ABSTRACT
Precise positioning of nucleosomes around regulatory sites is achieved by the action of chromatin remodelers, which use the energy of ATP to slide, evict or change the composition of nucleosomes. Chromatin remodelers act to bind nucleosomes, disrupt histone-DNA interactions and translocate the DNA around the histone core to reposition nucleosomes. Hence, remodeling is expected to involve nucleosomal intermediates with a structural organization that is distinct from intact nucleosomes. We describe the identification of a partially unwrapped nucleosome structure using methods that map histone-DNA contacts genome-wide. This alternative nucleosome structure is likely formed as an intermediate or by-product during nucleosome remodeling by the RSC complex. Identification of the loss of histone-DNA contacts during chromatin remodeling by RSC in vivo has implications for the regulation of transcriptional initiation.
KEYWORDS: chromatin remodeling, ChIP-seq, H4S47C cleavage mapping, MNase-seq, nucleosome, nucleosomal intermediate, RSC
The basic repeating unit of chromatin was described as DNA wrapped by an octameric core of histones around 4 decades ago,1 followed by the determination of the structures of the histone octamer2 and the nucleosome core particle (NCP).3 The NCP consists of a central H3-H4 tetramer flanked by 2 dimers of H2A-H2B wrapped by ∼1.7 turns of DNA. Each mirror-symmetric half of the nucleosome contains one H3-H4 dimer and one H2A-H2B dimer wrapping 73 bp of DNA. In parallel to the studies establishing the NCP as the steady-state configuration of chromatin, the structure of chromatin undergoing disruptive processes including transcription and replication had been proposed to depart from the NCP.4 To facilitate histone inheritance during replication, the nucleosome was envisioned to split in half, forming a hemisome, the symmetric half of the nucleosome, and each hemisome could be passed on to a daughter genome during replication, similar to formation of hemimethylated DNA during replication.5 Alternative nucleosome structures were also proposed in the context of active chromatin. Gene activation was shown to produce half-nucleosome footprints as mapped by DNase-I, hinting at routine formation of nucleosome intermediates during transcription.6 Although octameric nucleosomes are reconstituted in vitro with ≥150-bp DNA duplexes, stable hemisomes have been reconstituted by using ∼65-bp DNA duplexes.7,8
If nucleosomes are inherited as hemisomes during replication, we would expect an equal proportion of old and new histones making up nucleosomes after replication. However, many studies failed to observe nucleosome splitting at replication.9 This conclusion was confirmed in a recent study that tracked old and new histones through replication using differential isotope labeling and mass-spectroscopy, which clearly demonstrated that for the vast majority of the genome, the old H3-H4 tetramers did not split during replication.10 However, this study found that a significant fraction of tetramers with the H3.3 replacement histone variant did split. H3.3 is mostly enriched at active genes and enhancers, and subsequent mapping of H3.3 splitting events found them to be mainly occurring at enhancers.11 In budding yeast, which have only a single form of H3, active genes show splitting of H3-H4 tetramers.12 These observations of splitting of H3-H4 tetramers indicate that nucleosomes must go through intermediate structures at active genes and enhancers. These intermediate structures might feature loss of histones from the nucleosome or loss of specific histone-DNA contacts, or both.
