SUMMARY
What is the nature of evolutionary divergence of the jaw skeleton within the genus Oncorhynchus? How can two associated bones evolve new shapes and still maintain functional integration? Here, we introduce and test a ‘concordance’ hypothesis, in which an extraordinary matching of the evolutionary shape changes of the dentary and angular articular serves to preserve their fitting together. To test this hypothesis, we examined morphologies of the dentary and angular articular at parr (juvenile) stage, and at three levels of biological organization – between salmon and trout, between sister species within both salmon and trout, and among three types differing in life histories within one species, O. mykiss. The comparisons show bone shape divergences among the groups at each level; morphological divergence between salmon and trout is marked even at this relatively early life history stage. We observed substantial matching between the two mandibular bones in both pattern and amount of shape variation, and in shape covariation across species. These findings strongly support the concordance hypothesis, and reflect functional and/or developmental constraint on morphological evolution. We present evidence for developmental modularity within both bones. The locations of module boundaries were predicted from the patterns of evolutionary divergences, and for the dentary, at least, would appear to facilitate its functional association with the angular articular. The modularity results suggest that development has biased the course of evolution.
INTRODUCTION
Salmonids within the genus Oncorhynchus have been well studied (Groot et al. 2002; Quinn 2005). These fish, including salmon and trout of the Pacific Northwest of North America where the group likely originated, have been considered model organisms in evolutionary biology (Hendry and Sterns 2004; Noakes 2014). Oncorhynchus species show a variety of life history traits. Prominently, anadromy versus freshwater residency is widespread in the genus: O. mykiss trout famously evolve between fresh-water resident rainbow and ocean-going anadromous steelhead rapidly, and apparently independently in separate drainages (Docker and Heath 2003; McPhee et al. 2007; Christie et al. 2011; Pearse et al. 2014). Within-species variation also exists for the time of year when anadromous salmonids return to fresh water to breed – Spring-run versus Fall-run Chinook salmon, O. tshawytscha, and Summer versus Winter steelhead. Body morphology covaries with some of these behavioral differences (Carl and Healey 1984; Tiffan et al. 2000; Varian and Nichols 2010; Tiffan and Connor 2011; Billman et al. 2014). Body morphology is largely underlain by skeletal morphology, and the salmonid skeleton provides classical, textbook material in fish biology (Goodrich 1930; de Beer 1937). However, surprisingly little is known about likely life-history, evolutionary, or phenotypically plastic changes in the skeleton within Oncorhynchus. Our literature search revealed only a single comparative paper examining the Oncorhynchus skull. Qualitative in nature, it described the skull morphologies of adult salmon (Vladykov, 1962).
We are interested in the skull for the wealth of characters it provides in the morphologies of its many bones for studies of evolution and development (Kimmel 2014). A variety of adaptive life-history traits might well show up in skull morphologies. Understanding skull phenotypic variation would be expected to inform a variety of biological studies, including, for example, phenotypic plasticity and modularity in evolutionary developmental biology, ecology, and perhaps, considering salmonids particularly, conservation biology. Here we present quantitative morphological studies on the lower jaw, the mandible, of parr-stage juveniles of four Oncorhynchus species. Parr are advantageous for quantitative study in that they are relatively readily accessible in numbers required for accurately assessing within- and among-group variation. Further, irrespective of where and how adults may spend their lives – in the ocean, lakes or rivers – the natural environments of the juveniles of the different species are relatively uniform: the fish invariably deposit eggs within gravel beds of cold, well aerated, fast flowing streams, and after emergence from the gravel, the young parr of salmon and trout alike usually tend to remain for at least some weeks in their natal environments (Quinn 2005). Thus, because the parr are developing in a somewhat common environment, phenotypic differences likely are due to evolutionarily important genetic differences among the groups rather than plastic responses to environmental differences.
Our study of the mandible is motivated by a number of considerations. The teleost mandible includes two conjoined bones, the dentary and the angular articular. Evolution of the angular articular, and possibly the dentary as well, has involved evolutionary fusions of bones that are separate in osteichthyan outgroups (reviews, Patterson 1977; Jollie 1986). This history might bear on both functional morphology and development of the bones in modern fishes such as salmon and trout, as we consider in the Discussion. Trophic differences among Oncorhynchus species are likely to be reflected in evolutionary changes in jaw shapes and sizes, perhaps changes that are greater in extent than for other regions of the skeleton.
The two bones of the mandible differ in function – the dentary bears teeth and the angular articular houses the mandibular component of the jaw joint (Nelson 1973). Whereas most skull bones are extensively interconnected through sutures and articulations, the mandible connects only at localized articulations with the quadrate and the maxilla of the upper jaw. To maintain a perfect bite, the lower and upper jaw bones would be expected to evolve new shapes coordinately. However, the relative freedom of the lower jaw might suggest that its evolution could be somewhat independent of functional constraints arising from associations with other skull regions which might limit divergence. In contrast, the dentary and angular articular neatly and extensively fit together – as in the manner of a finger (angular articular) within a glove (dentary). The fit predicts that the pair of bones are integrated morphologically (Olson and Miller 1958), and that preservation of the fit would necessarily constrain their evolution. Yet, as we show here, the bones do evolve new shapes, and the evolutionary work-around that serves to circumvent constraint provides a focus for this report. We also address modularity, and how modular organization within the mandible might impact evolutionary change. The mammalian mandible, a single bone homologous to the teleost dentary, shows developmental modularity (Atchley and Hall 1991; Cheverud et al. 1997; Ehrich et al. 2003; Zelditch et al. 2008; Klingenberg 2009), but nothing is known about jaw modularity in fishes.
For our survey of lower jawbone morphologies we selected two salmon and two trout species, O. tshawytscha and O. kisutch (Chinook and Coho salmon), and O. mykiss and O. clarkii (rainbow/steelhead and cutthroat trout). Most phylogenetic studies of Oncorhynchus suggest that both pairs enjoy sister-group relationships (Fig. 1) (Oakley and Phillips 1999; Crespi and Fulton 2004; Crête-Lafrenière et al. 2012). This phylogenetic pattern predicts morphological differences to be more prominent between the salmon and trout groupings, than between the species pairs within these groupings. Our data verify this prediction. Furthermore we see clear differences in the bone shapes for both the O. tshawytscha – O. kisutch and the O. mykiss – O. clarkii species pairs. More minor differences are also present among three subgroups we examined within O. mykiss. Comparing how the two bones have diverged together reveals an unexpected matching between them, arguably serving as the basis for how they have preserved their fit in spite of extensive evolutionary change. We present this idea as a ‘concordance’ hypothesis. In support, we show that there is significant morphological integration between the two lower jawbones, apparently reflecting the way they fit together functionally. Moreover, we describe evidence for developmental modularity within both bones, and that the modular divisions reflect the patterns of evolutionary change within the genus.
Fig. 1.
Currently understood phylogenetic relationships among the four species in the study set. For citations, see text. Abbreviations used in figures and tables are indicated within the parentheses.
MATERIALS AND METHODS
Fish stocks and skeletal preparation
Table 1 summarizes collection data for the fish used in this study (n=182). All procedures were approved by the University of Oregon and/or Oregon State University Institutional Animal Care and Use Committees. Many of the parr were spawned from wild-captured parents and reared in the Oregon Hatchery Research Center (OHRC, Alsea, OR) or a facility at Oregon State University (OSU, Corvallis). Other hatchery-reared stocks were obtained from Oregon Department of Fish and Wildlife (ODFW) managed hatcheries. We obtained both hatchery and wild-captured O. clarkii, but only in small numbers, so we combined these two data sets for our analyses (n=25). The largest species data sets are for O. tshawytscha (n= 44, spring Chinook salmon) and O. mykiss (n=95, including hatchery and wild-captured resident freshwater rainbow trout as well as hatchery steelhead trout). Animals were euthanized by MS-222 overdose and preserved either in 4% formaldehyde solution, buffered to neutrality, for no longer than 4 hours, or, alternatively, in 70% ethanol. If the primary preservation was in ethanol they were later post fixed in very dilute 0.25% or 0.5% formaldehyde for 30 min. These precautions were necessary to avoid bone calcium and staining loss because salmonid bones are only very lightly mineralized at parr stages. Skeletal staining was by an acid-free combined Alcian Blue (for cartilage) and Alizarin Red (for bone) procedure, slightly modified from Walker and Kimmel (2007). Pigmentation was reduced by H2O2 bleaching. After clearing the preparations for several days in a solution of 0.1% KOH and 50% glycerol, individual bones were carefully dissected from the skulls, mounted between bridged coverslips and photographed at low magnification (2.5× objective lens) using Alizarin Red epifluorescence (Zeiss Axiophot microscope). Detailed procedures are available from the authors upon request.
Table 1.
