Abstract
BACKGROUND
The MUC1 heterodimeric oncoprotein is aberrantly overexpressed in human prostate cancers with more aggressive pathologic and clinical features. However, the signals that regulate MUC1 expression in prostate cancer cells are not well understood.
METHODS
MUC1 expression was studied in androgen-dependent and -independent prostate cancer cell lines by quantitative RT-PCR, immunoblotting and assessment of MUC1 promoter activation. Chromatin immunoprecipitation (ChIP) studies were performed to assess androgen receptor (AR) occupancy on the MUC1 promoter. Post-transcriptional regulation of MUC1 expression was assessed by miR-125b-mediated effects on activity of the MUC1 3′ untranslated region (3′UTR).
RESULTS
The present studies demonstrate that AR occupies a consensus AR element on the MUC1 promoter in androgen-dependent LNCaP, but not in androgen-independent DU145 and PC3, prostate cancer cells. The results further show that AR downregulates MUC1 gene transcription. Stable introduction of exogenous AR in PC3 (PC3/AR) cells and then silencing of AR confirmed AR-mediated repression of the MUC1 promoter. AR signaling has also been shown to drive miR-125b expression. The present studies further demonstrate that miR-125b suppresses MUC1 translation in LNCaP cells and that an anti-sense miR-125b upregulates expression of MUC1 protein. In addition, stable expression of miR-125b in DU145 cells resulted in decreases in MUC1 levels.
CONCLUSIONS
These findings demonstrate that AR signaling regulates MUC1 expression by transcriptional and posttranscriptional mechanisms in prostate cancer cells.
Keywords: prostate cancer, androgen-dependence, MUC1, androgen receptor, miR-125b
INTRODUCTION
The mucin 1 (MUC1) heterodimer localizes to the apical membrane of normal epithelial cells and is aberrantly overexpressed in diverse human carcinomas [1]. However, few insights are available regarding the mechanisms responsible for the regulation of MUC1 expression in carcinoma cells [1]. MUC1 is translated as a single polypeptide that is processed by autocleavage into two subunits that in turn form a heterodimer. The MUC1 N-terminal subunit (MUC1-N) contains highly glycosylated tandem repeats that are characteristic of mucin family members [1]. MUC1-N is positioned at the cell surface in a complex with the MUC1 C-terminal transmembrane subunit (MUC1-C) [1]. Release of MUC1-N from the heterodimer leaves MUC1-C as a potential transmembrane receptor for signaling stress to the interior of the cell [1]. In this context, the MUC1-C subunit associates with receptor tyrosine kinases at the cell membrane and localizes to the cytoplasm and nucleus [1]. In the nucleus, MUC1-C interacts with the transcription factors NF-κB p65 and STAT1/3, and promotes activation of the MUC1 gene in auto-inductive loops [2,3]. MUC1-C also interacts with the Wnt effector β-catenin and with p53, and thereby regulates expression of their target genes [4,5]. In carcinomas, MUC1-C contributes to the activation of specific gene families that are involved in oncogenesis and are associated with decreased patient survival [6–8]. In this capacity, MUC1-C overexpression is sufficient to confer anchorage-independent growth and tumorigenesis [4,9]. In addition to its induction at the transcriptional level, MUC1 expression is downregulated posttranscriptionally by miR-145 [10], miR-125b [11], and miR-1226 [12]. These findings have suggested that increases in MUC1 expression associated with progression of normal epithelia to carcinomas are controlled by both transcriptional and posttranscriptional mechanisms.
Overexpression of MUC1 in prostate cancer has been associated with more aggressive disease [13–18]. In this regard, MUC1 expression was detectable by immunohistochemical staining in ~90% of primary prostate cancers that were Gleason grade ≥7 or were metastatic to lymph nodes [13,14]. Gene expression profiling has also shown that MUC1 is highly expressed in prostate cancers with aggressive clinicopathologic characteristics and an increased risk of disease recurrence [19]. Prostate cancer cells are dependent on androgen receptor (AR) activation for their growth and survival [20]. Thus, treatment of androgen-dependent prostate cancer with surgical castration or leuteinizing hormone-releasing hormone (LHRH) agonists results in disease regression. However, these treatments to abrogate androgen action eventually fail as prostate cancer progresses to a castration-resistant or androgen-independent form of the disease. The progression to androgen-independent prostate cancer occurs in the presence of continued AR activation by mechanisms that include, among others, AR gene amplification and mutations, production of AR ligands by prostate cancer cells, and interactions with other signaling pathways [20–22]. Notably, recent work has shown that blocking MUC1-C function is associated with loss of androgen-independent, but not androgen-dependent, prostate cancer cell survival [23]. There is, however, no known direct interaction between AR and MUC1 signaling in androgen-dependent or -independent prostate cancer.
The present studies demonstrate that AR occupies the MUC1 promoter and downregulates MUC1 transcription. The results also demonstrate that AR decreases MUC1-C subunit protein levels by a miR-125b-mediated pathway. These findings support a model in which AR regulates MUC1 expression by both transcriptional and posttranscriptional mechanisms.
MATERIALS AND METHODS
Cell Culture
Human LNCaP, CWR22Rv1, PC3 and DU145 prostate cancer cells (ATCC) were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum, 100 μg/ml streptomycin, 100 units/ml penicillin and 2 mM L-glutamine. MDA PCa 2b prostate cancer cells (ATCC) were grown in Ham’s modified F12-K medium as described [23]. PC3/neo and PC3/AR cells were provided by Dr. Mien-Chie Hung (M.D. Anderson Cancer Center) [24]. The PC3/AR cells were transfected with a set of four SureSilencing shRNA plasmids targeting AR (ARshRNA) or a negative control shRNA (CshRNA) (SABiosciences), and then selected in hygromycin. LNCaP cells were infected with a control lentivirus (ZIP) or one expressing an anti-sense miR-125b (ZIP-125b) (System Biosciences) as described [11], and selected in puromycin. DU145 cells were infected with a control lentivirus (MIR) and one expressing miR-125b (MIR-125b) (System Biosciences), and then selected in puromycin.
