ABSTRACT
Herpesviruses are large DNA viruses that utilize the host nucleus for genome replication as well as capsid assembly. After maturation, these 125 nm large capsid assemblies must cross the nucleoplasm to engage the nuclear envelope and bud into the cytoplasm. Here we summarize our recent findings how this motility is facilitated. We suggest that herpesvirus induced nuclear remodeling allows capsids to move by diffusion in the nucleus and not by motor-dependent transport.
KEYWORDS: capsid, egress, herpes, nucleus, nuclear actin, ring-sheet
Introduction
Herpesvirus virions are enveloped and contain an icosahedral capsid carrying the doubled stranded DNA genome. The virions enter the host cell through fusion at the plasma membrane or in endocytic vesicles. Capsids are released into the cytoplasm where they engage molecular motors to move to the nuclear vicinity on microtubules. Here, capsids attach to nuclear pores and the viral genome is injected into the nucleus. The genome is then transcribed and replicated. New capsids form inside the nucleus and are filled with viral genomes. Afterwards, filled capsids leave the nucleus through a unique budding process. During this process, called nuclear egress, nuclear capsids first bud into the inner nuclear membrane, which results in an enveloped intermediate in the perinuclear space. This intermediate particle subsequently fuses with the outer nuclear membrane, releasing the capsid into the cytoplasm. Next, more viral proteins surround the capsid forming the tegument. This particle then acquires its final envelope by budding into the compartment of secondary envelopment that has markers of trans-Golgi membranes or endocytic membranes. Finally, the vesicle containing the mature enveloped particle fuses with the plasma membrane, releasing the mature virion in an exocytosis-like event.1-3
In the last 20 y or so it became very clear that both during entry as well as exit, herpesvirus particles use microtubule-associated, motor-dependent transport to cross the cytoplasmic space.4-6 However much less is known of how newly formed and packaged capsids cross the nuclear space to the reach the inner nuclear membrane for egress.
Nuclear capsid motility is not dependent on F-Actin
Shortly after the first herpesvirus expressing GFP-labeled capsids were constructed,7,8 it was apparent that capsids in the infected cell nucleus were highly motile. An early report suggested that this motility was based on a directed, ATP-dependent transport mechanism that relies on filamentous, nuclear actin (F-actin), possibly facilitating the transport of capsids to the nuclear envelope for egress. Myosin V was also implicated in this process as potential motor protein.9 Results from our lab also supported the hypothesis that herpesvirus infection induces nuclear F-actin at least in certain neurons and that Myosin V colocalizes with nuclear capsid accumulations.10 However, there was no direct evidence visualizing capsids transporting on F-actin. Instead, all evidence to this point was either based on inhibitors of F actin formation 9 or on Phalloidin staining in fixed cells.10
To test this model, we first attempted to visualize nuclear F-actin in living cells with the actin probe Lifeact.11 However we failed to detect nuclear F-actin in infected cells with Lifeact. Phalloidin staining in fixed cells gave the same results.12 We could only detect nuclear F-actin structures in infected superior cervical ganglion (SCG) neurons using Phalloidin as reported earlier, but not with Lifeact. We were therefore unable to directly visualize if nuclear F-actin structures support capsid motility because i) all cell types tested with the exception of primary cultures of SCG neurons did not induce detectable nuclear F-actin ii) Nuclear F-actin in SCG neurons could only be detected after fixation and staining with fluorescent phalloidin.
