Abstract
Mimicking non-covalent interaction based processes in nature has been an important goal of supramolecular chemistry. Here, we report on amphiphilic polypeptides that self-assemble to form nanoscale supramolecular assemblies and are programmed to disassemble in response to a specific protein. Benzenesulfonamide and carbonic anhydrase have been chosen as the ligand and protein respectively to demonstrate this possibility. Since the amphiphilic nanoassembly sequesters hydrophobic guest molecules, the protein-specific disassembly event provides a protein-sensitive molecular release as well. We envision that the binding induced disassembly and guest release might open up new opportunities for the next generation of supramolecular assemblies for protein-specific delivery and diagnostics.
Graphical abstract
The holy grail of supramolecular chemistry has been to design artificial molecules and molecular assemblies that mimic nature’s ability to execute specific processes through precisely engineered intermolecular forces.1 Nature has evolved specific interactions among macromolecules as an important recognition strategy to execute most of the biological processes. For example, proteins specifically interact with partner proteins to trigger biological events,2 with nucleic acids to transcribe the genetic code,3 and with carbohydrates to regulate cellular processes such as cell-cell communication.4 In most cases, the binding event causes a conformational change in the partner macromolecule to activate one of the binding partners to trigger a biological cascade. Inspired by these processes that are central to the existence of biological systems, we were interested in designing artificial peptide-based supramolecular assemblies that respond to a specific protein binding event and cause a discernible cascade of events.
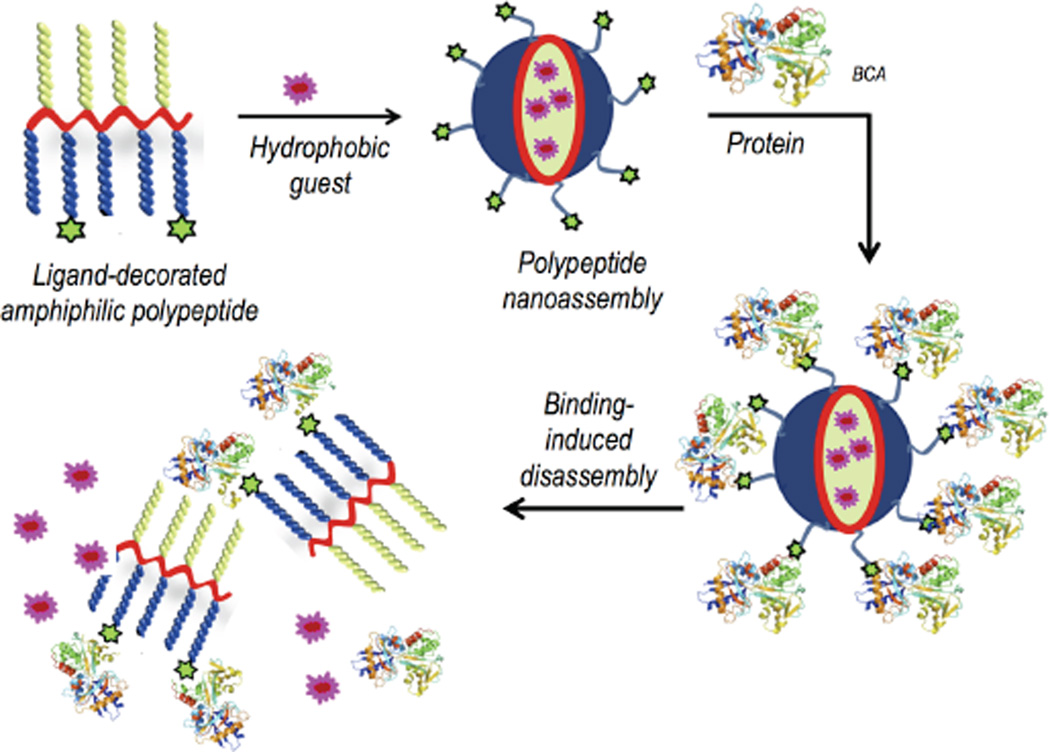
We chose peptide-based nanoassemblies, because they can be functionally diverse with precise functional group placements within the scaffold.5 As a first step, we were interested in a minimalist biomimetic design with peptide scaffolds. Accordingly, we have designed amphiphilic polypeptides, where the driving force for the nanoassembly formation is simply driven by hydrophobic forces. We also designed these peptide-based assemblies by derivatizing a peptide homopolymer with the requisite functionalities for achieving a nanoassembly that can specifically bind to a protein. In our molecular design, we utilize poly-L-gultamic acid, where a percentage of the carboxylic acid moieties are functionalized with hydrophobic units to render the polypeptide amphiphilic. Although this polymer itself is amphiphilic and self-assembles, we further derivatized the remaining carboxylic acid groups with oligoethyleneglycol (OEG) moieties in order to avoid non-specific, electrostatic interactions with proteins. To further insure specific binding with a targeted protein, a small percentage of the OEGs also contain protein-specific ligand moieties.
