ABSTRACT
Exploiting a C. elegans mutant (ncl-1) exhibiting nucleolar abnormalities, we recently identified the let-7/ncl-1/fib-1 genetic cascade underlying proper rRNA abundance and nucleolar size. These 3 factors, let-7 (a miRNA), NCL-1 (a member of the TRIM-NHL family), and fibrillarin (a nucleolar methyltransferase), are evolutionarily conserved across metazoans. In this article, we provide several lines of bioinformatic evidence showing that human and Drosophila homologues of C. elegans NCL-1, TRIM-71 and Brat, respectively, likely act as translational suppressors of fibrillarin. Moreover, since their 3′-UTRs contain putative target sites, they may also be under the control of the let-7 miRNA. We hypothesize that let-7, TRIM and fibrillarin contribute activities in concert, and constitute a conserved network controlling nucleolar size in eukaryotes. We provide an in-depth literature review of various molecular pathways, including the let-7/ncl-1/fib-1 genetic cascade, implicated in the regulation of nucleolar size.
KEYWORDS: C. elegans, DAO-5/Nopp140/Nolc1, fibrillarin, genetic cascade, membrane-less organelle, ribosome biogenesis, translational suppression, tumor suppressor
Introduction
The most prominent structure inside the nucleus is the nucleolus, or “small nucleus,” which was first described in 1835 and later coined in 1838. By 1953, the ultrastructure of this membrane-less organelle was revealed by electron microscopy to be comprised of 3 parts: the dense fibrillar component (DFC), the fibrillar center (FC) and the granular component (GC) (for a historical review, see 1). Since 1962, the nucleolus has been known to be the site of ribosome biogenesis or the “factory of the ribosome,” as evidenced by isotope-tracing of rRNA synthesis and the absence of new rRNA synthesis in Xenopus mutant embryos lacking nucleoli.2 In the last 3 decades, the nucleolus has been shown to be the site of multiple new functions, including signal recognition particle assembly, small RNA modification, cell cycle control, and cell stress sensing.3,4 In addition, it is now known to act as an “incubator” for more than 20 different types of viruses, facilitating their replication and propagation.5,6 The emerging role of the nucleolus as a harbor of regulatory non-coding RNAs (ncRNAs) has also been illustrated by various studies. Works by Audas et al. have revealed that ncRNA transcribed from the nucleolar intergenic spacer capture and immobilize proteins containing a nucleolar detention sequence (NoDS) within the nucleolus in response to diverse stimuli.7 Moreover, recent findings show that more than 10 microRNAs (miRNAs), including miR-664, miR-24, and miR-21—as well as some target mRNAs—reside in the nucleolus, further expanding the number of known nucleolar functions.8-10
Within the last 20 y, Lamond and his colleagues have also identified >700 nucleolar proteins by powerful mass spectrometric tools. Among them, 20% to 32% are associated with ribosomal proteins and/or ribosome biogenesis; however, 32% are novel and uncharacterized.11,12 Taken together, these data reveal that the nucleolus is a multifunctional organelle, and that additional functions may be identified as the >200 novel resident proteins are further characterized.
The nucleolus exhibits dynamic morphological changes when cells enter mitosis. A gradual disappearance of the nucleolus is observed in prophase, while nucleolus assembly (nucleologenesis) is seen at the beginning of telophase. The molecular control of nucleolus disassembly or assembly is associated with the inactivation or activation of rRNA transcription by Pol I, respectively. In this context, the Pol I transcriptional machinery and RNP processing complex are under the control of cell cycle checkpoint components CDK-1, cyclin B kinase, and PP1 phosphatases (for a review, see 13). During nucleolus disassembly, the nucleolar proteins for pre-RNA processing exit the DFC and GC prior to the cessation of rDNA transcription. By contrast, nucleologenesis entails several distinct steps. First, rRNA transcription is initiated in the rDNA cluster domains called nucleolar organizing regions (NOR) at the same time that early and late nucleolar processing proteins accumulate in the prenucleolar bodies (PNBs). Next, the PNBs are targeted to the NOR, the DFC and GC are assembled, and the PNBs become indistinguishable prior to the formation of a mature nucleolus.13
How is nucleolar size controlled?
