplicate samples. ***P < 0.001 (Student’s t test). (c) DNA methylation patterns detected in BCAT1 promoters 1 (left) and 2 (right) by MassARRAY analysis of bisulfite PCR amplicons A1–A7 in normal brain and astrocytic gliomas WHO grade 2–4. The asterisk indicates IDHwt anaplastic astrocytoma. (d–f) Extent of DNA methylation in NBr, IDHwt and IDHmut tumors in the average of all CpGs in promoter 2 (d; ***P < 0.0001), CpG1;2 (e; ***P = 0.0003) and CpG3 in promoter 1 (f; ***P < 0.0001). Statistical significance in d–f was calculated by Student’s t test. (g) Correlation of the average methylation grades of CpG1;2 and CpG3 and BCAT1 T1 expression.
We next explored the possibility that epigenetic silencing might account for BCAT1 transcript expression differences in tumors of patients with glioma. By analyzing patterns of BCAT1 promoter methylation with MassARRAY, we identified distinct methylation patterns in IDHwt and IDHmut (Fig. 3c). Promoter 2 showed the typical pattern of IDHmut-dependent methylation, as it was hypermethylated in IDHmut tumors but was mostly unmethylated in IDHwt tumors and normal brain (Fig. 3d). Hypermethylation in IDHmut tumors strongly correlated with BCAT1 downregulation. Notably, promoter 1 showed the opposite pattern, as it was hypermethylated in IDHwt tumors, particularly in the first three CpG dinucleotides of a small differentially methylated region (DMR) (Fig. 3e,f). The average degree of methylation of the three CpGs correlated with BCAT1 transcript T1 expression in IDHwt tumors, supporting its functional relevance (Fig. 3g). The observed CpG-specific methylation pattern suggests repressor binding to the unmethylated DMR, leading to the downregulation of the BCAT1 transcript T1 in IDHmut tumors. Consistent with HEY1 repressor activity, overexpression of HEY1 reduced transcript T1 expression, and chromatin immunoprecipitation analysis provided evidence for binding of HEY1 to the DMR in glioblastoma cells (Supplementary Fig. 5).
BCAT1 suppression reduces glutamate release and migration
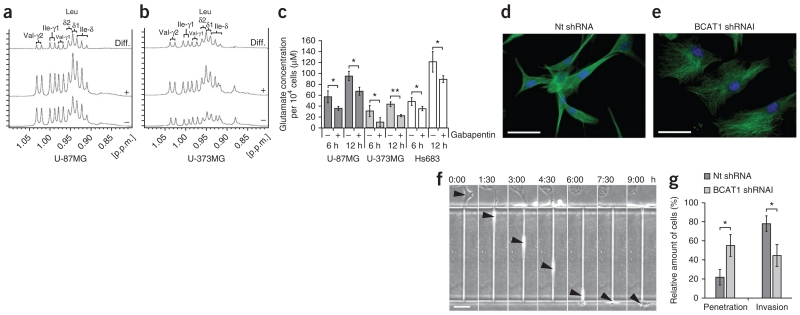
To gain additional insights into the functional role of BCAT1 in glioblastomas, we treated cells from the U-87MG and U-373MG lines with gabapentin, a leucine analog that specifically inhibits BCAT1 but not BCAT2 (ref. 41). 1H nuclear magnetic resonance (1H-NMR) spectroscopy of cell extracts that had been treated with 20 mM gabapentin for 20 h showed intracellular accumulation of BCAAs, which is consistent with BCAT1 inhibition (Fig. 4a,b). Glioblastomas release high concentrations of glutamate, leading to neuronal death by an excitotoxic mechanism42. Glutamate release was significantly reduced by inhibition of BCAT1 using gabapentin (Fig. 4c), indicating that BCAT1 is a major contributor to glutamate production through BCAA catabolism in IDHwt glioma. We analyzed the additional effects of BCAT1 suppression on amino acid metabolism by tandem mass spectrometry (MS/MS) after shRNA-mediated knockdown targeting all three BCAT1 transcripts (Supplementary Fig. 6). The observed changes in amino acid concentrations largely paralleled those reported in the plasma of Bcat2 knockout mice40. In Bcat2 knockout mice, inhibition of BCAA catabolism results in high concentrations of BCAAs and higher rates of protein synthesis in peripheral tissues through the mechanistic target of rapamycin (mTOR) signaling pathway and compensatory increased protein degradation (increased protein turnover). However, BCAT1 suppression did not cause increased phosphorylation of mTOR targets in glioma cells, perhaps because of the already high constitutive amounts of phosphorylation (Supplementary Fig. 7).
Figure 4.
BCAT1 suppression reduces glutamate release by glioma cells and limits glioblastoma cell invasion potential. (a,b) BCAA methyl group region of the 1H-NMR spectra obtained from extracts of U-87MG (a) and U-373MG (b) cells after treatment with solvent control (−) or 20 mM gabapentin (+) for 20 h. The horizontal axis represents the resonance frequency in p.p.m. relative to the trimethylsilyl propionate (TSP) reference. The vertical axis represents absolute signal intensities scaled to a constant cell count. Difference (diff.) spectra (treatment minus control) are shown at the top. After inhibitor treatment, the levels of valine (Val), leucine (Leu) and isoleucine (Ile) increased by factors of 1.09, 1.38, 1.19, respectively, in U-87MG cells and by factors of 1.83, 2.18 and 2.32, respectively, in U-373MG cells. δ values indicate chemical shifts, and γ values indicate gyromagnetic ratios. (c) Glutamate release by glioma cells at 6 and 12 h after the start of treatment with 20 mM gabapentin for BCAT1 inhibition (+) or solvent control (20 mM HEPES; −). Values represent the mean ± s.d. for n = 3 replicates. *P < 0.05, **P < 0.01 compared to the respective controls (Student’s t test). (d,e) Immunofluorescence labeling of tubulin (green) in control (d) and BCAT1 knockdown (e) U-87MG cells. Blue, DAPI. Scale bars, 50 μm. (f) Sequential images showing the permeation of a U-87MG cell (arrowheads) through a 5 μm × 11 μm × 300 μm microchannel over a period of 9 h. Scale bar, 100 μm. The labels above the images show the time elapsed as hours:minutes. (g) Effect of BCAT1 knockdown on the invasion potential of U-87MG cells compared to cells transduced with nontargeting shRNA. Results indicate the mean ± s.d. of three independent replicates (*P = 0.0146, Student’s t test).