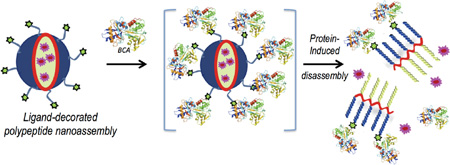
The key design hypothesis is that the amphiphilic polypeptide, mentioned above, would self-assemble to present a protein-specific ligand on the hydrophilic face of an amphiphilic nanoaggregate in water. We then envisage that the protein binding event would cause a rather large change in the self-assembled structure.6 Note that the protein binding causes a change in the hydrophilic face that presents the sulfonamide moiety to one that presents the hydrophilic surface of a rather large protein. This event should cause a significant change in the hydrophilic-lipophilic balance (HLB) of the amphiphile, resulting in supramolecular disassembly (Scheme 1). We chose carbonic anhydrase as the model protein, because of its disease relevance and because non-covalent ligands for carbonic anhydrase are well-established.7 Specifically, bovine carbonic anhydrase II (bCA-II) with a pI of 5.4 and a molecular weight of 30 kDa was used as the protein for this study.
Scheme 1.
Schematic presentation of the protein binding induced disassembly of a polypeptide nanoassembly
The motivations to explore polypeptides as the scaffolds for binding induced disassembly using carbonic anhydrase are multifold. Two of these factors emerge as rather significant ones. From the mechanistic possibility, prior work has shown that one needs multivalent protein-ligand interactions to cause the binding-induced disassembly.6a This requirement is rather limiting, as most biologically relevant proteins do not have multiple binding sites. Therefore, demonstration of binding induced disassembly with carbonic anhydrase, where the protein contains a single ligand binding site, would be a critical finding to expand the repertoire of this strategy. Second, we were interested in polypeptides, not only because they ultimately provide the opportunity to introduce diverse functional groups through sequencing, but also because its backbone is biodegradable.
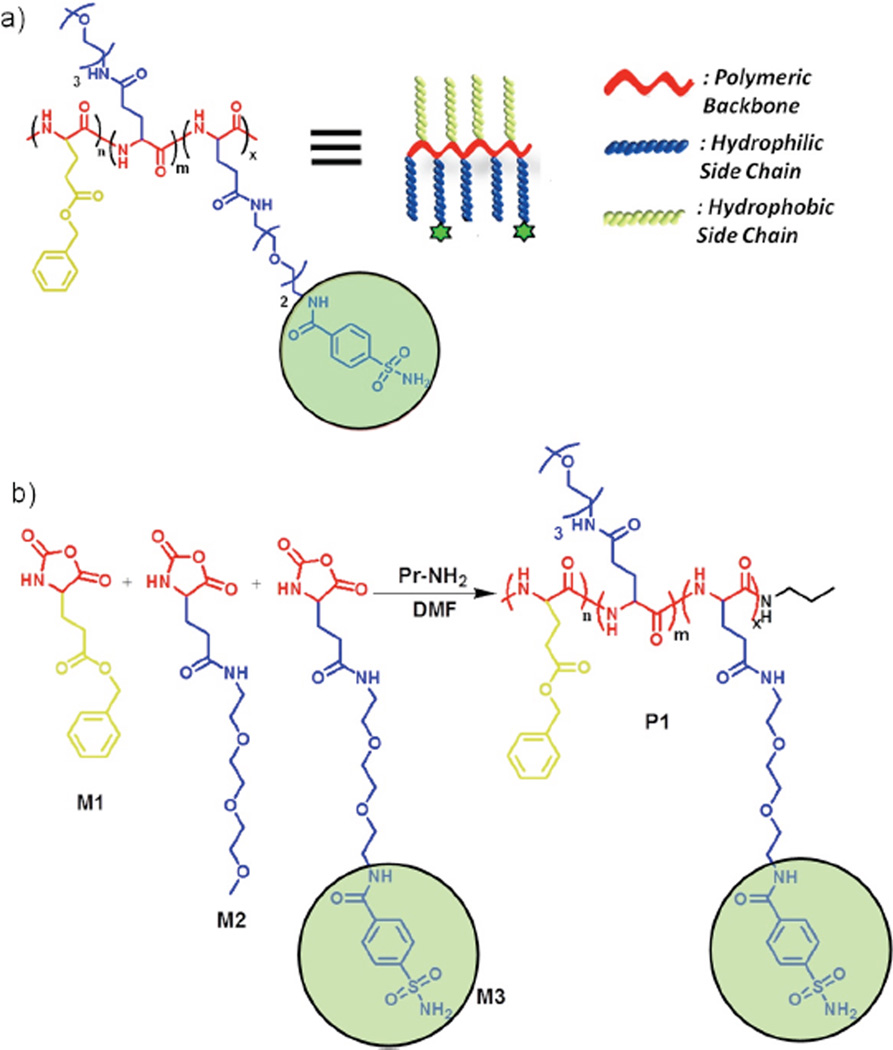
The targeted amphiphilic random co-polypeptide is made of three different substituted glutamic acid monomers (M1, M2 and M3, Scheme2). Benzene sulfonamide was chosen as ligand, because of its well-established, high binding affinity towards carbonic anhydrase.7 The polymer P1 was synthesized from NCA monomers as shown in Scheme 2. FTIR analysis of P1 showed complete disappearance of two characteristic peaks of the monomers at 1850 and 1782 cm−1 (Figure S1). NMR revealed that the experimental ratio of the monomers in P1 was about 0.48:0.37:0.15. Gel permeation chromatography (GPC) shows that Mn and dispersity were 3.2 kDa and 1.2 respectively (Figure S2).