As we failed to detect nuclear F-actin in all but SCG neurons after infection, we reanalyzed the original interpretations. The key finding in the original report9 was the inhibition of nuclear capsid motility by the F-actin depolymerizing drug Latrunculin A (LatA). We therefore repeated the original experiment using herpesviruses representing all 3 subfamilies and Lifeact expressing murine embryonic fibroblasts. This allowed us to monitor capsids as well as changes in actin dynamics in living cells. We indeed found that capsid motility stopped after addition of LatA and could quantify the effect using image correlation microscopy (ICM). However the block of motility was not due to the destruction of nuclear F-actin BUT due the induction of nuclear cofilin-actin rods to which capsids bound and were immobilized. Nuclear cofilin-actin rods are known to form in instances when high amounts of globular actin (G-actin) accumulate in the nucleus due the F-actin dissolving action of Latrunculin.13-15 These rods are most likely an artifact of Latrunculin action. Interestingly these rods have the capacity to bind herpesvirus capsids, which might have led to the original interpretation that nuclear F-Actin is involved in nuclear capsid motility as LatA stops this motility. In addition we could show that cells that do not induce actin rods upon LatA addition also show no difference in capsid motility and that 3 other F-actin depolymerizing drugs did not have an effect on bulk capsid motility as measured by ICM.12
Establishing a single particle tracking methodology that facilitates quantifying nuclear capsid motility
We concluded that nuclear F-Actin did not facilitate capsid motility. However, our ICM method only provided data on bulk capsid behavior and did not distinguish between directed and non-directed motility. We therefore wanted a method that enabled single particle tracking. Such a method would help determine if capsids were indeed actively transported as proposed originally. To quantify the data and avoid subjective biases, we implemented an automated tracking script. However as capsid motility is rapid and improved capsid-tagged mutants16 produced many more capsids in the nucleus, we needed a system that acquired images at high frame rates in the range of 30 and more frames per second (fps). Unfortunately, this speed of image capture would reduce the available signal dramatically so that commercial laser scanning confocal microscopy or spinning disk microscopy could not be used. Even the use of high sensitivity electron multiplying charged-coupled detectors (EM-CCD) on a spinning disk confocal microscope, was still not suitable for the task. Instead, we found that oblique illumination as a widefield method in combination with EM-CCD cameras overcame these problems and gave superior results. Oblique microscopy is a form of fluorescence dark field microscopy in which a collimated laser beam exits the objective at an angle that is does not lead to total internal reflection (TIRF) as used in TIRF microscopy. Instead the beam penetrates the sample at a relative flat angle almost as if the sample is illuminated by a horizontal light sheet. This results in an increased sectioning capability as out of focus fluorescence is reduced. In its classical implementation in which it is also called highly inclined and laminated sheet microscopy (HILO),17 the illuminating light enters the sample from one side only. This can lead to shading artifacts. Accordingly, we modified this setup by using a 2D galvonometer system that is capable of effectively rotating the direction of illumination several times per exposure. This way all shading artifacts as well as laser fringes are averaged out. As this technique rotates the oblique light sheet, we dubbed this method “Ring-Sheet.” This technology allowed us to image nuclear motility at very high speed and sensitivity, which resulted in superior data quality that allowed automated particle tracking. In addition, we added a cylindrical lens into the imaging path such that we could determine particle z-positions by the introduced astigmatism. For particle position fitting in x, y, and z, we used the ImageJ plugin Quickpalm and fed the resulting list into a Matlab script that uses SimpleTracker and MSD analyzer by Jean-Yves Tinevez.18 This methodology allowed us to localize mobile nuclear virus particles to about +/− 40 nm in all 3 dimensions.
Nuclear herpesvirus capsids diffuse in enlarged nuclear corrals
Analyzing the ensemble mean squared displacement (MSD) of thousands of particle tracks for 2 different herpesviruses revealed that nuclear capsids diffused almost freely early in infection and on timescale smaller than 1 second. We also analyzed each track separately and found that their anomalous diffusion exponents distributed normally (Gaussian) with a mean of 0.85+/− 0.26 with no recognizable subpopulations, again indicating that capsids diffusion over short timescales was essentially unobstructed.
