Abstract
In lymphatic tissues, chronic lymphocytic leukemia (CLL) cells are interspersed with CD68+ nurselike cells (NLCs), T cells, and other stromal cells that constitute the leukemia microenvironment. However, the mechanism regulating colocalization of CLL and these accessory cells are largely unknown. To dissect the molecular cross talk between CLL and NLCs, we profiled the gene expression of CD19-purified CLL cells before and after coculture with NLCs. NLC coculture induced high-level expression of B-cell maturation antigen and 2 chemoattractants (CCL3, CCL4) by CLL cells. CCL3/CCL4 induction in NLC cocultures correlated with ZAP-70 expression by CLL cells. High CCL3/CCL4 protein levels were found in CLL cocultures with NLCs, and CCL3/CCL4 induction was abrogated by R406, a Syk inhibitor, suggesting that NLCs induce these chemokines via B-cell receptor (BCR) activation. BCR triggering also caused robust CCL3/CCL4 protein secretion by CLL cells. High CCL3 and CCL4 plasma levels in CLL patients suggest that this pathway plays a role in vivo. These studies reveal a novel mechanism of cross talk between CLL cells and their microenvironment, namely, the secretion of 2 T-cell chemokines in response to NLC coculture and BCR stimulation. Through these chemokines, CLL cells can recruit accessory cells and thereby actively create a supportive microenvironment.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is characterized by the accumulation of monoclonal CD5+ B cells in blood, secondary lymphoid tissues, and the bone marrow.1 Most of the circulating leukemia cells are arrested in the G0/G1 phase of the cell cycle; therefore, the primary defect may be one of resistance to apoptosis rather than accelerated cell division.2 However, in vitro CLL cells undergo spontaneous apoptosis, suggesting that such ex vivo conditions lack factors necessary for leukemia-cell survival and that the resistance to apoptosis is not intrinsic to the leukemia B cell. In vitro apoptosis of CLL cells can be prevented by coculture with different accessory cells that are part of the CLL microenvironment, such as monocyte-derived nurselike cells (NLCs),3–6 mesenchymal marrow stromal cells (MSCs),3,7,8 or follicular dendritic cells,9 which provide survival signals to CLL cells. NLCs differentiate from monocytes into large, round, adherent cells that attract CLL cells and protect them from undergoing spontaneous or drug-induced cell death in a contact-dependent fashion.3,4,10 Because these cells share features in common with thymic nurse cells that nurture developing thymocytes,11 we designated these cells “nurselike cells.”3 NLCs can be found in the spleen and secondary lymphoid tissue of patients with CLL4 and thus represent a model for the microenvironment in secondary lymphatic tissues. High levels of CD68 make NLCs comparable with CD68+ lymphoma-associated macrophages in follicular lymphoma.12 Although still controversial, some studies demonstrated that a high CD68+ cell content in the microenvironment is associated with an aggressive clinical course and poor outcome in follicular lymphoma,12–14 suggesting that cell-cell interactions between the lymphoma cells and accessory cells of monocyte/macrophage lineage play a role in supporting neoplastic B-cell growth and drug resistance in lymphatic tissues. In addition, T cells are an integral part of the microenvironment in CLL. In CLL pseudofollicles (PFs), CLL cells are interspersed with T cells in proliferation clusters.15,16 PFs are a hallmark finding in CLL histopathology and are considered the proliferative compartment of this disease.17–19 In PFs, T cells are in intimate contact with CLL cells and express activation markers, such as CD40 ligand (CD154).15,20 Moreover, contact with activated CD4+ T cells induces Survivin15 and CD38 expression16 in CLL cells in PFs, suggesting that T cells promote CLL cell activation and proliferation in these areas.16,17 However, the factors promoting colocalization of T cells with CLL cells are largely unknown.16
Several molecules involved in cross talk between CLL cells and their microenvironment have recently been identified based on in vitro work and correlative studies on CLL tissue specimen. We characterized CXCL12 (SDF-1), a chemokine constitutively secreted by MSCs21 and NLCs,3 as a chemotactic and antiapoptotic factor for CLL cells, acting through its cognate receptor termed CXCR4, which is expressed at high levels on CLL cells.21,22 CXCR4 antagonists make CLL cells susceptible to chemotherapeutic drugs in stroma cocultures,10,23 and we therefore proposed the CXCL12-CXCR4 axis as a therapeutic target in CLL.3,10 However, our initial3 and a subsequent study6 indicated that the CXCR4-CXCL12 axis is not the only prosurvival pathway in CLL-NLC cross-talk.10,23 More recently, we reported that NLCs express another chemokine called CXCL13, which binds to CXCR5 chemokine receptors expressed at high levels on CLL cells.24,25 Similar to CXCL12, CXCL13 is another homeostatic chemokine constitutively secreted by stromal cells in B-cell follicles, which recruits circulating B cells to follicles.26,27 In addition to CXCL12 and CXCL13, NLCs express B cell–activating factor of the tumor necrosis factor (TNF) family (BAFF), a proliferation-inducing ligand (APRIL),6 CD31, and plexin-B1,5 which also protect CLL cells from apoptosis, indicating that NLCs use multiple distinct pathways to support CLL cell survival.
The aim of the present study was to analyze gene expression changes induced in purified CLL cells as a result of coculture with NLCs. First, we were interested in the feasibility of this experimental approach for dissecting the molecular cross talk between CLL cells and their microenvironment. Besides up-regulation of B-cell maturation antigen (BCMA), which appeared not particularly surprising in the context of previous studies on the importance of this molecule in CLL-NLC interactions,6,28 we found a robust up-regulation of 2 T-cell chemokines, CCL3 and CCL4, in CLL cells by coculture with NLCs. We also found high amounts of CCL3 and CCL4 protein in CLL cocultures with NLCs, which was inhibited by a specific Syk inhibitor. B-cell receptor (BCR) triggering also induced CCL3 and CCL4 secretion. These novel findings suggest that CLL cells play an active role in creating a supportive microenvironment and could explain why CLL cells are interspersed with T cells in tissue microenvironments, particularly in PFs.
Methods
Cell purification, flow cytometry, plasma samples, and CCL3 and CCL4 enzyme-linked immunosorbent assay
After informed consent was obtained in accordance with the Declaration of Helsinki, peripheral blood samples were collected from patients according to Institutional Review Board–approved protocols, fulfilling diagnostic and immunophenotypic criteria for B-cell CLL at the Leukemia Department, M. D. Anderson Cancer Center (Houston, TX). Peripheral blood mononuclear cells (PBMCs) were isolated via density gradient centrifugation over Ficoll Paque (GE Healthcare, Little Chalfont, United Kingdom) and used fresh or viably frozen in fetal calf serum plus 10% dimethyl sulfoxide (DMSO; Sigma-Aldrich, St Louis, MO) for storage in liquid nitrogen. ZAP-70 was detected in B-CLL cells by flow cytometry. A total of 106 mononuclear cells were incubated for 30 minutes at 4°C with saturating concentrations of the following monoclonal antibodies (mAbs): anti-CD3 phycoerythrin, anti-CD56 phycoerythrin, CD19 allophycocyanin (BD Biosciences PharMingen, San Diego, CA), and anti CD5 PerCP-Cy5.5 (BD Biosciences, San Jose, CA). The cells were washed twice and fixed with 4% paraformaldehyde in phosphate-buffered saline and then permeabilized with 0.1% saponin in Hank balanced salt solution for 5 minutes at 4°C. The cells were washed and then stained for 45 minutes at 4°C with a mAb anti-ZAP-70 Alexa-488 (Invitrogen, Carlsbad, CA). The cells were washed and analyzed by flow cytometry (FACSCalibur; BD Biosciences) and subsequent data analysis using the FlowJo software, version 8.5.2 (TreeStar, Ashland, OR). The expression of ZAP-70 was measured by calculating the percentage of CD19+CD3−CD56− cells that expressed ZAP-70 above the threshold. The threshold was set so that 0.1% of normal lymphocytes expressed ZAP-70. Normal lymphocytes were obtained from healthy donors and process in the same experiment as CLL samples. Indeed, CLL samples were considered positive when ZAP-70 expression was higher than 20%.29
Supernatants from CLL cells stimulated with anti-IgM mAbs for BCR stimulation were removed at the indicated time points and assayed for CCL3 and CCL4 protein by quantitative enzyme-linked immunosorbent assay (ELISA), according to the manufacturer's instructions (R&D Systems, Minneapolis, MN). CCL3 and CCL4 protein levels in blood plasma samples (n = 67) from patients with CLL at the Leukemia Department, M. D. Anderson Cancer Center and in normal blood plasma samples from healthy volunteers (n = 9) were also assayed by ELISA.
