Abstract
Background
The ingestion of probiotics to attempt to improve health is increasingly common, however quality control of some commercial products can be limited. Clinical practice is shifting toward the routine use of probiotics to aid in prevention of necrotizing enterocolitis in premature infants, and probiotic administration to term infants is increasingly common to treat colic and/or prevent atopic disease. Since bifidobacteria dominate the feces of healthy breast-fed infants, they are often included in infant-targeted probiotics.
Methods
We evaluated sixteen probiotic products to determine how well their label claims describe the species of detectable bifidobacteria in the product. Recently-developed DNA-based methods were used as a primary means of identification, and were confirmed using culture-based techniques.
Results
We found that the contents of many bifidobacterial probiotic products differ from the ingredient list, sometimes at a subspecies level. Only one of the sixteen probiotics perfectly matched its bifidobacterial label claims in all samples tested, and both pill-to-pill and lot-to-lot variation were observed.
Conclusion
Given the known differences between various bifidobacterial species and subspecies in metabolic capacity and colonization abilities, the prevalence of misidentified bifidobacteria in these products is cause for concern for those involved in clinical trials and consumers of probiotic products.
Introduction
Probiotics are dietary supplements containing non-pathogenic microbes that provide a health benefit to the host. A broad array of probiotics containing many different bacterial strains are commercially available, many of which were selected based on factors related to ease of production rather than identified mechanisms of protection. Well-designed studies of commercially available probiotics with established composition and purity are essential to establish safety and clinical efficacy, particularly in high-risk patients such as neonates and the immunocompromised. A thorough description of the components of the product is also crucial to understand the mechanism by which administration of a probiotic leads to desirable health outcomes. Factors to consider when establishing the efficacy of probiotic administration include the accurate identification and labeling of strains used, the viability of organisms administered, and consistency in product formulation over the time course of the study. Unfortunately, many commercial probiotics have been shown to fail at one or more of these criteria (1–8).
One area in which probiotics may have a strong benefit is in guiding the development of the microbial community of the gastrointestinal tract of the neonate. The development of the microbiota early in life has been shown to influence risk for susceptibility to infection and development of allergies and atopic disease (9,10). Perhaps the most compelling case for the use of probiotics is in the premature infant, where multiple randomized clinical trials have demonstrated a decreased risk of necrotizing enterocolitis in infants receiving probiotics (11,12). The hospital environment has been shown to be a likely source of inoculum for premature infants (13,14), and the gut microbiota of premature infants in the neonatal intensive care unit (NICU) was shown resemble that of NICU fomites (14). In order to combat this phenomenon, probiotics are increasingly administered to introduce alternative non-pathogenic species to colonize the gastrointestinal tract and occupy niches otherwise open to pathogens. Rare cases of infection from probiotic organisms or contaminants in premature infants underscore the importance of providing probiotic products with established composition and purity (15,16).
Bifidobacterium-containing products are often used in a NICU setting due to their status as generally recognized as safe (GRAS) microorganisms, their ubiquity in the gut of healthy breast-fed infants, and their observed health effects (17,18). Infants with bifidobacteria-dominated gastrointestinal tracts have improved responses to some vaccines, higher resistance to colonization by some pathogens, and better gut barrier function (19–22). Bifidobacteria aid the proper development of the infant’s acquired and innate immune systems, enhancing surveillance while reducing inflammation (22–25). Comparison studies between strains or species of bifidobactaria are limited. In a recent study, Bifidobacterium longum subsp. infantis was found to be a better colonizer of the premature gut than Bifidobacterium animalis subsp. lactis, especially in the presence of human milk (26). This advantage is likely due to the capacity of B. longum subsp. infantis to consume a wide spectrum of human milk oligosaccharides as a direct result of the extensive array of human milk oligosaccharide binding, transport and degrading enzymes encoded in its genome but not found in many species of Bifidobacterium including B. animalis subsp. lactis (27–30). B. longum subsp. infantis colonization is associated with improved responses to some vaccines; in addition B. longum subsp. infantis appears to decrease intestinal epithelia permeability, and to have anti-inflammatory effects in the premature intestine (20,22,31).