This finding was surprising, as previous reports had shown that the uninfected nucleus has a sponge-like structure consisting of chromatin and the interchromatin compartment. This sponge structure allows small proteins like GFP to diffuse unobstructed, while the motility of macromolecular complexes like RNP particles and capsid-sized latex beads is confined to corrals of about 300 nm in diameter 19-21 which corresponds to a volume of 0.014 µm3 for an idealized sphere. We could confirm this finding for non-infected cells by microrheology using microinjected, capsid-sized beads. Interestingly however, the corrals in infected cells had an about 27 fold larger volume (of 0.38 µm3 or 900 nm in diameter), using beads as well as virus capsids as probes. We could confirm this result also by staining host chromatin. In infected cells, chromatin was becoming more porous at 4 h and moved to the nuclear periphery with full marginalization at 6 h post infection as described earlier.22 It was clear that herpesvirus infection increases the nuclear corral size, which may allow capsids to diffuse freely.
Active diffusion allows nuclear capsids to hop between corrals
We found rare cases of viral particles moving with more directed motion, which we dubbed “capsid hopping” as it allowed particles to cross from one corral into neighboring corrals. This motility could be due to direct motor-driven transport of capsids or to the activity of unrelated energy-consuming processes that kick diffusing capsids in one direction (known as active diffusion).23 To distinguish between these 2 possibilities, we microinjected passivated beads into infected cells and compared their displacement probabilities to nuclear capsids. We found that the probabilities were indistinguishable, which let us conclude that rare capsid hopping must be due to active diffusion and not direct motor-dependent motility.
Viral remodeling of the nuclear architecture allows efficient capsid flux by diffusion in the absence of a motor-dependent transport mechanism
As described above, chromatin in infected cells was marginalized at the nuclear periphery at 6 hours post infection. Surprisingly, in Pseudorabies virus (PRV) infected cells, capsids did not fill up the chromatin depleted space in the nuclear center in infected cells. Instead, capsids were redistributed to the nuclear periphery. The nuclear interior instead was filled by the viral replication compartment (RC), the site of viral DNA replication as visualized by the viral single-stranded binding protein (a classic marker of replication compartments,24). We speculate that at least in the case of PRV the viral RC, filled with viral DNA and proteins might be so dense that it physically excludes capsids. As a result, capsids accumulate between the marginalized chromatin and the RC. This mechanism might be very advantageous for virus production as it concentrates capsids near the nuclear envelope such that a high percentage of capsids can access sites of egress by diffusion. However even without this concentration effect, around 50% of the nuclear volume is directly accessible to the nuclear membrane if we assume a nuclear volume of about 570 µm3 (as in our experimental system) and a space-filling array of corrals along the nuclear envelope with an average volume of 0.38 µm3 (for illustration see also Fig. 1b). In addition, hopping between corrals might allow an even higher proportion of particles to reach the inner nuclear membrane.
Figure 1.