Scheme 2.
a) Chemical structure of polymer P1 and corresponding cartoon. b) Schematic presentation of the final step of P1 synthesis.
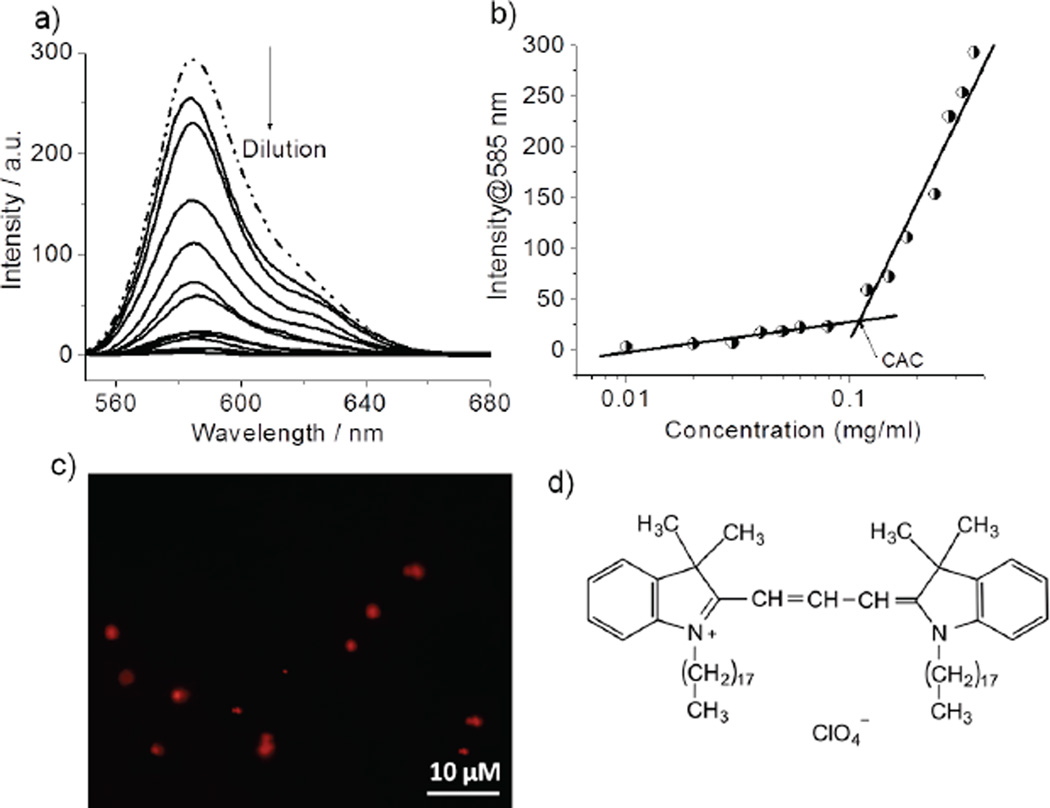
Next, we investigated the self-assembly features of the polypeptide P1 by evaluating its ability to act as host for a hydrophobic guest (DiI) in aqueous medium (Figure 1d). DiI is not soluble in water unless a hydrophobic pocket is provided. The emission from DiI increases with increase in P1 concentration, although initial DiI concentration was kept constant in all solutions (Figure 1a). The CAC of P1 was calculated to be about 31 µM (Figure 1b). Spectroscopic evidence of DiI encapsulation was also justified by OPM images, showing red-emitting spherical particles (Figure 1c).