Study sets of parr salmonids
| Group | Species/type | n | Age | SL ± SD | Rearing site (brood year) | Origin |
|---|---|---|---|---|---|---|
| ch06 |
O. tshawytscha Spring run |
12 | 202 | 49.0±2.0 | Hatchery, OHRC (2012) | No. Santiam stock |
| ch14 |
O. tshawytscha Spring run |
15 | 261 | 61.0±5.2 | Hatchery, OHRC (2012) | No. Santiam stock |
| ch31 |
O. tshawytscha Spring run |
17 | 182 | 46.6±2.0 | Lab, OCFWRU, OSU (2013) | No. Santiam stock |
| co30 | O. kitsutch | 15 | 146 | 42.2±1.5 | Big Creek Hatchery (2013) | Big Creek stock |
| ct25 |
O. clariii Coastal Freshwater resident |
10 | na | 44.7±6.7 | Wild, Trask River (2001) |
native |
| ct34 |
O. clarkii Coastal Freshwater resident |
15 | mixed | 43.9±3.0 | Oak Springs Hatchery (2013) | Hackleman strain |
| rr09 |
O. mykiss Freshwater resident |
15 | mixed | 43.0±3.4 | Roaring River Hatchery (2012) |
Cape Cod strain |
| rr15 |
O. mykiss Freshwater resident |
15 | mixed | 59.4±2.6 | Roaring River Hatchery (2012) |
Cape Cod strain |
| rr33 |
O. mykiss Freshwater resident |
15 | mixed | 43.0±2.0 | Roaring River Hatchery (2013) |
Cape Cod strain |
| rs42 |
O. mykiss Freshwater resident |
15 | na | 51.9±5.4 | Wild, Willamette River (2013) | ‘redside’ native |
| st18 |
O. mykiss Winter steelhead |
20 | 135 | 50.3±4.0 | Hatchery, OHRC (2012) | Alsea River stock |
| st40 |
O. mykiss Winter steelhead |
15 | 140 | 48.1±2.7 | Hatchery, OHRC (2013) | Alsea River stock |
Strain: spawned from hatchery maintained line. Stock: spawned from wild-captured parents.
Age: days post spawning, na: not available
SL±SD: Standard Length ± Standard Deviation (mm).
OCFWRU, Oregon Cooperative Fish and Wildlife Research Unit, Oregon State University, OHRC; Oregon Hatchery Research Center, Alsea OR; Big Creek Hatchery, Astoria OR; Oak Springs Hatchery; Maupin OR; Roaring River Hatchery, Scio, OR
Morphometrics
We principally used landmark-based geometric morphometrics to analyze shapes and sizes of the individual bones. The bones are quite flat, such that 2D rather than 3D landmarks sets are appropriate for their analysis. Landmarks were placed at positions identifiable along the bone edges (supporting information, Fig. S1) and digitized using tps Dig, version 2.12 (Rohlf 2008a). The digitized raw datasets were moved to MorphoJ (Klingenberg 2011) for Procrustes transformation, which removes size and orientation from the bone shape analysis. For the dentary we observed that upon Procrustes transformation three of the twelve landmarks (numbers 2, 9, and 10; supporting information, Fig. S1) showed high variation, with scatter oriented along the local contour of the edge. Treating these three as ‘sliding semi-landmarks’ during Procrustes alignment in tps RelW version 1.46 (Rohlf 2008b) eliminated this unwanted variation, and for the dentary, in addition to the raw digitized data, we also moved to MorphoJ the data that were Procrustes aligned with sliding of these semi-landmarks.
We analyzed shape variation, and quantified the variation of the individual bones among individuals and groups using MorphoJ procedures including principal component analysis (PCA), canonical variate analysis (CVA) and discriminant function analysis (DFA), all based upon the Procrustes shape coordinates. (Zelditch et al., 2012). Differences between groups were estimated from permutation analyses of Procrustes distances for CVA, and from the T-square statistic for DFA, equivalent to a test using Mahalanobis distance (Klingenberg, 2011). We examined covariance between the two bones by partial least squares, (PLS). PLS analyzes the covariance matrices derived from the Procrustes coordinates of the dentary and angular articular as separate blocks, and we used a pooled-within-group version to largely eliminate between group variation. Finally we used an ‘evaluate modularity’ procedure to test hypotheses that modularity exists within each of the two bones, the dentary and angular articular (Klingenberg 2011). The test uses PLS, and is based on shape variation; bone sizes and orientations are discarded. Covariance is measured by the RV coefficient, serving as a multivariate correlation coefficient. For the dentary in this modularity analysis we used Procrustes aligned data that did not involve sliding landmarks, thus avoiding introduced covariances coming from the sliding semi-landmark methodology. All tests of statistical significances in MorphoJ are based on permutation analyses.
In parallel with the landmark-based procedures we also used elliptical Fourier analysis (EFA), implemented in PAST (Hammer et al. 2001), with six elliptical Fourier modes for the analyses so as to avoid over-parameterization. The EFA procedure for shape analysis does not require placing landmarks on the bones, instead using outlines. We examined the lower jawbone shapes and also, from the same fish, and by EFA, opercles and subopercles (Fig. 2A). 2D outlines, in the form of lists of x, y coordinates, are readily obtained from the fluorescent images using tools in ImageJ (Rasband 2012). Identical starting positions and list lengths are required by the PAST routine, and we used custom software to initiate each list at the same position along the bone edge, and to prune the lists for each bone to identical lengths (greater than 300 x, y coordinates for each bone outline). PCA, CVA and DFA are available within the PAST environment to examine the EF-transformed data, as well as MANOVA-based tests of significance of the CVA results.
Fig. 2.
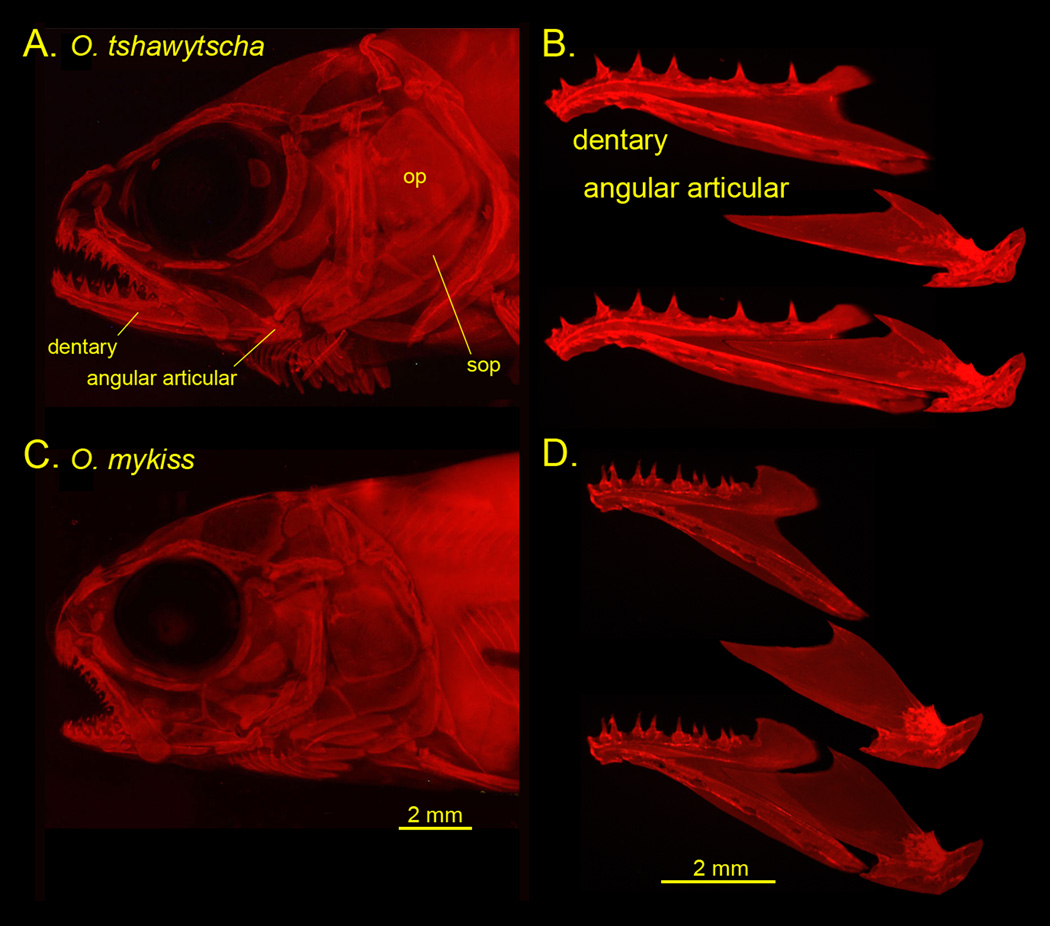
A, C, Parr skull preparations, and B, D dissected mandibular bones along with reconstructions of how they fit together. Alizarin Red staining and photographed by epifluorescence. op: opercle, sop: subopercle. The preparations are shown as left side views, with anterior to the left and dorsal up. We follow this convention in the diagrams shown in subsequent figures.