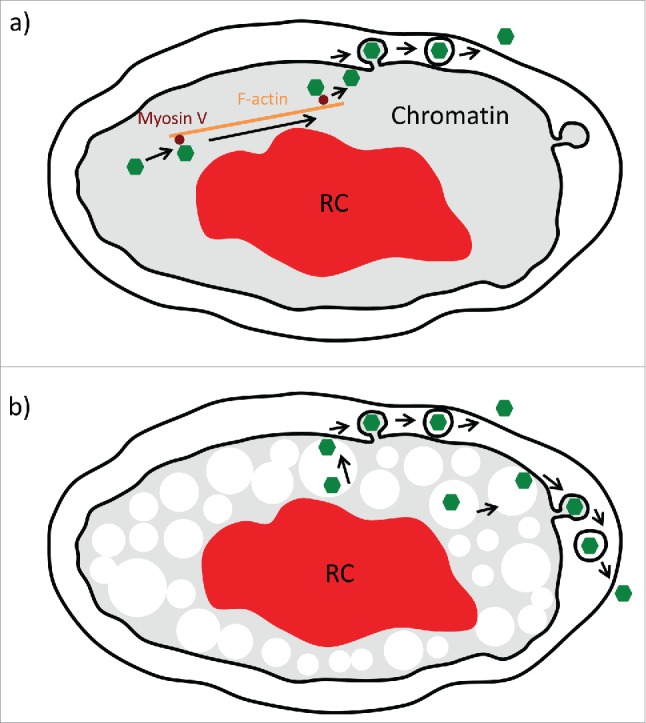
Models of nuclear capsid motility. a) Previous work suggested that herpesvirus infection induces nuclear F-actin (orange) that is used by viral capsids (green) for directed motility by recruiting Myosin V (brown), allowing transport through chromatin (gray) to the inner nuclear membrane (black inner circle). In this model, capsids attach and exit the nucleus by budding through the inner nuclear membrane and then fusing with the outer nuclear envelope (outer black circle). b) Our alternative model suggests that herpes infection enlarges nuclear corrals to an average of about 900 nm (white circles). Capsids form stochastically throughout the nucleus early in infection and can diffuse almost unobstructed in these enlarged corrals. In case of pseudorabies virus, the expansion of the viral replication compartment (RC, red) further concentrates capsids to the nuclear periphery. Capsids can reach the inner nuclear membrane by 2 pathways: Either diffusion in corrals located next to the membrane (left side) or by hopping between corrals by active diffusion (right side). In both cases, a high affinity/avidity binding system at the inner nuclear membrane is needed to provide a binding “sink” to capture the stochastic interactions for inner nuclear membrane budding.
These simple calculations argue for a model in which viral remodeling of nuclear architecture allows capsids to access the inner nuclear membrane from almost any starting position inside the nucleus without the need for a direct motor-dependent transport. This model might also help to understand the apparent overproduction of herpesviral particles in the nucleus. Based on a recent report, the number of nuclear capsids for a related herpesvirus is on the order of 3*104.25 However, the yield of mature virons is in the range of 2*103 particles per cell for the most productive viruses like PRV. These numbers mean that less than 10% of viral capsids produced in the nucleus will egress from the nucleus if the downstream morphogenesis steps have a reasonable efficiency. Overproduction of nuclear capsids might therefore insure that stochastic processes for nuclear egress like diffusion are not limiting.
Summary
Our results argue for a model in which the well-known herpesvirus-induced changes of the chromatin structure 22 enable capsids to escape the nucleus by Brownian motion. This model may be counterintuitive since long-range cytoplasmic transport of herpes particles requires motor-dependent motility. Our model offers a simple example showing that long-range capsid motility is not needed in the infected nucleus. First, to our knowledge, there is no mechanism described that concentrates immature capsid proteins or subassemblies in certain nuclear regions. Indeed, early FRAP results have shown that individual proteins have access to almost the whole nuclear space through rapid diffusion.26 This fact suggests that capsids will assemble stochastically throughout the nuclear space as long as the concentration of constituents is over a certain threshold. Further, we know from our work that capsids can move undisturbed in 0.38 µm3 corrals. A spacefilling array of on average 0.38 µm3 corrals around the nuclear envelope would include around 50% or more of the nuclear space depending on nuclear size and morphology as described above. This fact means that 50% or more of stochastically formed capsids have direct access to the inner nuclear membrane through passive diffusion. In addition, we show that capsids can hop between corrals, albeit with a much lower probability. As a result, even capsids that form in the center of the nucleus might be able to reach the nuclear envelope by a combination of hopping/active diffusion and diffusion.
What comes next?
There are at least 4 major question arising from our work:
How is chromatin remodeling achieved? Is it an active biological process like an antiviral host response or a biophysical phenomenon based on attraction depletion27 or phase separation28,29? Other viruses like parvo- and baculoviruses30,31 show similar phenotypes, arguing for a common mechanism.