Hydroxyacyl-CoA dehydrogenase (HADH) is downstream of BCAT1 in the BCAA catabolic pathway and is also an essential component of fatty-acid catabolism (beta oxidation) in the mitochondria. We found both pathways to be upregulated in IDHwt tumors relative to normal brain (Supplementary Fig. 1). After BCAT1 knockdown in glioblastoma cell lines, we observed decreased HADH protein concentrations (Supplementary Fig. 6e), providing additional evidence for the downregulation of the BCAA catabolic pathway, and concurrent accumulation of C4 short-chain fatty acids (Supplementary Fig. 6f), consistent with obstruction of beta oxidation through HADH suppression43.
We observed that BCAT1 knockdown strongly affected cellular morphology, resulting in a rounded, less extended appearance (Fig. 4d,e). To test whether these morphologies correlate with the ability of a tumor cell to invade adjacent tissues, we used a microchannel migration chip to simulate a three-dimensional environment (Fig. 4f). After BCAT1 knockdown, a majority of U-87MG cells (55%) penetrated only short distances into the microchannels and were unable to deform to completely invade the microchannels, whereas a vast majority of control cells (78%) completely invaded the channels (Fig. 4g and Supplementary Videos 1 and 2).
Glioblastoma growth is BCAT1 dependent
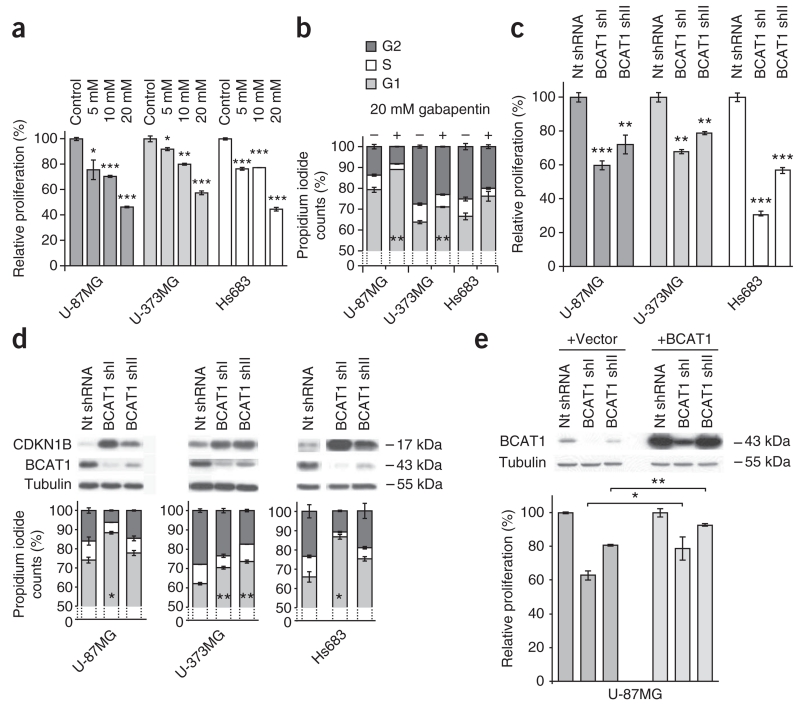
To further assess the impact of BCAT1 function on tumor cell proliferation, we performed BCAT1 inhibition and knockdown experiments. We found a concentration-dependent ~56% reduction of proliferation, estimated based on 5-ethynyl-2′-deoxyuridine (EdU) incorporation, when we treated glioma cells with the inhibitor gabapentin (Fig. 5a). Cell cycle analyses indicated that gabapentin treatment induced partial G1 arrest, as shown by an increased fraction of cells in the G1 phase along with concurrent decreases of the G2 and S fractions (Fig. 5b). shRNA-mediated BCAT1 knockdown elicited similar effects, decreasing proliferation by 20–70% in all three cell lines (Fig. 5c) and leading to G1 arrest and strong increases in cellular CDKN1B (also known as p27KIP1) concentrations (Fig. 5d). Notably, the degree of CDKN1B accumulation showed a positive correlation with the size of the G1 fraction (Fig. 5d). Cell death, as indicated by the fraction of cells in the sub-G1 phase, remained below 5%, except in the case of Hs683 cells, which showed a moderate increase (Supplementary Table 4). We observed comparable reductions of cell proliferation after BCAT1 knockdown in glioblastoma stem-like spheroid cultures in serum-free medium, although these cells showed a higher rate of apoptosis as determined by annexin V and 7-aminoactinomycin D (7-AAD) staining (Supplementary Fig. 8). Ectopic overexpression of BCAT1 in U-87MG BCAT1 knockdown cells significantly increased their proliferation rates and resulted in a partial rescue of the proliferation phenotype (Fig. 5e). When we induced cell cycle arrest using means other than BCAT1 knockdown, the metabolic effects (Supplementary Fig. 9) did not mirror the effects observed in the BCAT1 knockdown cells, indicating that the previously observed metabolic changes (Supplementary Fig. 6a–f) were specific effects of BCAT1 suppression and not secondary effects of cell cycle arrest.
Figure 5.
BCAT1 is essential for glioblastoma progression. (a–e) Cell proliferation using the Click-iT EdU cell proliferation assay and cell cycle analyses performed using DNA staining with propidium iodide of glioma cells after suppression of BCAT1 expression. The DNA distribution is shown for living cells. (a,b) Proliferation analysis (a) and cell cycle distribution (b) of glioma cell lines after BCAT1 inhibition with gabapentin for 20 h relative to control cells treated with solvent only. (c,d) Proliferation analysis (c) and cell cycle analysis (d) of glioma cells after knockdown of BCAT1 with two different shRNAs relative to cells treated with nontargeting shRNA. The western blots show protein expression of the G1 arrest marker CDKN1B, BCAT1 and the loading control tubulin. (e) Western blot and proliferation analyses of U-87MG cells treated with two BCAT1-targeting shRNAs or nontargeting shRNA after co-transduction with empty pLVX vector or the pLVX-BCAT1 vector, respectively. Values in the graphs (a–e) represent the mean ± s.d. for n = 3 replicates. *P < 0.05, **P < 0.01, ***P < 0.001 compared to the respective controls (Student’s t test).