RNA isolation from CLL B cells before and after NLC coculture
RNA was isolated from CD19-purified CLL cells from 9 different patients' PBMCs after Ficoll separation and subsequent purification with CD19 MicroBeads and the MACS technology according to the manufacturer's instructions (Miltenyi Biotec, Bergisch Gladbach, Germany). For comparison, the same CLL cell samples were cocultured for 14 days with NLCs (14d NLC). For coculture with NLCs, PBMCs from patients with CLL were suspended in complete RPMI medium (RPMI 1640 with 10% fetal calf serum, penicillin-streptomycin-glutamine; Invitrogen) to a concentration of 107/mL (total 20 mL) and incubated for 14 days in 75-cm2 tissue-culture flasks (Techno Plastic Products, Trasadingen, Switzerland) as described previously.3 Nonadherent lymphoid cells then were removed and the NLC layer was washed 2 times with phosphate-buffered saline. The complete removal of lymphocytes was verified by phase-contrast microscopy. The nonadherent cells together with the wash fractions were then used for RNA preparation. To purify the CLL B cells before RNA isolation, CLL PBMCs were passed through a 30-μm nylon mesh to obtain a single-cell suspension. Then CLL B cells were purified with CD19 MicroBeads. Using TRIzol (Invitrogen), B cells were lysed according to the manufacturer's instructions. Total RNA extraction then was performed with the PureLink Micro-to-Midi Total RNA Purification System as described by the manufacturer (Invitrogen). The RNA was quantified using a NanoDrop 1000 spectrophotometer (Fisher Scientific, Pittsburgh, PA).
Expression analysis
Gene expression studies were performed using HG U133 plus 2.0 oligonucleotide arrays from Affymetrix (Santa Clara, CA). The samples were processed following the protocol for eukaryotic samples in the expression analysis technical manual from Affymetrix (available at www.affymetrix.com). In detail, cDNA synthesis was performed with a starting amount of 1 to 2 μg total RNA using the One-cycle cDNA synthesis kit (Affymetrix) following the Affymetrix standard protocol. Double-stranded cDNA and cRNA were purified with the GeneChip sample cleanup module. Synthesis of biotin-labeled cRNA was performed using the IVT labeling kit from Affymetrix, followed by cleanup and fragmentation. After 16 hours of hybridization at 45°C, the arrays were washed, stained (Fluidics Station 450), and laser scanned (Affymetrix GC scanner 3000) following the Affymetrix manual. The array images were visually checked for hybridization irregularities. The average probe array signals were scaled to a target signal of 500 using the GeneChip operating software.
CCL3 and CCL4 protein expression in CLL-NLC cocultures
CLL cells from 20 patients (10 ZAP-70+ and 10 ZAP-70−) were placed in long-term cultures for generation of NLCs, as detailed in “RNA isolation from CLL B cells before and after NLC coculture.” Supernatant samples were obtained at 2, 7, and 14 days of coculture and assayed for CCL3 and CCL4 protein concentrations by ELISA. The patients' characteristics are displayed in Table S5 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
CLL-cell stimulation with anti-IgM: CLL cell viability, induction of CCL3 and CCL4 protein expression
BCR signaling is critical for the survival of normal and malignant mature B cells, such as CLL cells.30–32 To determine the efficacy of BCR stimulation of CLL cells, initially we analyzed CLL cell viability after stimulation with anti-IgM, or medium alone. CLL cells (107 cells/mL) were incubated in complete RPMI medium supplemented with 14 μg/mL or 42 μg/mL anti-IgM (polyclonal goat F(ab′)2 fragments to human IgM; MP Biomedicals, Irvine, CA), 10 μg/mL anti–human CD40 (clone LOB7/6; AbD Serotec, Oxford, United Kingdom), or complete medium alone. After 24 and 48 hours, CLL cell viability was determined by analysis of mitochondrial transmembrane potential by 3,3-dihexyloxocarbocyanine iodine (DiOC6, Invitrogen) and cell membrane permeability to propidium iodide (PI; Invitrogen), as described33 on a FACSCalibur (BD Biosciences). To determine the effect of different concentrations of anti-IgM on induction of CCL3 and CCL4 protein, CLL cells from 7 different patients were suspended in complete RPMI medium to a concentration of 107 cells/mL and incubated in 96-well plates for 24 hours at 37°C in 5% CO2 with anti-IgM at 0.5, 1, 5, 10, or 30 μg/mL anti-IgM. The same CLL samples were also incubated with 10 μg/mL of anti–human CD40 or RPMI medium alone. After 24 hours, the supernatants were removed and assayed for CCL3 and CCL4 protein by ELISA. To determine the time course of CCL3 and CCL4 protein secretion by CLL cells, CLL cells from 4 different patients were incubated with 30 μg/mL anti-IgM at a concentration of 107 cells/mL in complete medium for 48 hours at 37°C in 5% CO2. Supernatant samples were removed at different time points (0, 1, 2, 4, 6, 8, 24, and 48 hours), and CCL3 and CCL4 protein content was quantified by ELISA.
Effect of the Syk inhibitor R406 on secretion of CCL3 and CCL4 by CLL cells in CLL-NLC cocultures
The spleen tyrosine kinase (Syk) is a key mediator of immune receptor (T-cell receptor/TCR and BCR) signal transduction in lymphocytes. In B cells, Syk initiates downstream events and amplifies the initial BCR signal.34 R406 is a novel ATP-competitive, specific Syk inhibitor35 that induces apoptosis in non-Hodgkin lymphoma (NHL) cells,36 and its clinical activity in patients with NHL and CLL was recently reported.37 R406 was kindly provided by Rigel Pharmaceuticals (San Francisco, CA). A 100-mM stock solution, stored in aliquots at −80°C, was diluted in DMSO and added to the assay media to a final concentration of 5 μM. To examine whether R406 affects CCL3 and/or CCL4 secretion by CLL cells in cocultures of CLL cells with NLCs, CLL-NLC cocultures were established from 4 different patients. After 14 days, the CLL cells were removed from the NLCs by washing and incubated with 5 μM R406 or the respective concentration of DMSO (1%) for 30 minutes. Then, the CLL cell suspension was added back to the NLCs, and the supernatants of the (DMSO) controls and the R406-treated CLL cells were sampled at 24 and 48 hours for CCL3 and CCL4 protein detection by ELISA.
Data analysis, statistics
Results are shown as mean plus or minus SD or SEM of at least 3 experiments each. For statistical comparison between groups, the Student paired t test or Bonferroni t test was used. For correlations, the Pearson correlation coefficient was calculated. Analyses were performed using the Statistica 6.1 software (StatSoft, Tulsa, OK). Flow cytometry data were analyzed using the FlowJo software (TreeStar).
Results
Gene expression response of CLL cells after coculture with NLCs
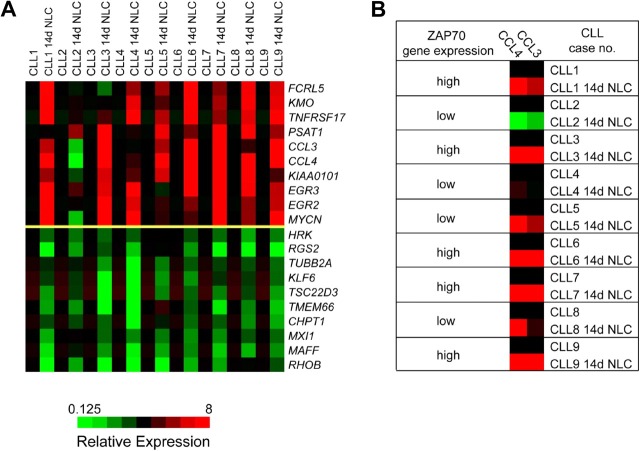
We observed a relatively homogeneous gene expression response of CLL cells after coculture with NLCs. Table 1 displays the 10 genes that were most highly up-regulated by NLC coculture, based on at least 3-fold up-regulation in at least 6 of the paired specimens and the overall mean up-regulation in all samples. Figure 1A displays the corresponding heatmaps that illustrate the top 10 up- and down-regulated genes in CLL cells after NLC coculture relative to the baseline expression immediately after isolation from the PB. As outlined in Table 1, the top 10 genes induced by NLC coculture are the CC chemokine CCL4, the BCMA (TNFRSF17), early growth response protein 3 gene (EGR3), the CC chemokine CCL3, phosphoserine aminotransferase 1 (PSAT1), N-myc proto-oncogene (MYCN), early growth response protein 2 gene (EGR2), the uncharacterized gene sequence KIAA0101, kynurenine 3-hydroxylase (KMO), and the Fc receptor-like 5 gene (FCRL5). Because of the high-level expression and because CCL3 and CCL4 have not previously been studied in CLL, we focused our subsequent studies on these 2 chemokines. The microarray data can be found in the Gene Expression Omnibus database under accession number GSE13811 (http://www.ncbi.nlm.nih.gov/geo/).
Table 1.