Figure 1.
a) Emission spectra of DiI encapsulated in the P1 micelle in varying concentration. b) Variation of λmax (585 nm) of DiI as a function of P1 concentration. c) Optical Fluorescence Microscopy (OPM) image of DiI encapsulated P1 micelle. d) Chemical structure of DiI dye molecule.
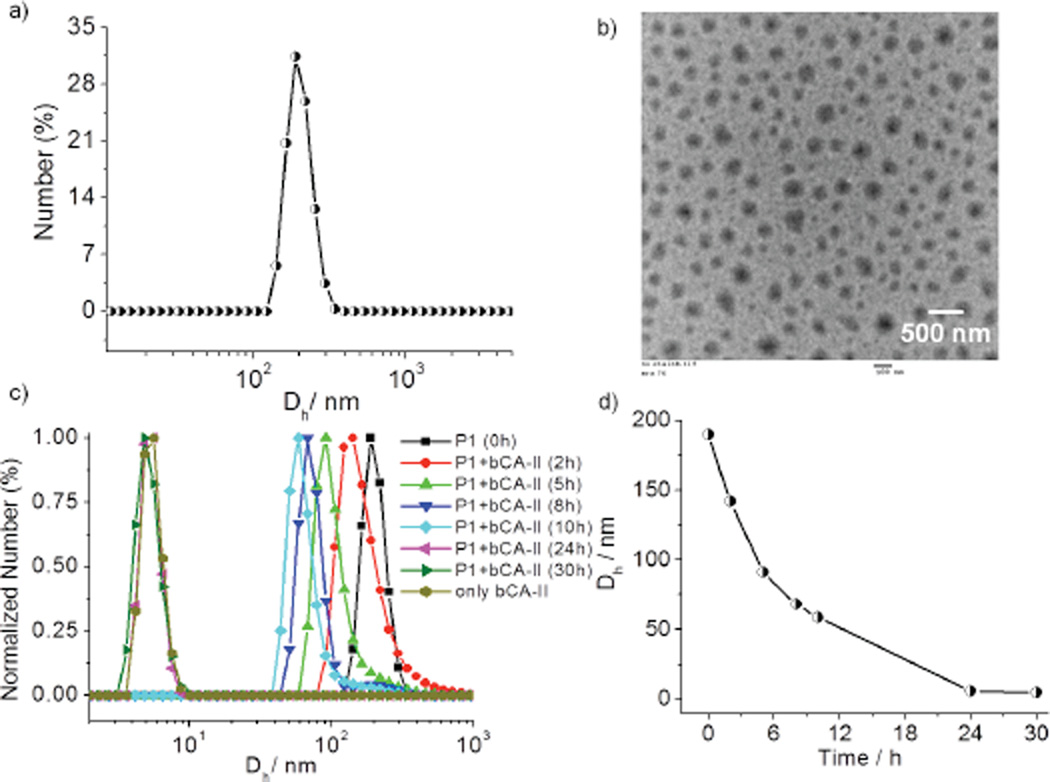
The formation of an amphiphilic aggregate with an average hydrodynamic diameter of 190–200 nm from P1 was confirmed using dynamic light scattering (DLS) (Figure 2a). Transmission electron microscopy (TEM) analysis showed the presence of spherical assemblies in the range of 170–180 nm (Figure 2b), which are slightly lower than those from DLS. This difference is likely due to the shrinkage of the particles in the dry state8 or due to overestimation of the size of the particles in DLS as it also includes hydration shells around the particles.
Figure 2.
a) DLS of P1 aggregates in water; (Concentration of P1 = 50 µM). b) TEM image of P1 aggregates. c) Time dependent DLS profile of P1 in presence of bCA-II protein. d) Size variation of the particle with time.