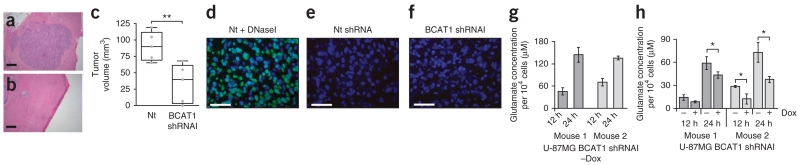
We evaluated the effect of BCAT1 knockdown on tumor growth in vivo using intracerebral transplantation of U-87MG cells into CD-1 nude (nu/nu) mice (Fig. 6a–c). Four weeks after transplantation of equal numbers of living cells, all six control mice transplanted with nontargeting shRNA–transduced cells but only one of six mice transplanted with BCAT1 shRNA–transduced cells showed neurologic symptoms such as lethargy or uncoordinated motor activities. H&E staining of mouse brain sections revealed large tumors in the mice transplanted with control cells (Fig. 6a), whereas we found markedly smaller tumors in the mice transplanted with BCAT1 knockdown cells (Fig. 6b). Quantitative analysis confirmed significant differences in tumor volume between the two groups (Fig. 6c; P = 0.0091). TUNEL assays did not detect DNA fragmentation in either control or BCAT1 knockdown tumors (Fig. 6d–f), indicating that the growth of BCAT1 knockdown tumors in vivo is limited mostly by reduced proliferation rather than increased apoptosis rates. We obtained equivalent results when we used a Tet-inducible BCAT1 shRNA vector for BCAT1 knockdown and rescue in vitro and for monitoring reduced growth of xenograft tumors in which we induced BCAT1 knockdown 10 d after implantation (Supplementary Fig. 10). We isolated cells from the tumors of mice transplanted with BCAT1 shRNA–transduced cells that had not been treated with doxycycline and removed contaminating mouse cells by puromycin selection. Analysis of glutamate concentrations in the supernatant showed that after passage through the mouse, glioblastoma cells still excreted high amounts of glutamate (Fig. 6g) in a BCAT1-dependent manner (Fig. 6h), strongly indicating that it was maintained in vivo.
Figure 6.
BCAT1 knockdown affects tumor growth in vivo. (a,b) Cross-sections of tumors 28 d after intracranial injection of U-87MG glioblastoma cells into CD-1 nu/nu mice. H&E staining is shown for mice injected with control nontargeting shRNA–transduced (a) or BCAT1 shRNAI–transduced (b) cells. Scale bars, 1 mm. (c) Quantification of tumor volumes (n = 5 mice for each group; **P = 0.0091, Student’s t test). (d–f) TUNEL staining of apoptotic cells (green) and nuclear counterstaining (blue) of representative orthotopic tumors from xenograft mice. (d) Brain sections treated with DNaseI, which served as positive control. (e,f) TUNEL staining of tumors that developed when mice were injected with control cells (e) or cells transduced with BCAT1 shRNAI (f). Scale bars, 50 μm. (g) Glutamate release by U-87MG cells 36 h after their explantation from xenograft tumors. Cells had been stably transduced with pLKO-Tet-On-BCAT1 shRNAI and were isolated 25 d after cranial implantation into CD1 nu/nu mice that did not receive doxycycline (−Dox). (h) Glutamate excretion by the same cells as those in g after 5 d in normal medium (−) or after doxycycline-induced knockdown of BCAT1 expression (+; 2 μg ml−1 doxycycline in cell culture medium). Values (g,h) represent the mean ± s.d. of quadruplicate samples. *P < 0.05 (Student’s t test).
DISCUSSION
Although it has been suggested that cancer cell proliferation is fueled primarily through the catabolism of glucose and glutamine10,44, the results presented here provide evidence that IDHwt glioma cells require catabolism of BCAAs to sustain an aggressive growth phenotype. Notably, with the exception of a small number of earlier reports45–49 and a recent in vitro metabolome analysis50, studies of BCAA metabolism have been restricted to nutrition science and glutamate neurotransmitter metabolism in the brain1,7,8. In the context of oncology research, the role of BCAT1 or BCAA catabolism in tumor growth has been largely overlooked.
The first crucial insight gleaned from our data is that although BCAT1 is clearly overexpressed in IDHwt gliomas, it is not expressed in the majority of IDHmut gliomas, implying a possible mechanistic link between these metabolic enzymes. IDH1 is a major source of α-KG39, which we showed to have a direct effect on the levels of BCAT1 expression. From this observation, one might be led to conclude that the observed BCAT1 downregulation in IDHmut cells could be explained in part by the depletion of α-KG in these cells. However, the partial reduction in the amounts of α-KG in IDHmut cells reported by others51 is insufficient to explain the near total absence of BCAT1 expression we observed in IDHmut tumors. Furthermore, we experimentally excluded direct inhibition of BCAT1 enzymatic activity by the oncometabolite 2-HG, as has been described for epigenetic modifiers32,33,35. This is consistent with the observation that hydroxy acids are not BCAT substrates52, as they cannot form a Schiff base with the vitamin B6 cofactor. We did, however, observe extensive DNA hypermethylation within the main promoter region of BCAT1 in IDHmut tumors that was lacking in IDHwt tumors or normal brain. These correlative data suggest that BCAT1 is silenced at least in part through IDH1mut-dependent DNA methylation.
In testing this hypothesis, we found that ectopic expression of IDH1mut in immortalized astrocytes38 resulted in a nearly 40% downregulation of BCAT1 expression, thus establishing a link between IDH1 mutation status and BCAT1 expression. However, it should be noted that IDHmut expression did not result in hypermethylation of the BCAT1 promoter in immortalized astrocytes, implying that DNA methylation might not have an initial regulatory role in BCAT1 suppression and might instead be a later event in BCAT1 silencing in IDHmut tumors.
In our study, suppression of BCAA catabolism by knockdown of BCAT1 resulted in reduced cell proliferation in vitro and in vivo. The mechanisms through which these metabolic changes affect glioblastoma cell proliferation are not known; however, we hypothesize that the blockage of BCAA and fatty acid catabolism (Supplementary Fig. 6g) might lead to reduced flow of acetyl-CoA and succinyl-CoA into the tricarboxylic acid cycle and thereby limit the ability of the cells to synthesize the macromolecules required for cell division. A recent study also demonstrated a clear link between amino acid metabolism and tumor aggressiveness in breast cancer, where growth of estrogen receptor (ER)− and human epidermal growth factor receptor 2 (HER2)− tumors was shown to be dependent on upregulation of the serine synthesis pathway53,54. Furthermore, higher expression of the glycine biosynthesis pathway has been found to be associated with greater mortality in patients with breast cancer55.
Although we have established the importance of BCAT1 for tumor growth, our finding that BCAT1 overexpression is a highly specific feature of IDHwt glioblastomas also makes BCAT1 a promising potential marker for diagnostic and prognostic assessment of gliomas, including the nearly 10% of IDHmut gliomas with mutations other than IDH1 R132H56 that are not amenable to IDH1 R132H diagnostic staining57.