RESULTS
Salmon versus trout: matching divergence in the shapes of the dentary and angular articular
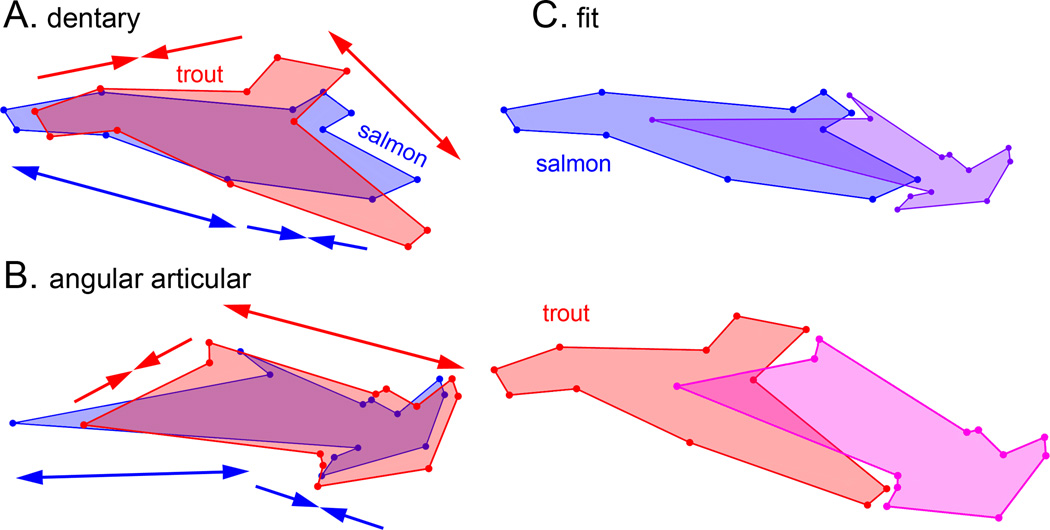
Examination of the skulls of salmon and trout parr reveals prominent differences in skeletal morphologies, particularly considering the jaws (Fig. 2). The snout of the salmon is more elongated, and the jaw joint extends behind the eye in this form, whereas in the trout at this stage the snout and jaw are relatively shortened. As revealed by geometric morphometrics, the overall shape disparities, summarized by Procrustes distances (PD), are considerable, and unexpectedly, are nearly of the same magnitude for the two bones; PD=0.19 for the dentary and 0.18 for the angular articular (Table 2). Since the number of landmarks is similar for each (11 for the dentary and 12 for the angular articular), this result suggests that the total amount of shape change between salmon and trout is comparable for the two bones. We examined the overall pattern of shape change by comparing the average Procrustes shapes of the two bones in salmon and trout, and found changes in the distinctive features of each bone (Fig. 3). For example, the dentary’s dorsal coronoid process is expanded in trout as compared with salmon. The most striking feature in these comparisons is a match-up in the pattern of divergence of the two bones. Relative to trout, the anterior regions of the salmon dentary and angular articular are both lengthened, and the posterior regions of both salmon bones are shortened. The deformations of the trout bones relative to salmon are the opposite – the anterior regions shortened and the posterior regions lengthened. We refer to this phenomenon as ‘matching’, and we propose a ‘concordance’ hypothesis that the apparent matching of how the bones have changed in shape is a feature of preservation of fitting together of the two bones. We explore this hypothesis further below.
Table 2.
Pairwise assignments of species by cross-validation, discriminant function analysis
| dentary | angular articular | |||
|---|---|---|---|---|
| Comparison | PD | Xval | PD | Xval |
| Salmon-Trout | 0.182 | 1/180 | 0.191 | 0/163 |
| ts – ki | 0.030 | 1/60 | 0.048 | 0/53 |
| ts – cl | 0.140 | 0/70 | 0.152 | 0/61 |
| ts – my | 0.197 | 0/130 | 0.211 | 0/128 |
| ki – cl | 0.135 | 0/40 | 0.137 | 0/38 |
| ki – my | 0.188 | 0/110 | 0.189 | 0/105 |
| cl – my | 0.064 | 7/120 | 0.068 | 6/113 |
Groupings: Species designations are the first two letters of the species name (see Table 1).
PD: Procrustes distance
Xval: number misclassified/total by the cross-validation test.
All pairwise discriminations are significant (P<0.0001) by T-square evaluation
Fig. 3.
Procrustes shapes for the salmon (dark) and trout (light) averages. A. dentary B. angular articular. In A and B each pair is in Procrustes alignment, such that the comparisons show shape deformations, not differences in orientations or sizes. The double headed arrows clarify the concordances in the shape changes: anterior regions (to the left) of both bones elongate for the salmon (darker arrows) and shrink for the trout (lighter arrows). Posterior regions (to the right) show the opposite shape changes. C. The bones are taken out of Procrustes alignment and presented as they approximately would fit together in both salmon and trout. For the salmon the size of the angular articular is reduced by 1/6th.
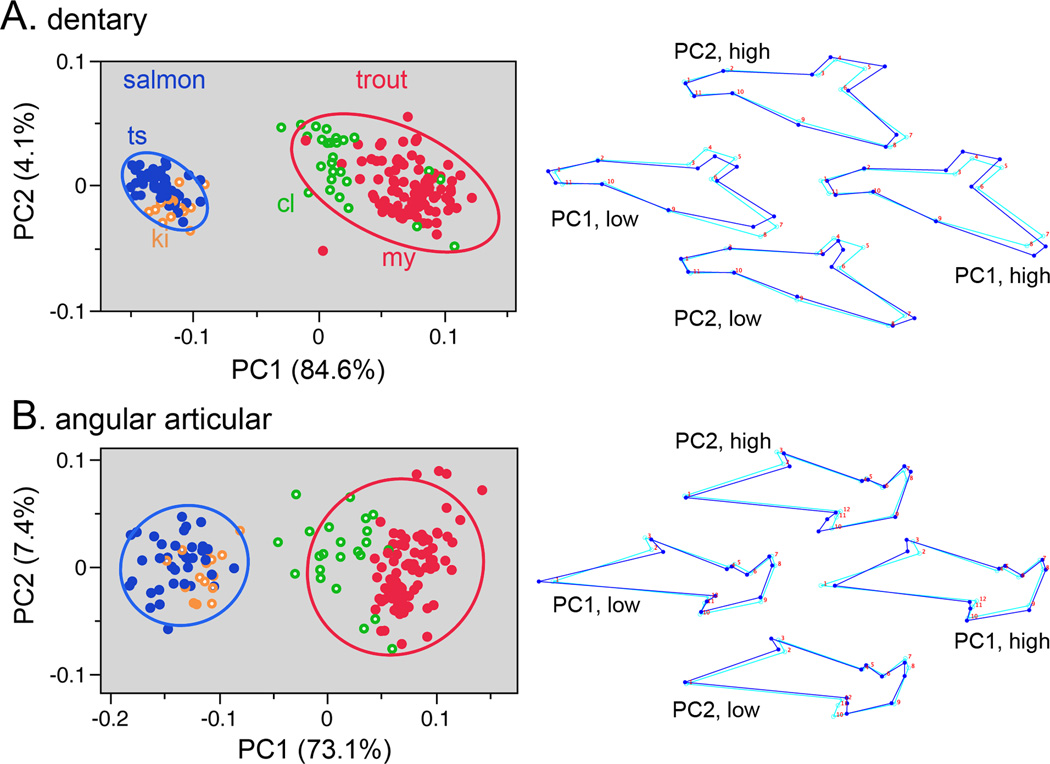
What is the bone shape variation among the individual samples in our study set? Using principal component analysis (PCA) and plotting the data on a PC1 by PC2 bivariate ‘shape space’ reveals non-overlapping and well-separated clusters of the data points representing salmon and trout (Fig. 4). Concerning the matching up of the shape changes of the two bones, for both the dentary and angular articular the separation between salmon and trout is along the leading eigenvector, PC1; there is essentially no separation along PC2. PC1 in both cases explains the large majority of the total shape variation, and explains similar amounts of the variation for both bones – 85% for the dentary and 73% for the angular articular. The magnitudes of the separation between salmon and trout along the PC1 axes are similar for both bones. Hence PCA shows the matching of divergence of the two bones between salmon and trout, and hence provides support for the concordance hypothesis.
Fig. 4.
Principal component analyses (PCAs) reveals shape divergences for the dentary (A) and angular articular (B) between salmon (2 species) and trout (2 species), and including 95% density ellipses. The data include all of the populations listed in Table 1. Species abbreviations as in Fig. 1. Wireframe diagrams to the right show landmark configurations representing the loadings on the PCs. For each, the light blue wireframes show the consensus configuration, identical for each diagram (at PC1=0, PC2=0), and the dark blue wireframes show the deformations at high and low scores for both PCs.
What differences in shape account for the PC1 divergences? These are shown by the wire-frame configurations in Fig. 4. Since PC1 captures most of the shape variation for both bones we would expect the PC1 deformations in Fig. 4 to match the total Procrustes deformations in Fig. 3. Indeed they do match (see legend to Fig. 4) revealing consistency in the methodology.
The different populations we collected for this study were comprised of parr of similar body sizes (standard length, SL; Table 1), but the groups are not perfectly size-matched. Hence, allometry, the covariation of size and shape, could be influencing the PC1 shape divergences shown in Fig. 4. To test this possibility we purposely included in our dataset a single sample set each of hatchery O. tshawytscha and hatchery O. mykiss (Spring Chinook salmon and resident rainbow trout) that were reared longer, resulting in a SL about 10 mm larger than the other samples (20% larger, groups ch14 and rr15, Table 1). We observed no significant change along PC1 for either bone between the younger and older fish (supporting information, Fig. S2). This finding suggests that variation in body size of the parr in our study sets is not markedly influencing the results shown in Fig. 4.