Studies to identify and characterize alternative nucleosome species have been rare compared to studies of canonical nucleosomes due to a dearth of techniques to identify dynamic nucleosomal intermediates. One of the most well-characterized alternative nucleosome structures is found at budding yeast centromeres.13 Yeast has a single nucleosome acting as a “point” centromere for each chromosome.14 The centromeric nucleosome has H3 replaced by the centromeric variant cenH3 (Cse4 in yeast, CENP-A in mammals). Unlike the canonical nucleosome that has a left-handed DNA wrap, the cenH3 nucleosome formed on the yeast centromeric sequence wraps DNA with right-handed chirality.15 Histone octamers support only left-handed wrapping, whereas histone tetramers can support either left-handed or right-handed wrapping, suggesting that a tetrameric complex forms the centromeric nucleosome. Furthermore, the cenH3 nucleosome is confined to an ∼80 bp region and contains H2A.13 Given the presence of cenH3 and H2A, their obligate partners H4 and H2B would also be expected to be part of the centromeric nucleosome. Furthermore, compared to the 147 bp of DNA that is protected by octameric nucleosomes, ∼80 bp would be expected to be protected only by a tetrameric complex of histones. These observations point to a hemisome that contains one each of cenH3, H4, H2A and H2B at yeast centromeres. In support of the in vivo observations, short centromeric sequences can be used to reconstitute stable cenH3 hemisomes.8
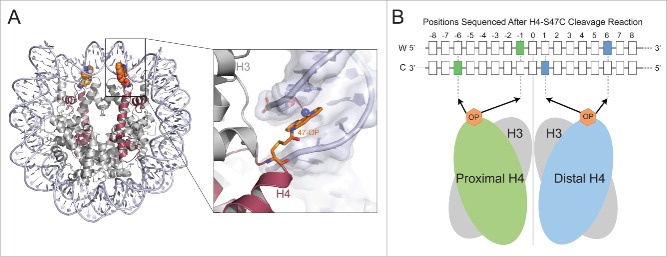
Hemisomes at budding yeast centromeres have been confirmed using H4S47C-anchored cleavage mapping, a technique originally developed to identify nucleosome positions genome-wide at base-pair resolution.16 In this method, yeast histone H4 is mutated at position 47 to carry cysteine instead of serine.17 Ex vivo labeling of cells with a phenanthroline ligand converts H4 into a site-specific DNA cleavage agent. H4S47C-phenanthroline chelates copper and in the presence of hydrogen peroxide cleaves nucleosomal DNA at highly specific positions (Fig. 1A). By preparing a sequencing library from the cleaved DNA and performing short read sequencing we found that the sequenced cleavage reactions correspond to nucleotide positions −6, −1, 0, +1 and +6 bp relative to the position of the dyad axis.17 Our structural model for the interaction of H4S47C-phenanthroline with DNA in the context of a nucleosome showed that the cleavages at −6 and −1 were due to the H4 upstream of the dyad, those at +1 and +6 were due to the H4 downstream of the dyad and that at the 0 position could be due to either H418 (Fig. 1B). Because the phenanthroline ligand must be within 4 Å of DNA for cleavage to occur, we can interrogate the status of DNA contacts formed by residues around H4-S47 for each H4 in the genome. By distinguishing cleavages due to each H4 at a given nucleosome position, we could ask if both H4s were contacting the DNA or just one. At the centromere, we found cleavage patterns that corresponded to the presence of only one H4 molecule.
Figure 1.
DNA cleavage by H4S47C-phenanthroline-Cu+ in vivo A) The position of 47-cysteine-orthophenanthroline (OP) in the nucleosome structure is shown (left). The magnified region (right) shows a copper ion (blue dot) bound to OP within 4 Å (dotted line) of the C1H atom of the deoxyribonucleotide near the dyad. OP binds in the minor groove of the nucleosomal DNA. B) The highlighted positions in the Watson (W) and Crick (C) strands around the nucleosome dyad are sequenced after OP cleavage. The positions cleaved by the proximal H4 are shown in green and the positions cleaved by the distal H4 are shown in blue.
We also characterized H4S47C-anchored cleavages based on their precise positions relative to the 16 functional centromere sequences. Cleavage positions that are near the dyad axis of an octameric nucleosome would be at the center of the DNA protected by an octameric nucleosome or a (Cse4/H4)2 tetrasome. In contrast, cleavage positions would be very near the end of the DNA protected by the hemisome. In the case of the yeast centromere, the cleavages were at the edge of the DNA protected by the nucleosome. Because of the tight binding of factors immediately on either side of the ∼80-bp region wrapped by the Cse4 nucleosome, these cleavages could not have been produced by octasomes or by (Cse4-H4)2 tetrasomes.19 Thus, H4S47C-anchored cleavage mapping confirms that hemisomes occupy yeast centromeres.