In addition, BCAT1 and BCAA metabolism could provide a basis for the development of new metabolism-based targeted approaches in glioma therapy. Blocking BCAA catabolism through selective inhibition of BCAT1 would, in addition to reducing tumor growth by interfering with tumor energy production and macromolecule synthesis, have the added benefit of reducing tumor glutamate excretion. This latter effect could potentially reduce neurotoxic damage in surrounding brain tissue42, alleviate tumor-associated epilepsy58 in patients with brain tumors and perturb the tumor redox balance by inhibiting glutathione production42. We believe that the results of this study should greatly encourage the development of BCAT1 inhibitors for tumor therapy.
ONLINE METHODS
RNA isolation and qRT-PCR
Total RNA was extracted using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen) according to the manufacturer’s instructions. FirstChoice Human Brain Reference Total RNA from Ambion served as the normal brain RNA pool (n = 23 donors). Total RNA (500 ng) was reverse transcribed using random primers and superscript II (Invitrogen) according to the manufacturer’s instructions. Each cDNA sample was analyzed in triplicate with the Applied Biosystems Prism 7900HT Fast Real-Time PCR System using Absolute SYBR Green ROX Mix (ABgene). The relative amount of specific mRNA was normalized to ARF1, B2M and TBP mRNA. Primer sequences are given in Supplementary Table 5.
Western blot analysis
All experiments involving the use of human tissues were carried out in line with the guidelines of the institutional review board of the Medical Faculty at Heinrich Heine University Düsseldorf. Fresh frozen human tumor tissue samples and a non-neoplastic brain tissue sample were lysed in guanidine isothiocyanate solution (4 M) using an ULTRA-TURRAX (IKA) and subjected to cesium chloride (CsCl) ultracentrifugation. Diafiltration of the obtained protein fractions was performed using Amicon Ultra-0.5 centrifugal filter devices (Millipore; 3-kDa cutoff) essentially as described59. Total protein of cell lines was extracted using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen). Ten micrograms of protein was separated by 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Millipore). The membrane was blocked in blocking solution (5% BSA in TBS and 0.1% Tween 20) and incubated with primary antibodies overnight at 4 °C. Horseradish peroxidase (HRP)-conjugated secondary antibodies were incubated for 1 h at room temperature before chemiluminescent detection of protein (ECL kit, GE Healthcare).
The following antibodies were used: monoclonal mouse antibody to α-tubulin (1:2,000, clone DM1A, #T9026, Sigma-Aldrich), monoclonal mouse antibody to BCAT1 (1:2,000, ECA39, clone 51, #611270, BD Biosciences), monoclonal rat antiserum to IDH1 (1:2, generously provided by A. von Deimling), monoclonal mouse antibody to HADH (1:500, #H00003033-M01, Abnova), monoclonal rabbit antibody to CDKN1B (1:1,000, p27Kip1, #3686, Cell Signaling Technology), polyclonal rabbit antibody to p70 S6 kinase (1:1,000, #9202, Cell Signaling Technology), polyclonal rabbit antibody to phosphorylated p70 S6 kinase (Thr389) (1:1,000, #9205, Cell Signaling Technology), polyclonal rabbit antibody to 4E-BP1 (1:1,000, #9452, Cell Signaling Technology), polyclonal rabbit antibody to phosphorylated 4E-BP1 (Thr37/46) (1:1,000, #9459, Cell Signaling Technology), monoclonal rabbit antibody to phosphorylated S6 ribosomal protein (Ser235/236) (1:1,000, #4858, Cell Signaling Technology), HRP-conjugated antibody to rat IgG (1:10,000, generously provided by A. von Deimling), HRP-conjugated horse antibody to mouse IgG (1:5,000, #7076, Cell Signaling Technology) and HRP-conjugated goat antibody to rabbit IgG (1:2,000, #7074, Cell Signaling Technology).
Immunohistochemical staining
Tumor sections were deparaffinized using xylol and rehydrated in decreasing concentrations of ethanol. Antigen retrieval was performed by heating for 40 min in a steamer (90–95 °C) in 10 mM sodium citrate buffer (pH 6.0). Endogenous peroxidase was inactivated by incubating the tumor sections in 3% hydrogen peroxide for 10 min. Sections were incubated overnight with primary mouse monoclonal antibody to BCAT1 (ECA39, clone 51, #611270, BD Biosciences) diluted 1:500 or mouse antibody to human R132H IDH1 (generously provided by A. von Deimling) diluted 1:20 in Dako REAL antibody diluent (Dako). Staining for the detection of bound antibody was performed according to standard protocols using the EnVision detection system (peroxidase/DAB+, rabbit/mouse) (Dako), and subsequent counterstaining was done using hematoxylin.
Mutation analysis
The BCAT1 gene (ENSG00000060982) was investigated for mutations in 20 tumors using the primers shown in Supplementary Table 5. All 11 exons and 1,000 bp of the promoter region of the BCAT1 transcript variant T1 (ENST00000261192) plus 1,700 bp of the promoter region of the transcript isoform T6 (ENST00000538118), including the two alternative exons (1b and 1c of the transcript isoforms T4 and T6), were amplified by PCR using GoTaq polymerase (Promega). PCR products were treated with ExoSAP (ExoSAP-IT, GE Healthcare) and sequenced in both directions using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems).
DNA methylation analysis
DNA methylation was analyzed by the MassARRAY technique as previously described60. Briefly, 500 ng of genomic DNA was chemically modified with sodium bisulfite using the EZ methylation Kit (Zymo Research) according to the manufacturer’s instructions. The bisulfite-treated DNA was PCR amplified with primers generating four amplicons (A1–A4) from −990 bp to +612 bp around the transcription start site of the BCAT1 transcript T1 (ENST00000261192) and three amplicons (A5–A7) of the promoter of the BCAT1 transcripts T4 (ENST00000539282) and T6 (ENST00000538118) from −198 bp to +63 bp. The amplicons were transcribed by T7 polymerase, followed by thymidine-specific cleavage using RNase A. The digested fragments were quantified by mass spectrometry. The primer sequences are given in Supplementary Table 5. DNA methylation standards (0%, 20%, 40%, 60%, 80% and 100% methylated genomic DNA) were used to confirm the unbiased amplification of the amplicons.