How is efficient nuclear egress facilitated? If nuclear capsid transport is not actively directed toward sites of nuclear egress but instead is stochastic, the sites of nuclear egress must act as high affinity or avidity sinks for nuclear capsids to enable efficient capsid export. The use of specifically labeled viral mutants and the recently described structure of the PRV nuclear egress complex might enable this.32,33 However our early results suggests that a thorough quantification of capsid fate is only possible if we use imaging methodologies that use the available photon budget very efficiently. Developments in light sheet microscopy might ultimately allow us to reach this goal, resulting in a whole cell, “DynOmics” view of capsid dynamics.
Who is kicking? Or better what are the sources of active diffusion? In general, every energy-consuming process would be able to “kick” particles.23 Therefore it might be unlikely that certain processes are the sole source. But likely candidates might be chromatin remodeling and viral polymerase activity. Another option, might be a very recently described form of nuclear actin,34 which plays a role in DNA double stranded break (DSB) repair. As herpesvirus replication compartments recruit various proteins involved in DSB repair, possibly for recombination-dependent replication35 it might be that this new form of actin plays a role in kicking viral particles. It is however not known if this new form of nuclear actin can be disturbed by the panel of drugs used in our recent ICM study.12 Future work using single particle tracking is required to evaluate this idea, possibly using further advanced particle localization algorithms that can detect single particles even at high densities.36
Why do PRV capsids accumulate around the viral RC while HSV-1 capsids accumulate in the RC? We hypothesize that compartment density plays a role. Microrheology using capsid-sized beads might allow to measure RC density later in infections, however technical challenges must be solved first.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank all of our co-authors of our 2 recent papers in which we re-evaluated nuclear herpes motility for their year-long support.
Funding
This work was supported by National Institutes of Health (NIH) Grants NS033506 and NS060699 (to L.W.E.). J.B.B. was supported by a DFG fellowship during the work described here (BO 4158/1-1) and is currently supported by a DFG reintegration grant (BO 4158/2-1). We apologize to all colleagues whose work could not be cited due to space restrictions.
References
- [1].Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: an update. Virus Res 2009; 143:222-34; PMID:19651457; http://dx.doi.org/ 10.1016/j.virusres.2009.03.018 [DOI] [PubMed] [Google Scholar]
- [2].Mettenleiter TC, Müller F, Granzow H, Klupp BG. The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol 2012; 15:170-8; PMID:23057731 [DOI] [PubMed] [Google Scholar]
- [3].Hogue IB, Bosse JB, Hu J-R, Thiberge SY, Enquist LW. Cellular mechanisms of alpha herpesvirus egress: live cell fluorescence microscopy of pseudorabies virus exocytosis. PLoS Pathog 2014; 10:e1004535; PMID:25474634; http://dx.doi.org/ 10.1371/journal.ppat.1004535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Radtke K, Döhner K, Sodeik B. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Microbiol 2006; 8:387-400; PMID:16469052; http://dx.doi.org/ 10.1111/j.1462-5822.2005.00679.x [DOI] [PubMed] [Google Scholar]