Among species shape variation: a second kind of matching
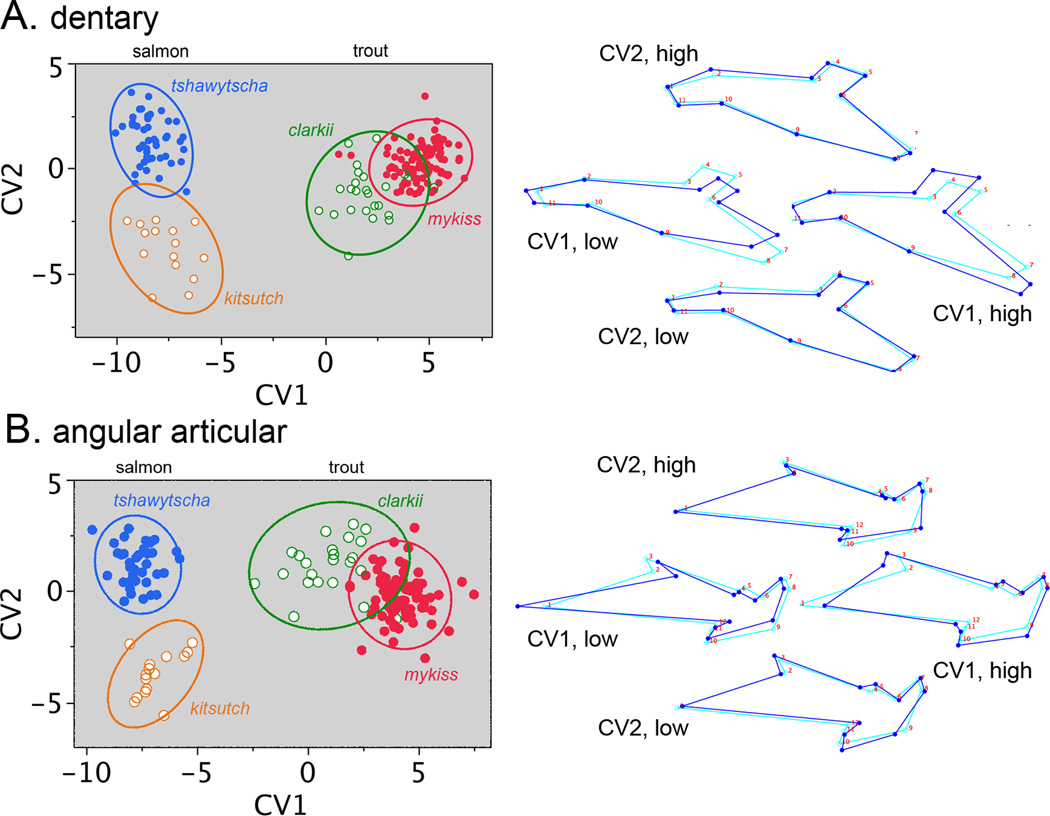
The results described above suggest that there is concordance in pattern and scale in the shape evolution of the two bones of the lower jaw between salmon and trout. Does concordance extend to the species level? Canonical variate analysis (CVA, Fig. 5) separates species better than PCA (Fig. 4). CV1 in the CVA yields a similar distribution to PC1 in the PCA for both bones, prominently separating trout and salmon species, and showing similar deformations of the landmark configurations. Two trout species O. clarkii and O. mykiss almost completely separate along CV1, showing that O. clarkii has evolved a more salmon-like lower jaw than O. mykiss, and also signaling that the salmon-trout pattern of matching up of the shape divergences of the two jawbones has occurred between these two trout species as well, but at much reduced scale. Matching is also apparent in the distribution of points between the two CVA scatter plots in Fig. 5, just as we described above for the PCA scatter plots (Fig. 4).
Fig. 5.
Canonical variate analyses (CVAs) reveal among-species differences in bone shapes. Presentation as in Fig. 4. The data points show individual measurements (same Procrustes transformed data as Fig. 4) grouped according to species. The plots include 95% density ellipses. Trout and salmon separate along CV1, and the two trout species, O. mykiss and O. clarkii, are also largely separated by CV1. The two salmon species O. tshawytscha and O. kisutch completely separate along CV2. To the right, the CV1 wireframe diagrams show the changes between trout and salmon (essentially identical in detail to the PC1 wireframes in Fig. 3), and the similar changes, but to a lesser degree, between O. mykiss and O. clarkii. The CV2 wireframes show the changes between O. tshawytscha and O. kisutch.
CV2 separates the two salmon species O. tshawytscha and O. kisutch into completely non-overlapping clusters of data points, and in a similar manner for the two bones (Fig. 5). However, considering the wire-frame diagrams in this figure, we do not detect any apparent one to one matching of the dentary and angular articular shape divergences along CV2. So how does evolution provide for functional integration between the two bones – their fitting together? Only small shape occurs in the posterior region of the dentary and in the anterior region of the angular articular. These are the regions where the bones associate with one another. Hence, instead of a matching up of evolutionary divergences, local conservative shape change in these specific regions seems to have accomplished the matching.
The scatter plots in Figs. 4 and 5 clearly reveal that for both mandibular bones shape divergence being much more prominent between salmon and trout than between the two salmon species and the two trout species. This pattern is predicted by the phylogeny (‘phylogenetic signal’), as currently understood (Fig. 1). Morphology has diverged to a larger extent among more distantly related species than between more closely related species. Evaluation of PDs and pairwise discriminant function analyses reinforce our conclusions, and provide statistical support (Table 2).
Elliptical Fourier investigation of among-species bone shape divergences
As a check on the landmark analysis, we obtained canonical variates using elliptical Fourier analysis (EFA), a procedure using bone outlines and not requiring landmarking. Our EFA examined the shapes of the same mandibular bones (i.e., the same bones from the same images) that we used for the landmark-based analyses, and we observed essentially the same results as those in the landmark analyses (supporting information, Fig. S3A,B, Table S1). These data support that our findings are due to biology, they are not simply a feature of the analytical method we used to examine bone shapes.
How does variation of mandibular bones compare with that in other regions of the skull? We used EFA to examine the shape variation of two opercular region bones, the opercle and subopercle, and observed considerably more overlap among species for the opercular region bones than for the mandibular region bones (supporting information, Fig. S3C, D). This difference might mean that mandibular divergences have been the more prominent, but more analyses will be required to learn if this is indeed the case, for increased within-group variation in the opercular region is an alternative explanation for the result.
Within-O. mykiss variation in bone shape
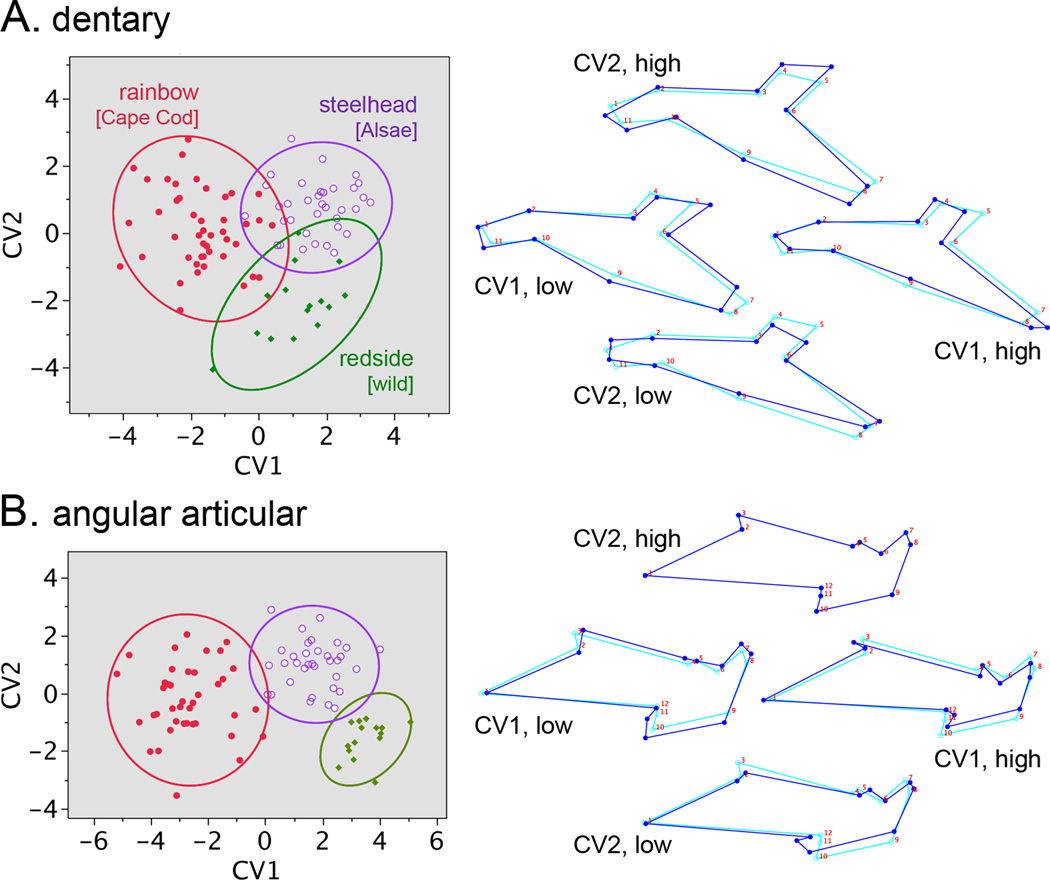
Our dataset includes samples of three classes of O. mykiss that differ in life history – hatchery reared freshwater-resident rainbow trout, hatchery-reared steelhead trout, and wild-captured freshwater-resident rainbow ‘redside’ trout. Pairwise PDs among these groups are small, and pairwise comparisons by DFA resulted in a number of incorrect assignments in the cross-validation tests. Nevertheless differences among the groups are highly significant (Table 3; p<0.0001, T-square analysis), and the three groups separate reasonably well in CV1 versus CV2 scatter plots (Fig. 6). Replicated samples representing different brood years for the hatchery fish (Table 1) show that even though subtle, the rainbow-steelhead separation is reproducible (supporting information, Fig. S4).