Cell culture
The human glioma cell lines U-87MG (HTB-14), LN-229 (CRL-2611), Hs683 (HTB-138) and U-373MG (HTB-17) were cultured in DMEM containing 4.5 g l−1 glucose (DMEM) supplemented with 10% FCS (Biochrom #S0115) and a 1% penicillin and streptomycin mix. For all cells transduced with Tet (tetracycline)-inducible vectors, doxycycline-free serum (Clonetech) was used. Cell lines were authenticated by short tandem repeat analysis. Glioblastoma stem-like (NCH421k) cells were established at the Division of Experimental Neurosurgery, University of Heidelberg, Germany. They were characterized genotypically and phenotypically in a previous study61. NCH421k cells were grown as floating aggregates (neurospheres) on uncoated tissue culture dishes in DMEM and F-12 medium containing 20% BIT serum-free supplement, basic fibroblast growth factor and epidermal growth factor at a concentration of 20 ng ml−1 each (all from Provitro). HEK293T and HEK293 cells were maintained as monolayer cultures in DMEM containing 10% FCS without antibiotics. IHAs expressing wild-type IDH1 or R132H mutant IDH1 were provided by T.A.C. and prepared as previously described62. They were cultured in DMEM supplemented with 10% FCS, 0.5 μg ml−1 G418 and 1% penicillin and streptomycin mix.
All cell lines were cultivated at 37 °C in a humidified incubator with 5% CO2. For hypoxia experiments, cells were cultured at 1% O2 in a nitrogen-supplied C16 hypoxia incubator for 24 h (Labotect).
Cloning of lentiviral vectors
To generate inducible shRNA-expressing lentiviral constructs, double-stranded oligonucleotides encoding the desired shRNAs were cloned into the single vector–inducible shRNA construct pLKO-Tet-On using the AgeI and EcoRI restriction sites. Briefly, reconstituted sense and antisense oligonucleotides were mixed with 10× Oligo Annealing Buffer (Invitrogen), incubated at 95 °C for 4 min and left in the turned off heat block overnight. Double-stranded oligonucleotides (0.1 nmol μl−1) were cloned into the predigested and purified pLKO-Tet-On vector using the Roche Rapid Ligation Kit following the manufacturer’s instructions. The sense and antisense oligonucleotide sequences are given in Supplementary Table 5.
For overexpression of BCAT1, the insert was generated using PCR with the primers shown in Supplementary Table 5 and subsequently cloned into the XbaI and XhoI sites of the pLVX-puro lentiviral vector (Clontech).
Virus production and transduction
Lentiviral particles were produced by cotransfection of HEK293T cells with the psPAX2 (Addgene 12260, Didier Trono; packaging plasmid), pMD2.G (Addgene 12259, Didier Trono; envelope plasmid) and respective pLKO.1 shRNA constructs (Sigma-Aldrich), inducible pLKO-Tet-On shRNA constructs or the pLVX-puro overexpression vector (Clontech). Transfections were carried out using TransIT-LT1 (Mirus Bio LLC), and viruses was harvested at 48 and 72 h after transfection and combined.
Infection of glioma cells with virus at a multiplicity of infection of 2 was carried out in the presence of 8 μg ml−1 of polybrene (Chemicon). Virus-containing supernatant was removed after 24 h, and cells were split 3 d, 5 d and 7 d after transduction.
Two shRNA constructs (TRCN0000005907 and TRCN0000005909) targeting different regions of human BCAT1 and two shRNA constructs (TRCN0000027298 and TRCN0000027289) targeting different regions of human IDH1 mRNA transcripts were used. Nontargeting shRNA (Mission SHC002) was used as control.
Inducible BCAT1 knockdown cells were established by infecting U-87MG and U-373MG cells with pLKO-Tet-On nontargeting shRNA and pLKO-Tet-On BCAT1 shRNAI lentiviral particles, respectively, and selecting with puromycin (1 μg ml−1) for 7 d. Induction was achieved by adding 2 μg ml−1 doxycycline to the medium for at least 5 d. This inducible lentiviral system was previously described63.
Quantification of knockdown or overexpression was assessed by qRT-PCR and western blot analysis.
Treatment with dimethyl–α-KG and gabapentin
Fifty thousand cells per well were seeded in 24-well plates in a total volume of 500 μl cell culture medium. The medium was removed 16 h after seeding the cells and replaced with 500 μl medium containing 1 mM, 5 mM or 10 mM dimethyl–α-KG (dimethyl 2-oxoglutarate; Sigma-Aldrich) or 5 mM, 10 mM or 20 mM gabapentin (1-(aminomethyl)-cyclohexane; Sigma-Aldrich). Corresponding volumes of 200 mM HEPES buffer (pH 7.4) were added to the respective control wells.
Cell cycle analysis and detection of apoptosis
Cell cycle analysis was performed 20 h after treatment with gabapentin and 6 d after lentiviral transduction. Nicoletti buffer (0.1% sodium citrate, pH 7.4, 0.05% Triton X-100 and 50 μg ml−1 propidium iodide) was added to the wells containing both dead and living cells. After 4 h in the dark at 4 °C, DNA content was analyzed by flow cytometry using a FACS Canto II (BD Biosciences). FACS Diva software was used to quantify the distribution of cells in each cell cycle phase: sub-G1 (dead cells), G1, S and G2/M.
For investigation of apoptotic activity after BCAT1 lentiviral knockdown, NCH421k cells were incubated with annexin V–phycoerythrin (PE) and 7-AAD (BD Biosciences) for 15 min in the dark, which was immediately followed by flow cytometry.
Proliferation analysis
To assess the proliferation of glioma cells after treatment with gabapentin or after lentiviral knockdown, the Click-iT EdU cell proliferation assay (Invitrogen) was used following the manufacturer’s instructions. The cells were incubated with 10 μM of the nucleoside analog EdU for 10–16 h. Quantification of cells that incorporated EdU was performed using a FACS Canto II (BD Biosciences).
Synchronization of cells by double thymidine block
U-87MG cells were synchronized at the G1/S boundary by culturing them in the presence of 2 mM thymidine (Sigma-Aldrich) in DMEM plus 10% FCS for 16 h. They were washed with PBS and allowed to recover in complete growth medium that lacked thymidine for 11 h and then propagated again in 2 mM thymidine for an additional 14 h.
Glutamate quantification
Glutamate concentration in the supernatants of cells was determined using the Glutamine and Glutamate Determination Kit (GLN-1, Sigma-Aldrich) according to the manufacturer’s instructions. The reaction volumes were scaled down to 100 μl total volume, and absorbance was measured in triplicate in a microplate (Corning 96 Well Clear Flat Bottom UV-Transparent Microplate) using a Tecan Infinite M200 plate reader (Tecan). The data were normalized to the number of cells per well.