Up-regulated genes after coculture of CLL cells with NLCs
| Gene name | No. of cases with more than 3-fold up-regulation | Mean up-regulation in 9 samples, fold change |
|---|---|---|
| CCL4 | 7 | 10.16 |
| TNFRSF17 | 7 | 6.72 |
| EGR3 | 7 | 5.97 |
| CCL3 | 6 | 6.09 |
| PSAT1 | 6 | 4.98 |
| MYCN | 6 | 4.96 |
| EGR2 | 6 | 4.84 |
| KIAA0101 | 6 | 4.39 |
| KMO | 6 | 3.93 |
| FCRL5 | 6 | 3.31 |
This table displays, in descending order, the top 10 genes up-regulated in CLL cells after coculture with NLCs. The left column displays the gene names. The middle column displays the number of CLL paired specimens that displayed a more than 3-fold gene up-regulation (of a total of 9 cases). The right column indicates the mean up-regulation of the genes in all 9 cases.
CCL4 indicates CC chemokine; TNFRSF17, B-cell maturation antigen (BCMA); EGR3, early growth response protein 3 gene; CCL3, CC chemokine; PSAT1, phosphoserine aminotransferase 1; MYCN, N-myc proto-oncogene; EGR2, early growth response protein 2 gene; KIAA0101, uncharacterized gene sequence; KMO, kynurenine 3-hydroxylase; and FCRL5, Fc receptor-like 5.
Figure 1.
DNA microarray analysis identifies a homogeneous gene expression response of CLL cells after coculture with NLCs. (A) These heatmaps depict the top 10 genes that are up-regulated (top 10 rows of the graph) or down-regulated (bottom 10 rows) in 9 different CLL cell samples before and after 14 days of coculture with NLCs (14d NLC). The changes in gene expression are depicted for each gene relative to its expression level immediately after Ficoll isolation and CD19 purification and before initiation of NLC cultures (black squares). Shades of red and green indicate up- or down-regulation, respectively, of a given gene according to the color scheme displayed below the heatmaps. Displayed are the expression responses in 9 different CLL patient samples, as indicated on the top horizontal axis. (B) To illustrate the correlation between ZAP-70 expression and CCL3 and CCL4 induction, ZAP-70 gene expression in individual CLL cases was classified as high or low, as determined by microarray analysis and correlated with CCL3 and CCL4 expression before and after NLC coculture. In CLL cases with high ZAP-70 gene expression (cases 1, 3, 6, 7, and 9), NLC coculture uniformly induced a robust expression of CCL3 and CCL4, whereas in cases with low ZAP-70 gene expression the response was more heterogeneous (no induction of CCL3 and CCL4 in case 2; intermediate induction in cases 4, 5, and 8).
Induction of CCL3 and CCL4 correlates with ZAP-70 expression of the CLL cells
Because BCR activation and downstream signaling via the ζ-associated protein of 70 kD (ZAP-70) are discussed as a central mechanism for CLL cell survival in tissue microenvironments,1,38 we analyzed whether ZAP-70 expression, as determined in our array studies, correlated with the expression of up-regulated genes in NLC cocultures. Figure 1B illustrates the correlation between ZAP-70 gene expression and induction of CCL3 and CCL4. For example, in cases with increased ZAP-70 expression (cases 1, 3, 6, 7, and 9), NLC coculture uniformly induced a robust expression of CCL3 and CCL4; whereas in cases with low ZAP-70 expression, the response was more heterogeneous or absent. To determine the statistical significance of this correlation, we calculated the Pearson correlation between the induced CCL3 or CCL4 expression (CCL3 or CCL4 in CLL-NLC cocultures minus the respective baseline CCL3 or CCL4 expression at day 0) and the mean ZAP-70 expression by CLL cells in CLL-NLC cocultures (Figure S1A,B). We found a significant correlation between ZAP-70 expression and CCL3 expression (Pearson correlation coefficient r = 0.798, P = .005) and between ZAP-70 expression and CCL4 expression by the CLL cells (r = 0.733, P = .012).
High levels of CCL3 and CCL4 protein expression in supernatants from CLL-NLC cocultures
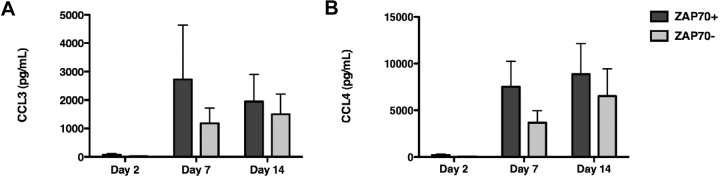
To determine whether our observation of CCL3 and CCL4 gene expression by microarray analysis is associated with an increased production and secretion of the respective proteins, we assayed supernatants from CLL cells placed in long-term cultures with outgrowth of NLCs for CCL3 and CCL4 protein by ELISA. After 2 days of culture (Figure 2), the CCL3 and CCL4 levels generally were low, with higher levels and more heterogeneity in case of CCL4 (Figure 2C). In contrast, we detected high levels of both chemokines after 7 and 14 days of culture, which coincided with the outgrowth of NLCs. We reported earlier that NLCs can be observed in CLL PBMC cultures as early as 3 to 4 days after culture initiation and the number of NLC plateaus after 7 days.3 Figure 2 illustrates the kinetics and variability of CCL3 and CCL4 protein expression in supernatants of CLL cells from 20 different patients, illustrating the mean CCL3 and CCL4 levels at the different time points (Figure 2A) and the individual patients' protein levels for CCL3 (Figure 2B) and CCL4 (Figure 2C) in ZAP-70+ and ZAP-70− patients. The exact protein concentrations corresponding to these figures are detailed in Tables S1 and S2. CLL-NLC cocultures from ZAP-70+ patients generally displayed higher levels of CCL3 (Figure 3A) and CCL4 (Figure 3B) than ZAP-70− patients. These differences, however, were not statistically significant (Tables S3, S4).
Figure 2.
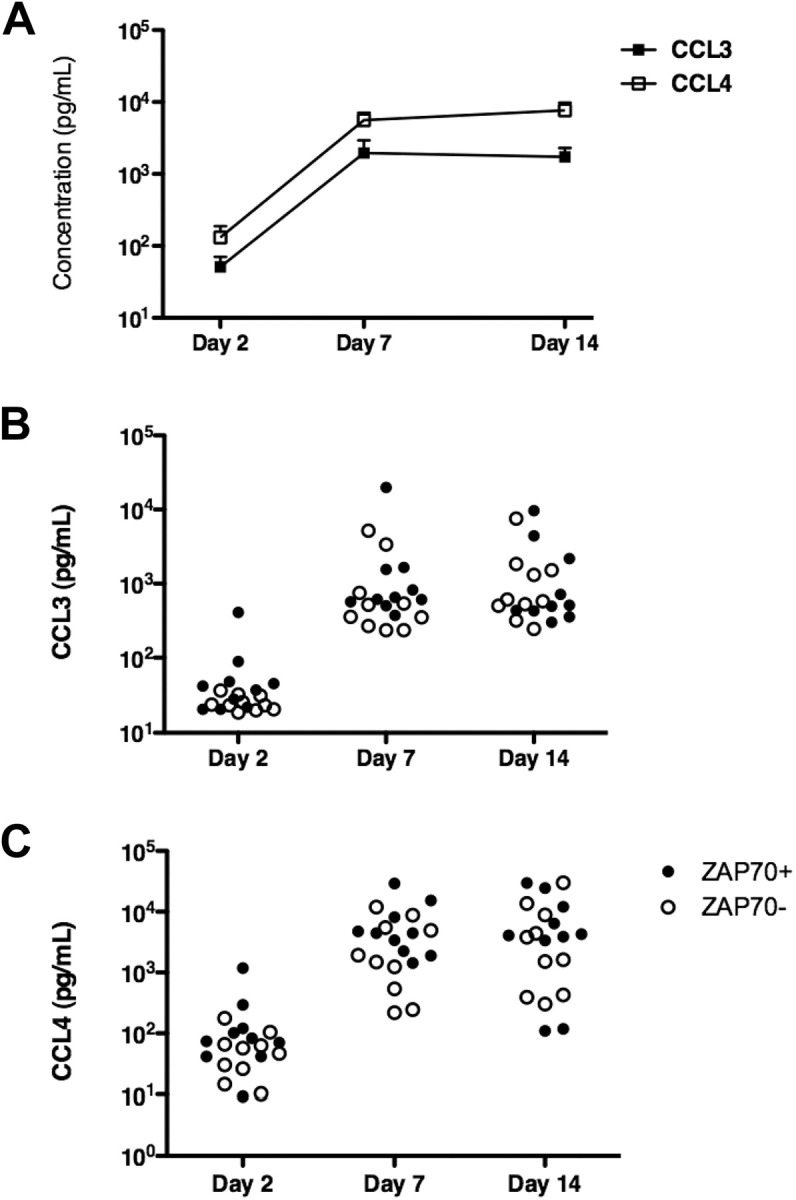
Induction of CCL3 and CCL4 protein expression in CLL-NLC cocultures. CLL PBMCs were placed in long-term cultures with outgrowth of NLCs, as described in “RNA isolation from CLL B cells before and after NLC coculture.” To determine the kinetics of induction of CCL3 and CCL4 protein expression, we analyzed supernatant samples at the time points indicated on the horizontal axis by ELISA. (A) The different symbols that are connected by the lines represent the mean CCL3 and CCL4 concentrations in 20 different CLL patients' samples at the indicated time points. In accordance with the microarray data, we found higher CCL4 protein expression compared with CCL3. (B,C) The individual protein expression data for CCL3 and CCL4 in the 20 CLL cases are displayed in dot plot diagrams. ZAP-70+ cases (•) can be distinguished from ZAP-70− cases (○).