Immunoblot Analysis
Lysates from subconfluent cells were prepared as described [25]. Soluble proteins were subjected to immunoblotting with anti-AR (H-280; Santa Cruz Biotechnology), anti-MUC1-C (Labvision) and anti-β-actin (Sigma). Immune complexes were detected with horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (GE Healthcare Biosciences).
RT-PCR
Total RNA was isolated from cells using an RNeasy Minikit (Qiagen). cDNAs were synthesized with 1 μg of RNA using the first-strand cDNA synthesis kit (Invitrogen). Expression of MUC1 and β-actin mRNA was analyzed with 1 μl of cDNA using the Taq DNA polymerase (Qiagen). Primers used for RT-PCR are listed in Supplemental Table I.
Quantitative RT-PCR
For real time qRT-PCR, cDNA synthesis was performed with 1 μg of total RNA using the Superscript-III One-step RT-PCR system (Invitrogen). The SYBR green qPCR assay kit (Applied Biosystems) was used with 1 μl of cDNA diluted 25-fold from each sample and the samples were amplified with the ABI Prism 7000 Sequence Detector (Applied Biosystems). Primers used for qRT-PCR are listed in Supplemental Table II. Fold enrichment was calculated as described [26] and the results are expressed as the mean ±SD of triplicate values for each sample.
MUC1 Promoter-Luciferase Assays
Cells were transfected with pGL3, pGL3-pMUC1-Luc [27] or pGL3-pMUC1(mARE)-Luc and, as an internal control, SV-40-Renilla-Luc (Promega) in the presence of Lipofectamine 2000. After 48 hr, the cells were lysed in passive lysis buffer. The lysates were analyzed for firefly and Renilla luciferase activities using the dual luciferase assay kit (Promega).
Chromatin Immunoprecipitation (ChIP) Assays
Soluble chromatin was prepared as described [28] and precipitated with anti-AR or a control non-immune IgG. For PCR, 2 μl from a 50 μl DNA extraction were used with 25–35 cycles of amplification. The primers used for PCR are listed in Supplemental Table III. For real time ChIP qPCR, the SYBR green Champion ChIP qPCR assay kit (SA Biosciences) was used and the samples were amplified with the ABI Prism 7000 Sequence Detector (Applied Biosystems). The primers used for qPCR are listed in Supplemental Table III. Relative fold enrichment was calculated as described [26]. The results are expressed as the mean ±SD of triplicate values for each sample.
Analysis of miR-125b Expression
Total RNA was isolated from cells using the RNeasy total RNA isolation kit (Qiagen). Expression of miR-125b was assessed using a small RNA specific RT-PCR kit (System Biosciences) with a universal reverse primer and forward primers specific for miR-125b or for human U6 small RNA as a control.
Luciferase-MUC1 3′UTR Reporter Assays
The MUC1 3′UTR was cloned into the pMIR-LUC vector (Applied Biosystems). The wild type MUC1 3′UTR (MUC1-3′UTR) was mutated (MUC1-mut3′UTR) or deleted (MUC1-del3′UTR) at the CAGGG sequence. Cells were cotransfected with the pMIR-Luc vectors and SV-40-Renilla-Luc (Promega) in the presence of Superfect (Qiagen). After 48 hr, the cells were lysed in passive lysis buffer. The lysates were analyzed for firefly and Renilla luciferase activities using the dual luciferase assay kit (Promega).
RESULTS
MUC1-C Expression Is Suppressed in Androgen-Dependent Prostate Cancer Cells
Immunoblot analysis of the androgen-dependent LNCaP, androgen-sensitive MDA PCa 2b [29] and androgen-responsive CWR22Rv1 [30] prostate cancer cells demonstrated readily apparent AR expression and low to undetectable levels of MUC1-C protein (Fig. 1A). By contrast, MUC1-C was clearly detectable in the androgen-independent DU145 and PC3 prostate cancer cells, which have undetectable levels of AR protein (Fig. 1A). Based on these results, RT-PCR was performed to determine whether the differences in MUC1-C expression are regulated at the mRNA level. MUC1 transcripts were detectable in all of the prostate cancer cell lines and highest in DU145 cells (Fig. 1B, left). Similar results were obtained when MUC1 mRNA levels were analyzed by qRT-PCR (Fig. 1B, right). PC3 cells transfected to stably express exogenous AR have been shown to regain androgen responsiveness [24]. Compared to PC3/neo cells, MUC1-C protein was downregulated in the PC3/AR cells (Fig. 1C). Introduction of exogenous AR in PC3 cells was also associated with decreases in MUC1 transcripts as detected by RT-PCR (Fig. 1D, left) and qRT-pCR (Fig. 1D, right). These findings indicated that AR signaling regulates MUC1-C expression at the transcriptional and translational levels.
Fig. 1.