Table 3.
Pairwise assignments of groupings within O. mykiss by cross-validation: Discriminant Function Analysis
| dentary | angular articular | |||
|---|---|---|---|---|
| Comparison | PD | Xval | PD | X val |
| rb – rs | 0.040 | 9/60 | 0.059 | 1/56 |
| rb – st | 0.035 | 9/80 | 0.048 | 9/76 |
| rs – st | 0.029 | 6/50 | 0.034 | 4/49 |
Groupings (see Table 1): rb: rainbow trout, hatchery Cape Cod strain, rs: ‘redside’ trout, wild-captured freshwater resident, st: steelhead trout, hatchery Alsea stock.
PD: Procrustes distance
Xval: number misclassified/total by the cross-validation test.
All pairwise discriminations are significant (P<0.0001) by T-square evaluation
Fig. 6.
Within-species CVAs reveals bone shape divergences among three groupings of O. mykiss trout; freshwater resident rainbow (Cape Cod strain Roaring River Hatchery-reared), wild-captured freshwater resident ‘redside’ from the upper Willamette River, OR, and steelhead obtained from wild-captured Alsea River OR parents and reared at the OHRC. The data points show individual measurements and the plots include 95% density ellipses. The rainbow dataset is a pool of three collections (two brood years) shown in Table 1, and the steelhead dataset is a pool of two collections (two brood years).
To the point of matching divergences between the dentary and angular articular, the scatter plots for the two bones in Fig. 6 are similar in scale and are roughly similar in how the three groups distribute. Matching is difficult to discern from the wireframe diagrams, but may be present nevertheless (see the next section, on PLS).
Comparisons within the O. mykiss dataset show the differences among groups are small and complex, but tend to signal more streamlined and delicate bone shapes in steelhead and redside. It seems clear that the dentaries and angular articulars have diverged to similar extents among these groups.
Shapes of the dentary and angular articular bones covary
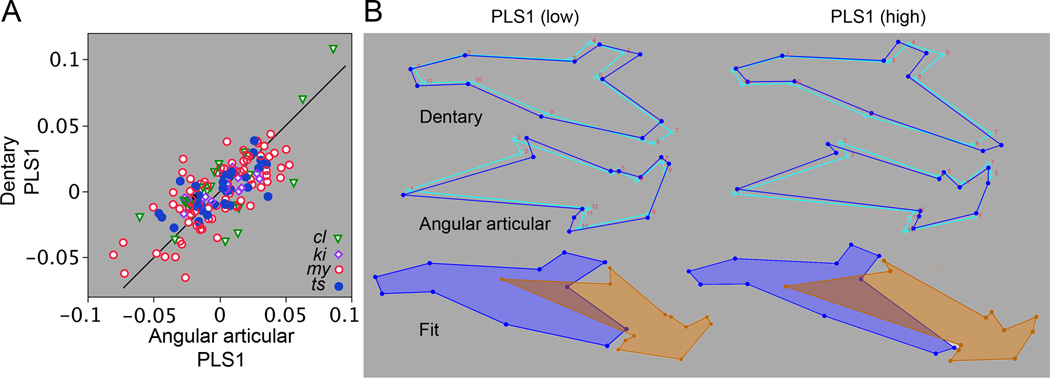
The foregoing results have demonstrated a matching between the evolutionary shape divergences of the dentary and angular articular, supporting the concordance hypothesis. The hypothesis predicts shapes of the two bones covary, and we tested this prediction directly by partial least squares analysis (PLS). We find a highly significant RV coefficient of 0.32 (P<0.0001 by permutation analysis), thus showing substantial covariation, as predicted. PLS1, the leading eigenvector in the analysis, accounts for most of the total covariance (71%, significant by permutation analysis at P<0.0001). Plotting the PLS1 scores for the dentary against the PLS1 scores for the angular articular graphically demonstrates the covariance (Fig. 7A). We include on this plot a 45° line, and notably the data points fall close to this line. Although performing a simple bivariate regression analysis would be inappropriate (because the PLS1 scores are not simple variables), the apparent slope of the relationship being close to 1:1 suggests that the variation in the scores of the two bones among samples is similar in scale, another demonstration of matching, and thus agreeing with conclusions from the PCAs and CVAs described above. The deformations of the PLS1 wireframe configurations representing the two bones also exhibit a clear matching (Fig. 7B) – the deformations toward low PLS scores similar to the changes we observed by PCA and CVA for salmon, and the deformations toward high PLS scores similar to those for trout. This result is most interesting, considering the PLS we used is a pooled within group analysis, expected to be focused on covariation shared by the groups rather than covariation among the groups.
Fig. 7.
Partial least squares (PLS) pooled within group analyses reveal significant covariance between the dentary and angular articular. Our entire within-genus Oncorhynchus dataset is included, pooling is according to species. By pooling within group, each analysis examines covariance within the dataset collectively, species differences are eliminated by mean-centering. (A) RV coefficient = 0.32, p<0.0001 (permutation analysis). A. The scatter plot shows individual PLS1 scores for one bone against the other, and includes a 45° diagonal line (see text for explanation). Abbreviations are the first two letters of the species names (Fig. 1). PLS1 accounts for 71% of the total covariance, p<0.0001. B. Wireframe diagrams representing shape deformations from the mean shapes (light blue wireframes) in the positive (high) and negative (low) directions along the PLS1 axis (dark blue). The bottom diagrams illustrate the approximate fitting together of the two bones at high and low PLS1 scores.
We also examined dentary-angular articular shape covariance using the within O. mykiss data subset, and obtained results agreeing with the PLS bone shape results just described for our full among-species dataset – reasonably high and significant covariance (RV coefficient=0.26; p<0.0001). However, the covariance is quantitatively not so prominent as for the within-genus analysis, and the plot of PLS1 scores looking more scattered for the O. mykiss data (supporting information, Fig. S5A). Differences between the within-genus and within-species analyses might be due to the smaller sample size for the latter, and especially to the relatively very low shape variation we observed for this dataset, for presence of covariation depends on having variation among the samples (Hallgrímsson et al. 2009).
Notably, the PLS1 shape changes for the within O. mykiss analysis (supporting information, Fig. S5B) are similar to those obtained for the complete dataset, and also preserve the fitting together of the two bones. Because PLS1 explains most of the covariation for both datasets, this observation suggests that the covariance structure of the lower jaw is similar within O. mykiss as it is for the genus as a whole. Overall, the PLS findings strongly support the concordance hypothesis.
Evolutionary change between salmon and trout predicts developmental modularity
Whereas the divergences we have described are likely due to Darwinian adaptations, development may also impact evolutionary change. Skull development is modular (Cheverud 1982; Hallgrímsson et al. 2007; Drake and Klingenberg 2010; Jamniczky and Hallgrímsson 2011; Cvijanović et al. 2014) where modules are local regions that possess high internal covariance and are interconnected by substantially lower covariances (Wagner 1996; Klingenberg 2009). The low covariance module boundaries provide for evolutionary dissociability – i.e., for adjacent modules having the potential to diverge from one another in shape largely independently. Hence we can hypothesize that dissociability due to modular organization could bias the way the mandible evolves. To test this hypothesis we examine our shape divergence data for regions within each of the bones that show prominent dissociation from one another, and then test whether these specific regions behave statistically as modules.