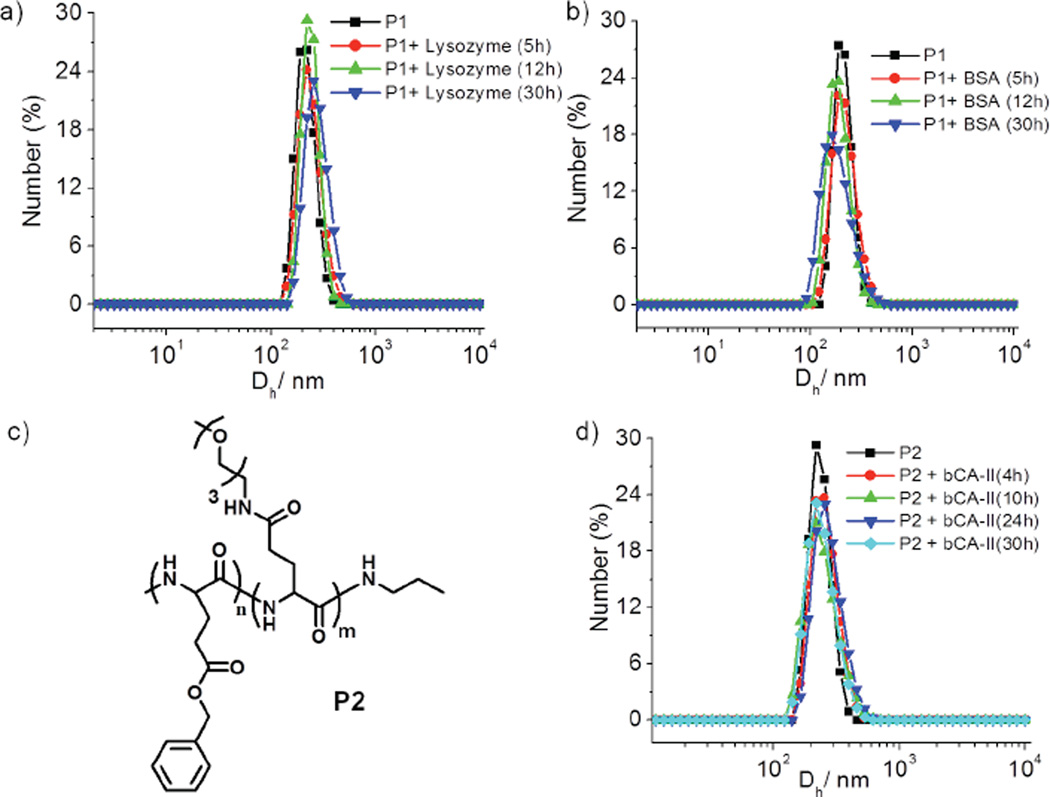
To investigate whether the specific interaction between the ligand and the complementary protein would cause significant changes to the aggregation state of the assembly from P1, an aqueous solution of P1 (50 µM) was treated with bCA-II (30 µM). The size evolution of the aggregates was monitored over 30 h by DLS. Upon addition of protein, the size decreased from ~200 nm to ~5 nm, which is close to the size of the bCA-II protein by itself (Figure 2c and 2d). On the other hand, no size change was observed over the same time period in the absence of protein (Figure S3). These data provided the indication that is consistent with our binding induced disassembly hypothesis, illustrated in Scheme 1. It is however important to show that this presumed binding-induced disassembly is indeed due to a specific ligand-protein interaction. For this purpose, we used noncomplementary proteins such as pepsin, bovine serum albumin (BSA) and lysozyme. These proteins were selected as non-complementary proteins for their diversity in pI values, since this is often the source of nonspecific interactions (pI = 1.0 (pepsin), 4.8 (BSA) and 11 (lysozyme)). Indeed, we noticed that there was no change in size of the aggregates in solution over 30h for any of these proteins (Figure 3a, 3b and S4). To further validate that the observed disassembly is due to specific protein-ligand interactions, we synthesized a control polypeptide P2, which does not contain the complementary sulfonamide ligand (Figure 3c). P2 also forms assemblies similar to those from the ligand-bearing polymer P1 (Figure S5). The size of the aggregates from P2 was found to be ~140 nm by both DLS and TEM. When an aqueous solution of P2 is treated with bCA-II, the size did not change over 30 h (Fig. 3d), again supporting the hypothesis that the observed size change is due to binding-induced disassembly caused by a specific ligand-protein interaction.
Figure 3.
DLS of P1 aggregates in presence of a) lysozyme. b) BSA (Concentration of P1 = 50 µM). c) Chemical structure of P2. d) Time dependent DLS of P2 in presence of bCA-II protein.
In addition, the results with the control polypeptide P2 also rules out a potential alternate explanation for disassembly in response to carbonic anhydrase. It is known that carbonic anhydrase has some esterase activity.9 Therefore, it is possible that the observed disassembly in P1 is due to the cleavage of the ester moiety in the hydrophobic part of the amphiphilic peptide. In this case, the disassembly is anticipated due to the resultant change in the hydrophilicity of the self-assembling peptide. The fact that the structurally identical polymer P2, except for the ligand presence, did not disassemble in the presence of bCA-II rules out this alternate hydrolysis based disassembly pathway (Figure 3d).