B. longum has two subspecies found in humans that historically have been challenging to distinguish, B. longum subsp. longum and B. longum subsp. infantis. Previous studies indicate B. longum subsp. longum and B. longum subsp. infantis possess different suites of glycolytic enzymes (27,29,32). While closely related, the two subspecies are not distinguishable using common methods which focus on analysis of the 16S rRNA gene (27,29,32,33). One commonly used probiotic product whose label lists two subspecies, B. longum subsp. longum and B. longum subsp. infantis, as ingredients recently reclassified the included strains as B. animalis subsp. lactis. This unfortunately has led to confusion, as there is a history of academics publishing results using this product with these strains and listing them as B. longum subsp. longum and B. longum subsp. infantis (34). The risk of species and subspecies misidentification is high, especially given the recently refined definition of these two B. longum subspecies through genome sequencing (27,32). Motivated by the potentially unfortunate consequences of species and subspecies confusion in clinical trials, we surveyed several Bifidobacterium-containing probiotic products to evaluate their label claims with regard both to these two B. longum subspecies and to other bifidobacterial species. To facilitate this effort, we developed a reliable and inexpensive PCR-based method for rapid identification of B. longum subsp. infantis at the subspecies level (20,35). To verify that these DNA-based methods give data that accurately reflect input DNA, we also validated our methods against a series of artificial mock communities.