Immunofluorescence staining
Cells were plated on glass coverslips 5 d after lentiviral transduction. Cells were fixed in 4% formaldehyde, rinsed twice in PBS and permeabilized in PBS containing 0.2% Triton X-100. After rinsing with PBS, cells were incubated in 10% goat serum for 30 min at room temperature. Samples were incubated for 1 h with the primary antibody (mouse antibody to α-tubulin, 1:200, clone DM1A, #T9026, Sigma-Aldrich) and for 1 h at room temperature with the secondary antibody (FITC-conjugated goat mouse-specific antibody, 1:100, ab6785, Abcam) after mounting with DAPI-containing Vectashield mounting medium (Vector Laboratories). For fluorescence imaging, images were taken using a ×40 objective lens on a Zeiss Axioplan microscope.
Three-dimensional microchannel migration assay
For migration assays, we used poly(dimethylsiloxane) (PDMS)-based microchannel chips (provided by R. Kemkemer). Microfabricated channel structures with the dimensions 5 μm × 11 μm × 300 μm (width × height × length) were biofunctionalized by incubation with a 50 μg ml−1 fibronectin solution before use. The chip was fixed on a Teflon holder, and 2 × 105 cells were seeded on the chip in close proximity to the channels. After cells were attached on the chip, live-cell imaging was performed for 25 h. During the experiments, no chemical gradient or flow inside the channels was applied. Phase-contrast time-lapse pictures of multiple positions were captured every 10 min with an automated inverted microscope (Zeiss Cell Observer, Carl Zeiss) equipped with an air-humidified and heated chamber. Images were recorded and processed with Zeiss AxioVision and ImageJ software. Cell behaviors were analyzed and categorized as reported previously64,65.
Animal experiments
Animal work for the orthotopic brain tumor model was approved by the governmental authorities (Regierungspraesidium Karlsruhe, Germany) and supervised by institutional animal protection officials in accordance with the US National Institutes of Health guidelines Guide for the Care and Use of Laboratory Animals. Protocols for animal use for the 2-HG inhibition studies were approved by the Animal Care Use Committee of Virginia Polytechnic Institute in accordance with the US National Institutes of Health guidelines.
Orthotopic brain tumor model
A total of 2 × 105 U-87MG cells with BCAT1 shRNAI knockdown or nontargeting shRNA were stereotactically implanted into the brains of six female 7- to 9-week-old athymic mice (CD-1 nu/nu; Charles River Laboratories). Four weeks after implantation, mice were euthanized, and their brains were removed and cryosectioned. Brain sections were stained with H&E, and tumor volume was calculated using ImageJ.
Inducible orthotopic brain tumor model
Two-hundred thousand U-87MG cells stably transduced with inducible pLKO-Tet-On-BCAT1 shRNAI were stereotactically implanted into the brains of 12 female 7- to 9-week-old CD-1 nu/nu mice. Ten days after cranial implantation, six mice received water containing 2 mg ml−1 doxycycline and 1.25% sucrose, and six mice were fed with water containing 1.25% sucrose only. After MRI at 18 and 25 d after implantation, mice were euthanized, and their brains were removed and cryosectioned, or tumor cells were isolated for quantification of glutamate release.
MRI
MRI scans were performed using a custom-developed transmit and receive small-animal coil in a conventional whole-body 1.5 T MRI scanner (Symphony, Siemens). T1-weighted imaging was performed using a high-resolution spin echo sequence (echo delay time, 600 ms; repetition time, 14 ms; field of view, 40 mm × 40 mm; matrix, 192; voxel size, 0.2 × 0.2 × 1 mm3) 5 min after intraperitoneal administration of 80 μl gadopentetic acid (Gd-DTPA) contrast agent (0.5 mmol ml−1; Magnevist, Bayer Schering Pharma).
Isolation of cells from xenograft tumors
After mice were euthanized, resected tumor pieces were immediately stored in sterile, cold PBS. Tumor pieces were washed thoroughly and minced in cold PBS. Tissue pieces were incubated in 1 ml trypsin at 37 °C and suspended in DMEM supplemented with 10% FCS and a 1% penicillin and streptomycin mix after 10 min. After collecting the pieces by centrifugation, this step was repeated twice, followed by seeding of cells into six-well plates. After 10 h of incubation at 37 °C, DMEM containing 2 μg ml−1 puromycin was added to eliminate contaminating mouse tissue.
TUNEL staining of brain sections
Fluorescent TUNEL staining was performed using the In situ Cell Death Detection Kit (Roche Diagnostic GmbH) according to the manufacturer’s instructions. Frozen brain sections were fixed in 4% paraformaldehyde for 20 min at room temperature, rinsed three times in PBS and permeabilized in 0.1% citrate buffer (pH 6.0) containing 0.1% Triton X-100 for 2 min on ice. After washing in PBS, the sections were incubated with a mixture of terminal deoxynucleotidyl transferase (TdT) enzyme solution and FITC-labeled deoxyuridine triphosphate (dUTP) solution in a humidified chamber in the dark at 37 °C for 60 min. After washing, the slides were mounted with DAPI-containing Vectashield mounting medium (Vector Laboratories). Slides were examined with a Zeiss Axioplan microscope, and images were taken using a ×40 objective lens. Negative controls were obtained by replacing the TdT solution with distilled water, and positive controls were obtained by incubation with 200 U ml−1 DNaseI for 10 min at room temperature before TdT solution treatment.
Perchloric acid cell extraction and 1H-NMR spectroscopy
For quantification of branched-chain amino acids after treatment with gabapentin or solvent, control glioma cells were harvested by trypsinization and washed once with ice-cold PBS, and the cell pellets were frozen at −80 °C. Typically, 1 × 108 harvested cells were subjected to perchloric acid extraction followed by KOH neutralization according to published protocols66. 1H-NMR spectra of the sample extracts and a calibration solution were acquired on a Bruker Avance AV-600 spectrometer (Bruker BioSpin GmbH) operating at a proton frequency of 600 MHz. A detailed description of the procedure is given in the Supplementary Methods.
Analysis of acylcarnitines and amino acids
For quantification of acylcarnitines and amino acids, glioma cells were harvested by trypsinization 8 d after lentiviral BCAT1 knockdown, washed with cold PBS and stored at −80 °C. Cell pellets were diluted in 100 μl water and homogenized by sonification (Ultrasonic device; 3–5 × 20 cycles, output 80%; Branson Sonifier 450). Protein concentration was determined according to the methods of Lowry et al.67 with the modifications of Helenius and Simons68 using BSA as a standard. The final protein concentrations of the homogenates were in a range of 2–4 mg ml−1.