Given the ability of H4S47C-anchored cleavage mapping to identify the status of H4-DNA contacts near the dyad genome-wide, we asked if there are alternative nucleosome structures outside of the centromeres. We identified a subset of positions (5% of total positions genome-wide) that were conspicuously asymmetric with respect to H4S47C-anchored cleavages, although the asymmetry is an order of magnitude less than that of centromeric nucleosomes.
At centromeric nucleosome positions, asymmetric H4S47C cleavages could be explained only by hemisomes, which leads to the possibility of hemisomes at asymmetric H4S47C-anchored cleavages outside of centromeres. A hemisome would protect only ∼70–80 bp of DNA whereas an octasome protects ∼150 bp of DNA. The amount of DNA protected by a protein complex can be determined by treating chromatin with Micrococcal Nuclease (MNase) followed by sequencing the isolated DNA (MNase-seq). MNase is an endo-exonuclease that preferentially chews on linker DNA and stops when it encounters a protein-DNA contact. By performing paired-end sequencing of fragments protected by MNase, we can estimate not only the position in the genome that is protected, but also the length of DNA that is protected. At asymmetric nucleosome positions as well as nucleosome positions genome-wide, we found the expected protection of ∼150 bp. We also observed a protection of ∼73 bp on either side of the dyad. This protection indicated nucleosome splitting at the dyad, making the DNA around the dyad accessible to MNase. If the asymmetric H4S47C-anchored cleavages represent hemisomes, we would expect the distribution of the ∼73 bp fragments to also be asymmetric. However, we observed these fragments to be symmetric around the dyad. This means that both halves of the nucleosome are present at asymmetric positions and the asymmetric S47C-anchored cleavages represent partial loss of histone-DNA contacts without loss of histones.
We identified asymmetric particles throughout the genome, but they were especially enriched around transcription start sites (TSSs). Ordered nucleosome arrays are found flanking TSSs, where genic nucleosome positions are numbered +1, +2 and so on starting from the first nucleosome position downstream of the TSS. TSSs are usually depleted of nucleosomes, and the nucleosome positions upstream of the promoter are numbered −1, −2 and so on. The +1 position was most enriched in asymmetric nucleosomes, with significant enrichment also at the −1 position. Interestingly, asymmetric nucleosomes were also recently identified at +1 nucleosome positions by ChIP-exo mapping of histones (Fig. 2A–B) and validated by MNase-seq (Fig. 2C).20 However, we find that these particles are not at the same positions as those at the +1 position that we identified by H4S47C-anchored cleavage mapping (data not shown). Furthermore, asymmetric particles identified by histone ChIP-exo show symmetric H4-S47C cleavage patterns characteristic of octasomes (Fig. 2D). It is likely that the asymmetric particles identified by histone ChIP-exo represent a distinct class of nucleosomal intermediates.
Figure 2.
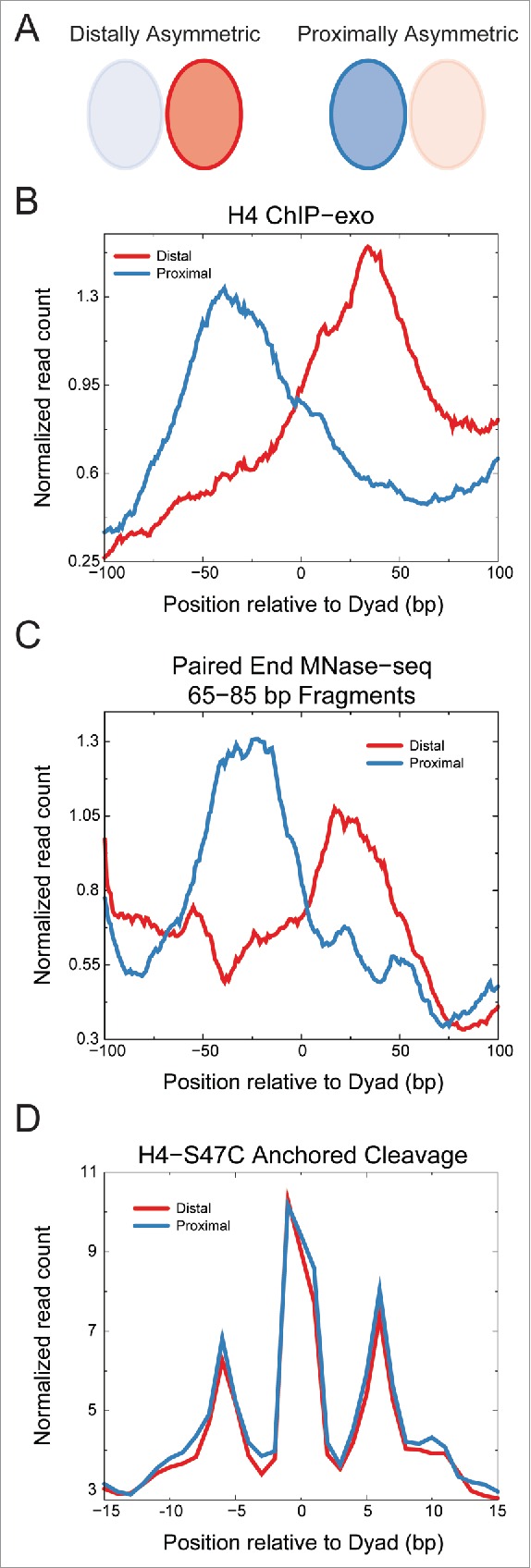