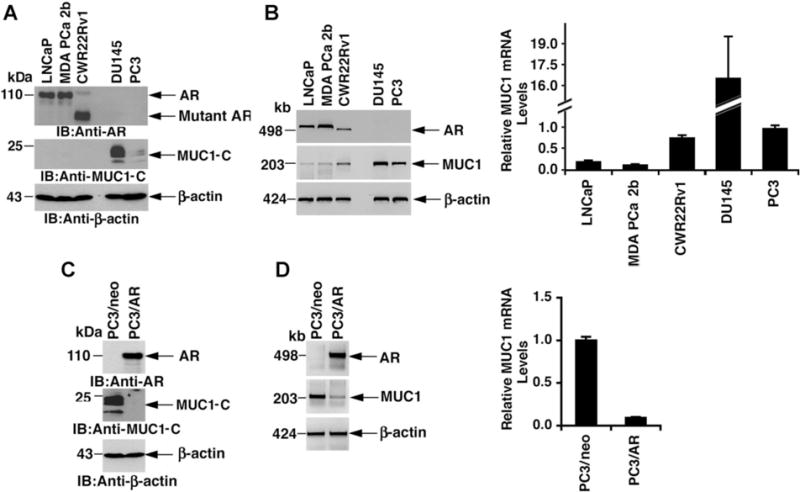
Expression of AR and MUC1 in prostate cancer cell lines. A: Lysates from LNCaP, MDA PCa 2b, CWR22Rv1, DU145 and PC3 cells were immunoblotted with the indicated antibodies. B: Total RNA from the indicated cells was analyzed for AR, MUC1and β-actin mRNA levels by RT-PCR (left). Total RNA was also analyzed for MUC1mRNA levels by qRT-PCR with the results (mean SD of three determinations) expressed as relative to that obtained for PC3 cells (assigned a value of 1) (right). C: Lysates from PC3/neo and PC3/AR cells were immunoblotted with the indicated antibodies. D: Total RNA from the indicated cells was analyzed for (i) AR, MUC1 and β-actin mRNA levels (left) and (ii) MUC1 mRNA levels by qRT-PCR with the results (mean ± SD of three determinations) expressed as relative to that obtained for PC3/neo cells (assigned a value of1) (right).
Downregulation of MUC1 Transcription by a Consensus AR Binding Site in the MUC1 Promoter
Analysis of the MUC1 promoter using TESS (transcription element search software) identified a consensus AR binding site (AGAACA) located at position −194 to −189 upstream to the transcription initiation site (Fig. 2A). To determine whether this putative AR element (ARE) in the MUC1 promoter regulates MUC1 transcription, a MUC1 promoter-Luciferase (pMUC1-Luc) reporter was mutated at that site (AGAACA - → ATAGTA; mARE). Transfection of LNCaP cells demonstrated that the wild-type MUC1 promoter in pMUC1-Luc is activated about sevenfold compared to that obtained with the pGL3 vector (Fig. 2A). Moreover, comparison of pMUC1-Luc with the pMUC1 (mARE)-Luc reporter demonstrated that mutation of the ARE increases activation of the MUC1 promoter (Fig. 2A). Similar results were obtained with CWR22Rv1 cells (Fig. 2B), indicating that repression of the MUC1 promoter is attenuated by mutation of the AGAACA site. In DU145 cells, the wild-type MUC1 promoter was activated over 20-fold compared to pGL3 and mutation of the ARE had little if any effect (Fig. 2C). Repression of the MUC1 promoter was also more pronounced in PC3/AR as compared to PC3/neo cells (Fig. 2D). Moreover, the extent of activation was similar in PC3/neo and PC3/AR cells when transfected with the pMUC1 (mARE)-Luc mutant (Fig. 2D). These findings indicated that activation of the MUC1 promoter is suppressed through a consensus AR binding site.
Fig. 2.
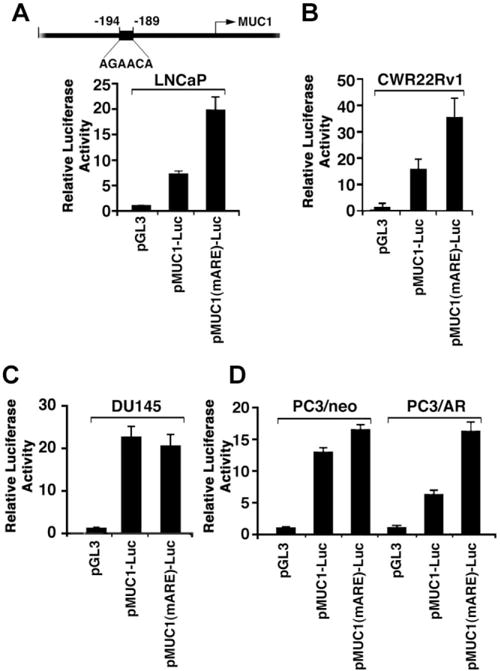
Activation of the MUC1 promoter is regulated by mutation of a putative AR element (ARE). A: Schema of the MUC1 promoter region with positioning of the putative AR binding site. A–D: The indicated cells were transfected with the empty pGL3, pMUC1-Luc or pMUC1 (mARE)-Luc and, as an internal control, the SV-40-Renilla-Luc plasmid. Luciferase activity was measured at 48 hr after transfection. The results are expressed as relative luciferase activity (mean ± SD from three separate experiments) compared to that obtained with cells transfected with an empty pGL3 vector (assigned a value of 1).
AR Occupies the MUC1 Promoter
To determine if AR occupies the putative ARE, we performed ChIP assays using soluble chromatin from LNCaP cells. Immunoprecipitation of the MUC1 promoter region (−348 to −46) containing the ARE with anti-AR was analyzed by semiquantitative PCR. The results indicated that AR occupies this region of the MUC1 promoter (Fig. 3A, left). As a control, there was no detectable signal in immunoprecipitates performed with nonimmune IgG (Fig. 3A, left). There was also no detectable occupancy of a control region (CR; +4596 to +4817) of the MUC1 gene downstream to the ARE (Fig. 3A, left). These results were confirmed by qPCR (Fig. 3A, right). By contrast, there was no detectable AR occupancy of the MUC1 promoter in DU145 cells by ChIP-PCR (Fig. 3B, left) or qPCR (Fig. 3B, right). AR occupancy of the MUC1 ARE was also not detectable in PC3/neo cells (Fig. 3C). However, stable AR expression in PC3/AR cells was associated with the detection of AR occupancy on the ARE (Fig. 3D).
Fig. 3.