Figure 3.
CCL3 and CCL4 protein expression in ZAP-70+ and ZAP-70− CLL cases. Displayed are the mean and SEM for CCL3 (A) and CCL4 (B) protein concentrations in supernatants from ZAP-70+ CLL samples ( , n = 10) or ZAP-70− CLL samples (
, n = 10) or ZAP-70− CLL samples ( , n = 10). In ZAP-70+ CLL samples, we found higher levels of both CCL3 and CCL4 at all time points. However, these differences did not reach statistical significance.
, n = 10). In ZAP-70+ CLL samples, we found higher levels of both CCL3 and CCL4 at all time points. However, these differences did not reach statistical significance.
Anti-IgM stimulation protects CLL cells from undergoing apoptosis
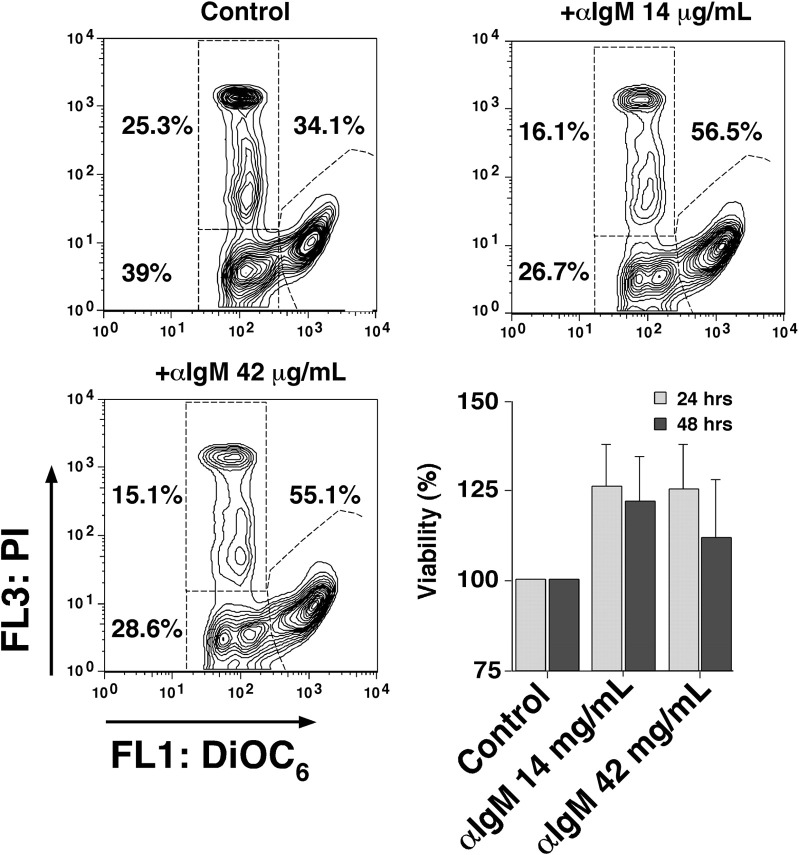
Self- or auto-antigens can continuously stimulate the BCR on CLL cells, which, along with other signals from the microenvironment, appear to be critical for promoting CLL cell survival and growth.1 Cross-linking the BCR with anti-IgM mimics this pathway in vitro, although not all CLL cases (only ∼ 50%) appear to respond to BCR stimulation, with a preferential response to anti-IgM in nonmutated, ZAP-70+ CLL cases.1,39,40 Based on the published studies, survival,31,41 rather than apoptosis,42 seems the more common response of CLL cells to anti-IgM stimulation, using anti-IgM concentrations between 10 and 20 μg/mL.31,41 In our patient samples, we noticed low to moderate increases (Figure 4) in viability after 24 and 48 hours after stimulation with anti-IgM, compared with control CLL cells in medium alone (100%). The mean viability after 24 hours was 125.8% plus or minus 23.4% of controls (mean ± SD, n = 4) with 14 μg/mL and 124.9% plus or minus 25.1% with 42 μg/mL anti-IgM (mean ± SD, n = 4). After 48 hours, the mean viability with 14 μg/mL anti-IgM was 121.8% plus or minus 24.3% and 111.6 plus or minus 31.9% with 42 μg/mL anti-IgM.
Figure 4.
BCR engagement protects CLL cells from undergoing apoptosis. Presented are contour maps of CLL B cells from one patient defining the relative green (DiOC6) and red (PI) fluorescence intensities of CLL cells on the horizontal and vertical axes, respectively. The viability was determined after 24 hours of culture in medium alone (control) or medium supplemented with anti-IgM mAbs at the concentrations indicated above each of the contour maps. This stain allows us to gate on the vital cell population (DiOC6bright, PIexclusion), apoptotic cells (DiOC6dim, PIexclusion), and the dead cells (DiOC6dim, PIpositive). The percentage of each cell population is indicated next to each of these gates. In this case, anti-IgM stimulation protected approximately 20% of the cells from undergoing apoptosis, resulting in a viable population of 56.5% at 14 μg/mL anti-IgM and 55.1% at 42 μg/mL anti-IgM. The bar diagram on the lower right displays an increased viability of CLL cells after culture with anti-IgM in 4 different CLL samples. CLL cells were cultured in medium alone (control) or medium supplemented with anti-IgM mAbs at the indicated concentrations, and the viability was assessed after 24 and 48 hours by staining with DiOC6 and PI. Displayed are the means plus or minus SD of 4 different patient samples.
IgM stimulation induces CCL3 and CCL4 protein secretion by CLL cells
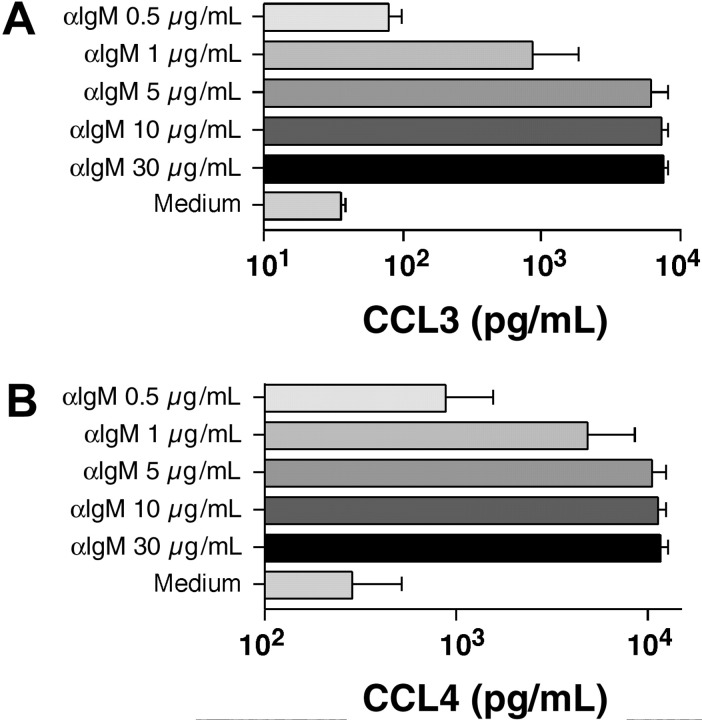
Here we incubated CLL cells with increasing concentrations of anti-IgM and tested CLL supernatants after 24 hours for CCL3 and CCL4 protein by ELISA. As displayed in Figure 5, incubation of CLL cells for 24 hours with anti-IgM induced a dose-dependent induction of CCL3 and CCL4 between 0.5 and 10 μg/mL anti-IgM, which reached a plateau at concentrations greater than 10 μg/mL. CLL samples in medium alone or supplemented with anti-CD40 did not show any significant levels of CCL3 or CCL4 (Tables S6, S7).
Figure 5.
Induction of CCL3 and CCL4 protein expression by CLL cells after BCR engagement: anti-IgM dose-response. CLL cells were placed in culture medium supplemented with different concentrations of anti-IgM, as indicated on the vertical axis. CLL cells cultured with medium alone were used as controls. After 24 hours, supernatants were removed and assayed for CCL3 (A) and CCL4 (B) protein expression by ELISA. Displayed are the mean (± SD, n = 3) CCL3 and CCL4 concentrations, as indicated on the horizontal axis. Whereas anti-IgM induced a dose-dependent induction of CCL3 and CCL4, CLL cells cultured in medium alone did not express significant amounts of CCL3 and CCL4.