The mechanism by which the size and scale of organisms, cells, or organelles are controlled is a longstanding and fundamental question in biology. Numerous studies in different model systems have provided important pieces to this puzzle. In 1970, Miller and Gurdon demonstrated that the size of the nucleolus is smaller in Xenopus mutants carrying a haploid rDNA gene.14 Nucleolar size has also been reported to be influenced by the environment or nutrient availability. For example, the nucleolar size of liver cells is changed when rats are subjected to partial hepatoectomy or fed a methionine-free diet.15 Notably, most cancer cells share the feature of enlarged nucleoli. Abnormal nucleolar size and number can be used by pathologists as a cancer marker and to classify malignancy degree.16 Since enlarged nucleoli reflect increased ribosome biogenesis and nuclear DNA synthesis, cells with a larger nucleolus should have larger volumes of the nucleus and cytoplasm. Using the fission yeast model, 2 groups independently demonstrated that nuclear size is determined by the amount of cytoplasm, following a nucleus to cytoplasm (N/C) ratio.17,18 Neumann and Nurse further extended their study to determine a nucleolus to nucleus (No/N) ratio that governs the size of the nucleolus.18 This appears to be true in the case of human sensory ganglia neurons in which the sizes of nuclei and nucleoli increase with cell size.19
For both membrane-enclosed and membrane-less organelles, being proportional in size to the overall dimensions of the cell is called “organelle size scaling.”20 Chan and Marshall proposed the existence of size-sensing control mechanisms either involving direct measurements by sensor molecules or indirect functional readouts.21 Fibrillarin, a nucleolar methyltranferase conserved from archaea to eukaryotes,22 likely serves as one such molecular sensor for regulating nucleolar size.23 Brangwynne and his colleagues introduced the notion that membrane-less nuclear bodies, including the nucleolus, behave like “liquid-phase droplets” and that “phase separation” is a general mechanism for their assembly.24 By using fibrillarin-fused green fluorescence protein (FIB-1::GFP) as a reporter, they demonstrated that nucleolar assembly is controlled by a concentration-dependent phase transition in C. elegans embryos.25 In addition, they showed that rRNA transcription is important to create a thermodynamically favorable condition for the nucleation of nucleolar components at the NOR.26 The finding that fibrillarin can methylate histone H2A at the rDNA loci,27 thereby promoting Pol I transcriptional activity, is in line with the scenario that concentration of fibrillarin plays an important role in regulation of nucleolar activity and size.
Molecules linked to the assembly of membrane-less nuclear bodies are hypothesized to contain a domain of “low sequence complexity” (LCS), such as RG, QN and YG amino acid repeats, for interaction with multiple associating proteins and/or RNAs.28 The LCS domain, a signature feature of intrinsic disorder, is indeed found in many nucleolar proteins, including nucleolin and Collin.29 Using the PONDR program, we discovered LCS domains in C. elegans and human fibrillarins as well as in C. elegans DAO-5/Nopp140, a nucleolar protein to facilitate Pol I transcription activity and its human homolog Nolc1 30 (Fig. 1). Hepatitis D viral antigen (HDAg), which is constantly observed in the nucleolus upon infection, has also been shown to be an intrinsically disordered protein.31 Interestingly, HDAg is perfectly co-localized with fibrillarin in HeLa cells but not in C. elegans intestinal cells, possibly reflecting distinct nucleolar constituents or other host differences between worms and humans.32
Figure 1.
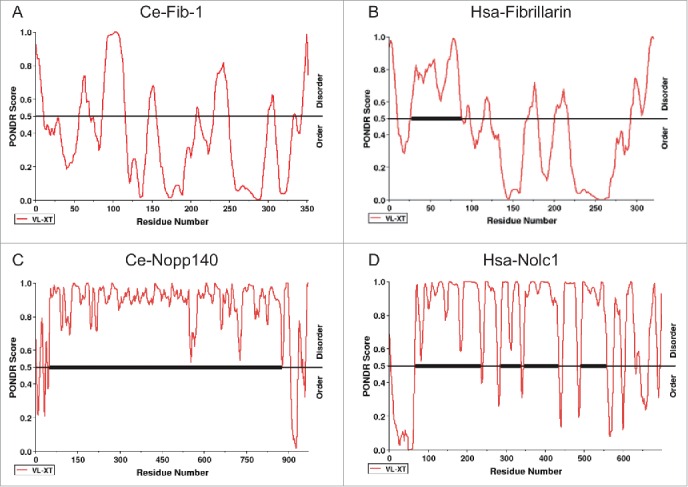
The predicted distribution of the intrinsically disordered regions in 2 major nucleolar proteins, FIB-1/fibrillarin (A and B) and Nopp140/Nolc1 (C and D), in C. elegans (A and C, indicated as Ce-Fib-1 and Ce-Nopp140) and human (B and D, indicated as Hsa-Fibrillarin and Hsa-Nolc1). Results reveal that the thermodynamic patterns and tendencies between the 2 homologous gene sets are conserved. Higher PONDR scores reflect a stronger tendency toward intrinsic structural disorder. Notably, the central region of Nopp140, with a tandem pair of acidic and basic regions,30 significantly coincides with the low-complexity region. Scoring was based on the VX-LT algorithm. The sequence prediction analyses were determined by PONDR (www.pondr.com).