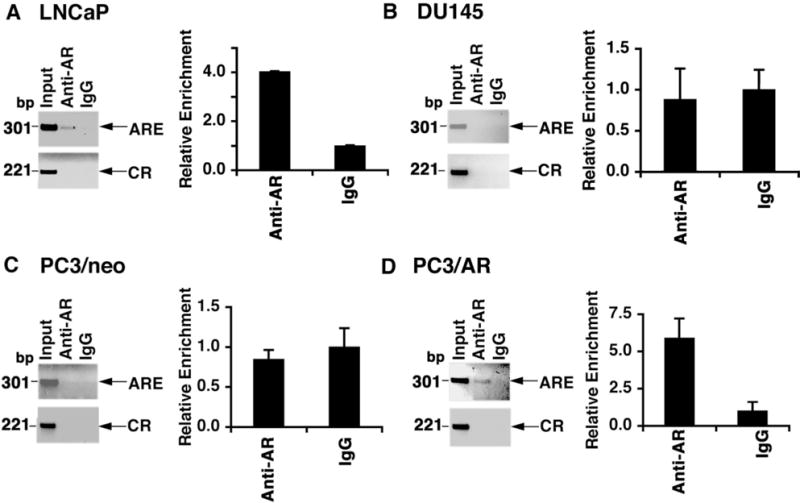
AR occupies the MUC1 promoter. A–D: Soluble chromatin from the indicated cells was immunoprecipitated with anti-AR or a control IgG. The final DNA extractions were amplified by PCR (left panels) and by qPCR with the results (mean ± SD from three determinations) expressed as relative enrichment compared to that obtained with the control IgG (assigned a value of1) (right panels).
AR Occupancy Suppresses MUC1 Transcription
To determine whether AR occupancy suppresses the MUC1 promoter, PC3/AR cells were stably transfected to express a control shRNA or an AR shRNA. Immunoblot analysis of the transfected cells demonstrated that AR silencing is associated with upregulation of MUC1-C expression (Fig. 4A). In concert with these results, silencing AR was also associated with increases in MUC1 mRNA levels (Fig. 4B). ChIP analysis of the MUC1 promoter further demonstrated that, compared to PC3/AR cells expressing the CshRNA (Fig. 4C, left), silencing AR results in decreased occupancy of AR on the ARE (Fig. 4C, right). In addition, activation of the pMUC1-Luc reporter was increased by AR silencing (Fig. 4D). These findings confirmed that AR occupancy of the MUC1 promoter suppresses MUC1 transcription.
Fig. 4.
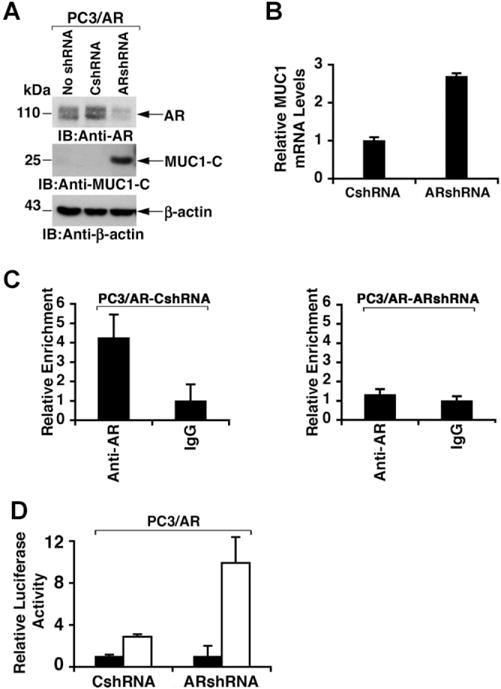
Silencing AR in PC3/AR cells relieves downregulation of MUC1expression. A: Lysates from PC3/AR cells left untransfected and stably transfected with control (CshRNA) or AR shRNA plasmids were immunoblotted with the indicated antibodies. B: Total RNA isolated from PC3/AR cells expressing CshRNA or ARshRNA was analyzed for MUC1 mRNA levels by qRT-PCR. The results (mean ± SD from three determinations) are expressed as relative MUC1 mRNA levels as compared to that obtained from the CshRNA expressing cells (assigned a value of 1). C: Soluble chromatin from PC3/AR cells expressing CshRNA (left) or ARshRNA (right) was precipitated with anti-AR or IgG. The final DNA extractions were analyzed by qPCR with the results (mean SD from three determinations) expressed as relative enrichment compared to that obtained with the control IgG (assigned a value of 1). D: The indicated PC3/AR cells were transfected with the empty pGL3 (solid bars) or pMUC1-Luc (open bars) and the Renilla-Luc plasmid. Luciferase activity was measured at 48 hr after transfection. The results are expressed as the relative luciferase activity (mean ± SD from three separate experiments) compared to that obtained with cells transfected with an empty pGL3 vector (assigned a value of 1).
AR Signaling Regulates MUC1 Expression by a miR-125b-Dependent Mechanism
Other studies have shown that (i) AR signaling is associated with increased miR-125b expression [31], and (ii) miR-125b downregulates MUC1 translation by binding to the MUC1-3′UTR at a UCAGGG site (Fig. 5A) [11]. Consequently, we asked if AR regulates MUC1 expression by a miR-125b-mediated mechanism in prostate cancer cells. RT-PCR analysis of RNA from LNCaP cells demonstrated expression of miR-125b (Fig. 5A). Notably, however, miR-125b levels were undetectable in DU145 cells (Fig. 5A). Moreover, miR-125b expression was increased in PC3/AR, as compared to PC3/neo, cells (Fig. 5A), consistent with findings that AR induces miR-125b expression [31]. The MUC1 3′UTR was cloned into the pMIR-Luc reporter (pMIR-Luc/MUC1-3′UTR) [11] (Fig. 5B). In addition, the miR-125b binding motif 5′-CAGGG-3′ in the MUC1 3′UTR was mutated to 5′-AGUUU-3′ (pMIR-Luc/MUC1-mut3′UTR) or deleted (pMIR-Luc/MUC1-del3′UTR) (Fig. 5B). Activation of pMIR-Luc/MUC1-3′UTR in LNCaP cells was substantially downregulated compared to that obtained with the pMIR-Luc vector (Fig. 5B). Moreover, this downregulation of pMIR-Luc/MUC1-3′UTR activity was reversed in part by transfection of pMIR-Luc/MUC1-mut3′UTR and pMIR-Luc/MUC1-del3′UTR (Fig. 5B). Whereas these results indicated that miR-125b suppresses MUC1 3′UTR-mediated expression, we infected LNCaP cells with a control lentivirus (LNCaP/ZIP) or one that expresses an antisense miR-125b (LNCaP/ZIP-125b). Immunoblot analysis of the infected LNCaP cells demonstrated upregulation of MUC1-C protein (Fig. 5C). qRT-PCR analysis of these cells further demonstrated little if any effect of miR-125b on MUC1 mRNA levels (Fig. 5D), consistent with the finding that miR-125b suppresses MUC1 translation, and not MUC1 mRNA levels [11].