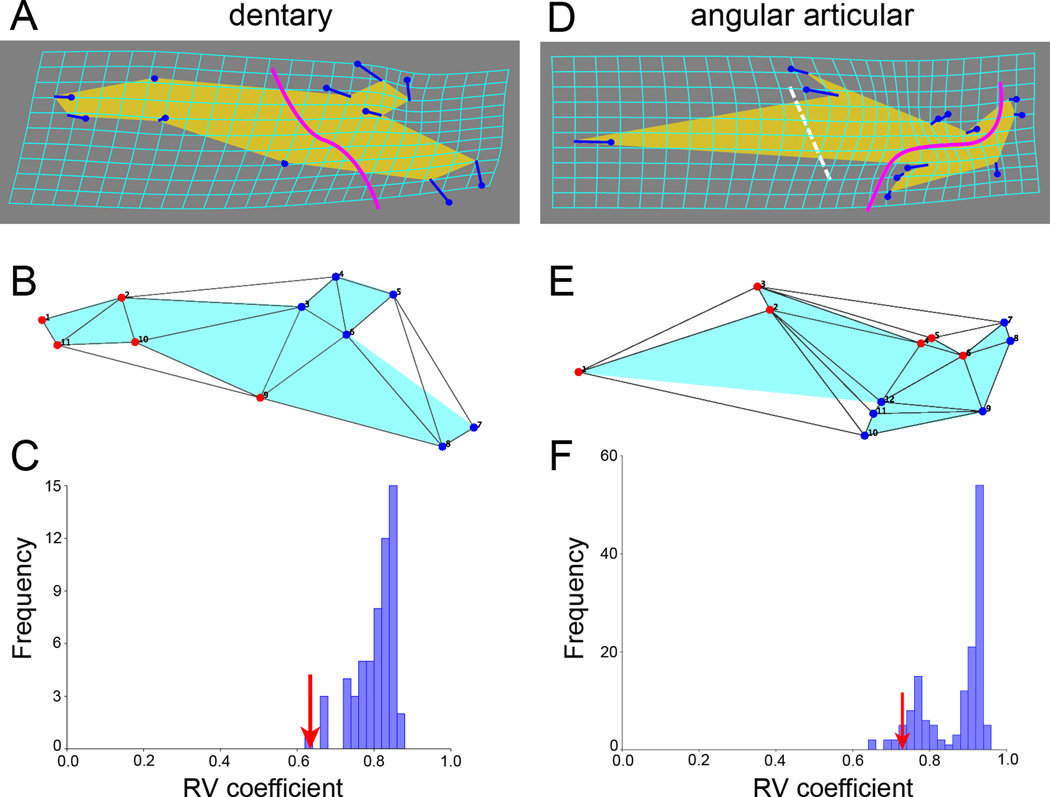
A clear hypothesis of modularity arises from the matching pattern of evolutionary change we have been addressing: namely the anterior and posterior regions of each bone reshape distinctively, hence displaying dissociation from one another. For the dentary a hypothetical module boundary would then separate an anterior tooth-bearing region from a posterior angular articular-associated region (Fig. 8A, bold curving line). The proposed anterior dentary module includes landmarks 1, 2, and 9–11 (Fig. 8B). The proposed posterior dentary module includes landmarks 3–8.
Fig. 8.
Shape diversity among Oncorhynchus species successfully predicts modularity, suggesting that modularity has influenced the pattern of evolution within the genus. A–C: dentary, D–F: angular articular. A, D: Thin-plate spline transformation grids, equivalent to the wire frame diagrams in Fig 2, and showing the deformation of the averaged salmon dataset. Dissociations to be examined by the modularity evaluation procedures are shown by the bold curving lines crossing the bones. The dashed line crossing the angular articular shows the location of a predicted boundary concordant with the hypothetical boundary across the dentary. B, E: Setting up the test of the modularity hypotheses with the ‘evaluate modularity’ procedure (Klingenberg, 2009). The landmark configurations are partitioned into two blocks (hypothetical modules, red versus blue landmarks). The lines connecting the landmarks were obtained by DeLaunay triangulation (the default procedure) and show the adjacency profiles used to determine contiguity of the blocks. C, F: Testing the hypotheses. RV coefficients are determined that describe the covariance between the two blocks. These values (one for each bone, red arrows) are compared with RV coefficients for all other partitions of the configurations that separate spatially contiguous blocks of the same sizes as the hypothetical ones (blocks of 5 and 6 landmarks for the dentary, and blocks of 6 landmarks each for the angular articular). The frequency histograms show the RV coefficients for all of the possible partitions. The relatively low RV coefficients observed for the hypothetical modules supports modularity since by definition modules are interconnected by low covariance. PL: proportion lower, approximating a P value (see text).
We cannot test a corresponding boundary for the angular articular (dashed white line, Fig. 8D), because the putative anterior module only includes a single landmark; its internal covariance cannot be assessed. However, the pattern of landmark displacement within the angular articular suggests the location of a separate boundary, located at the posterior-most end of the bone near the angular articular’s contribution to the jaw joint (Fig. 8D, curving line). We hypothesize the boundary separates a dentary-associated angular articular module that includes landmarks 1–6 from a joint region angular articular module that includes landmarks 7–12 (Fig. 8E).
Klingenberg (2009) has provided an elegant way to test these hypotheses, and implemented the test in MorphoJ software. Covariance between the proposed modules is examined by PLS and quantified by the RV coefficient. This value is compared with RV coefficients for the complete set of other comparable partitions of the configuration (see legend to Fig. 8). For the dentary the predicted boundary yields the lowest covariance of all (Fig. 8C; proportion lower = 0/58). For the angular articular only 7 out of a total of 143 partitions had lower RV coefficients than the proposed module pair (Fig. 8F, proportion lower = 0.049). Both results are unlikely to be obtained by chance alone (the values for ‘proportion lower’ can be used as probabilities; Klingenberg 2009), for they are less than 0.05. These findings suggest that the dissociations we observe in the evolutionary changes within the mandibular bones are influenced by developmental modularity. For the dentary, at least, modular organization could facilitate the way evolution has circumvented functional constraint to provide for shape change without wrecking the fitting together of the two bones.
DISCUSSION
We have examined morphological variation and covariation of the two bones comprising the mandibular skeleton within Oncorhynchus. Comparisons at three levels of biological organization – between salmon and trout, among two salmon species and two trout species, and among three life history forms within O. mykiss show shape divergence among the groups at each level. We also quantified shape covariance between the dentary and angular articular, and observed it to be substantial across species. This result indicates that mandibular structure is highly integrated. However, morphological integration is not uniform throughout the mandible: Rather, we find evidence for modular organization within each of the two bones. Furthermore, the modular patterning – a feature that must be due to processes underlying jaw development – is related to the patterning of evolutionary change of jaw morphology within the genus. This connection provides evidence for a positive role of development in biasing the course of adaptive evolution of the jaw.
Levels of shape diversification within Oncorhynchus
Mandibular morphology varies hierarchically: Differences between salmon and trout are much greater than are species differences within these two groups, and, in turn, as we examined for O. mykiss, within-species variation is lower still. Between salmon and trout we observe slimmer, more streamlined and elongated dentaries and angular articulars for the salmon. The trout dentary is more angled as compared with the same bone in salmon. It is evident that the long dentary, including where the two bones are overlapped, makes up most of the length of the lower jaw (in fact about 70% of the total length in both salmon and trout parr; data not shown), and it is clear that most of the increased length of the lower jaw of salmon (evident by visual inspection of the skulls; Fig. 2) is due to the morphological evolution of the dentary.
Within trout, O. clarkii deviates toward the salmon morphology, compared with O. mykiss. This result is not surprising because a relatively long jaw is a field mark of the cutthroat parr (Quinn 2005, p. 29). Within O. mykiss one might propose that the jawbones of steelhead would appear more like salmon-like than those of freshwater resident rainbow, because steelhead and salmon are both anadromous. However, the steelhead bones don’t deform toward the salmon shape, but even so, they are, like salmon, relatively more streamlined than the hatchery-reared resident rainbow. Within salmon, O. kisutch deviates from O. tshawytscha primarily by having thinner, more delicate processes – notably the anterior (tooth-bearing) process of the dentary and in the posterior (jaw-joint) region of the angular articular.
Such differences in morphology might be adaptive, but we note caveats to this interpretation. Changes could be due to neutral processes and drift. Furthermore, some possible causes of variation were uncontrolled: our hatchery steelhead O. mykiss derive from wild-captured parents, whereas our hatchery freshwater resident O. mykiss come from a long maintained hatchery-bred ‘Cape-Cod’ strain (Williamson et al. 2010), likely adapted to hatchery rearing and possibly bottlenecked. The salmon-trout comparison is better designed – including replicate populations from separate brood years of O. tshawytscha and steelhead O. mykiss from wild-captured parents and reared in the same research-oriented hatchery facility, the Oregon Hatchery Research Center. Here we have essentially a common-garden experiment suggesting that the differences are genetic, rather than being due to phenotypic plasticity, and likely have been selected for.
If morphological differences we observe do indeed have an adaptive basis, we consider that although we have examined parr-stage juveniles, the differences in morphologies possibly are foreshadowing adaptations for later life history stages, when habitats differ, and behaviors, including trophic behaviors, are likely more divergent between the different forms than they are at parr stage. One might speculate that the more elongate, streamlined jaws we found for both salmon and steelhead trout are be adaptive for the oceanic habitat at the adult stage. Modeling how jaw shape influences trophic behaviors could be a productive approach to explore the ecological and adaptive significances of our findings.
Functional integration and modularity within the lower jaw
Our finding significant covariation between the shapes of dentary and angular articular, predicted by the concordance hypothesis, is expected because the two bones function together as a lower jaw ‘mandibular unit’. This consideration prompts interpretation that the covariance reflects functional integration within the lower jaw (sensu Armbruster et al. 2014), where integration can be defined as the coordinated variation of developmentally and/or functionally related morphological features (Olson and Miller 1958; Hallgrimsson et al. 2009). Considering that within the genus both the dentary and angular articular have evolved different shapes suggests further that the mandible is exhibiting evolutionary as well as functional integration, where evolutionary integration refers to the disposition for them to evolve jointly (Lewontin 1978; Armbruster et al. 2014).