In an attempt to systematically map the species-specific genetic networks that regulate nucleolar size and influence Pol I-mediated transcription, Neumuller et al. recently employed D. melanogaster and S. cerevisiae models to identify conserved or non-conserved molecular complexes.33 Their data indicated that the TRAMP complex (Trf4/Air2/Mtr4p polyadenylation complex) as well as other molecular complexes participating in histone acetylation; poly(A)+ mRNA export; or ER-to Golgi vesicle-mediated transport are evolutionarily conserved regulators of nucleolar size.33 In a genome-wide RNAi screen in Drosophila, they also found that loss of rpl23, sip3a and brat led to enlarged nucleoli whereas loss of tif1a, nopp140 and fib reduced nucleolar size. The mechanisms employed by brat and fibrillarin to regulate size were not identified. Our recent work extended their findings by revealing that NCL-1, a C. elegans homolog of Drosophila Brat, is in fact a translational suppressor of fibrillarin, and its expression is regulated by the let-7 miRNA.23
A genetic cascade of let-7/ncl-1/fib-1 regulates nucleolar size in C. elegans
There are several advantages to using C. elegans as a model for studying nucleolar size regulation. The adult worms are composed of ∼1000 somatic cells with well-characterized cell types and lineages.34 They have a transparent body and can be easily cultured in laboratories. Furthermore, their cells display various sizes of nucleoli ranging from less than 1 to ∼5–7 microns. Intriguingly, nucleoli are undetectable in the −1 oocyte, which is adjacent to the spermatheca and blastomeres of early embryos.32 A previously identified ncl-1 mutant with enlarged nucleoli in nearly all cells 32,35 could serve as a valuable tool for determining the mechanism of nucleolar size control as well as for identifying upstream and downstream genes involved in the process.
In our recent work, we showed that the amount of C. elegans fibrillarin (FIB-1) is correlated with nucleolar size, and that fib-1 acts directly downstream of ncl-1. In wild-type embryos with no observable nucleoli, a smaller amount of FIB-1 was observed compared to ncl-1 mutant embryos, in which nucleoli were clearly detected (Fig. 2A and B). When C. elegans ncl-1 mutants were fed with dsRNA-expressing bacteria to knockdown fib-1 expression, nucleolar size in embryos decreased (Fig. 2A and C), similar to observations in a Drosophila cell culture–based RNAi screen.33 In addition, we demonstrated that NCL-1 cooperates with 2 RNA-binding post-transcriptional regulators, Pumillio (PUM or PUF) and NANOS, to suppress FIB-1 translation by binding to the fib-1 3′-UTR. When the PUF (PUM) binding sequences in the fib-1 3′-UTR were mutated, FIB-1::GFP expression was up-regulated approximately 3-fold.23
Figure 2.
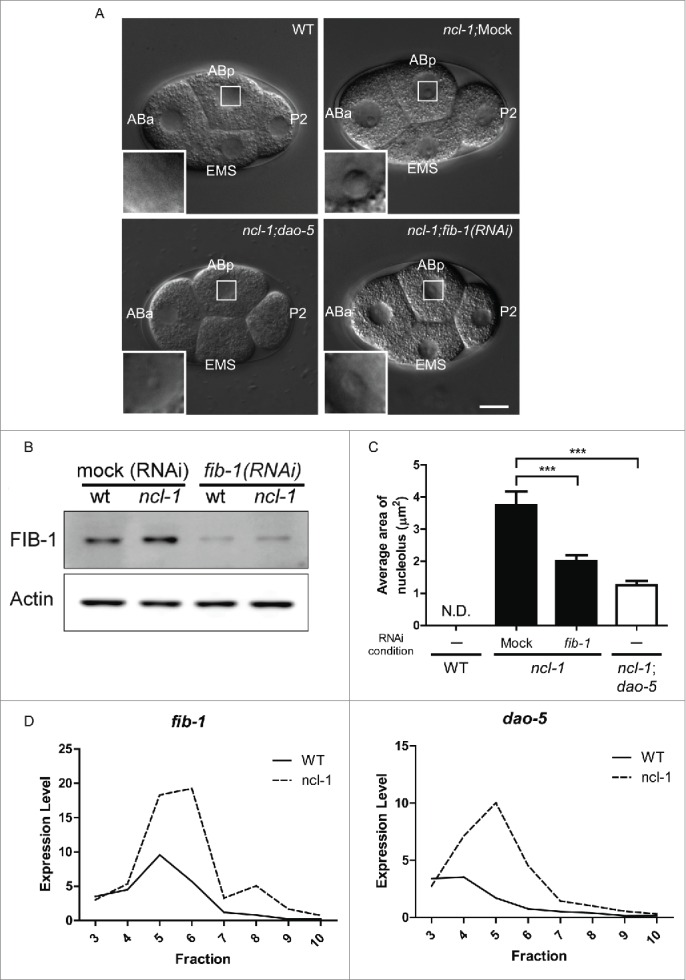
ncl-1 is an upstream translational suppressor of fib-1 and dao-5. The amount of FIB-1 and DAO-5 present can be correlated with the size and function of the nucleolus. (A) Comparison of nucleolar size in embryos of various genetic backgrounds at the 4–cell stage. The nucleolus of ABp cell is highlighted and magnified in the insets. Scale bar, 20 μm. (B) Western blot analysis shows the amount of fibrillarin (FIB-1), which is correlated with the nucleolar size shown in (A). Actin serves as the loading control. (C) Quantitative representation of the nucleolar size from (A). Figures (A–C) are partially adapted and modified from Yi et al., (2015)23 with permission of PLoS Genetics. (D) Total RNAs were extracted from embryos of wild-type (N2) and ncl-1 mutants and fractionated by centrifugation in a sucrose gradient for polysome profile analysis. The amounts of fib-1 and dao-5 mRNA in each fraction were then determined by RT-qPCR and indicated by solid line (N2) or dotted line (ncl-1). The ribosome fractions from low to high sucrose gradients are indicated from left to right, representing a transition from monosomes to polysomes.