Fig. 5.
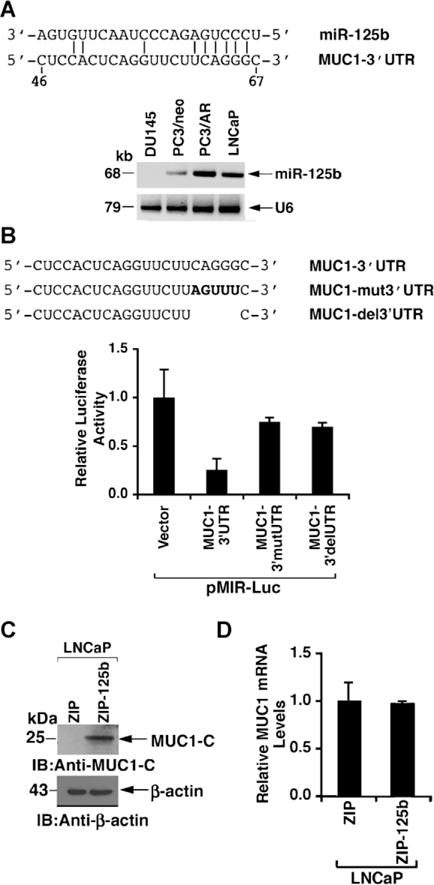
AR induced miR-125b suppressess MUC1 translation. A: Sequence of miR-125b with six bases in the seed region that are complementary to the MUC1 3′UTR. Expression of miR-125b and, as a control, U6 in the indicated cell lines was determined by RT-PCR. B: The wild-type MUC13′UTR was cloned into the pMIR-luciferase vector (pMIR-Luc/MUC1-3′UTR). The indicated MUC1 sequence was then mutated (MUC1-mut3′UTR) or deleted (MUC1-del3′UTR). LNCaP cells were transfected with the indicated pMIR-Luc vectors and Renilla-Luc. Luciferase activity was measured at 48 hr after transfection. The results (mean SD of three determinations) are expressed as relative luciferase activity as compared to that obtained with the pMIR-Luc vector (assigned a value of 1). C: Lysates from LNCaP cells stably infected with a control lentivirus vector (MIR-ZIP) or one expressing an anti-sense for miR-125b (MIR-ZIP-125b) were immunoblotted with the indicated antibodies. D: Total RNA isolated from the LNCaP/ZIP and LNCaP/ZIP-125b cells was analyzed for MUC1mRNA levels by qRT-PCR. The results (mean ± SD of three determinations) are expressed as relative MUC1 mRNA levels as compared to that obtained from LNCaP/ZIP cells (assigned a value of 1).
To confirm the effects of miR-125b, DU145 cells were stably infected with a control lentivirus (DU145/MIR) or one expressing miR-125b (DU145/MIR-125b). Immunoblot analysis of the DU145/miR-125b cells demonstrated a marked suppression of MUC1-C protein (Fig. 6A). As determined by qRT-PCR, there was no effect of exogenous miR-125b on MUC1 mRNA levels (Fig. 6B), again consistent with miR-125b-mediated inhibition of MUC1 translation [11]. Transfection of the DU145/vector cells with the pMIR-Luc vectors demonstrated no apparent suppression of the MUC1 3′UTR or an effect of mutating or deleting the miR-125b binding motif (Fig. 6C, left). By contrast, the MUC1-3′UTR was suppressed in DU145/MIR-125b cells and this effect was reversed with pMIR-Luc/MUC1-mut3′UTR and pMIR-Luc/MUC1-del3′UTR (Fig. 6C, right). These findings collectively indicated that AR regulates MUC1 expression by a miR-125b-mediated inhibition of MUC1 translation.
Fig. 6.
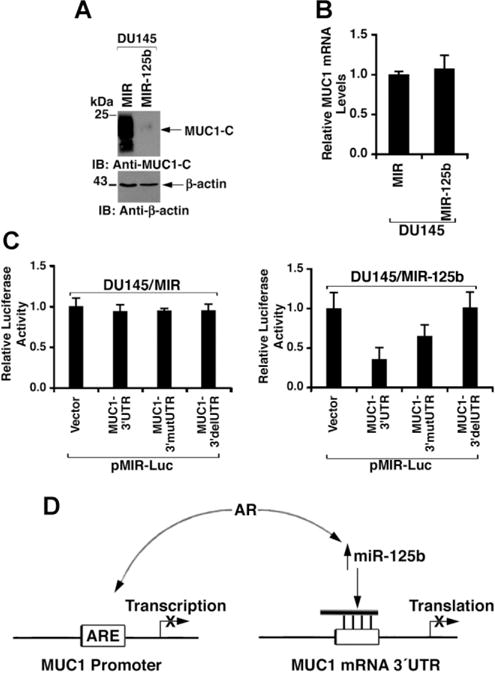
Expression of exogenous miR-125b in DU145 cells suppresses MUC1 translation. A: Lysates from DU145 cells stably infected with a control lentivirus (MIR) or one expressing miR-125b (MIR-125b) were immunoblotted with the indicated antibodies. B: Total RNA isolated from DU145/MIR and DU145/MIR-125b cells was analyzed for MUC1 mRNA levels by qRT-PCR. The results (mean SD of three determinations) are expressed as relative MUC1 mRNA levels as compared to that obtained from DU145/MIR cells (assigned a value of 1). C: DU145/MIR (left) and DU145/MIR-125b (right) cells were transfected with the indicated pMIR-Luc vectors and Renilla-Luc. Luciferase activity was measured at 48 hr after transfection. The results (mean SD of three determinations) are expressed as relative luciferase activity as compared to that obtained with the pMIR-Luc vector (assigned a value of 1). D: Schema depicting AR-induced suppression of MUC1expression by downregulating MUC1 promoter activation and MUC1 mRNA translation.