Results
Method Validation Using Mock Community
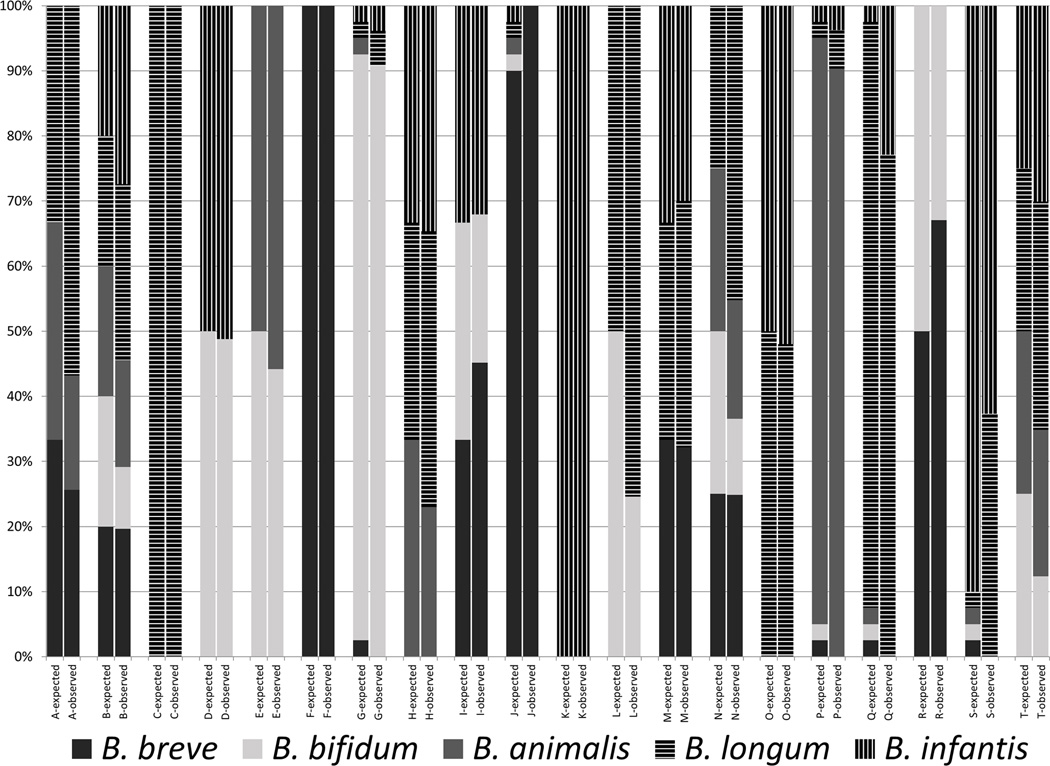
We have previously developed bifidobacterial terminal restriction length polymorphism (Bif-TRFLP) analysis to readily differentiate common bifidobacterial species and Bifidobacterium longum/infantis ratio analysis (BLIR) to differentiate B. longum subspecies (20,33). In order to evaluate commercial probiotic products we first examined if the Bif-TRFLP/BLIR correctly differentiates a number of mock communities containing different combinations of bifidobacterial strain DNA. The Bif-TRFLP/BLIR results generally reflected the known mock communities of input DNA from common bifidobacteria probiotic species (Figure 1). All five tested species were observable and distinguishable from each other, and there was no specific bias against any species or subspecies. Communities containing 2, 3, 4, or 5 different strains in approximately equal levels were identified as containing the correct mix of strains. However, when multiple strains were present each in levels under 5% of total DNA, some minority strains were not observed in the output data, a known limitation of the TRFLP-based technique (33). Specifically, no B. animalis and B. breve were observed in mixture G, mixture J appeared as containing only B. breve despite all 5 species being added, mixture P was missing B. breve and B.bifidum, and mixtures Q and S were missing B. breve, B. bifidum, and B. animalis.
Figure 1.
Mock community composition and measurement. The expected values (as defined by the ratios of input DNA initially measured by A260) and observed values for each of the 20 different mock communities assayed are shown here.