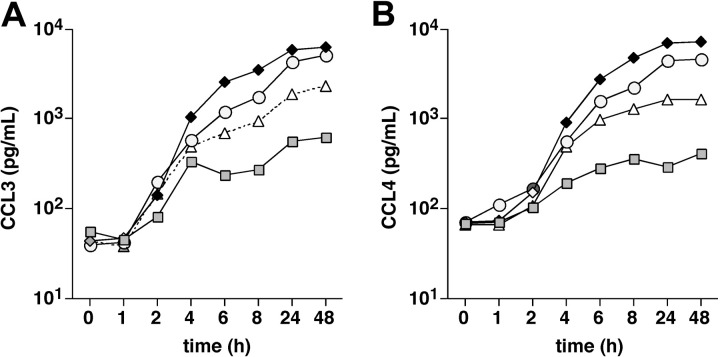
Time course of CCL3 and CCL4 protein secretion by CLL cells in response to BCR stimulation
In this experiment, we stimulated CLL cells for various times with 15 μg/mL of anti-IgM and determined CCL3 and CCL4 protein levels in the CLL supernatants at the indicated time points (Figure 6). The first increases in CCL3 and CCL4 levels in the supernatants were detected 2 hours after initiation of the cultures, and the levels continuously increased over the following hours up to the 24-hour time point (Figure 5). After the 24-hour time point, CCL3 and CCL4 levels plateaued, suggesting that the optimal response was reached at 24 hours.
Figure 6.
Time course of CCL3 and CCL4 protein expression by CLL cells after BCR engagement. CLL cells were placed in culture medium and stimulated with 15 μg/mL anti-IgM. Supernatants were removed at the time points indicated on the horizontal axis and assayed for CCL3 (A) and CCL4 (B) protein expression by ELISA. The different symbols that are connected by the lines represent the CCL3 and CCL4 concentrations in 4 different CLL samples at the indicated time points.
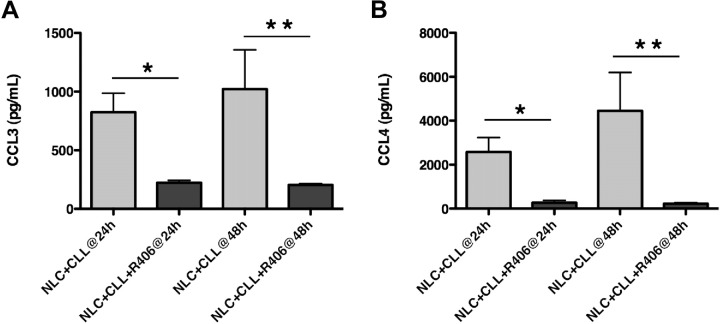
The Syk inhibitor R406 inhibits CCL3 and CCL4 secretion in CLL cell cocultures with NLCs
Incubation of CLL cells with the specific Syk inhibitor R406 almost completely abrogated the induction of CCL3 and CCL4 secretion by CLL cells in cocultures with NLCs. R406 at 24 and 48 hours significantly reduced the detectable CCL3 and CCL4 levels in supernatants from CLL-NLC cocultures (P < .05, n = 4), as displayed in Figure 7, suggesting that the induction of CCL3 and CCL4 secretion in CLL-NLC cocultures is Syk-dependent and therefore could involve the BCR.
Figure 7.
The Syk inhibitor R406 abrogates induction of CCL3 and CCL4 secretion by CLL cells in cocultures with NLCs. The bars represent the mean (±SEM) CCL3 and CCL4 concentration in CLL cell supernatants from 4 different patients incubated for 24 or 48 hours with NLCs in the presence or absence of the Syk inhibitor R406, as indicated on the horizontal axis. Treatment with R406 significantly inhibited the induction of CCL3 (A) and CCL4 (B) secretion by the CLL cells, as indicated by the asterisks, with P values less than .05 at 24 hours (*) and 48 hours (**). No significant CCL3 or CCL4 concentrations were detected in supernatants of NLCs alone (data not shown).
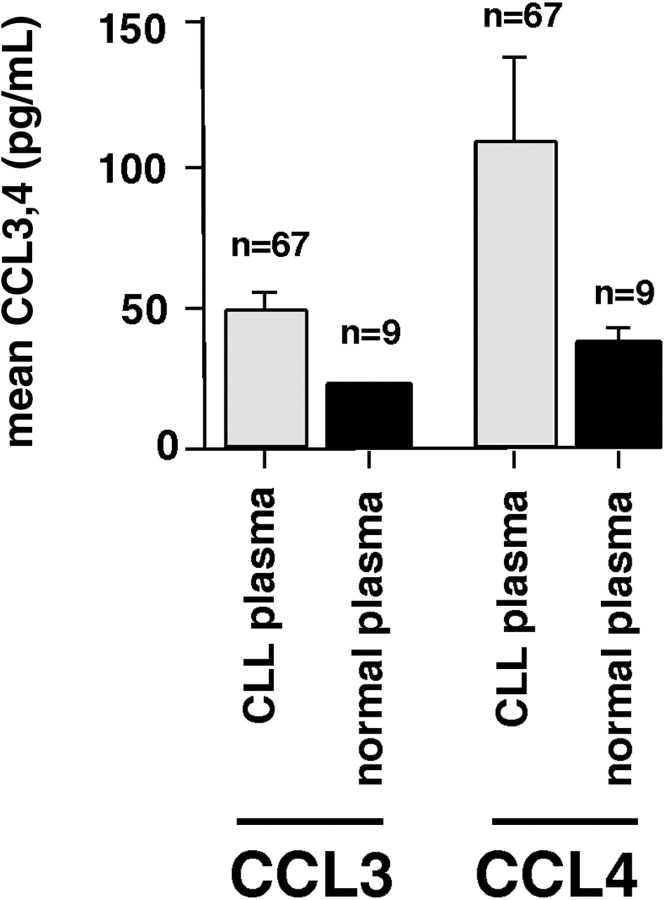
Increased levels of CCL3 and CCL4 in blood plasma from CLL patients
CLL cells may produce CCL3 and CCL4 in response to BCR stimulation (or other stimuli in the microenvironment) in vivo; consequently, elevated levels of these chemokines in CLL patients would be expected. Therefore, we tested blood plasma samples from CLL patients and healthy volunteers for CCL3 and CCL4 protein by ELISA. In CLL patients, the mean CCL3 blood plasma level was 47.2 plus or minus 7 ng/mL (mean ± SEM, n = 67), compared with 20.5 plus or minus 0.3 ng/mL (mean ± SEM, n = 9) in healthy controls. The mean CCL4 blood plasma level was 107.7 plus or minus 29 ng/mL (mean ± SEM, n = 67) in CLL patients, compared with 35.7 plus or minus 5.1 ng/mL (mean ± SEM, n = 9) in healthy controls (Figure 8). However, the differences between plasma levels in CLL patients and controls did not reach statistical significance. CCL3 and CCL4 levels were higher in the plasma of CLL patients who had adverse prognostic features, in particular, a β-2 microglobulin of more than 4 mg/dL, a nonmutated status of the IgVH genes, or more than 20% CD38+ CLL cells (data not shown).
Figure 8.
CCL3 and CCL4 protein levels in blood plasma from CLL patients and healthy volunteers. This bar diagram displays the mean (± SEM) concentrations of CCL3 and CCL4 in 67 plasma samples from CLL patients and normal donors (n = 9), as displayed on the horizontal axis. CLL plasma displayed higher levels of both CCL 3 and CCL4 compared with normal blood plasma, suggesting that secretion of these chemokines by CLL cells occurs in vivo.
Discussion
Even though in the last few years increasing emphasis has been placed on exploring the tumor microenvironment as a potential therapeutic target, we are currently faced with difficulties finding suitable methods to dissect the complex cellular and molecular interactions between tumor cells and the host. In this study, we cocultured CLL cells and NLCs to model interactions between CLL cells and CD68+ NLCs in vivo. Despite the limitation that an in vitro model is never going to be able to fully recapitulate the in vivo microenvironment, several recent studies suggest that this model is a close approximation to the in vivo microenvironment in tissue compartments, such as the secondary lymphoid tissues. Fully differentiated NLCs are detected in lymphoid tissues from CLL patients,4 and several molecular pathways that are essential for normal B-lineage cell biology are operational in this system. We therefore decided to use this model for a systematic gene expression analysis in CD19-purified CLL cells before and after coculture with NLCs to discover new molecular pathways of interaction between CLL cells and their microenvironment.
First, this study demonstrates the feasibility of this approach, as indicated by a relatively homogeneous gene expression profile with induction and down-regulation of several interesting genes that could be involved in microenvironment interactions and/or survival of the CLL cells (Figure 1A). Based on the published literature, the induction of 3 genes, CCL3, CCL4, and BCMA, appeared particularly important for CLL cell interactions with the microenvironment. The importance of BCMA for CLL cell interaction with NLCs already has been highlighted by several recent studies,6,28 and we therefore focused on the expression, induction, and possible function of the chemokines CCL3 and CCL4.