CONCLUSION
AR Regulates MUC1 Gene Transcription
MUC1 is overexpressed in certain prostate cancers [13–18]. However, the basis for dysregulation of MUC1 expression in prostate cancer is not known. The present studies demonstrate that AR occupies a consensus binding sequence in the MUC1 promoter. The results also indicate that AR occupancy of the MUC1 promoter suppresses MUC1 transcription (Fig. 6D). Overexpression of MUC1 in carcinoma cells has been attributed, at least in part, to activation of the MUC1 promoter through NF-κB and STAT3 [2,32]. The NF-κB pathway is constitutively activated in prostate cancer cells [33,34]. In addition, MUC1-C binds to NF-κB p65 and promotes activation of NF-κB target genes, including the MUC1 gene itself in an auto-inductive loop [2]. In concert with the expression of MUC1-C, NF-κB is constitutively activated in MUC1-positive DU145 and PC3 cells, but not in the MUC1-negative LNCaP cells [35,36]. Thus, AR-mediated suppression of the MUC1 promoter may override the inductive effects of a constitutively activated NF-κB pathway. STAT3 also activates MUC1 gene transcription by occupancy of the MUC1 promoter [32]. Moreover, like NF-κB, STAT3 is constitutively activated in DU145 and PC3, but not LNCaP, cells [37]. AR binds directly to STAT3 and thus could interfere with STAT3-dependent activation of MUC1 expression [38]. Alternatively, AR may suppress the MUC1 promoter by a dominant mechanism of repression over that conferred by STAT3 activation. AR represses gene transcription in part through the recruitment of corepressors, such as NCoR and SMRT, which act in concert with histone deacetylases to induce chromatin modification and condensation [39]. Corepressors can also function by inhibiting co-activator recruitment and by blocking interactions of the AR N- and C-terminal domains [39]. Thus, further studies will be needed to more precisely define how AR occupancy of the MUC1 promoter suppresses MUC1 transcription.
AR Regulates MUC1Expression by a miR-125b-Mediated Mechanism
MiR-125b is upregulated by AR signaling in prostate cancer cell lines and is overexpressed in certain human prostate cancers [31]. Targeting of the pro-apoptotic p53, PUMA and Bak1 genes by miR-125b has been associated with the promotion of prostate cancer cell growth [40]. The present results demonstrate that miR-125b also suppresses expression of MUC1 in prostate cancer cells (Fig. 6D). Thus, in LNCaP cells that express miR-125b, stable expression of an anti-sense-miR-125b was associated with upregulation of MUC1-C levels. In addition, expression of exogenous miR-125b in DU145 cells, which have low to undetectable endogenous miR-125b levels, resulted in downregulation of the MUC1-C protein. In concert with previous studies in breast cancer cells [11], miR-125b decreased MUC1 translation in DU145 cells in the absence of a detectable effect on MUC1 mRNA levels. These results indicate that AR suppresses MUC1 expression by promoter repression and by inhibiting MUC1 translation (Fig. 6D). In this model, AR-induced promoter repression is incomplete as evidenced by the detection of MUC1 transcripts in androgen-dependent prostate cancer cells. AR-induced increases in miR-125b thus represent a second level for MUC1 regulation by a post-transcriptional mechanism. MiR-125b induces androgen-independent growth of prostate cancer cells and, as such, has been associated with an oncogenic function [31,40]. The demonstration that miR-125b levels are decreased in other human cancers has provided support for a role as a tumor suppressor [41–43]. These findings have indicated that cell context dictates whether miR-125b contributes to oncogenesis or functions as a tumor suppressor.
MUC1 Is Overexpressed in Aggressive Prostate Cancers
MUC1 is overexpressed in prostate cancers with higher Gleason grades and is associated with an increased risk of recurrence [13–18], suggesting that MUC1 is of potential importance to more aggressive prostate cancers. Indeed, recent studies with a MUC1-C oncoprotein inhibitor have shown that DU145 and PC3 cells are dependent on MUC1-C for their growth and survival in vitro and in tumor xenograft models [23]. By contrast, LNCaP, MDA PCa 2b and CWR22Rv1 cells, which express AR and undetectable levels of MUC1-C were unaffected by the MUC1-C inhibitor [23]. In addition, the MUC1-C inhibitor had no effect on PC3 cells expressing exogenous AR that downregulates MUC1-C levels [23]. These results invoked the possibility that prostate cancers in which AR is effective in downregulating MUC1-C are dependent on AR and not MUC1-C for growth and survival. By extension, prostate cancers in which AR signaling is not sufficient to downregulate MUC1-C could progress with dependence on MUC1-C function. In that line of reasoning, androgen-independent prostate cancers have been linked to constitutive activation of the IKK → NF-κB pathway [21,44,45], and MUC1-C interacts with both IKKβ and RelA in the constitutive activation of the NF-κB pathway [2,46]. MUC1-C could also interact directly or indirectly with AR signaling in androgen-independent prostate cancers. For example, MUC1-C stabilizes β-catenin [4], which is a coactivator of AR-mediated transcription [47–49]. Progression of prostate cancers to androgen-independence could thus be associated with upregulation of MUC1-C and thereby dependence on this oncoprotein for growth and survival. Of interest in this regard, other studies have demonstrated that activation of MUC1-C in prostate cancer cells is associated with downregulation of AR expression and dependence on MUC1-C for survival (unpublished data). Therefore, an agent, such as GO-203, that targets MUC1-C and is entering Phase I clinical trials may be effective in the treatment of androgen-independent prostate cancers that express this oncoprotein.