Asymmetric nucleosome positions identified by ChIP-exo show symmetric H4S47C-anchored cleavages. A) Schematic of ChIP-exo asymmetry relative to the transcription start site (TSS). Ratio of H4-ChIP-exo counts between proximal and distal sides of the dyad axis relative to the TSS from Rhee et al.20 was used to identify asymmetric nucleosome positions. The H4-ChIP-exo ratios were obtained from Table S4 of Rhee et al. and a cutoff of greater than 2 or less than 0.5 was used. B) Averages of H4 ChIP-exo data for the distally (n = 264) and proximally (n = 293) asymmetric nucleosome positions. The running average over a 15 bp window is plotted. C) Averages of centers of fragments from MNase-seq30 are plotted for distally and proximally asymmetric nucleosome positions identified by H4-ChIP-exo. The running average over a 15 bp window is plotted for fragments of 65–85 bp in length, which would include half-nucleosome protections. Enrichment is observed on the distal side of the dyad for the distally asymmetric positions and on the proximal side of the dyad for the proximally asymmetric positions. D) Average read counts of the ends of fragments generated by H4S47C-anchored cleavage16,18 are plotted for distally and proximally asymmetric nucleosome positions identified by H4-ChIP-exo. We observe no asymmetry in the H4S47C-anchored cleavage data for both distally and proximally asymmetric positions identified by H4-ChIP-exo.
H4S47C-anchored cleavage mapping reports on only one out of 6 histone-DNA contact sites on each half of the nucleosome. How do we determine the status of all histone-DNA contact sites at a given nucleosome position? We turned to an aspect of MNase-seq data that has received little attention. MNase stops digesting DNA when it encounters a protein-DNA contact, so that histone-DNA contact positions in the genome would be more protected compared to DNA positions that are exposed. The ends of the sequenced fragments would then reveal the positions where MNase stopped upon encountering a protein-DNA contact. Around the nucleosome dyad, we observed a 10 bp periodicity in MNase cut frequency, coinciding with the 10 bp periodicity of histone-DNA contacts. This indicates that the MNase cut frequency is reporting on histone-DNA contacts at nucleosome positions genome-wide. At asymmetric nucleosome positions, we found loss of enrichment of fragment ends on the same side of the dyad that lost H4S47C-anchored cleavages. The contacts that were lost are the ones right next to the dyad. Thus, asymmetric nucleosome positions show a loss of a subset of histone-DNA contacts on one side of the dyad. Loss of specific histone-DNA contacts could be due to the action of chromatin remodelers, which disrupt histone-DNA contacts to reposition nucleosomes. This can be confirmed by asking whether cleavage asymmetry is found at positions found to have highly dynamic nucleosomes. Most dynamic nucleosomes can be found in regions around TSSs due to disruption and reassembly of nucleosomes caused by pervasive binding of chromatin remodelers, transcription factors and the transcription machinery. When we looked at the enrichment of asymmetric nucleosomes genome-wide, they were highly enriched at ±1 positions but not at promoters or genic positions, indicating that asymmetric nucleosomes could be formed due to nucleosome dynamics.