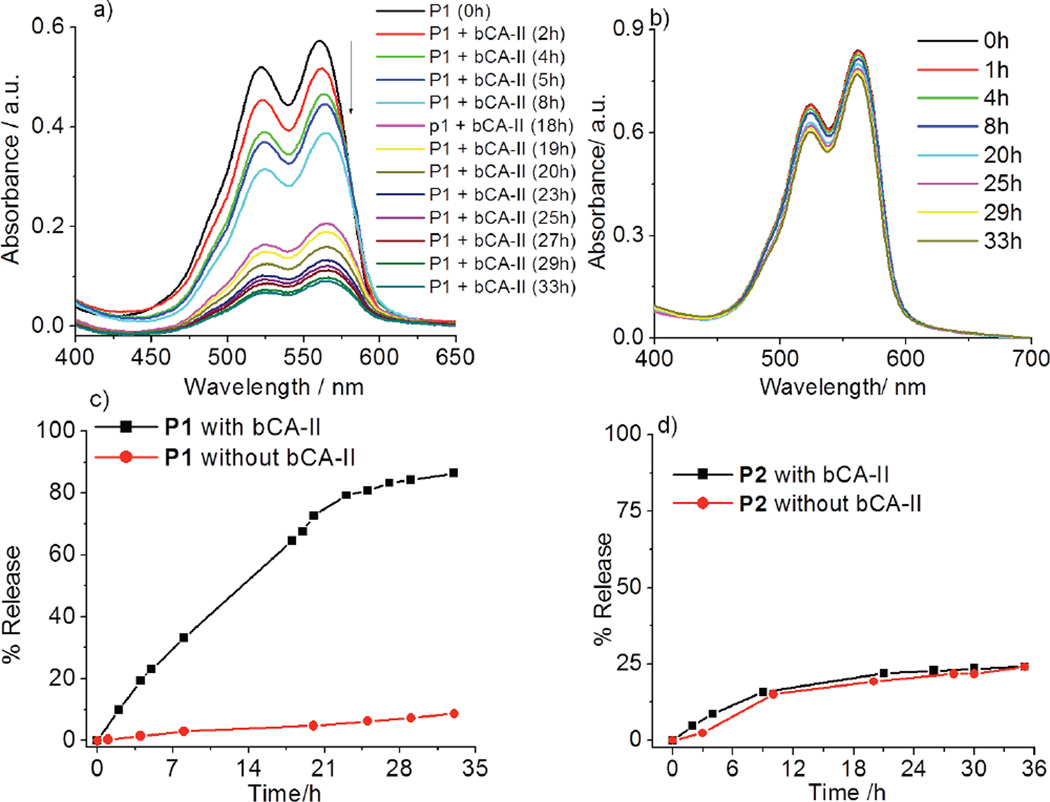
Since the protein binding causes a disassembly, it is possible that we can utilize this process to cause the guest molecules to be released. Since the hydrophobic guest is insoluble in water, it is likely that it will simply precipitate out of solution. To test this hypothesis, DiI-encapsulated P1 (50 µM) was treated with bCAII (30 µM) and the possible guest release of the DiI was monitored by absorption spectroscopy. Indeed, the absorbance of DiI decreased overtime in presence of bCA-II, while there was no such change in the absence of bCA-II (Figure 4a and 4b). These observations indicate that the observed process is a protein-specific molecular release (~85% release over 33h, Figure 4c). To further test whether the process is specific to bCA-II, in a control experiment, we exposed the same solution to non-complementary proteins with different surface charges, viz. pepsin, BSA and lysozyme. The guest release was found to be relatively insignificant (Figure S6). Similarly, we also monitored the possible guest release from P2 aggregate and it was <20%. More importantly, the extent of guest release was found to be identical to that without the protein from this assembly over the same amount of time (Figure 4d and S7). These results indicate that the guest release from the P1 assembly is indeed due to specific ligand-protein interaction induced disassembly.
Figure 4.
DiI release from P1 micelle a) in presence and b) in absence of bCA-II protein (Concentration of P1 = 50 µM); c) Plot of % release with time; d) Plot of % release of DiI from control polymer P2 in presence and absence of bCA-II.
Finally, since the degradable components of this polypeptide are considered biocompatible, we envisaged that the amphiphilic polypeptide itself might be biocompatible, i.e. not cytotoxic. To test this, we studied in vitro cell viability using an Alamar blue assay with HeLa cell lines and found the cells to be ~80% viable even at 250 µg/ mL of polymer solution (Figure S8).