Supplementary Material
Acknowledgments
Grant sponsor: Department of Defense Prostate Cancer Idea Award; Grant number: W81XWH-08-1-0093.
Abbreviations
- MUC1
mucin 1
- MUC1-N
MUC1 N-terminal subunit
- MUC1-C
MUC1 C-terminal subunit
- AR
androgen receptor
- ARE
AR element
- ChIP
chromatin immunoprecipitation
- Luc
luciferase
Footnotes
Additional Supporting Information may be found in the online version of this article.
D. Kufe holds equity in Genus Oncology and is a consultant to the company.
References
- 1.Kufe D. Mucins in cancer: Function, prognosis and therapy. Nature Reviews Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad R, Raina D, Joshi MD, Kawano T, Kharbanda S, Kufe D. MUC1-C oncoprotein functions as a direct activator of the NF-kappaB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khodarev N, Ahmad R, Rajabi H, Pitroda S, Kufe T, McClary C, Joshi MD, MacDermed D, Weichselbaum R, Kufe D. Cooperativity of the MUC1 oncoprotein and STAT1 pathway in poor prognosis human breast cancer. Oncogene. 2010;29(6):920–929. doi: 10.1038/onc.2009.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks GSK3beta-mediated phosphorylation and degradation of beta-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 5.Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7:167–178. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 6.Khodarev N, Pitroda S, Beckett M, MacDermed D, Huang L, Kufe D, Weichselbaum R. MUC1-induced transcriptional programs associated with tumorigenesis predict outcome in breast and lung cancer. Cancer Res. 2009;69:2833–2837. doi: 10.1158/0008-5472.CAN-08-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pitroda S, Khodarev N, Beckett M, Kufe D, Weichselbaum R. MUC1-induced alterations in a lipid metabolic gene network predict response of human breast cancers to tamoxifen treatment. Proc Natl Acad Sci USA. 2009;106:5837–5841. doi: 10.1073/pnas.0812029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacDermed DM, Khodarev NN, Pitroda SP, Edwards DC, Pelizzari CA, Huang L, Kufe DW, Weichselbaum R. MUC1-associated proliferation signature predicts outcomes in lung adenocarcinoma patients. BMC Med Genomics. 2010;3:16. doi: 10.1186/1755-8794-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3/MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–6110. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010;70(1):378–387. doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajabi H, Jin C, Ahmad R, McClary C, Kufe D. Mucin 1 oncoprotein expression is suppressed by the miR-125b oncomir. Genes Cancer. 2010;1(1):62–65. doi: 10.1177/1947601909357933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin C, Rajabi H, Kufe D. miR-1226 targets expression of the mucin 1 oncoprotein and induces cell death. Int J Oncol. 2010;37(1):61–69. doi: 10.3892/ijo_00000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirschenbaum A, Itzkowitz SH, Wang JP, Yao S, Eliashvili M, Levine AC. MUC1 expression in prostate carcinoma: Correlation with grade and stage. Mol Urol. 1999;3(3):163–168. [PubMed] [Google Scholar]
- 14.Cozzi PJ, Wang J, Delprado W, Perkins AC, Allen BJ, Russell PJ, Li Y. MUC1, MUC2, MUC4, MUC5AC and MUC6 expression in the progression of prostate cancer. Clin Exp Metastasis. 2005;22(7):565–573. doi: 10.1007/s10585-005-5376-z. [DOI] [PubMed] [Google Scholar]
- 15.Arai T, Fujita K, Fujime M, Irimura T. Expression of sialylated MUC1 in prostate cancer: Relationship to clinical stage and prognosis. Int J Urol. 2005;12(7):654–661. doi: 10.1111/j.1442-2042.2005.01112.x. [DOI] [PubMed] [Google Scholar]
- 16.Andrén O, Fall K, Andersson SO, Rubin MA, Bismar TA, Karlsson M, Johansson JE, Mucci LA. MUC-1 gene is associated with prostate cancer death: A 20-year follow-up of a population-based study in Sweden. Br J Cancer. 2007;97(6):730–734. doi: 10.1038/sj.bjc.6603944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garbar C, Mascaux C, Wespes E. Expression of MUC1 and sialyl-Tn in benign prostatic glands, high-grade prostate intraepithelial neoplasia and malignant prostatic glands: A preliminary study. Anal Quant Cytol Histol. 2008;30(2):71–77. [PubMed] [Google Scholar]
- 18.Rabiau N, Dechelotte P, Guy L, Satih S, Bosviel R, Fontana L, Kemeny JL, Boiteux JP, Bignon YJ, Bernard-Gallon D. Immunohistochemical staining of mucin 1 in prostate tissues. In Vivo. 2009;23(2):203–207. [PubMed] [Google Scholar]
- 19.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, Ekman P, DeMarzo AM, Tibshirani R, Botstein D, Brown PO, Brooks JD, Pollack JR. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101(3):811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: Directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23(32):8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 21.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1(1):34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 22.Taplin ME, Balk SP. Androgen receptor: A key molecule in the progression of prostate cancer to hormone independence. J Cell Biochem. 2004;91(3):483–490. doi: 10.1002/jcb.10653. [DOI] [PubMed] [Google Scholar]
- 23.Joshi MD, Ahmad R, Raina D, Rajabi H, Bubley G, Kharbanda S, Kufe D. MUC1 oncoprotein is a druggable target in human prostate cancer cells. Mol Cancer Ther. 2009;8:3056–3065. doi: 10.1158/1535-7163.MCT-09-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cha T, Qiu L, Chen C, Wen Y, Hung M. Emodin down-regulates androgen receptor and inhibits prostate cancer cell growth. Cancer Res. 2005;65(6):2257–2295. doi: 10.1158/0008-5472.CAN-04-3250. [DOI] [PubMed] [Google Scholar]