ATP-dependent chromatin remodelers bind nucleosomes and disrupt histone-DNA interactions to slide, evict or change nucleosome composition. The enrichment of asymmetric nucleosome positions around sites of constant nucleosome turnover and remodeler binding strongly suggested that asymmetric nucleosomes might be forming due to remodeler action. To ask which of the remodelers in yeast could be responsible for this phenomenon, we determined the enrichment of remodelers at asymmetric ±1 nucleosome positions using ChIP-seq data. Out of 7 yeast remodelers, we found only RSC to be enriched at asymmetric ±1 nucleosome positions. That RSC action is associated with asymmetric nucleosomes was further supported by the fact that RSC depletion causes higher nucleosome loss at asymmetric ±1 nucleosome positions compared to all ±1 nucleosome positions. We also found that RSC depletion resulted in much lower expression of genes with asymmetric +1 nucleosomes compared to all genes. Finally, the presence of asymmetric +1 nucleosomes correlated with lower RNA polymerase II stalling at +1 nucleosomes. These functional correlations indicate that RSC function at asymmetric ±1 nucleosome positions is essential for the proper expression of genes containing asymmetric nucleosomes.
RSC has been studied in detail using structural and biochemical tools. Hence, we can ask if known mechanisms of RSC interaction with nucleosomes could explain the formation of asymmetric nucleosomes. RSC is a large remodeling complex (1.3 MDa), whose structural characterization has been limited to electron microscopy (EM).21,22 Negative stain EM images of RSC alone revealed a huge cavity, which is complementary to the nucleosome surface.21 Cryoelectron microscopy of the RSC-nucleosome complex revealed RSC tightly enveloping the nucleosome.22 The densities in the binding cavity could account for the entire histone octamer and a part of the ∼150 bp nucleosomal DNA, leading to the speculation that RSC binding partially unwraps the nucleosome. Three binding sites of RSC to the nucleosome could be discerned, the major one being close to the dyad. The RSC densities were directly contacting the octamer at the site near the dyad and no DNA densities were observable. The local unwrapping of DNA near the dyad as observed by cryoEM suggests that RSC binding could lead to asymmetric nucleosome formation in vivo.
The structural alterations to the nucleosome induced by RSC as seen by cryoEM are strongly supported by in vitro experiments that have characterized RSC action on nucleosomes. DNAse footprinting of nucleosomes bound by RSC in the absence of ATP shows overall protection of the nucleosomal DNA compared to free nucleosomes. However, there is a site 2 turns from the dyad that is specifically footprinted only in the presence of RSC.23 In the absence of ATP, RSC unwraps nucleosomes right up to the dyad while remaining bound to both the histone octamer and the nucleosomal DNA.24 Upon addition of ATP, nucleosomal DNA is highly susceptible to nuclease attack, indicating mobilization of the DNA by RSC. Comparing the in vitro observations with properties of asymmetric nucleosomes observed in vivo, we infer that asymmetric nucleosomes detected by H4S47C-anchored cleavage mapping result from a RSC-nucleosome complex in the absence of ATP. The presence of ATP would activate RSC to mobilize the nucleosomal DNA and possibly evict the histone octamer, enabling transcriptional activation. This model can explain how RSC function is required for robust expression of genes with asymmetric nucleosomes at their +1 position.