Interestingly, absence of Nopp140/dao-5 expression in ncl-1 embryos also led to a reduced nucleolar size (Fig. 2A and C), suggesting that NCL-1 may regulate size by suppressing both fib-1 and dao-5 expression. To test whether NCL-1 is a translational suppressor of fib-1 and dao-5, we performed polysome profile analysis36 of wild-type and ncl-1 embryonic lysates and analyzed fractionation of fib-1 and dao-5 mRNAs. As shown in Fig. 2D, higher levels of fib-1 and dao-5 mRNA were detected in the lighter fractions for wild-type embryos, which represent monosomes and messenger ribonuclear proteins. The distribution of fib-1 or dao-5 mRNAs shifted to the heavier fractions for ncl-1 mutant embryos, strongly indicating that NCL-1 is a translational suppressor of fib-1 and dao-5 expression.
While searching for upstream factors that regulate ncl-1 expression, our bioinformatic analyses uncovered target sequences for 2 miRNAs, let-7 and mir-49, in the ncl-1 3′-UTR.23 We examined the expression of GFP reporters fused with ncl-1 3′-UTRs carrying wild-type or mutated let-7 binding sequences and detected a higher GFP level in animals with the latter. Fluorescence microscopic examination confirmed these GFP reporter expression patterns in seam cells and the vulva, which are known to express higher levels of let-7. Further confirmation of the possibility that ncl-1 is regulated by let-7 was obtained by assessing nucleolar size in vulva cells of a temperature-sensitive hypomorphic let-7(n2853) mutant. Compared to animals grown at the permissive temperature (15°C), we observed a 25% decrease in nucleolar size at the non-permissive temperature (25°C). No discernible difference in nucleolar size was detected when a let-7; ncl-1 double mutant was used for the assay (Fig. 3A and B). Fibrillarin was also expressed at consistently higher levels in let-7(n2853) worms grown at 15°C compared to those grown at the restrictive temperature (25°C), but only a minor difference in expression was seen in the double mutants grown at each temperature (Fig. 3C). Collectively, our findings showed that let-7, ncl-1, and fib-1 function together as a novel genetic pathway to regulate nucleolar size in C. elegans.
Figure 3.
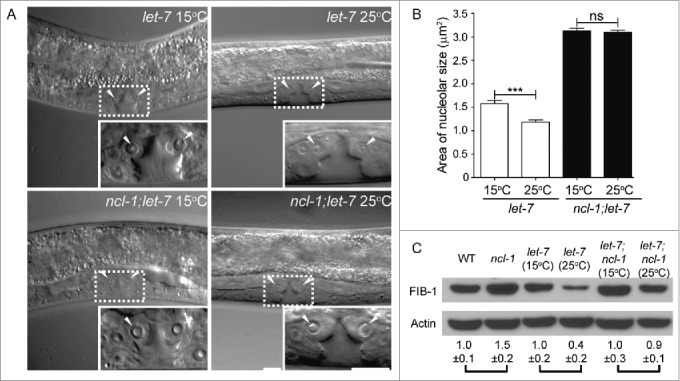
let-7 regulates nucleolar size in vulva cells by controlling fibrillarin expression. (A) DIC microscopy of the vulva cells of let-7(n2853) and let-7(n2853); ncl-1(e1942) worms, at the permissive (15°C) or non-permissive (25°C) temperatures. Insets represent enlarged images of the boxed regions in the corresponding figures. Arrowheads point to the nucleoli of the vulva cells. Scale bar, 10 μm. (B) Quantitative representation of the results shown in (A), illustrating the nucleolar sizes in the vulva. Asterisks signify the difference in sizes observed at the indicated temperatures, where ***P < 0.001, n = 22–36 for let-7(n2853). NS = not significant, n = 100–110 for ncl-1(e1942); let-7(n2853). (C) Western blot analysis of FIB-1 and an Actin control in the indicated strains. Numbers below represent the relative levels of FIB-1 protein expression (normalized to the control sample of each pair-wise comparison). Adapted from Yi et al., (2015)23 with permission from PLoS Genetics.