In summary, we have designed and synthesized a polypeptide, the amphiphilic nature of which provides a nanoscale supramolecular assembly that can stably encapsulate hydrophobic guest molecules in aqueous media. The polypeptide is engineered to present a protein-specific ligand in its hydrophilic face. We show that the binding interaction between the ligand moiety and the complementary protein causes the assembly to fall apart. This binding-induced disassembly has been shown to be specific to bCAII and to cause release of guest molecules. The extent of guest release in response to protein binding was found to be substantial (~85%). This feature, along with the simplicity of the synthetic route, highlights the utility of peptide-based assemblies for protein-induced supramolecular disassembly. Although responsive molecular assemblies have been consistently targeted for applications such as delivery and diagnostics,10 systems that respond to protein activity are very limited. These are interesting, because aberrant protein activity is the basis for all genetic diseases. While there have been significant efforts on systems that respond to enzymatic activity variations,11 assemblies that respond to non-enzymatic proteins are very limited. The polypeptides, outlined here, are poised to make a significant impact in this area with their biocompatible, biodegradable, and high fidelity responsive disassembly characteristics.
Supplementary Material
Acknowledgments
We thank the NIGMS of the National Institutes of Health (GM-065255) for support.
Footnotes
Supporting Information
Detailed synthetic procedures and characterizations of the polymers. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Gao Y, Zhao F, Wang Q, Zhanga Y, Xu B. Chem. Soc. Rev. 2010;39:3425–3433. doi: 10.1039/b919450a. [DOI] [PubMed] [Google Scholar]; (b) Breslow R. Acc. Chem. Res. 1995;28:146–153. [Google Scholar]; (c) Zhao H, Foss FW, Breslow R., Jr J. Am. Chem. Soc. 2008;130:12590–12591. doi: 10.1021/ja804577q. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Bruns CJ, Stoddart JF. Acc. Chem. Res. 2014;47:2186–2199. doi: 10.1021/ar500138u. [DOI] [PubMed] [Google Scholar]; (e) Coskun A, Banaszak M, Astumian RD, Stoddart JF, Grzybowski BA. Chem. Soc. Rev. 2012;41:19–30. doi: 10.1039/c1cs15262a. [DOI] [PubMed] [Google Scholar]
- 2.(a) Schreiber G, Haran G, Zhou H-X. Chem. Rev. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Keskin O, Gursoy A, Ma B, Nussinov R. Chem. Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]; (c) Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Proc. Natl. Acad. 2000;97:1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Ramírez-Tapia LE, Martin CT. J. Biol. Chem. 2012;287:37352–37361. doi: 10.1074/jbc.M112.370643. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Papantonis A, Cook PR. Chem. Rev. 2013;113:8683–8705. doi: 10.1021/cr300513p. [DOI] [PubMed] [Google Scholar]; (c) Michaelis J, Treutlein B. Chem. Rev. 2013;113:8377–8399. doi: 10.1021/cr400207r. [DOI] [PubMed] [Google Scholar]
- 4.(a) Wong P, Hampton B, Szylobryt E, Gallagher AM, Jaye M, Burgess WH. J. Biol. Chem. 1995;270:25805–25811. doi: 10.1074/jbc.270.43.25805. [DOI] [PubMed] [Google Scholar]; (b) Spivak-Kroizman T, Lemmon MA, Dikic I, Ladbury JE, Pinchasi D, Huang J, Jaye M, Crumley G, Schlessinger J, Lax I. Cell. 1994;79:1015–1024. doi: 10.1016/0092-8674(94)90032-9. [DOI] [PubMed] [Google Scholar]; (c) Powell AK, Fernig DG, Turnbull JEJ. Biol. Chem. 2002;277:28554–28563. doi: 10.1074/jbc.M111754200. [DOI] [PubMed] [Google Scholar]
- 5.(a) Zhou M, Smith AM, Das AK, Hodson NW, Collins RF, Ulijn RV, Gough JE. Biomaterials. 2009;30:2523–2530. doi: 10.1016/j.biomaterials.2009.01.010. [DOI] [PubMed] [Google Scholar]; (b) Cui H, Muraoka T, Cheetham AG, Stupp SI. Nano Lett. 2009;9:945–951. doi: 10.1021/nl802813f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) O'Leary LER, Fallas JA, Bakota EL, Kang MK, Hartgerink JD. Nature Chem. 2011;3:821–828. doi: 10.1038/nchem.1123. [DOI] [PubMed] [Google Scholar]