- [5].Lyman MG, Enquist LW. Herpesvirus interactions with the host cytoskeleton. J Virol 2009; 83:2058-66; PMID:18842724; http://dx.doi.org/ 10.1128/JVI.01718-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Koyuncu OO, Hogue IB, Enquist LW. Virus infections in the nervous system. Cell Host Microbe 2013; 13:379-93; PMID:23601101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol 1998; 72:7563-8; PMID:9696854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Smith GA, Gross SP, Enquist LW. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc Natl Acad Sci 2001; 98:3466-70; PMID:11248101; http://dx.doi.org/ 10.1073/pnas.061029798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Forest T, Barnard S, Baines JD. Active intranuclear movement of herpesvirus capsids. Nat Cell Biol 2005; 7:429-31; PMID:15803134; http://dx.doi.org/ 10.1038/ncb1243 [DOI] [PubMed] [Google Scholar]
- [10].Feierbach B, Piccinotti S, Bisher M, Denk W, Enquist LW. Alpha-herpesvirus infection induces the formation of nuclear actin filaments. PLoS Pathog 2006; 2:e85; PMID:16933992; http://dx.doi.org/ 10.1371/journal.ppat.0020085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al.. Lifeact: a versatile marker to visualize F-actin. Nat Methods 2008; 5:605-7; PMID:18536722; http://dx.doi.org/ 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bosse JB, Virding S, Thiberge SY, Scherer J, Wodrich H, Ruzsics Z, Koszinowski UH, Enquist LW. Nuclear herpesvirus capsid motility is not dependent on f-actin. mBio 2014; 5:e01909-14-e01909-14; PMID:25293761; http://dx.doi.org/ 10.1128/mBio.01909-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pendleton A, Pope B, Weeds A, Koffer A. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem 2003; 278:14394-400; PMID:12566455; http://dx.doi.org/ 10.1074/jbc.M206393200 [DOI] [PubMed] [Google Scholar]
- [14].Domazetovska A, Ilkovski B, Cooper ST, Ghoddusi M, Hardeman EC, Minamide LS, Gunning PW, Bamburg JR, North KN. Mechanisms underlying intranuclear rod formation. Brain 2007; 130:3275-84; PMID:17928315; http://dx.doi.org/ 10.1093/brain/awm247 [DOI] [PubMed] [Google Scholar]
- [15].de Lanerolle P, Serebryannyy L. Nuclear actin and myosins: life without filaments. Nat Cell Biol 2011; 13:1282-8; PMID:22048410; http://dx.doi.org/ 10.1038/ncb2364 [DOI] [PubMed] [Google Scholar]
- [16].Hogue I, Bosse J, Engel E, Scherer J, Hu J-R, del Rio T, Enquist L. Fluorescent protein approaches in alpha herpesvirus research. Viruses 2015; 7:5933-61; PMID:26610544; http://dx.doi.org/ 10.3390/v7112915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Meth 2008; 5:159-61; PMID:18176568; http://dx.doi.org/ 10.1038/nmeth1171 [DOI] [PubMed] [Google Scholar]
- [18].Tarantino N, Tinevez J-Y, Crowell EF, Boisson B, Henriques R, Mhlanga M, Agou F, Israël A, Laplantine E. TNF and IL-1 exhibit distinct ubiquitin requirements for inducing NEMO-IKK supramolecular structures. The Journal of Cell Biology 2014; 204:231-45; PMID:24446482; http://dx.doi.org/ 10.1083/jcb.201307172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Görisch SM, Wachsmuth M, Ittrich C, Bacher CP, Rippe K, Lichter P. Nuclear body movement is determined by chromatin accessibility and dynamics. Proc Natl Acad Sci USA 2004; 101:13221-6; PMID:15331777; http://dx.doi.org/ 10.1073/pnas.0402958101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Görisch SM, Lichter P, Rippe K. Mobility of multi-subunit complexes in the nucleus: accessibility and dynamics of chromatin subcompartments. Histochem Cell Biol [Internet] 2005; 123:217-28. Available from: http://www.springerlink.com/index/10.1007/s00418-005-0752-y; PMID:15830242; http://dx.doi.org/ 10.1007/s00418-005-0752-y [DOI] [PubMed] [Google Scholar]
- [21].Tseng Y, Lee JSH, Kole TP, Jiang I, Wirtz D. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J Cell Sci [Internet] 2004; 117:2159-67. Available from: http://jcs.biologists.org/cgi/doi/10.1242/jcs.01073; PMID:15090601; http://dx.doi.org/ 10.1242/jcs.01073 [DOI] [PubMed] [Google Scholar]