The disruption of histone-DNA interactions at the dyad by RSC implies that these contacts represent barriers against nucleosome disruption. If this were the case, mutations in histones that weaken these histone-DNA interactions would alleviate the requirement of remodelers for nucleosome disruption and in turn for gene activation. RSC is required for various functions including chromosome segregation25 and is essential for survival, precluding identification of mutants that alleviate the loss of RSC. However, the homolog of RSC, the SWI/SNF complex is not essential for survival and it is required for expression of a significant fraction of genes in yeast. Based on the evolutionary conservation between RSC and SWI/SNF, we expect that the 2 complexes have similar mechanisms of action on nucleosomes. SWI/SNF independence (SIN) mutations, which overcome the loss of SWI/SNF function,26 can provide clues to the mechanism of SWI/SNF action. Several SIN mutants harbor point mutations in H3 and H4 at residues near the dyad axis.27,28 Crystal structures of these SIN mutants reveal the destabilization of histone-DNA interactions near the dyad axis, which implies that SWI/SNF functions in part by destabilizing histone-DNA contacts near the dyad axis. These findings support our model whereby asymmetric nucleosomes result from disruption of histone-DNA interactions by RSC.
RSC binding that results in partial unwrapping of the nucleosome represents the first in vivo demonstration of a nucleosome remodeling intermediate. RSC and SWI/SNF are evolutionarily conserved from yeast to humans, and the human and fly homologs (BRM and BRG1 respectively) have essential roles in development. Several mutations in human remodeling complex subunits with BRM and BRG1 have been identified as cancer driver mutations.29 However, the mechanisms of BRM/BRG1 action are difficult to study because combinations of subunits give rise to numerous distinct complexes. Our paradigm of understanding remodeler action through identification of alternate nucleosome structures presents a new way to understand remodeler function that can override the combinatorial complexity of remodelers in metazoans.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Thomas JO, Kornberg RD. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A 1975; 72:2626-30; PMID:241077; http://dx.doi.org/ 10.1073/pnas.72.7.2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci U S A 1991; 88:10148-52; PMID:1946434; http://dx.doi.org/ 10.1073/pnas.88.22.10148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997; 389:251-60; PMID:9305837; http://dx.doi.org/ 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- [4].van Holde KE, Lohr DE, Robert C. What happens to nucleosomes during transcription? J Biolog Chem 1992; 267:2837-40; PMID:1310672 [PubMed] [Google Scholar]
- [5].Weintraub H, Worcel A, Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell 1976; 9:409-17; PMID:991271; http://dx.doi.org/ 10.1016/0092-8674(76)90085-4 [DOI] [PubMed] [Google Scholar]
- [6].Lee MS, Garrard WT. Transcription-induced nucleosome ‘splitting’: an underlying structure for DNase I sensitive chromatin. EMBO J 1991; 10:607-15; PMID:2001676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tatchell K, Van Holde KE. Nucleosome reconstitution: effect of DNA length on nuclesome structure. Biochemistry 1979; 18:2871-80; PMID:476061; http://dx.doi.org/ 10.1021/bi00580a031 [DOI] [PubMed] [Google Scholar]
- [8].Furuyama T, Codomo CA, Henikoff S. Reconstitution of hemisomes on budding yeast centromeric DNA. Nucleic Acids Res 2013; 41:5769-83; PMID:23620291; http://dx.doi.org/ 10.1093/nar/gkt314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Annunziato AT. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem 2005; 280:12065-8; PMID:15664979; http://dx.doi.org/ 10.1074/jbc.R400039200 [DOI] [PubMed] [Google Scholar]
- [10].Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 2010; 328:94-8; PMID:20360108; http://dx.doi.org/ 10.1126/science.1178994 [DOI] [PubMed] [Google Scholar]