Whereas function is likely the ultimate cause of an integrated jaw structure, development also seems to be a critical determinant. Our evidence suggests both bones of the lower jaw are modularly patterned. That the anterior module of the dentary bears teeth, and posterior module associates with the angular articular, indicates that functionality of the bone may be served by modular design. Similarly, a boundary partitions the angular articular near its extreme posterior end, separating a jaw-joint region module from a dentary-associated module.
The module boundary crossing the angular articular is in the vicinity of what is thought to be an ancient fusion of separate angular and articular bones that existed in ancestors of the salmonids (Nelson 1973; Patterson 1977; Jollie 1986). Possibly the boundary is a persistent remnant of the location of the fusion, with the low covariance across it reflecting separate ancestral developmental origins, e.g., in embryonic mesenchymal condensations (Hall 2005). Indeed, Jollie (1984) reported separate early ossifications for the angular and articular in young salmon parr that later appeared fused into the angular articular. If Jollie’s interpretation is correct, the two ossifications could be developmental remnants of the ancestral condition.
In the same vein we also note that Jollie (1986) has argued that the teleost dentary should be termed a ‘dentosplenial’ because of an ancient dentary-splenial fusion. However In this case the location of region of the would-be ancient fusion is obscure in teleosts, so imagining it to persist as a module boundary is poorly supported. However dentary modularity originated in evolution, the position of the boundary we discovered is striking considering how modularity may have influenced the maintenance of the dentary’s fit with the angular articular during evolutionary divergence within Oncorhynchus.The concordant matching up of the shape changes of the two bones requires dissociation between the expanding and shrinking regions of the bones, and, at least for the dentary, modularity seems to provide for this dissociability. There could be a corresponding module boundary within the angular articular but the landmark configuration we used is not suitable to show it. However, it is evident from the nature of the landmark displacements that dissociation is present at the predicted position in the angular articular (Fig. 8D, dashed line).
These findings suggest that developmental modularity has facilitated the observed evolutionary shape changes within the salmonid lower jaw. The logic behind the analysis and findings match previous work in which we demonstrated modularity in the opercular region of the skulls of threespine stickleback, Gasterosteus aculeatus (Kimmel et al. 2012; Jamniczky et al. 2013; Kimmel 2014). In the stickleback work and in this study we predicted the locations of module boundaries from observed dissociations in evolutionary shape changes. That this strategy of detecting modularity appears to work supports a role of development in “serving to structure the expression of phenotypic variation on which natural selection acts” (Hallgrímsson et al. 2009. p. 355).
Conclusion
Dissociability has emerged as a theme, not only in our study of salmonid jaw reported here, but also in our recent stickleback analyses. Wagner and Altenberg (1996) use ‘variability’ as a dispositional term, meaning the propensity to vary, as contrasted with ‘variation’ – the latter term meaning realized variation as one can measure directly. Following this terminology (see also Armbruster et al. 2014), we feel it useful to contrast ‘dissociability’ as a dispositional term versus ‘dissociation’. Modularity provides for dissociability, and, as we interpret, in the jaw we see the result – dissociation, possibly because modularity positively influences the nature of adaptive change that is under selection. A sophisticated and modular developmental genetic basis for building bone shapes (review: Kimmel 2014) may well be providing for flexibility of adaptive evolution of the jaw.
Supplementary Material
Acknowledgments
We thank John Dowd, and Bonnie Ullmann for technical assistance. Assistance and collections of parr from Oregon Department of Fish and Game Hatcheries were courtesy of Luke Allen, Lyle Curtis, Rob Dietrichs, Joseph O’Neill, and Jeff Ziller. A collection from Oregon State University was courtesy of Carl Schreck. Heather Jamniczky and Brian Hall provided critical comments on drafts of the manuscript. The research was funded by US National Science Foundation grant IOS-0818738 and National Institutes of Heath grant DE013834.
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information maybe be found in the online version of this article.
Figure S1. Landmark positions, located at local maximum curvatures along of the dentary (A) and angular articular (B). Fig. 2B and D in the main text shows these same bones without the landmarks, permitting examination of the detailed structure (e.g. the small protuberance shows at angular articular landmark 5). For the dentary the assignment of the positions of curvature maxima were easy to identify for most landmarks (e.g. dentary landmarks #1, 4, 7), but were difficult for dentary landmarks #2, #3 and #9, as determined from replicates. Hence experimental error likely accounts for high variation encountered for these landmarks and motivated our assigning them as sliding semil and marks (see Methods in the main text).
Figure S2. Allometry, meaning variation in bone shape as a function of body size, is modest over the size ranges of the parr in our study set (grand mean SL = 48.7 mm). A. dentary, B: angular articular. The bars show mean PC1 scores (with 95% confidence intervals shown by the error bars) as measures of bone shapes. Included are two collections of O. tshawytscha (O ts; Chinook salmon) from the same brood stock but differing in SL by a mean of 12 mm (ch06, ch14; Table 1) and two collections of O. mykiss (O my; rainbow trout from the same brood stock but differing in SL by a mean of 16 mm (rr09, rr15; Table 1). Shape differences in both comparisons between the smaller (S) and larger (L) fish are insignificant (P>0.05, Tukey-Kramer test).
Figure S3. Among-species CVAs using elliptical Fourier transformation of the bone outlines rather than landmarks to examine bone shapes. CV1 versus CV2 plots for the dentary, angular articular, opercle, and subopercle. The same samples (same initial bone images) were used for both the landmark and elliptical Fourier analyses. The scatter plots for the dentary and angular articular may be compared with those in Fig. 5 of the main text; the similarities in the plots are evident in spite of one method using landmark displacements and the other using deformation of the entire bone outline. Matching the landmark analysis, here the trout and salmon groups completely resolved along the CV1 axis. The scatter plots for the opercular region bones show more among-species overlaps than those for the mandibular region bones. We did not carry out landmark analyses of the opercular region bones.
Figure S4. Within-O. mykiss CVAs for the two groups for which we have replicate datasets, (replicates collected from the same hatcheries, but for different brood years). The group means are computed identically to Fig. 6 in the main text, but here the 95% ellipses and colors of the data points show individual collections (listed in Table 1 in the main text). rainbow: blue and red circles, collections rr09 and rr33, steelhead: lavender and orange circles, collections st18 and st40. There is extensive overlap between replicates of both rainbow and steelhead, showing reproducibility, and supporting that the differences between the between the rainbow and steelhead groupings are not due simply to their being from separate collections.
Figure S5. Partial least squares (PLS) pooled within group analyses reveal significant covariance between the dentary and angular articular for the O. mykiss dataset. Presentation as in Fig. 7 in the main text, The dataset is as in Fig. 6 in the main text. RV coefficient = 0.26, p<0.0001.
A. PLS1 scatter plot including a 45° diagonal. Abbreviations: rb: rainbow, rs: redside, st: steelhead. PLS1 accounts for 47.5% of the total covariance, p<0.0001. B. Shape deformations and approximate fits between the two bones.
Table S1. Pairwise assignments among species by cross-validation: Discriminant function analysis for the elliptical Fourier dataset.