Conservation of genetic pathways in regulation of nucleolar size
While the fibrillarin gene is structurally unique in most metazoans, the homologues of NCL-1 are divergent from each other in various organisms. NCL-1 homologous proteins belong to the TRIM/RBCC/NHL (NCL-1, HT2A, and LIN-41) family characterized by the presence of a RING domain, a B-box zinc finger, and a coil-coiled domain.37,38 Proteins in the NHL family are divided into 2 groups (groups 1 and 2) based on their C-terminal domain sequences. The TRIM proteins in group 2 possess a C-terminal SPRY domain and are absent in invertebrates, whereas the proteins in group 1 possess a variety of C-terminal domains, including NHL, PHD, and MATH, and are present in both vertebrates and invertebrates.39 The NHL domain folds into 6-bladed β propellers, a structure well known for protein binding. Many TRIM-NHL proteins have now been found to bind to single-stranded RNA.40 A Brat (Drosophila homolog of worm NCL) binding sequence motif (UCGUUG designated as an NHL site) that is distinct from the Pumillio (PUM) binding sequence (UGUAUAUA designated as a PUM site) was recently identified,41,42 providing initial evidence that Brat/NCL-1 can directly target mRNA transcripts to suppress gene expression.
To this end, we identified adjacent NHL and PUM sites in the C. elegans fib-1 3′-UTR (Fig. 4A) as well as in the Drosophila and human fib-1 3′-UTRs. We also discovered multiple NHL sites and a PUM site in the C. elegans dao-5 3′-UTR and in the Drosophila Nopp140 and human Nolc1 3′-UTRs (Fig. 4B). These results suggest that dao-5/nopp140 may also be direct targets of NCL-1, expanding the potential role of NCL-1 in translational regulation across species. To further explore the possible parallels between the C. elegans let-7/ncl-1/fib-1 cascade and related genes in Drosophila and human, we analyzed the brat and TRIM-71 3′-UTRs and found let-7 binding sites in both sequences (Fig. 4C). Taken together, we propose that let-7/trim/fib-1 constitutes a conserved genetic network to control nucleolar size.
Figure 4.
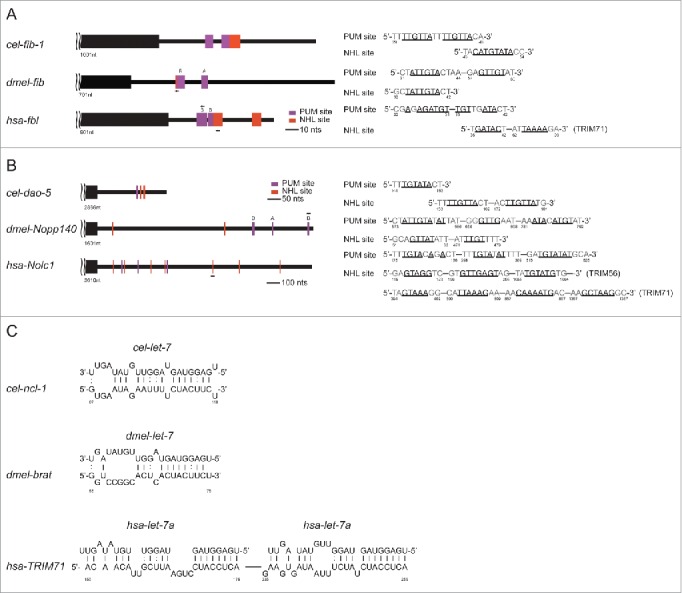
Evolutionary conservation among C. elegans, Drosophila and human of the regulatory cascade including let-7, TRIM/NHL, and fibrillarin as well as dao-5/nopp140/Nolc1. (A and B) The presence of PUM and NHL sites in fib-1/fib/fbl 3′-UTR (A) and in dao-5/nopp140/Nolc1 3′-UTR (B) is depicted. (C) The let-7 binding sites in cel-ncl-1, dmel-brat and hsa-TRIM71 are indicated. let-7 binding sites on hsa-TRIM71 have also been reported previously.46
Interestingly, this pathway is reportedly linked to animal development, lending further support to its functional significance and conservation. During early embryonic development, LIN-28, which is an essential regulator of let-7 biogenesis and a pluripotency factor, mediates nucleolus maturation.43 The pathway has also been implicated in tumor formation. Phylogenic tree analysis revealed that Drosophila brat is most closely related to C. elegans ncl-1 and then to its ortholog Mei-P26.41 Flies without brat exhibit enlargement of nucleoli and are prone to brain tumor formation. However, while the ncl-1 loss-of-function mutant phenocopies the nucleoli enlargement defect and can be rescued by brat, it shows no extra cell proliferation. Based on this observation, we speculate that brat's effect on nucleolus enlargement takes place prior to tumor formation, or that these 2 events are uncoupled. However, within the context of nucleolar size and cell proliferation, brat and p53 share many important functional attributes. Enlargement of nucleoli is the hallmark of many human tumors and can be primarily attributed to the loss of p53,44 and brat and p53 are each known as tumor suppressor genes. Perhaps more importantly, both proteins converge on the regulation of fib-1 expression, with p53 reportedly repressing at the level of transcription 45 and Brat suppressing translation. Collectively, these findings imply that the complex process of nucleolar size control is governed by multiple conserved genetic networks, which likely crosstalk to each other to ensure proper cell proliferation.