- [11].Huang C, Zhang Z, Xu M, Li Y, Li Z, Ma Y, Cai T, Zhu B. H3.3-H4 tetramer splitting events feature cell-type specific enhancers. PLoS Genetics 2013; 9:e1003558; PMID:23754967; http://dx.doi.org/ 10.1371/journal.pgen.1003558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Katan-Khaykovich Y, Struhl K. Splitting of H3-H4 tetramers at transcriptionally active genes undergoing dynamic histone exchange. Proc Natl Acad Sci U S A 2011; 108:1296-301; PMID:21220302; http://dx.doi.org/ 10.1073/pnas.1018308108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Krassovsky K, Henikoff JG, Henikoff S. Tripartite organization of centromeric chromatin in budding yeast. Proc Natl Acad Sci U S A 2012; 109:243-8; PMID:22184235; http://dx.doi.org/ 10.1073/pnas.1118898109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci U S A 2007; 104:14706-11; PMID:17804787; http://dx.doi.org/ 10.1073/pnas.0706985104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell 2009; 138:104-13; PMID:19596238; http://dx.doi.org/ 10.1016/j.cell.2009.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Henikoff S, Ramachandran S, Krassovsky K, Bryson TD, Codomo CA, Brogaard K, Widom J, Wang JP, Henikoff JG. The budding yeast Centromere DNA Element II wraps a stable Cse4 hemisome in either orientation in vivo. eLife 2014; 3:e01861; PMID:24737863; http://dx.doi.org/ 10.7554/eLife.01861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Brogaard K, Xi L, Wang JP, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature 2012; 486:496-501; PMID:22722846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ramachandran S, Zentner GE, Henikoff S. Asymmetric nucleosomes flank promoters in the budding yeast genome. Genome Res 2015; 25:381-90; PMID:25491770; http://dx.doi.org/ 10.1101/gr.182618.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 2007; 129:1153-64; PMID:17574026; http://dx.doi.org/ 10.1016/j.cell.2007.04.026 [DOI] [PubMed] [Google Scholar]
- [20].Rhee HS, Bataille AR, Zhang L, Pugh BF. Subnucleosomal structures and nucleosome asymmetry across a genome. Cell 2014; 159:1377-88; PMID:25480300; http://dx.doi.org/ 10.1016/j.cell.2014.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Leschziner AE, Saha A, Wittmeyer J, Zhang Y, Bustamante C, Cairns BR, Nogales E. Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc Natl Acad Sci U S A 2007; 104:4913-8; PMID:17360331; http://dx.doi.org/ 10.1073/pnas.0700706104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chaban Y, Ezeokonkwo C, Chung WH, Zhang F, Kornberg RD, Maier-Davis B, Lorch Y, Asturias FJ. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat Struct Mol Biol 2008; 15:1272-7; PMID:19029894; http://dx.doi.org/ 10.1038/nsmb.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol 2005; 12:747-55; PMID:16086025; http://dx.doi.org/ 10.1038/nsmb973 [DOI] [PubMed] [Google Scholar]
- [24].Lorch Y, Maier-Davis B, Kornberg RD. Mechanism of chromatin remodeling. Proc Natl Acad Sci U S A 2010; 107:3458-62; PMID:20142505; http://dx.doi.org/ 10.1073/pnas.1000398107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hsu JM, Huang J, Meluh PB, Laurent BC. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol Cell Biol 2003; 23:3202-15; PMID:12697820; http://dx.doi.org/ 10.1128/MCB.23.9.3202-3215.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kruger W, Peterson CL, Sil A, Coburn C, Arents G, Moudrianakis EN, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev 1995; 9:2770-9; PMID:7590252; http://dx.doi.org/ 10.1101/gad.9.22.2770 [DOI] [PubMed] [Google Scholar]
- [27].Muthurajan UM, Bao Y, Forsberg LJ, Edayathumangalam RS, Dyer PN, White CL, Luger K. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J 2004; 23:260-71; PMID:14739929; http://dx.doi.org/ 10.1038/sj.emboj.7600046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Flaus A, Rencurel C, Ferreira H, Wiechens N, Owen-Hughes T. Sin mutations alter inherent nucleosome mobility. EMBO J 2004; 23:343-53; PMID:14726954; http://dx.doi.org/ 10.1038/sj.emboj.7600047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv 2015; 1:e1500447; PMID:26601204; http://dx.doi.org/ 10.1126/sciadv.1500447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci U S A 2011; 108:18318-23; PMID:22025700; http://dx.doi.org/ 10.1073/pnas.1110731108 [DOI] [PMC free article] [PubMed] [Google Scholar]