Macrophage inflammatory protein-1 (MIP-1) initially was purified from supernatants of endotoxin (lipopolysaccharide)-stimulated macrophages.43 Two distinct but highly related proteins, MIP-1α and MIP-1β, were subsequently characterized; and according to the current chemokine nomenclature, these members of the CC chemokine subfamily are now termed CCL3 and CCL4.44 CCL3 and CCL4 expression can be induced in most mature hematopoietic cells. B cells and dendritic cells produce CCL3 and CCL4 in response to various stimuli, such as BCR triggering, CD40 ligand, lipopolysaccharide, or TNF-α.45 In Boyden chamber chemotaxis assays, CCL3 and CCL4 are potent chemoattractants for monocytes46 and lymphocytes,47,48 even though the activity of these chemokines on distinct T-cell subsets remains controversial.43 Compared with CXCL12 (SDF-1), CCL3 and CCL4 are more potent chemoattractants, that is, they require lower concentrations for maximal activity (1-10 ng/mL).47,48 CCL3 signals through the chemokine receptors CCR1 and CCR5, whereas CCL4 signals only through the CCR5 chemokine receptor.44 Besides its physiologic function as a chemokine receptor on T cells and monocytes/macrophages, CCR5 also functions as a coreceptor for M-tropic HIV-1 strains.49 CCL3 and CCL4 have been implicated in several human diseases, such as rheumatoid arthritis,50 inflammatory and autoimmune central nervous system diseases such as multiple sclerosis,51 or inflammatory diseases of the respiratory tract.43 In multiple myeloma (MM), CCL3 enhances osteoclast formation and promotes MM cell migration and survival.52 CCL3 levels in marrow plasma from patients with active MM are elevated,52 and CCL3 expression by the MM cells, as determined by microarray studies, correlated with disease activity.53 These studies conclude that local (over)expression of CCL3 and CCL4 induces recruitment of accessory cells, particularly lymphocytes, and also may provide growth signals to neoplastic cells.
A more specific function of CCL3 and CCL4 in the lymphatic tissues has been suggested by recent in vitro54 and in vivo55 studies. Antigen binding to the BCR selectively induces CCL3 and CCL4 in naive, memory, and germinal center B cells.54 The kinetics of CCL3 and CCL4 protein release after BCR stimulation in normal B cells are very similar to our observations in CLL cells with a rapid increase over the first 24 hours and a subsequent plateau (Figure 6). In addition, Krzysiek et al reported that the induction of CCL3 and CCL4 secretion by B cells was restricted to BCR stimulation, whereas anti-CD40 mAbs and interleukin-4 (IL-4) stimulation had no effect.54 Because of the observation that CD4+ CD45RO+ T cells migrated along a chemotactic gradient formed by BCR-stimulated B cells in a CCL3- and CCL4-dependent fashion, Krzysiek et al suggested that this mechanism allows for recruitment and cognate interactions between Ag-specific T and B cells.54 More recently, the in vivo importance of CCL3 and CCL4 expression in lymph nodes for T-cell recruitment to sites of antigen-presenting cell (APC) T-cell interaction during the induction of specific immune responses was highlighted by Castellino et al.55 In an elegant vaccination model, they demonstrated that CD8+ T cells up-regulate CCR5, which then allows these T cells to migrate to sites of CD4+ T-cell/dendritic-cell interaction in response to CCL3 and CCL4.
The role of T cells in the pathogenesis of CLL remains controversial. CLL cells can respond to signals delivered by T cells, such as CD40 ligand (CD40L), which in turn can promote CLL cell survival and proliferation.15,56 In addition, there are considerable numbers of CD4+ T cells in CLL pseudofollicles,20,57,58 the proliferative compartment of this disease.17 In this context, Ghia et al reported the induction of another T-cell chemokine, CCL22, by CLL cells in response to CD40 ligation.20 As such, CLL cells can secrete at least 3 different T-cell chemokines depending on the mode of stimulation. Given the homogeneous CCL3 and CCL4 induction in our model, the elevated plasma levels of these chemokines in CLL patients, and the in vivo evidence that CCL3 and CCL4 are critical for T-cell recruitment to lymphoid tissues,55 it appears that CCL3 and CCL4 have a dominant role in T-cell recruitment for CLL–T-cell interactions.
ZAP-70 is a prognostic marker in CLL and plays an important role in BCR signaling in CLL.40 When analyzing the microarray data, we noticed a correlation between ZAP-70 expression by the CLL cells and the induction of CCL3 and CCL4 RNA in NLC cocultures (Figures 1B and S1). We also noticed a trend for higher levels of CCL3 and CCL4 protein in supernatants from ZAP-70+ CLL-NLC cocultures (Figure 3), which, however, did not reach statistical significance. This set of data suggests that ZAP-70 labels CLL cases that are more BCR-responsive, which in turn can respond to BCR stimulation with a more pronounced CCL3 and CCL4 response. On the other hand, the lack of significance when correlating ZAP-70 and CCL3/CCL4 protein induction by anti-IgM indicates that ZAP-70 may not be directly involved, and signals for CCL3/CCL4 could be transduced through other BCR signaling components, such as Syk, rather than ZAP-70, as suggested by our data using the Syk inhibitor R406.
In the context of our finding that BCR stimulation with anti-IgM induces CCL3/CCL4 secretion (Figures 5,6), it is tempting to speculate that CCL3 and CCL4 induction in NLC cocultures may be a consequence of CLL cell activation through the BCR in response to antigen(s) presented by NLCs. This hypothesis is strengthened by our finding that R406, a specific Syk inhibitor, significantly inhibited the induction of CCL3 and CCL4 protein secretion by CLL cells in CLL-NCL cocultures (Figure 7). Because Syk (along with ZAP-70) is a key molecule in BCR signaling, this finding suggests that indeed BCRs become activated by CLL-NCL interactions. However, additional experiments need to be performed to validate this hypothesis and to elucidate the mechanism and possible antigen(s) involved in this interaction. One approach could be to determine whether or not the expression profile induced in CLL cells by NCL coculture, and in particular, the induction of CCL3 and CCL4, is a NLC-specific finding. In preliminary experiments, we found that coculture with MSCs induced a distinct expression profile in CLL cells that was different from the expression profile induced by NLCs. More specifically, and in contrast to NLCs, MSCs did not induce the expression of CCL3 and CCL4 in CLL cells (data not shown). Ongoing experiments in our group now focus on the characterization of the expression profile induced in CLL cells by CLL-MSC cocultures.
NLCs express BAFF and APRIL,6 which normally are expressed by monocytes, macrophages, and dendritic cells. BAFF and APRIL are potent regulators of normal B-cell development and function59,60 and also protect neoplastic B cells, such as CLL cells28,61 or MM cells62 from undergoing apoptosis. BCMA, a receptor of the TNF superfamily, binds BAFF and APRIL and is preferentially expressed on plasma cells and tonsillar memory B cells.63 The specific signals that regulate BCMA expression are largely unknown. During BAFF-induced differentiation of memory B cells to Ig-secreting B cells, CD40 and BAFF-R are down-regulated, whereas BCMA is up-regulated,63,64 suggesting that BAFF promotes survival of Ig-secreting B cells via BCMA up-regulation. In addition, B-cell stimulation with CpG/IL-2, and IL-15 can induce BCMA expression.63 As such, it is tempting to speculate that coculture with NLCs induces a similar activation/differentiation response in CLL cells, leading to the BCMA induction that we observed after NLC coculture (Table 1, Figure 1).
Collectively, our data demonstrate CCL3 and CCL4 secretion by CLL cells in response to NLC coculture and BCR stimulation. Secretion of these chemokines by the leukemia B cells can foster productive CLL–T-cell interactions within the lymph node microenvironment, and therefore may participate, along with other signals from the microenvironment, in the propagation of the CLL clone. The high efficacy of the Syk inhibitor R406 in inhibiting the induction of CCL3 and CCL4 in CLL-NLC cocultures provides a rationale to further explore the possibility of BCR activation in CLL-NLC cocultures. In addition, the secretion of T-cell chemokines by CLL cells indicates that CLL cells are not passive “seed” but rather active players in creating a favorable microenvironment.
Acknowledgments
The authors thank Marina Henneberg and Matthias Niedermeier for expert technical assistance and Yasumichi Hitoshi and Ann Lowe from Rigel Pharmaceuticals for valuable suggestions and for providing R406.