REFERENCES
- Armbruster WS, Pélabon C, Bolstad GH, Hansen TF. Integrated phenotypes: understanding trait covariation in plants and animals. Phil. Trans. R. Soc. B. 2014;369 doi: 10.1098/rstb.2013.0245. 20130245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Hall BK. A model for development and evolution of complex, morphological structures. Biol. Rev. 1991;66:101–157. doi: 10.1111/j.1469-185x.1991.tb01138.x. [DOI] [PubMed] [Google Scholar]
- Billman EJ, E J, Whitman LD, Schroeder RK, Sharpe CS, Noakes DLG, Schreck CB. Body morphology differs in wild juvenile Chinook salmon Oncorhynchus tshawytscha that express different migratory phenotypes in the Willamette River, Oregon, U.S.A. J. Fish Biol. 2014;85:1097–1110. doi: 10.1111/jfb.12482. [DOI] [PubMed] [Google Scholar]
- Carl LM, Healey MC. Differences in enzyme frequency and body morphology among three juvenile life history types of Chinook salmon (Oncorhynchus tshawytscha) in the Nanaimo River, British Columbia. Canadian Journal of Fisheries and Aquatic Sciences. 1984;41:1070–1077. [Google Scholar]
- Christie MR, Marine ML, Blouin MS. Who are the missing parents? Grandparentage analysis identifies multiple sources of gene flow into a wild population. Molecular Ecology. 2011 doi: 10.1111/j.1365-294X.2010.04994.x. [DOI] [PubMed] [Google Scholar]
- Cheverud JM. Phenotypic, genetic, and environmental morphological integration in the cranium. Evolution. 1982;36:499–516. doi: 10.1111/j.1558-5646.1982.tb05070.x. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Routman EJ, Irschick DJ. Pleiotropic effects of individual gene loci on mandibular morphology. Evolution. 1997;51:2006–2016. doi: 10.1111/j.1558-5646.1997.tb05122.x. [DOI] [PubMed] [Google Scholar]
- Crespi BJ, Fulton MJ. Molecular systematics of Salmonidae: combined nuclear data yields a robust phylogeny. Mol. Phyologenet. Evol. 2004;31:658–679. doi: 10.1016/j.ympev.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Crête- Lafrenière A, Weir LK, Bernatchez L. Framing the Salmonidae family phylogenetic portrait: A more complete picture from increased taxon sampling. PLoS ONE. 2012;7:e46662. doi: 10.1371/journal.pone.0046662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvijanović M, Ivanović A, Miloš L, Kalezic L, Miriam L, Zelditch ML. The ontogenetic origins of skull shape disparity in the Triturus cristatus group. Evol. Dev. 2014;16:306–317. doi: 10.1111/ede.12093. [DOI] [PubMed] [Google Scholar]
- de Beer GR. The Development of the Vertebrate Skull. London: Oxford University Press; 1937. [Google Scholar]
- Docker M, Heath D. Genetic comparison between sympatric anadromous steelhead and freshwater resident rainbow trout in British Columbia, Canada. Conservation Genetics. 2003;4:227–231. [Google Scholar]
- Drake AG, Klingenberg CP. Large-Scale Diversification of Skull Shape in Domestic Dogs: Disparity and Modularity. Am. Nat. 2010;175:289–301. doi: 10.1086/650372. [DOI] [PubMed] [Google Scholar]
- Ehrich TH, Vaughn TT, Koreishi SF, Linsey RB, Pletscher LS, Cheverud JM. Pleiotropic effects on mandibular morphology I. Developmental morphological integration and differential dominance. J. Exp. Zool. 2003;296B:58–79. doi: 10.1002/jez.b.9. [DOI] [PubMed] [Google Scholar]
- Goodrich ES. Structure and Development of Vertebrates. London: Macmillan; 1930. [Google Scholar]
- Groot C, Margolis L, Clark WC. Physiological Ecology of Pacific Salmon. Vancouver: University of British Columbia Press; 2002. [Google Scholar]
- Hall BK. Bones and Cartilage: Developmental and Evolutionary Skeletal Biology. Amsterdam: Elsevier Academic Press; 2005. [Google Scholar]
- Hallgrímsson B, Lieberman DE, Young NM, Parsons TE, Wat S. Evolution of covariance in the mammalian skull. Novartis Found. Symp. 2007;284:164–185. doi: 10.1002/9780470319390.ch12. discussion 185–190. [DOI] [PubMed] [Google Scholar]
- Hallgrímsson B, Jamniczky H, Young NM, Rolian C, Parsons TE, Boughner JC, Marcucio RS. Deciphering the Palimpsest: Studying the Relationship Between Morphological Integration and Phenotypic Covariation. Evo. Bio. 2009;36:355–376. doi: 10.1007/s11692-009-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electronica. 2001;4(1):9. [Google Scholar]
- Hendry AP, Sterns SC, editors. Evolution Illuminated. Oxford: Oxford University Press; 2004. [Google Scholar]
- Jamniczky HA, Hallgrímsson B. Modularity in the skull and cranial vasculature of laboratory mice: implications for the evolution of complex phenotypes. Evol. Dev. 2011;13:28–37. doi: 10.1111/j.1525-142X.2010.00453.x. [DOI] [PubMed] [Google Scholar]
- Jamniczky HA, Harper EE, Garner R, Cresko WA, Wainwright PC, Hallgrímsson B, Kimmel CB. Association between integration structure and functional evolution in the opercular four- bar apparatus of the threespine stickleback, Gasterosteus aculeatus (Pisces, Gasterosteidae) Biol. J. Linn. Soc. Lond. 2014;111:375–390. [Google Scholar]
- Jollie M. Development of the head skeleton and pectoral girdle in salmons, with a note on scales. Can. J. Zool. 1984;62:1757–1778. [Google Scholar]
- Jollie M. A primer of bone names for the understanding ot the actinopterygioan head and pectoral girdle skeletons. Can. J. Zool. 1986;64:365–379. [Google Scholar]
- Kimmel CB. Skull developmental modularity: a view from a single bone – or two. J. Appl. Ichthyol. 2014;30:600–607. doi: 10.1111/jai.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Hohenlohe PA, Ullmann B, Currey M, Cresko WA. Developmental dissociation in morphological evolution of the stickleback opercle. Evol. Dev. 2012;14:326–337. doi: 10.1111/j.1525-142X.2012.00551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP. Morphometric integration and modularity in configurations of landmarks: tools for evaluating a priori hypotheses. Evol. Dev. 2009;11:405–421. doi: 10.1111/j.1525-142X.2009.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP. MorphoJ: an integrated software package for geometric morphometrics. Molecular Ecology Resources. 2011;11:353–357. doi: 10.1111/j.1755-0998.2010.02924.x. [DOI] [PubMed] [Google Scholar]
- McPhee MV, Utter F, Stanford JA, Kuzishchin KV, Savvaitova KA, Pavlov DS, Allendorf FW. Population structure and partial anadromy in Oncorhynchus mykiss from Kamchatka: relevance for conservation strategies around the Pacific Rim. Ecol. Freshw. Fish. 2007;16:539–547. [Google Scholar]
- Nelson GJ. Relationships of clupeomorphs, with remarks on the structure of the lower jaw in fishes. In: Greenwood PH, Miles RS, Patterson C, editors. Interrelationships of Fishes. Zool. J. Linn. Soc. Suppl. 1. Vol. 53. 1973. pp. 333–349. [Google Scholar]
- Noakes DLG. Behavior and genetics of salmon. In: Woo PTK, Noakes DJ, editors. Salmon: Biology, Ecological Impacts and Economic Importance. Vol. 1. New York: Nova Science Publishers; 2014. pp. 195–222. [Google Scholar]
- Oakley TH, Phillips RB. Phylogeny of salmonine fishes based on growth hormone introns: Atlantic (Salmo) and Pacific (Oncorhynchus) salmon are not sister taxa. Mol. Phyologenet. Evol. 1999;11:381–393. doi: 10.1006/mpev.1998.0599. [DOI] [PubMed] [Google Scholar]
- Olson EC, Miller RL. Morphological Integration. Chicago: University of Chicago Press; 1958. [Google Scholar]
- Patterson C. Cartilage bones, dermal bones and membrane bones, or the exoskeleton versus the endoskeleton. In: Andrews SM, Miles RS, Walker AD, editors. Problems in Vertebrate Evolution. London: Academic Press; 1977. pp. 77–122. [Google Scholar]
- Pearse DE, Miller MR, Abadía-Cardoso A, Garza JC. Rapid parallel evolution of standing variation in a single, complex, genomic region is associated with life history in steelhead/rainbow trout. Proc. R. Soc. B. 2014;281 doi: 10.1098/rspb.2014.0012. 20140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn TP. The Behavior and Ecology of Pacific Salmon and Trout. Bethesda MD: American Fisheries Society; Seattle: University of Washington Press; 2005. [Google Scholar]
- Rasband WS. ImageJ. Bethesda: U.S. National Institutes of Health; 2012. imagej.nih.gov/ij/ [Google Scholar]
- Rohlf F. tpsDig2, digitize landmarks and outlines, version 2.12 (computer software) Department of Ecology and Evolution, State Univ. of New York at Stony Brook; 2008a. [Google Scholar]
- Rohlf F. tpsRelw, relative warps analysis, version 1.46 (computer software) Department of Ecology and Evolution, State Univ. of New York at Stony Brook; 2008b. [Google Scholar]
- Tiffan KF, Connor WP. Distinguishing between natural and hatchery Snake River fall Chinook salmon subyearlings in the field using body morphology. Trans. Am. Fish. Soc. 2011;140:21–30. [Google Scholar]
- Tiffan KF, Rondorf DW, Rodney DG, Verhey PA. Identification of juvenile fall versus spring Chinook salmon migrating through the Lower Snake River based on body morphology. Trans. Am. Fish. Soc. 2000;129:1389–1395. [Google Scholar]
- Vladykov VD. Osteological studies on Pacific salmon of the genus Oncorhynchus. Ottawa: Fisheries Research Board of Canada; 1962. (no. 136) [Google Scholar]
- Varian A, Nichols KM. Heritability of morphology in brook trout with variable life histories. PLoS ONE. 2010;5:e12950. doi: 10.1371/journal.pone.0012950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP. Homologues, natural kinds and the evolution of modularity. Am. Zool. 1996;36:36–43. [Google Scholar]
- Wagner G, Altenberg L. Complex adaptations and the evolution of evolvability. Evolution. 1996;50:967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- Walker MB, Kimmel CB. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 2007;82:23–28. doi: 10.1080/10520290701333558. [DOI] [PubMed] [Google Scholar]
- Williamson K, DeHaan P, Hawkins D. Genetic origin of Oncorhynchus mykiss collected from the Upper Willamette River Basin, OR. United States Fish & Wildlife Service Abernathy Fish Technology Center Report; 2010. [Google Scholar]
- Zelditch ML, Swiderski DL, Sheets HD. Geometric Morphometrics for Biologists: A Primer. 2nd. San Diego: Elsevier Academic Press; 2012. [Google Scholar]
- Zelditch ML, Wood AR, Bonett RM, Swiderski DL. Modularity of the rodent mandible: Integrating bones, muscles, and teeth. Evol. Dev. 2008;10:756–768. doi: 10.1111/j.1525-142X.2008.00290.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.