- 6.(a) Amado Torres D, Garzoni M, Subrahmanyam A, Pavan GM, Thayumanavan SJ. Am. Chem. Soc. 2014;136:5385–5399. doi: 10.1021/ja500634u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Azagarsamy MA, Yesilyurt V, Thayumanavan S. J. Am. Chem. Soc. 2010;132:4550–4551. doi: 10.1021/ja100746d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Takaoka Y, Sakamoto T, Tsukiji S, Narazaki M, Matsuda T, Tochio H, Shirakawa M, Hamachi I. Nat. Chem. 2009;1:557–561. doi: 10.1038/nchem.365. [DOI] [PubMed] [Google Scholar]; (d) Anees P, Sreejith S, Ajayaghosh A. J. Am. Chem. Soc. 2014;136:13233–13239. doi: 10.1021/ja503850b. [DOI] [PubMed] [Google Scholar]; (e) Yamaguchi N, Zhang L, Chae B, Palla CS, Furst EM, Kiick KL. J. Am. Chem. Soc. 2007;129:3040–3041. doi: 10.1021/ja0680358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Krishnamurthy VM, Kaufman GK, Urbach AR, Gitlin I, Gudiksen KL, Weibel DB, Whitesides GM. Chem. Rev. 2008;108:946–1051. doi: 10.1021/cr050262p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Alterio V, Di Fiore A, D’Ambrosio K, Supuran CT, De Simone G. Chem. Rev. 2012;112:4421–4468. doi: 10.1021/cr200176r. [DOI] [PubMed] [Google Scholar]
- 8.(a) Moughton AO, O’Reilly RK. Chem. Commun. 2010;46:1091–1093. doi: 10.1039/b922289h. [DOI] [PubMed] [Google Scholar]; (b) Qian C, Zhang S, Li J, Zuo B, Wang X. Soft Matter. 2014;10:1579–1590. doi: 10.1039/c3sm52761a. [DOI] [PubMed] [Google Scholar]
- 9.Şentürk M, Gülçin I, Beydemir Ş, Küfrevioğlu Öİ, Supuran CT. Chem Biol Drug Des. 2011;77:494–499. doi: 10.1111/j.1747-0285.2011.01104.x. [DOI] [PubMed] [Google Scholar]
- 10.(a) Gillies ER, Jonsson TB, Frechet JMJ. J. Am. Chem Soc. 2004;126:11936–11943. doi: 10.1021/ja0463738. [DOI] [PubMed] [Google Scholar]; (b) Ryu J-H, Chacko R, Jiwpanich S, Bickerton S, Babu RP, Thayumanavan S. J. Am. Chem. Soc. 2010;132:17227–17235. doi: 10.1021/ja1069932. [DOI] [PubMed] [Google Scholar]; (c) Zhang Q, Clark JCG, Wang M, Remsen EE, Wooley KL. Nano Lett. 2002;2:1051–1054. [Google Scholar]; (d) Nishiyama N, Iriyama A, Jang WD, Miyata K, Itaka K, Inoue Y, Takahashi H, Yanagi Y, Tamaki Y, Koyama H, Kataoka K. Nat. Mater. 2005;4:934–941. doi: 10.1038/nmat1524. [DOI] [PubMed] [Google Scholar]; (e) Nam J-M, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]; (f) Dan K, Ghosh S. Angew. Chem. Int. Ed. 2013;52:7300–7305. doi: 10.1002/anie.201302722. [DOI] [PubMed] [Google Scholar]; (g) Azagarsamy MA, Alge DL, Radhakrishnan SJ, Tibbitt MW, Anseth KS. Biomacromolecules. 2012;13:2219–2224. doi: 10.1021/bm300646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Amir RJ, Zhong S, Pochan DJ, Hawker CJ. J. Am. Chem. Soc. 2009;131:13949–13951. doi: 10.1021/ja9060917. [DOI] [PubMed] [Google Scholar]; (b) Shamis M, Lode HN, Shabat D. J. Am. Chem. Soc. 2004;126:1726–1731. doi: 10.1021/ja039052p. [DOI] [PubMed] [Google Scholar]; (c) Azagarsamy MA, Sokkalingam P, Thayumanavan S. J. Am. Chem. Soc. 2009;131:14184–14185. doi: 10.1021/ja906162u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Guo J, Zhuang J, Wang F, Raghupathi KR, Thayumanavan S. J. Am. Chem. Soc. 2014;136:2220–2223. doi: 10.1021/ja4108676. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Zhu L, Kate P, Torchilin VP. ACS Nano. 2012;6:3491–3498. doi: 10.1021/nn300524f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Gao Y, Kuang Y, Guo Z-F, Guo Z, Krauss IJ, Xu B. J. Am. Chem. Soc. 2009;131:13576–13577. doi: 10.1021/ja904411z. [DOI] [PubMed] [Google Scholar]; (g) Thornton PD, Mart RJ, Ulijn RV. Adv. Mater. 2007;19:1252–1256. [Google Scholar]; (h) Bacinello D, Garanger E, Taton D, Chiu Tam K, Lecommandoux S. Biomacromolecules. 2014;15:1882–1888. doi: 10.1021/bm500296n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.