- 25.Ren J, Agata N, Chen D, Li Y, Yu W-H, Huang L, Raina D, Chen W, Kharbanda S, Kufe D. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anti-cancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19(5):631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 27.Yin L, Kufe D. Human MUC1 carcinoma antigen regulates intracellular oxidant levels and the apoptotic response to oxidative stress. J Biol Chem. 2003;278(37):35458–35464. doi: 10.1074/jbc.M301987200. [DOI] [PubMed] [Google Scholar]
- 28.Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor a. Mol Cell. 2006;21(2):295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 29.Navone N, Olive M, Ozen M, Davis R, Troncoso P, Tu S, Johnston D, Pollack A, Pathak S, von Eschenbach A, Logothetis C. Establishment of two human prostate cancer cell lines derived from a single bone metastasis. Clin Cancer Res. 1997;3(12 Pt 1):2493–2500. [PubMed] [Google Scholar]
- 30.Tepper CG, Boucher DL, Ryan PE, Ma AH, Xia L, Lee LF, Pretlow TG, Kung HJ. Characterization of a novel androgen receptor mutation in a relapsed CWR22 prostate cancer xenograft and cell line. Cancer Res. 2002;62(22):6606–6614. [PubMed] [Google Scholar]
- 31.Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, Tepper CG, Evans CP, Kung HJ, deVere White RW. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci USA. 2007;104(50):19983–19988. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaemers IC, Vos HL, Volders HH, van der Valk SW, Hilkens J. A STAT-responsive element in the promoter of the episialin/MUC1 gene is involved in its overexpression in carcinoma cells. J Biol Chem. 2001;276(9):6191–6199. doi: 10.1074/jbc.M009449200. [DOI] [PubMed] [Google Scholar]
- 33.Suh J, Rabson AB. NF-kappaB activation in human prostate cancer: Important mediator or epiphenomenon? J Cell Biochem. 2004;91(1):100–117. doi: 10.1002/jcb.10729. [DOI] [PubMed] [Google Scholar]
- 34.Jin RJ, Lho Y, Connelly L, Wang Y, Yu X, Saint Jean L, Case TC, Ellwood-Yen K, Sawyers CL, Bhowmick NA, Blackwell TS, Yull FE, Matusik RJ. The nuclear factor-kappaB pathway controls the progression of prostate cancer to androgen-independent growth. Cancer Res. 2008;68(16):6762–6769. doi: 10.1158/0008-5472.CAN-08-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palayoor ST, Youmell MY, Calderwood SK, Coleman CN, Price BD. Constitutive activation of IkappaB kinase alpha and NF-kappaB in prostate cancer cells is inhibited by ibuprofen. Oncogene. 1999;18(51):7389–7394. doi: 10.1038/sj.onc.1203160. [DOI] [PubMed] [Google Scholar]
- 36.Parrondo R, de las Pozas A, Reiner T, Rai P, Perez-Stable C. NF-kappaB activation enhances cell death by antimitotic drugs in human prostate cancer cells. Mol Cancer. 2010;9:182. doi: 10.1186/1476-4598-9-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mora LB, Buettner R, Seigne J, Diaz J, Ahmad N, Garcia R, Bowman T, Falcone R, Fairclough R, Cantor A, Muro-Cacho C, Livingston S, Karras J, Pow-Sang J, Jove R. Constitutive activation of Stat3 in human prostate tumors and cell lines: Direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 2002;62(22):6659–6666. [PubMed] [Google Scholar]
- 38.Ueda T, Bruchovsky N, Sadar MD. Activation of the androgen receptor N-terminal domain by interleukin-6 via MAPK and STAT3 signal transduction pathways. J Biol Chem. 2002;277(9):7076–7085. doi: 10.1074/jbc.M108255200. [DOI] [PubMed] [Google Scholar]
- 39.Burd CJ, Morey LM, Knudsen KE. Androgen receptor corepressors and prostate cancer. Endocr Relat Cancer. 2006;13(4):979–994. doi: 10.1677/erc.1.01115. [DOI] [PubMed] [Google Scholar]
- 40.Shi XB, Xue L, Ma AH, Tepper CG, Kung HJ, White RW. miR-125b promotes growth of prostate cancer xenograft tumor through targeting pro-apoptotic genes. Prostate. 2010 doi: 10.1002/pros.21270. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 42.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, Tavare S, Caldas C, Miska EA. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8(10):R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Sawyers C, Scher H. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol. 2008;8(4):440–448. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gasparian A, Yao Y, D K, Lyakh L, Karseladze A, Slaga T, Budunova I. The role of IKK in constitutive activation of NF-κB transcription factor in prostate carcinoma cells. J Cell Sci. 2002;115(Pt 1):141–151. doi: 10.1242/jcs.115.1.141. [DOI] [PubMed] [Google Scholar]
- 46.Ahmad R, Raina D, Trivedi V, Ren J, Rajabi H, Kharbanda S, Kufe D. MUC1 oncoprotein activates the I|B kinase β complex and constitutive NF-|B signaling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G, Wang J, Sadar MD. Crosstalk between the androgen receptor and beta-catenin in castrate-resistant prostate cancer. Cancer Res. 2008;68(23):9918–9927. doi: 10.1158/0008-5472.CAN-08-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Truica CI, Byers S, Gelmann EP. Beta-catenin affects androgen receptor transcriptional activity and ligand specificity. Cancer Res. 2000;60(17):4709–4713. [PubMed] [Google Scholar]
- 49.Yang F, Li X, Sharma M, Sasaki CY, Longo DL, Lim B, Sun Z. Linking beta-catenin to androgen-signaling pathway. J Biol Chem. 2002;277(13):11336–11344. doi: 10.1074/jbc.M111962200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