This work was supported by a Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research (J.A.B.), an ASCO Career Development Award (J.A.B.), and CLL Global Research Foundation grants (J.A.B., A.R.). and A.R. and E.H are supported by the Interdiscinplinary Center for Clinical Research, University of Würzburg, Würzburg, Germany.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.A.B. designed the research, supervised the study, analyzed the data, and wrote the paper; M.P.Q. performed NLC cocultures and analyzed CCL3 and CCL4 protein induction and expression and the effect of R406; E.H. and A.B. performed microarray studies and analysis and provided CCL3 and CCL4 serum data; W.G.W. and M.J.K. provided patients' samples, analyzed data, and reviewed the manuscript; and A.R. designed and supervised the microarray studies and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan A. Burger, Department of Leukemia, Unit 428, University of Texas M. D. Anderson Cancer Center, PO Box 301402, Houston, TX 77230-1402; e-mail: jaburger@mdanderson.org.
References
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Munk Pedersen I, Reed J. Microenvironmental interactions and survival of CLL B-cells. Leuk Lymphoma. 2004;45:2365–2372. doi: 10.1080/10428190412331272703. [DOI] [PubMed] [Google Scholar]
- 3.Burger JA, Tsukada N, Burger M, Zvaifler NJ, Dell'Aquila M, Kipps TJ. Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell-derived factor-1. Blood. 2000;96:2655–2663. [PubMed] [Google Scholar]
- 4.Tsukada N, Burger JA, Zvaifler NJ, Kipps TJ. Distinctive features of “nurselike” cells that differentiate in the context of chronic lymphocytic leukemia. Blood. 2002;99:1030–1037. doi: 10.1182/blood.v99.3.1030. [DOI] [PubMed] [Google Scholar]
- 5.Deaglio S, Vaisitti T, Bergui L, et al. CD38 and CD100 lead a network of surface receptors relaying positive signals for B-CLL growth and survival. Blood. 2005;105:3042–3050. doi: 10.1182/blood-2004-10-3873. [DOI] [PubMed] [Google Scholar]
- 6.Nishio M, Endo T, Tsukada N, et al. Nurselike cells express BAFF and APRIL, which can promote survival of chronic lymphocytic leukemia cells via a paracrine pathway distinct from that of SDF-1alpha. Blood. 2005;106:1012–1020. doi: 10.1182/blood-2004-03-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panayiotidis P, Jones D, Ganeshaguru K, Foroni L, Hoffbrand AV. Human bone marrow stromal cells prevent apoptosis and support the survival of chronic lymphocytic leukaemia cells in vitro. Br J Haematol. 1996;92:97–103. doi: 10.1046/j.1365-2141.1996.00305.x. [DOI] [PubMed] [Google Scholar]
- 8.Lagneaux L, Delforge A, Bron D, De Bruyn C, Stryckmans P. Chronic lymphocytic leukemic B cells but not normal B cells are rescued from apoptosis by contact with normal bone marrow stromal cells. Blood. 1998;91:2387–2396. [PubMed] [Google Scholar]
- 9.Pedersen IM, Kitada S, Leoni LM, et al. Protection of CLL B cells by a follicular dendritic cell line is dependent on induction of Mcl-1. Blood. 2002;100:1795–1801. [PubMed] [Google Scholar]
- 10.Burger M, Hartmann T, Krome M, et al. Small peptide inhibitors of the CXCR4 chemokine receptor (CD184) antagonize the activation, migration and antiapoptotic responses of CXCL12 in chronic lymphocytic leukemia B cells. Blood. 2005;106:1824–1830. doi: 10.1182/blood-2004-12-4918. [DOI] [PubMed] [Google Scholar]
- 11.Wekerle H, Ketelsen UP. Thymic nurse cells: Ia-bearing epithelium involved in T-lymphocyte differentiation? Nature. 1980;283:402–404. doi: 10.1038/283402a0. [DOI] [PubMed] [Google Scholar]
- 12.Farinha P, Masoudi H, Skinnider BF, et al. Analysis of multiple biomarkers shows that lymphoma-associated macrophage (LAM) content is an independent predictor of survival in follicular lymphoma (FL). Blood. 2005;106:2169–2174. doi: 10.1182/blood-2005-04-1565. [DOI] [PubMed] [Google Scholar]
- 13.Dave SS, Wright G, Tan B, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 14.Byers RJ, Sakhinia E, Joseph P, et al. Clinical quantitation of immune signature in follicular lymphoma by RT-PCR based gene expression profiling. Blood. 2008;111:4764–4770. doi: 10.1182/blood-2007-10-115915. [DOI] [PubMed] [Google Scholar]
- 15.Granziero L, Ghia P, Circosta P, et al. Survivin is expressed on CD40 stimulation and interfaces proliferation and apoptosis in B-cell chronic lymphocytic leukemia. Blood. 2001;97:2777–2783. doi: 10.1182/blood.v97.9.2777. [DOI] [PubMed] [Google Scholar]
- 16.Patten PE, Buggins AG, Richards J, et al. CD38 expression in chronic lymphocytic leukemia is regulated by the tumor microenvironment. Blood. 2008;111:5173–5181. doi: 10.1182/blood-2007-08-108605. [DOI] [PubMed] [Google Scholar]
- 17.Caligaris-Cappio F. Role of the microenvironment in chronic lymphocytic leukaemia. Br J Haematol. 2003;123:380–388. doi: 10.1046/j.1365-2141.2003.04679.x. [DOI] [PubMed] [Google Scholar]
- 18.Soma LA, Craig FE, Swerdlow SH. The proliferation center microenvironment and prognostic markers in chronic lymphocytic leukemia/small lymphocytic lymphoma. Hum Pathol. 2006;37:152–159. doi: 10.1016/j.humpath.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 19.Stein H, Bonk A, Tolksdorf G, Lennert K, Rodt H, Gerdes J. Immunohistologic analysis of the organization of normal lymphoid tissue and non-Hodgkin's lymphomas. J Histochem Cytochem. 1980;28:746–760. doi: 10.1177/28.8.7003001. [DOI] [PubMed] [Google Scholar]
- 20.Ghia P, Strola G, Granziero L, et al. Chronic lymphocytic leukemia B cells are endowed with the capacity to attract CD4+, CD40L+ T cells by producing CCL22. Eur J Immunol. 2002;32:1403–1413. doi: 10.1002/1521-4141(200205)32:5<1403::AID-IMMU1403>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 21.Burger JA, Burger M, Kipps TJ. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood. 1999;94:3658–3667. [PubMed] [Google Scholar]
- 22.Mohle R, Failenschmid C, Bautz F, Kanz L. Overexpression of the chemokine receptor CXCR4 in B cell chronic lymphocytic leukemia is associated with increased functional response to stromal cell-derived factor-1 (SDF-1). Leukemia. 1999;13:1954–1959. doi: 10.1038/sj.leu.2401602. [DOI] [PubMed] [Google Scholar]
- 23.Zeng Z, Samudio IJ, Munsell M, et al. Inhibition of CXCR4 with the novel RCP168 peptide overcomes stroma-mediated chemoresistance in chronic and acute leukemias. Mol Cancer Ther. 2006;5:3113–3121. doi: 10.1158/1535-7163.MCT-06-0228. [DOI] [PubMed] [Google Scholar]
- 24.Burkle A, Niedermeier M, Schmitt-Graff A, Wierda WG, Keating MJ, Burger JA. Overexpression of the CXCR5 chemokine receptor, and its ligand, CXCL13 in B-cell chronic lymphocytic leukemia. Blood. 2007;110:3316–3325. doi: 10.1182/blood-2007-05-089409. [DOI] [PubMed] [Google Scholar]
- 25.Durig J, Schmucker U, Duhrsen U. Differential expression of chemokine receptors in B cell malignancies. Leukemia. 2001;15:752–756. doi: 10.1038/sj.leu.2402107. [DOI] [PubMed] [Google Scholar]
- 26.Gunn MD, Ngo VN, Ansel KM, Ekland EH, Cyster JG, Williams LT. A B-cell-homing chemokine made in lymphoid follicles activates Burkitt's lymphoma receptor-1. Nature. 1998;391:799–803. doi: 10.1038/35876. [DOI] [PubMed] [Google Scholar]
- 27.Ansel KM, Ngo VN, Hyman PL, et al. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 2000;406:309–314. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]
- 28.Endo T, Nishio M, Enzler T, et al. BAFF and APRIL support chronic lymphocytic leukemia B-cell survival through activation of the canonical NF-kappaB pathway. Blood. 2007;109:703–710. doi: 10.1182/blood-2007-04-081786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 30.Gururajan M, Jennings CD, Bondada S. Cutting edge: constitutive B cell receptor signaling is critical for basal growth of B lymphoma. J Immunol. 2006;176:5715–5719. doi: 10.4049/jimmunol.176.10.5715. [DOI] [PubMed] [Google Scholar]