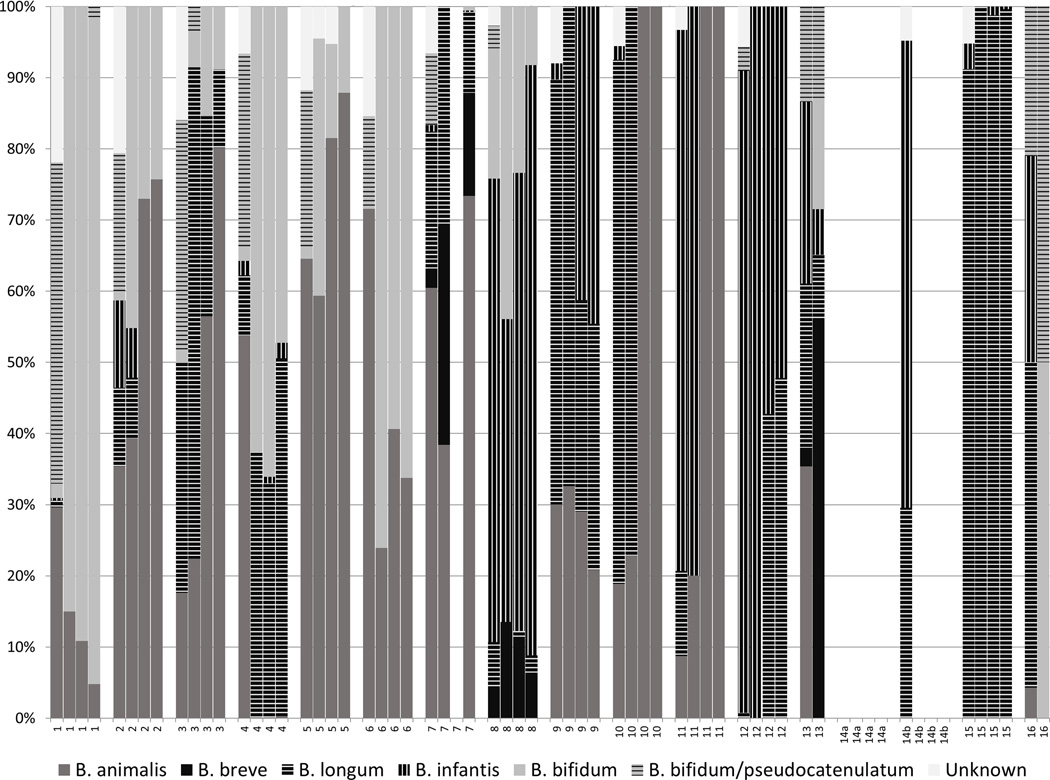
Bif-TRFLP/BLIR of Commercial Probiotics
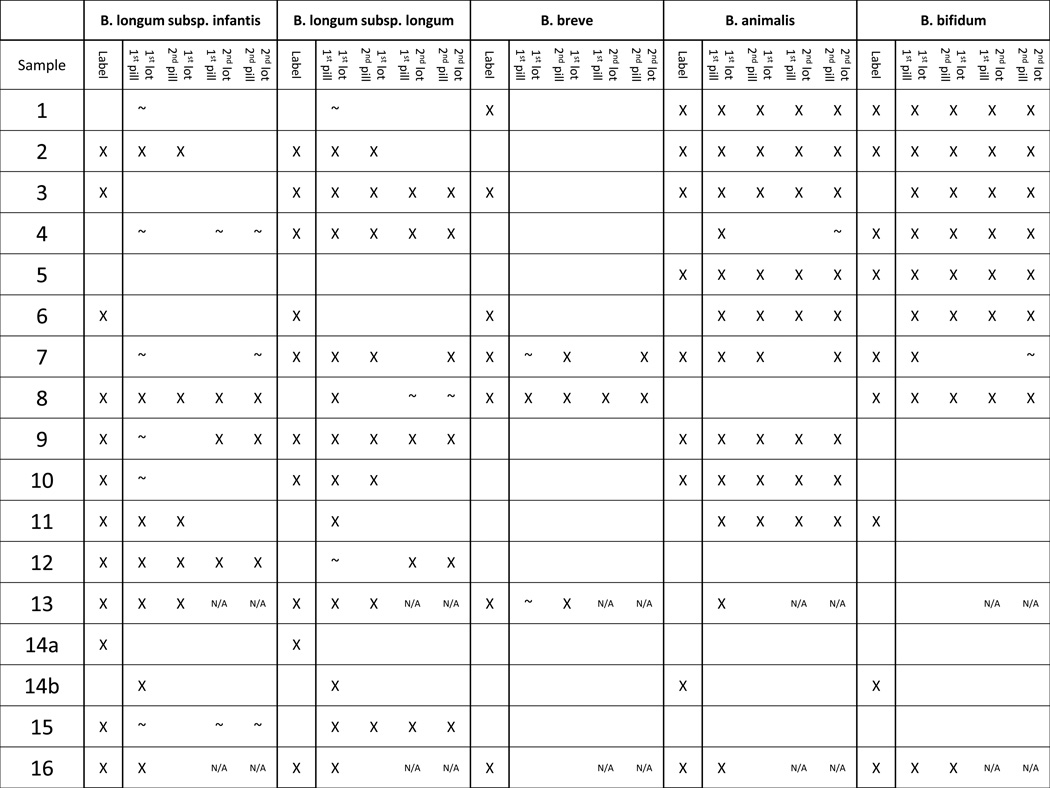
After successful validation of the identification methods, they were used to describe the bifidobacterial contents of the selected probiotic products. Species observed in these probiotics include B. longum (both subspecies), B. bifidum, B. breve, and B animalis subsp. lactis. Ambiguous peaks that match both B. bifidum and B. pseudocatenulatum were observed, but were assumed to be B. bifidum due to the probiotic product context. The measured contents of the probiotic products were often consistent across both pills and lots (Figure 2). The analysis showed pill-to-pill variation (within a lot) in probiotic 16, and lot-to-lot variation in probiotics 2, 10, and 11. Multiple probiotic products either contained unlisted species and/or did not contain the species their label claimed (Table 1). Only one probiotic (#5) perfectly matched its label claims in all four samples assayed, with no missing or additional bifidobacterial species. Amplification of bifidobacterial DNA was obtained from every sample except for probiotic 7 lot 2 pill 1, all probiotic 14a pills, and all probiotic pill 14b pills except lot 1 pill 1. It is unknown whether this represents a lack of bifidobacteria in the samples or potential PCR inhibitors that co-eluted with the extracted DNA. Attempting to reduce the concentration of potential PCR inhibitors by a 1:50 dilution of the genomic DNA from these failed samples, did not result in positive amplification (data not shown).