The amounts of acylcarnitines and amino acids were determined in cell homogenates by electrospray ionization MS/MS (ESI-MS/MS) according to a modified method as previously described69 using a Quattro Ultima triple quadrupole mass spectrometer (Micromass) equipped with an electrospray ion source and a Micromass MassLynx data system. Therefore, 4 μl of the cell homogenates (total protein of 8–16 μg) were placed on a 4.7-mm filter paper punch, dried at room temperature overnight and extracted with 100 μl of deuterium-labeled standard solution in methanol69.
Preparation of mouse brain extracts and BCAT1 assay
Brain extracts were prepared from Bcat2 knockout mice70 essentially as described71 with modifications. The extraction buffer contained additional protease inhibitors, 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride (AEBSF), 800 nM aprotinin, 50 μM bestatin, 15 μM E-64, 10 μM pepstatin A and no di-isopropylfluorophosphate. After homogenization, the extract was centrifuged for 10 min at 13,000 r.p.m., and the brain supernatant was used to assay BCAT1 activity under non–maximal activity (Vmax) conditions72. The time was adjusted so that ≤25% of the substrate was consumed during the assay. The substrates branched-chain KIV and branched-chain isoleucine were used at concentrations of 0.1 mM and 1.2 mM, respectively. Competing α-keto acid substrates (KIC and α-KG) and the hydroxyl acid of α-KG (2-HG) concentrations ranged from 1 to 10 mM.
Statistical analyses
The relationship between IDH1 or IDH2 mutation status and BCAT1 protein expression was tested with the two-sided Fisher’s exact test. Student’s t test (two-tailed, unpaired) was used for all other statistical comparisons. *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Tödt for bioinformatic expertise, M. Kirchgäßner, K. Pfleger, E. Wieland, A.-C. Klein, S. Emmerich and the DKFZ Light Microscopy Facility for excellent technical support, C. Schmidt (DKFZ) for providing the HEY1-pDest-vectors and A. von Deimling (University of Heidelberg) for providing the IDH1-specific antibodies. This work was supported by the German Federal Ministry of Education and Research (BMBF) within the National Genome Research Network NGFNplus (01GS0883 and 01GS0884) to B.R., P.L. and G.R., the German Cancer Aid (Deutsche Krebshilfe, grant number 108456) to B.R. and P.L. and the intramural funding program of the National Tumor Center Heidelberg. Y.J.P. was supported by the Roman Herzog research fellowship from the Hertie Foundation, Germany, and the Korean Health Technology R&D Project, Ministry of Health and Welfare, Republic of Korea (A120071-1211-0000200).
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
M.T. and S.B. developed the concept, designed and performed experiments, analyzed data and wrote the manuscript. Y.J.P. and A.M.L. conducted the promoter methylation analysis, chromatin immunoprecipitation analysis and HEY1 knockdown. W.W. conducted dimethyl–α-KG and hypoxia experiments. M.S. assisted with experiments. S.V.P. conducted lentiviral knockdown in NCH421k cells. A.H.C.B. assisted with microchannel experiments. D.K. conducted western blot analysis of patient samples. R.M.P. and R. König mapped metabolic pathways. J.F. performed IDH1 mutation studies and immunohistochemical analyses. D.L. performed animal experiments. I.W., V.H., S.T., T.A.C., D.S., H.W. and S.M.P. assisted with the methylation analysis. A.A. and S.M.H. performed and interpreted BCAT1 inhibition experiments. C.G.R. and R. Kemkemer synthesized the microchannel chips. B.C. and C.H.-M. analyzed tissue microarray data. K.S. conducted the MS/MS and gas chromatography–mass spectrometry. W.-E.H. performed the 1H-NMR spectroscopy. M.J. performed magnetic resonance imaging. C.P. was involved in the methylation analysis. J.G.O. analyzed the mass spectrometry data. G.R. performed histological and immunohistochemical analyses and contributed to writing the manuscript. P.L. oversaw all research phases and contributed to writing the manuscript. B.R. developed the concept, supervised the project and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Additional methods
Detailed methodology is described in the Supplementary Methods.
References
- 1.Lieth E, et al. Nitrogen shuttling between neurons and glial cells during glutamate synthesis. J. Neurochem. 2001;76:1712–1723. doi: 10.1046/j.1471-4159.2001.00156.x. [DOI] [PubMed] [Google Scholar]
- 2.Hutson SM. The case for regulating indispensable amino acid metabolism: the branched-chain α-keto acid dehydrogenase kinase-knockout mouse. Biochem. J. 2006;400:e1–e3. doi: 10.1042/BJ20061506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ichihara A, Koyama E. Transaminase of branched chain amino acids. I. Branched chain amino acids–α-ketoglutarate transaminase. J. Biochem. 1966;59:160–169. doi: 10.1093/oxfordjournals.jbchem.a128277. [DOI] [PubMed] [Google Scholar]
- 4.Taylor RT, Jenkins WT. Leucine aminotransferase. II. Purification and characterization. J. Biol. Chem. 1966;241:4396–4405. [PubMed] [Google Scholar]
- 5.García-Espinosa MA, Wallin R, Hutson SM, Sweatt AJ. Widespread neuronal expression of branched-chain aminotransferase in the CNS: implications for leucine/glutamate metabolism and for signaling by amino acids. J. Neurochem. 2007;100:1458–1468. doi: 10.1111/j.1471-4159.2006.04332.x. [DOI] [PubMed] [Google Scholar]
- 6.Hall TR, Wallin R, Reinhart GD, Hutson SM. Branched chain aminotransferase isoenzymes. Purification and characterization of the rat brain isoenzyme. J. Biol. Chem. 1993;268:3092–3098. [PubMed] [Google Scholar]
- 7.Sweatt AJ, et al. Branched-chain amino acid catabolism: unique segregation of pathway enzymes in organ systems and peripheral nerves. Am. J. Physiol. Endocrinol. Metab. 2004;286:E64–E76. doi: 10.1152/ajpendo.00276.2003. [DOI] [PubMed] [Google Scholar]
- 8.Hutson SM, Sweatt AJ, Lanoue KF. Branched-chain [corrected] amino acid metabolism: implications for establishing safe intakes. J. Nutr. 2005;135(suppl. 6):1557S–1564S. doi: 10.1093/jn/135.6.1557S. [DOI] [PubMed] [Google Scholar]
- 9.Eagle H. Nutrition needs of mammalian cells in tissue culture. Science. 1955;122:501–514. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 10.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seltzer MJ, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70:8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C, et al. Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 2009;69:7986–7993. doi: 10.1158/0008-5472.CAN-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase–dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. USA. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balss J, et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 19.Ichimura K, et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro-oncol. 2009;11:341–347. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennan C, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS ONE. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colman H, et al. A multigene predictor of outcome in glioblastoma. Neuro-oncol. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 26.Toedt G, et al. Molecular signatures classify astrocytic gliomas by IDH1 mutation status. Int. J. Cancer. 2011;128:1095–1103. doi: 10.1002/ijc.25448. [DOI] [PubMed] [Google Scholar]