- 31.Bernal A, Pastore RD, Asgary Z, et al. Survival of leukemic B cells promoted by engagement of the antigen receptor. Blood. 2001;98:3050–3057. doi: 10.1182/blood.v98.10.3050. [DOI] [PubMed] [Google Scholar]
- 32.Deglesne PA, Chevallier N, Letestu R, et al. Survival response to B-cell receptor ligation is restricted to progressive chronic lymphocytic leukemia cells irrespective of Zap70 expression. Cancer Res. 2006;66:7158–7166. doi: 10.1158/0008-5472.CAN-06-0085. [DOI] [PubMed] [Google Scholar]
- 33.Zamzami N, Marchetti P, Castedo M, et al. Reduction in mitochondrial potential constitutes an early irreversible step of programmed lymphocyte death in vivo. J Exp Med. 1995;181:1661–1672. doi: 10.1084/jem.181.5.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudd CE. Adaptors and molecular scaffolds in immune cell signaling. Cell. 1999;96:5–8. doi: 10.1016/s0092-8674(00)80953-8. [DOI] [PubMed] [Google Scholar]
- 35.Braselmann S, Taylor V, Zhao H, et al. R406, an orally available spleen tyrosine kinase inhibitor blocks fc receptor signaling and reduces immune complex-mediated inflammation. J Pharmacol Exp Ther. 2006;319:998–1008. doi: 10.1124/jpet.106.109058. [DOI] [PubMed] [Google Scholar]
- 36.Chen L, Monti S, Juszczynski P, et al. SYK-dependent tonic B-cell receptor signaling is a rational treatment target in diffuse large B-cell lymphoma. Blood. 2008;111:2230–2237. doi: 10.1182/blood-2007-07-100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedberg J, Sharman J, Schaefer-Cutillo J, et al. Tamatinib fosdium (TAMF), an oral SYK inhibitor, has significant clinical activity in B-cell non-Hodgkin's lymphoma (NHL): Proceedings of the 10th International Conference on Malignant Lymphoma, Lugano, Switzerland. 2008. Ann Oncol. 2008;19:116. [Google Scholar]
- 38.Chen L, Apgar J, Huynh L, et al. ZAP-70 directly enhances IgM signaling in chronic lymphocytic leukemia. Blood. 2005;105:2036–2041. doi: 10.1182/blood-2004-05-1715. [DOI] [PubMed] [Google Scholar]
- 39.Lanham S, Hamblin T, Oscier D, Ibbotson R, Stevenson F, Packham G. Differential signaling via surface IgM is associated with VH gene mutational status and CD38 expression in chronic lymphocytic leukemia. Blood. 2003;101:1087–1093. doi: 10.1182/blood-2002-06-1822. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Widhopf G, Huynh L, et al. Expression of ZAP-70 is associated with increased B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2002;100:4609–4614. doi: 10.1182/blood-2002-06-1683. [DOI] [PubMed] [Google Scholar]
- 41.Longo PG, Laurenti L, Gobessi S, Sica S, Leone G, Efremov DG. The Akt/Mcl-1 pathway plays a prominent role in mediating antiapoptotic signals downstream of the B-cell receptor in chronic lymphocytic leukemia B cells. Blood. 2008;111:846–855. doi: 10.1182/blood-2007-05-089037. [DOI] [PubMed] [Google Scholar]
- 42.Zupo S, Cutrona G, Mangiola M, Ferrarini M. Role of surface IgM and IgD on survival of the cells from B-cell chronic lymphocytic leukemia. Blood. 2002;99:2277–2278. doi: 10.1182/blood-2001-11-0126. [DOI] [PubMed] [Google Scholar]
- 43.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13:455–481. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 44.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 45.Sallusto F, Palermo B, Lenig D, et al. Distinct patterns and kinetics of chemokine production regulate dendritic cell function. Eur J Immunol. 1999;29:1617–1625. doi: 10.1002/(SICI)1521-4141(199905)29:05<1617::AID-IMMU1617>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 46.Uguccioni M, D'Apuzzo M, Loetscher M, Dewald B, Baggiolini M. Actions of the chemotactic cytokines MCP-1, MCP-2, MCP-3, RANTES, MIP-1 alpha and MIP-1 beta on human monocytes. Eur J Immunol. 1995;25:64–68. doi: 10.1002/eji.1830250113. [DOI] [PubMed] [Google Scholar]
- 47.Taub DD, Conlon K, Lloyd AR, Oppenheim JJ, Kelvin DJ. Preferential migration of activated CD4+ and CD8+ T cells in response to MIP-1 alpha and MIP-1 beta. Science. 1993;260:355–358. doi: 10.1126/science.7682337. [DOI] [PubMed] [Google Scholar]
- 48.Schall TJ, Bacon K, Camp RD, Kaspari JW, Goeddel DV. Human macrophage inflammatory protein alpha (MIP-1 alpha) and MIP-1 beta chemokines attract distinct populations of lymphocytes. J Exp Med. 1993;177:1821–1826. doi: 10.1084/jem.177.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Littman DR. Chemokine receptors: keys to AIDS pathogenesis? Cell. 1998;93:677–680. doi: 10.1016/s0092-8674(00)81429-4. [DOI] [PubMed] [Google Scholar]
- 50.Patel DD, Zachariah JP, Whichard LP. CXCR3 and CCR5 ligands in rheumatoid arthritis synovium. Clin Immunol. 2001;98:39–45. doi: 10.1006/clim.2000.4957. [DOI] [PubMed] [Google Scholar]
- 51.Balashov KE, Rottman JB, Weiner HL, Hancock WW. CCR5(+) and CXCR3(+) T cells are increased in multiple sclerosis and their ligands MIP-1alpha and IP-10 are expressed in demyelinating brain lesions. Proc Natl Acad Sci U S A. 1999;96:6873–6878. doi: 10.1073/pnas.96.12.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi SJ, Cruz JC, Craig F, et al. Macrophage inflammatory protein 1-alpha is a potential osteoclast stimulatory factor in multiple myeloma. Blood. 2000;96:671–675. [PubMed] [Google Scholar]
- 53.Magrangeas F, Nasser V, Avet-Loiseau H, et al. Gene expression profiling of multiple myeloma reveals molecular portraits in relation to the pathogenesis of the disease. Blood. 2003;101:4998–5006. doi: 10.1182/blood-2002-11-3385. [DOI] [PubMed] [Google Scholar]
- 54.Krzysiek R, Lefevre EA, Zou W, et al. Antigen receptor engagement selectively induces macrophage inflammatory protein-1 alpha (MIP-1 alpha) and MIP-1 beta chemokine production in human B cells. J Immunol. 1999;162:4455–4463. [PubMed] [Google Scholar]
- 55.Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 2006;440:890–895. doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- 56.Furman RR, Asgary Z, Mascarenhas JO, Liou HC, Schattner EJ. Modulation of NF-kappa B activity and apoptosis in chronic lymphocytic leukemia B cells. J Immunol. 2000;164:2200–2206. doi: 10.4049/jimmunol.164.4.2200. [DOI] [PubMed] [Google Scholar]
- 57.Pizzolo G, Chilosi M, Ambrosetti A, Semenzato G, Fiore-Donati L, Perona G. Immunohistologic study of bone marrow involvement in B-chronic lymphocytic leukemia. Blood. 1983;62:1289–1296. [PubMed] [Google Scholar]
- 58.Lampert IA, Wotherspoon A, Van Noorden S, Hasserjian RP. High expression of CD23 in the proliferation centers of chronic lymphocytic leukemia in lymph nodes and spleen. Hum Pathol. 1999;30:648–654. doi: 10.1016/s0046-8177(99)90089-8. [DOI] [PubMed] [Google Scholar]
- 59.Schiemann B, Gommerman JL, Vora K, et al. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science. 2001;293:2111–2114. doi: 10.1126/science.1061964. [DOI] [PubMed] [Google Scholar]
- 60.Mackay F, Schneider P, Rennert P, Browning J. BAFF AND APRIL: a tutorial on B cell survival. Annu Rev Immunol. 2003;21:231–264. doi: 10.1146/annurev.immunol.21.120601.141152. [DOI] [PubMed] [Google Scholar]
- 61.Novak AJ, Bram RJ, Kay NE, Jelinek DF. Aberrant expression of B-lymphocyte stimulator by B chronic lymphocytic leukemia cells: a mechanism for survival. Blood. 2002;100:2973–2979. doi: 10.1182/blood-2002-02-0558. [DOI] [PubMed] [Google Scholar]
- 62.Moreaux J, Legouffe E, Jourdan E, et al. BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone. Blood. 2004;103:3148–3157. doi: 10.1182/blood-2003-06-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darce JR, Arendt BK, Wu X, Jelinek DF. Regulated expression of BAFF-binding receptors during human B cell differentiation. J Immunol. 2007;179:7276–7286. doi: 10.4049/jimmunol.179.11.7276. [DOI] [PubMed] [Google Scholar]
- 64.Avery DT, Kalled SL, Ellyard JI, et al. BAFF selectively enhances the survival of plasmablasts generated from human memory B cells. J Clin Invest. 2003;112:286–297. doi: 10.1172/JCI18025. [DOI] [PMC free article] [PubMed] [Google Scholar]