Figure 2.
Bifidobacterial composition of probiotic products by PCR-based methods. Each product was assayed four times, shown here in order grouped by product, lot 1 pill 1, lot 1 pill 2, lot 2 pill 1, lot 2 pill 2. Blank plot area not between two different products indicates no amplicon was detected.
Table 1.
Bifidobacterial Isolation
For purposes of content comparison, bifidobacterial isolates were obtained and identified (by MALDI Biotyper and BLIR for B. longum group isolates) from each of the probiotic products. Not all species listed species were isolated from the probiotics (Supplemental Table S1) likely a consequence of differing viability after storage, as well as differences in culturability. B. animalis subsp. lactis was the most commonly-isolated organism, and was frequently the only species isolated from the products. Indeed, B. animalis subsp. lactis was the only species not listed on the product labels that was found by isolation. In 3 out of 4 cases where Bif-TRFLP found B. animalis subsp. lactis where it was not listed on the label, the species was also successfully isolated (Probiotics 4, 6, and 11, but not 13).
Genome Sequencing to Confirm Results of BLIR Analysis
The BLIR method is based on unique genetic loci found in the genomes of multiple strains of each B. longum subspecies. Several potentially mislabeled products containing B. longum subsp. longum and B. longum subsp. infantis were observed using this method. To further confirm the results of the BLIR method, the genomes of two isolates from Product 15, where the label claimed B. longum subsp. infantis and BLIR indicated the contents were B. longum subsp. longum, were sequenced for comparison. 2.5 million reads were obtained for each isolate and the average genome size was 2.239 Mb. This predicted genome size is noticeably smaller than that of B. longum subsp. infantis ATCC 15697 (2.832 Mb) (27). Reads from the Product 15 isolate did not map to key loci in the B. longum subsp. infantis ATCC15697 genome, including the subspecies-stereotypic HMO cluster described by Sela et al (27,29). While average read coverage for this genome was 272-fold, the average read coverage for the 40kb HMO cluster was zero (Supplementary Figure S1). Conversely, the Product 15 sequence aligned well with B. longum subsp. longum DJO10A (36), indicating that the Product 15 isolates were B. longum subsp. longum strains rather than the advertised B. longum subsp. infantis, (although strain-level differences were observed, as indicated by small gaps in coverage in supplemental Figure S1A). This is consistent with the classification given by the BLIR method.
Discussion
Consistent with previous studies, we present further evidence that the advertised content of many probiotic products containing bifidobacteria vary significantly from the actual content (1–8). Indeed, only 1 of the 16 products tested exactly matched the bifidobacterial species claims on the label in every sample tested. Some products were not internally consistent as both pill-to-pill and lot-to-lot variation were observed. We note that many of these products also contained non-bifidobacterial species contents, which, for the purposes of this study, were not evaluated. These results suggest that quality control of probiotics is lacking. In order for clinical trials to provide meaningful data about the benefits of specific probiotic strains and enable clinicians to make informed decisions about prescribing or recommending probiotics, increased standards of strain identification are needed, particularly given the lack of regulatory oversight for certification of probiotics in the United States.
Debate continues regarding probiotic administration to premature infants, with several authors arguing for the routine prophylactic use of probiotics for the prevention of necrotizing enterocolitis (37,38). Which probiotic strain and whether a single strain or a combination product is superior are open questions requiring further trials. There is evidence supporting the use of B. longum subsp. infantis as a potential candidate species for infants, especially when provided in combination with breast milk (26,39). However, the current study shows that B. longum subsp. infantis is commonly misidentified in commercial probiotics, some of which are marketed towards infants. To this end, we present and validate potentially useful tools for evaluating probiotic contents, including a method to distinguish B. longum subsp. infantis from B. longum subsp. longum, a subspecies differentiation not possible on the basis of traditional 16S ribosomal RNA sequencing alone.