Conclusion and perspectives
Recent studies by our group and others have reported numerous conserved complexes and networks with implications in nucleolar size control, providing important insights into the underlying molecular determinants and further reinforcing the complexity of this process. Genome-wide screens in lower eukaryotic models have facilitated the identification of relevant regulators and should be further exploited for in-depth, mechanistic understanding of its regulation. Although we have provided additional evidence to illustrate the evolutionary conservation of regulatory molecules and networks governing nucleolar size, control mechanisms are likely much more complicated in higher eukaryotes given the greater number of cell types and developmental pathways. Understanding at this level thus poses a daunting challenge, but recent advances in genome editing techniques and imaging tools should expedite our understanding of how different networks connect to one another to establish the size of nucleoli across distinct tissues and functions.
Although many new roles of the nucleolus have been explored, ribosome production and assembly have long been regarded as the most critical function of this organelle. The size of the nucleolus closely correlates with Pol I transcription activity and ribosome biogenesis, and is therefore tightly coordinated with metabolic demands of the cell. Consequently, nucleolar size is impinged on by a number of cellular growth and proliferation signal pathways, including oncogenes, such as Ras, Myc and PI3K, and tumor suppressor genes, such as p53, Rb and PTEN.44 However, whether nucleolar size is the driving force for tumor formation or rather a consequence of dis-regulation of cellular signaling or Pol I is still unresolved, and should be thoroughly addressed. Intriguingly, while both NCL-1 and Brat are suppressors of nucleolar size, NCL-1 is distinct in its lack of effect on cellular proliferation, as observed with the Drosophila homolog Brat. To further dissect the implications of this functional separation, genetic screens based on the ncl-1 mutant should be performed to identify putative enhancers of proliferation. Ultimately, a better understanding of the role of the nucleolus in maintaining and/or promoting tumorigenesis could have far-reaching implications with regard to the development of new anti-cancer drugs.
Abbreviations
- NHL
a family protein of NCL-1, HT-2A and LIN-41 in C. elegans
- TRIM-NHL
tripartite motif protein with a C-terminus of NHL- domain
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Katherine Olsson-Carter for critically reviewing the manuscript.
Funding
This work was supported by grants from the Ministry of Education (EMRPD1A043, EMRPD1B0081, and EMRPD1C0051), the Ministry of Science and Technology (MOST 102-2311-B-182-005 and MOST 103-231-B-182-003) and the Chang Gung Memorial Hospital (BMRP742) to SJL; the National Health Research Institute (NHRI-EX105-10321S1), the Ministry of Science and Technology (MOST 104-2320-B-182-029-MY3) and the Chang Gung Memorial Hospital (CMRPDIC0843 and CMRPD3E0152) to BCMT; the National Science Council (NSC 100-2311-B-002-006-MY3), the National Health Research Institute (NHRI-EX102-10151SI) and the National Taiwan University (10R71602A4) to SPC; and the Ministry of Science and Technology (MOST 103-2811-B-182-010 and MOST 104-2811-B-182-021) to YHY.
References
- [1].Lo SJ, Lee CC, Lai HJ. The nucleolus: reviewing oldies to have new understandings. Cell Res 2006; 16:530-8; PMID:16775624; http://dx.doi.org/ 10.1038/sj.cr.7310070 [DOI] [PubMed] [Google Scholar]
- [2].Brown DD, Gurdon JB. Absence of ribosomal RNA synthesis in the anucleolate mutant of Xenopus laevis. Proc Natl Acad Sci U S A 1964; 51:139-46; PMID:14106673; http://dx.doi.org/ 10.1073/pnas.51.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pederson T, Powell K. Thoru Pederson: Spotting novel roles for the nucleolus. J Cell Biol 2015; 208:384-5; PMID:25688131; http://dx.doi.org/ 10.1083/jcb.2084pi [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tsai RY, Pederson T. Connecting the nucleolus to the cell cycle and human disease. FASEB J 2014; 28:3290-6; PMID:24790035; http://dx.doi.org/ 10.1096/fj.14-254680 [DOI] [PubMed] [Google Scholar]
- [5].Pederson T. “Compact” nuclear domains: reconsidering the nucleolus. Nucleus 2010; 1:444-5; PMID:21326828; http://dx.doi.org/ 10.4161/nucl.1.5.13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rawlinson SM, Moseley GW. The nucleolar interface of RNA viruses. Cell Microbiol 2015; 17:1108-20; PMID:26041433; http://dx.doi.org/ 10.1111/cmi.12465 [DOI] [PubMed] [Google Scholar]