- 27.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartmann C, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120:707–718. doi: 10.1007/s00401-010-0781-z. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki M, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26:2038–2049. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki M, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang L, Jin S, Su SM. IDH mutations in glioma and acute myeloid leukemia. Trends Mol. Med. 2010;16:387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate–dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schramm G, et al. PathWave: discovering patterns of differentially regulated enzymes in metabolic pathways. Bioinformatics. 2010;26:1225–1231. doi: 10.1093/bioinformatics/btq113. [DOI] [PubMed] [Google Scholar]
- 37.Gravendeel LA, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 38.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J. Natl. Cancer Inst. 2010;102:932–941. doi: 10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.She P, et al. Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab. 2007;6:181–194. doi: 10.1016/j.cmet.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goto M, et al. Structural determinants for branched-chain aminotransferase isozyme-specific inhibition by the anticonvulsant drug gabapentin. J. Biol. Chem. 2005;280:37246–37256. doi: 10.1074/jbc.M506486200. [DOI] [PubMed] [Google Scholar]
- 42.Sontheimer H. A role for glutamate in growth and invasion of primary brain tumors. J. Neurochem. 2008;105:287–295. doi: 10.1111/j.1471-4159.2008.05301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasquali M, Monsen G, Richardson L, Alston M, Longo N. Biochemical findings in common inborn errors of metabolism. Am. J. Med. Genet. C. Semin. Med. Genet. 2006;142C:64–76. doi: 10.1002/ajmg.c.30086. [DOI] [PubMed] [Google Scholar]
- 44.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 45.de Bont JM, et al. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro-oncol. 2008;10:648–660. doi: 10.1215/15228517-2008-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goto M, Shinno H, Ichihara A. Isozyme patterns of branched-chain amino acid transaminase in human tissues and tumors. Gann. 1977;68:663–667. [PubMed] [Google Scholar]
- 47.Weggen S, et al. Identification of amplified genes from SV40 large T antigen–induced rat PNET cell lines by subtractive cDNA analysis and radiation hybrid mapping. Oncogene. 2001;20:2023–2031. doi: 10.1038/sj.onc.1204287. [DOI] [PubMed] [Google Scholar]
- 48.Yoshikawa R, et al. ECA39 is a novel distant metastasis-related biomarker in colorectal cancer. World J. Gastroenterol. 2006;12:5884–5889. doi: 10.3748/wjg.v12.i36.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou W, et al. Functional evidence for a nasopharyngeal carcinoma-related gene BCAT1 located at 12p12. Oncol. Res. 2007;16:405–413. doi: 10.3727/000000007783980873. [DOI] [PubMed] [Google Scholar]
- 50.Reitman ZJ, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc. Natl. Acad. Sci. USA. 2011;108:3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao S, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davoodi J, et al. Overexpression and characterization of the human mitochondrial and cytosolic branched-chain aminotransferases. J. Biol. Chem. 1998;273:4982–4989. doi: 10.1074/jbc.273.9.4982. [DOI] [PubMed] [Google Scholar]
- 53.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jain M, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartmann C, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 57.Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol. 2009;118:599–601. doi: 10.1007/s00401-009-0595-z. [DOI] [PubMed] [Google Scholar]
- 58.Buckingham SC, et al. Glutamate release by primary brain tumors induces epileptic activity. Nat. Med. 2011;17:1269–1274. doi: 10.1038/nm.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grzendowski M, et al. Simultaneous extraction of nucleic acids and proteins from tissue specimens by ultracentrifugation: a protocol using the high-salt protein fraction for quantitative proteome analysis. Proteomics. 2009;9:4985–4990. doi: 10.1002/pmic.200800902. [DOI] [PubMed] [Google Scholar]
- 60.Ehrich M, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. USA. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Campos B, et al. Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin. Cancer Res. 2010;16:2715–2728. doi: 10.1158/1078-0432.CCR-09-1800. [DOI] [PubMed] [Google Scholar]
- 62.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiederschain D, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8:498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 64.Bai AH, et al. MicroRNA-182 promotes leptomeningeal spread of non-sonic hedgehog-medulloblastoma. Acta Neuropathol. 2012;123:529–538. doi: 10.1007/s00401-011-0924-x. [DOI] [PubMed] [Google Scholar]
- 65.Rolli CG, Seufferlein T, Kemkemer R, Spatz JP. Impact of tumor cell cytoskeleton organization on invasiveness and migration: a microchannel-based approach. PLoS ONE. 2010;5:e8726. doi: 10.1371/journal.pone.0008726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sze DY, Jardetzky O. Determination of metabolite and nucleotide concentrations in proliferating lymphocytes by 1H-NMR of acid extracts. Biochim. Biophys. Acta. 1990;1054:181–197. doi: 10.1016/0167-4889(90)90240-e. [DOI] [PubMed] [Google Scholar]
- 67.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 68.Helenius A, Simons K. The binding of detergents to lipophilic and hydrophilic proteins. J. Biol. Chem. 1972;247:3656–3661. [PubMed] [Google Scholar]
- 69.Sauer SW, et al. Intracerebral accumulation of glutaric and 3-hydroxyglutaric acids secondary to limited flux across the blood-brain barrier constitute a biochemical risk factor for neurodegeneration in glutaryl-CoA dehydrogenase deficiency. J. Neurochem. 2006;97:899–910. doi: 10.1111/j.1471-4159.2006.03813.x. [DOI] [PubMed] [Google Scholar]
- 70.She P, et al. Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab. 2007;6:181–194. doi: 10.1016/j.cmet.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suryawan A, et al. A molecular model of human branched-chain amino acid metabolism. Am. J. Clin. Nutr. 1998;68:72–81. doi: 10.1093/ajcn/68.1.72. [DOI] [PubMed] [Google Scholar]
- 72.Hutson SM, et al. Role of branched-chain aminotransferase isoenzymes and gabapentin in neurotransmitter metabolism. J. Neurochem. 1998;71:863–874. doi: 10.1046/j.1471-4159.1998.71020863.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.