- [22].Monier K, Armas JC, Etteldorf S, Ghazal P, Sullivan KF. Annexation of the interchromosomal space during viral infection. Nat Cell Biol 2000; 2:661-5; PMID:10980708; http://dx.doi.org/ 10.1038/35023615 [DOI] [PubMed] [Google Scholar]
- [23].Brangwynne CP, Koenderink GH, MacKintosh FC, Weitz DA. Cytoplasmic diffusion: molecular motors mix it up. J Cell Biol 2008; 183:583-7; PMID:19001127; http://dx.doi.org/ 10.1083/jcb.200806149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].de Bruyn Kops A, Knipe DM. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 1988; 55:857-68; PMID:2847874; http://dx.doi.org/ 10.1016/0092-8674(88)90141-9 [DOI] [PubMed] [Google Scholar]
- [25].Villinger C, Neusser G, Kranz C, Walther P, Mertens T. 3D analysis of HCMV induced-nuclear membrane structures by FIB/SEM tomography: insight into an unprecedented membrane morphology. Viruses 2015; 7:5686-704; PMID:26556360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Görisch SM, Lichter P, Rippe K. Mobility of multi-subunit complexes in the nucleus: accessibility and dynamics of chromatin subcompartments. Histochem Cell Biol 2005; 123:217-28; PMID:15830242; http://dx.doi.org/ 10.1007/s00418-005-0752-y [DOI] [PubMed] [Google Scholar]
- [27].Marenduzzo D, Finan K, Cook PR. The depletion attraction: an underappreciated force driving cellular organization. J Cell Biol 2006; 175:681-6; PMID:17145959; http://dx.doi.org/ 10.1083/jcb.200609066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhu L, Brangwynne CP. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol 2015; 34:23-30; PMID:25942753; http://dx.doi.org/ 10.1016/j.ceb.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. J Cell Biol 2013; 203:875-81; PMID:24368804; http://dx.doi.org/ 10.1083/jcb.201308087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ihalainen TO, Niskanen EA, Jylhävä J, Paloheimo O, Dross N, Smolander H, Langowski J, Timonen J, Vihinen-Ranta M. Parvovirus induced alterations in nuclear architecture and dynamics. PLoS One 2009; 4:e5948; PMID:19536327; http://dx.doi.org/ 10.1371/journal.pone.0005948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Laakkonen JP, Kaikkonen MU, Ronkainen PHA, Ihalainen TO, Niskanen EA, Häkkinen M, Salminen M, Kulomaa MS, Ylä-Herttuala S, Airenne KJ, et al.. Baculovirus-mediated immediate-early gene expression and nuclear reorganization in human cells. Cell Microbiol 2008; 10:667-81; PMID:18042259; http://dx.doi.org/ 10.1111/j.1462-5822.2007.01074.x [DOI] [PubMed] [Google Scholar]
- [32].Hagen et al. Structural Basis of Vesicle Formation at the Inner Nuclear Membrane. Cell 2015; 163(7):1692-1701; http://dx.doi.org/10.1016/j.cell.2015.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zeev-Ben-Mordehai et al. Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights into Inner Nuclear Membrane Remodeling. Cell Report 2015; 13(12):2645-2652; http://dx.doi.org/10.1016/j.celrep.2015.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Belin BJ, Lee T, Mullins RD, Lappalainen P. DNA damage induces nuclear actin filament assembly by Formin-2 and Spire-1/2 that promotes efficient DNA repair. eLife 2015; 4:e07735; PMID:26287480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Weller SK, Coen DM. Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harbor Perspect Biol 2012; 4:a013011-1; PMID:22952399; http://dx.doi.org/ 10.1101/cshperspect.a013011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chenouard N, Smal I, de Chaumont F, Maška M, Sbalzarini IF, Gong Y, Cardinale J, Carthel C, Coraluppi S, Winter M, et al.. Objective comparison of particle tracking methods. Nat Meth 2014; 11:281-9; PMID:24441936; http://dx.doi.org/ 10.1038/nmeth.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]