- [7].Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell 2012; 45:147-57; PMID:22284675; http://dx.doi.org/ 10.1016/j.molcel.2011.12.012 [DOI] [PubMed] [Google Scholar]
- [8].Politz JC, Hogan EM, Pederson T. MicroRNAs with a nucleolar location. RNA 2009; 15:1705-15; PMID:19628621; http://dx.doi.org/ 10.1261/rna.1470409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bai B, Liu H, Laiho M. Small RNA expression and deep sequencing analyses of the nucleolus reveal the presence of nucleolus-associated microRNAs. FEBS Open Bio 2014; 4:441-49; PMID:24918059; http://dx.doi.org/ 10.1016/j.fob.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Reyes-Gutierrez P, Ritland Politz JC, Pederson T. A mRNA and cognate microRNAs localize in the nucleolus. Nucleus 2014; 5:636-42; PMID:25485975; http://dx.doi.org/ 10.4161/19491034.2014.990864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol 2007; 8:574-85; PMID:17519961; http://dx.doi.org/ 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- [12].Lam YW, Evans VC, Heesom KJ, Lamond AI, Matthews DA. Proteomics analysis of the nucleolus in adenovirus-infected cells. Mol Cell Proteomics 2010; 9:117-30; PMID:19812395; http://dx.doi.org/ 10.1074/mcp.M900338-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hernandez-Verdun D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2011; 2:189-94; PMID:21818412; http://dx.doi.org/ 10.4161/nucl.2.3.16246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Miller L, Gurdon JB. Mutations affecting the size of the nucleolus in Xenopus laevis. Nature 1970; 227:1108-10; PMID:5451103; http://dx.doi.org/ 10.1038/2271108a0 [DOI] [PubMed] [Google Scholar]
- [15].Bailey RP, Vrooman MJ, Sawai Y, Tsukada K, Short J, Lieberman I. Amino acids and control of nucleolar size, the activity of RNA polymerase I, and DNA synthesis in liver. Proc Natl Acad Sci USA 1976; 73:3201-5; PMID:1067612; http://dx.doi.org/ 10.1073/pnas.73.9.3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Derenzini M, Montanaro L, Trere D. What the nucleolus says to a tumour pathologist. Histopathology 2009; 54:753-62; PMID:19178588; http://dx.doi.org/ 10.1111/j.1365-2559.2008.03168.x [DOI] [PubMed] [Google Scholar]
- [17].Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B. The size of the nucleus increases as yeast cells grow. Mol Biol Cell 2007; 18:3523-32; PMID:17596521; http://dx.doi.org/ 10.1091/mbc.E06-10-0973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Neumann FR, Nurse P. Nuclear size control in fission yeast. J Cell Biol 2007; 179:593-600; PMID:17998401; http://dx.doi.org/ 10.1083/jcb.200708054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Berciano MT, Novell M, Villagra NT, Casafont I, Bengoechea R, Val-Bernal JF, Lafarga M. Cajal body number and nucleolar size correlate with the cell body mass in human sensory ganglia neurons. J Structural Biol 2007; 158:410-20; http://dx.doi.org/ 10.1016/j.jsb.2006.12.008 [DOI] [PubMed] [Google Scholar]
- [20].Chan YH, Marshall WF. Scaling properties of cell and organelle size. Organogenesis 2010; 6:88-96; PMID:20885855; http://dx.doi.org/ 10.4161/org.6.2.11464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chan YH, Marshall WF. How cells know the size of their organelles. Science 2012; 337:1186-9; PMID:22955827; http://dx.doi.org/ 10.1126/science.1223539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rodriguez-Corona U, Sobol M, Rodriguez-Zapata LC, Hozak P, Castano E. Fibrillarin from Archaea to human. Biol Cell / Under Auspices European Cell Biol Organization 2015; 107:159-74; http://dx.doi.org/ 10.1111/boc.201400077 [DOI] [PubMed] [Google Scholar]
- [23].Yi YH, Ma TH, Lee LW, Chiou PT, Chen PH, Lee CM, Chu YD, Yu H, Hsiung KC, Tsai YT, et al.. A Genetic Cascade of let-7-ncl-1-fib-1 Modulates Nucleolar Size and rRNA Pool in Caenorhabditis elegans. PLoS Genetics 2015; 11:e1005580; PMID:26492166; http://dx.doi.org/ 10.1371/journal.pgen.1005580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. J Cell Biol 2013; 203:875-81; PMID:24368804; http://dx.doi.org/ 10.1083/jcb.201308087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Weber SC, Brangwynne CP. Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr Biol 2015; 25:641-6; PMID:25702583; http://dx.doi.org/ 10.1016/j.cub.2015.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci U S A 2015; 112:E5237-5245; PMID:26351690; http://dx.doi.org/ 10.1073/pnas.1509317112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tessarz P, Santos-Rosa H, Robson SC, Sylvestersen KB, Nelson CJ, Nielsen ML, Kouzarides T. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 2014; 505:564-8; PMID:24352239; http://dx.doi.org/ 10.1038/nature12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhu L, Brangwynne CP. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol 2015; 34:23-30; PMID:25942753; http://dx.doi.org/ 10.1016/j.ceb.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al.. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 2015; 57:936-47; PMID:25747659; http://dx.doi.org/ 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lee CC, Tsai YT, Kao CW, Lee LW, Lai HJ, Ma TH, Chang YS, Yeh NH, Lo SJ, et al.. Mutation of a Nopp140 gene dao-5 alters rDNA transcription and increases germ cell apoptosis in C. elegans. Cell Death Dis 2014; 5:e1158; PMID:24722283; http://dx.doi.org/ 10.1038/cddis.2014.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Alves C, Cheng H, Roder H, Taylor J. Intrinsic disorder and oligomerization of the hepatitis delta virus antigen. Virology 2010; 407:333-40; PMID:20855099; http://dx.doi.org/ 10.1016/j.virol.2010.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lee LW, Lee CC, Huang CR, Lo SJ. The nucleolus of Caenorhabditis elegans. J Biomed Biotech 2012; 2012:601274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Neumuller RA, Gross T, Samsonova AA, Vinayagam A, Buckner M, Founk K, Hu Y, Sharifpoor S, Rosebrock AP, Andrews B, et al.. Conserved regulators of nucleolar size revealed by global phenotypic analyses. Sci Signal 2013; 6:ra70; PMID:23962978; http://dx.doi.org/ 10.1126/scisignal.2004145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 1983; 100:64-119; PMID:6684600; http://dx.doi.org/ 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- [35].Frank DJ, Roth MB. ncl-1 is required for the regulation of cell size and ribosomal RNA synthesis in Caenorhabditis elegans. J Cell Biol 1998; 140:1321-9; PMID:9508766; http://dx.doi.org/ 10.1083/jcb.140.6.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lai MC, Sun HS, Wang SW, Tarn WY. DDX3 functions in antiviral innate immunity through translational control of PACT. FEBS J 2016; 283:88-101; PMID:26454002; http://dx.doi.org/ 10.1111/febs.13553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 2000; 5:659-69; PMID:10882102; http://dx.doi.org/ 10.1016/S1097-2765(00)80245-2 [DOI] [PubMed] [Google Scholar]
- [38].Slack FJ, Ruvkun G. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem Sci 1998; 23:474-5; PMID:9868369; http://dx.doi.org/ 10.1016/S0968-0004(98)01299-7 [DOI] [PubMed] [Google Scholar]
- [39].Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al.. The tripartite motif family identifies cell compartments. EMBO J 2001; 20:2140-51; PMID:11331580; http://dx.doi.org/ 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Loedige I, Stotz M, Qamar S, Kramer K, Hennig J, Schubert T, Löffler P, Längst G, Merkl R, Urlaub H, et al.. The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev 2014; 28:749-64; PMID:24696456; http://dx.doi.org/ 10.1101/gad.236513.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Loedige I, Jakob L, Treiber T, Ray D, Stotz M, Treiber N, Hennig J, Cook KB, Morris Q, Hughes TR, et al.. The Crystal Structure of the NHL Domain in Complex with RNA Reveals the Molecular Basis of Drosophila Brain-Tumor-Mediated Gene Regulation. Cell Reports 2015; 13:1206-20; PMID:26527002; http://dx.doi.org/ 10.1016/j.celrep.2015.09.068 [DOI] [PubMed] [Google Scholar]
- [42].Laver JD, Li X, Ray D, Cook KB, Hahn NA, Nabeel-Shah S, Kekis M, Luo H, Marsolais AJ, Fung KY, et al.. Brain tumor is a sequence-specific RNA-binding protein that directs maternal mRNA clearance during the Drosophila maternal-to-zygotic transition. Genome Biol 2015; 16:94; PMID:25962635; http://dx.doi.org/ 10.1186/s13059-015-0659-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Vogt EJ, Meglicki M, Hartung KI, Borsuk E, Behr R. Importance of the pluripotency factor LIN28 in the mammalian nucleolus during early embryonic development. Development 2012; 139:4514-23; PMID:23172912; http://dx.doi.org/ 10.1242/dev.083279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Woods SJ, Hannan KM, Pearson RB, Hannan RD. The nucleolus as a fundamental regulator of the p53 response and a new target for cancer therapy. Biochimica Et Biophysica Acta 2015; 1849:821-9; PMID:25464032; http://dx.doi.org/ 10.1016/j.bbagrm.2014.10.007 [DOI] [PubMed] [Google Scholar]
- [45].Marcel V, Ghayad SE, Belin S, Therizols G, Morel AP, Solano-Gonzàlez E, Vendrell JA, Hacot S, Mertani HC, Albaret MA, et al.. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 2013; 24:318-30; PMID:24029231; http://dx.doi.org/ 10.1016/j.ccr.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lin YC. Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Mol Biol Evolution 2007; 24:2525-34; http://dx.doi.org/ 10.1093/molbev/msm195 [DOI] [PubMed] [Google Scholar]