While the clear congruence between the known mock community input and resultant Bif-TRFLP/BLIR data was encouraging from a methodological standpoint, the results from the products tested was concerning. There were numerous mismatches between label claims and our results. Probiotic 11, for example, was advertised as containing B. bifidum and B. longum subsp. infantis, but Bif-TRFLP/BLIR indicated its contents were B. animalis subsp. lactis and B. longum subsp. infantis, with some B. longum subsp. longum present in one pill. The second lot of Probiotic 11 had the same stock keeping unit (SKU) number as the first lot, was produced by the same manufacturer, and was even purchased from the same store. However, the product label from the second lot differed from the first lot. It now listed a B. animalis subsp. lactis strain (with the same trade name as the previous B. bifidum), no B. longum subsp. infantis, and a Lactobacillus strain. The Bif-TRFLP data from lot 2 of this product agrees with the new label claim, and the relabeling of the B. bifidum-named strain as B. animalis subsp. lactis. Such an unannounced change may or may not be noticed and included in the analysis of the results of a clinical trial using this product.
Probiotic 15 was consistently listed as a containing only a B. longum subsp. infantis strain, while it clearly contained B. longum subsp. longum by Bif-TRFLP/BLIR. Genomic comparisons of two isolates from the same product lot showed it was missing large genomic sections present in the B. longum subsp. infantis type strain (ATCC 15697) including gene clusters dedicated to catabolism of human milk oligosaccharides and urea—genetic loci specific to the B. longum subsp. infantis subspecies (27,29). Conversely, the genome sequence of Product 15 aligned very closely with B. longum subsp. longum DJO10A thus clearly showing membership within that subspecies. Products 3, 9, and 10 also had misidentified B. longum subspecies contents in some samples. These results demonstrate the difficulty of distinguishing between these two B. longum subspecies. BLIR analysis is an inexpensive tool for quality control screening by clinical labs or the probiotics industry to rapidly differentiate B. longum product subspecies that would otherwise only be possible via whole genome sequencing (27) or multilocus sequence typing (29).
The contents of probiotic 6 were especially different from its labeling, as it did not appear to contain any of the three label species (B. longum subsp. longum, B. longum subsp. infantis, and B. breve) and contained two non-label species (B. bifidum and B. animalis). However further investigation of the Probiotic 6 label revealed a footnote indicating that the manufacturers reclassified both their B. longum subsp. longum and B. longum subsp. infantis strains as B. animalis subsp. lactis, which is in agreement with the Bif-TRFLP/BLIR data developed in this study. However it is unclear if such subtle footnoted-label changes, in lieu of actual relabeling, will be fully understood and recognized by clinicians and consumers.
While our use of the Bif-TRFLP and BLIR techniques has revealed numerous inconsistencies of product labeling with product contents, there are limitations to the methods used in this study. It is possible for a mutation causing an rRNA gene restriction site to change a cognate Bif-TRFLP fragment length and cause an incorrect (or unknown) species identification. In addition, while our terminal restriction fragment database is extensive, it does not contain every known bifidobacterial strain. Moreover, in products containing a mixture of strains, one strain might be present but not detected due to it being below the limit of detection in a mixed community, a known limitation of the technique (33). The absence of any detectable bifidobacterial amplicon in some samples was concerning, however, the possibility remains that there may have been PCR inhibitors in the sample which resulted in a false negative.
In conclusion, the state of species and subspecies quality control for many bifidobacteria-containing probiotic products remains inconsistent. In our study, we found that probiotic products often do not correctly identify the bifidobacterial species they list on their labels. Probiotic products can shift their contents over time without warning, even while keeping the same SKU. Differentiating the two subspecies B. longum subsp. longum from B. longum subsp. infantis appeared to be of particular difficulty.
Clinicians face a number of challenges when administering probiotics to patients. While not addressed in our study, the issues of strain viability upon dosing and potential contamination by adventitous microorganisms (and even pathogens) are also concerns for those planning clinical trials. Despite the GRAS (generally recognized as safe) status of probiotic supplements, the strain identity issues discussed here must be taken into account when planning and conducting clinical trials to ensure the proper interpretation the data generated. Misidentified probiotics are not likely to pose a danger to patients, however they certainly cloud clinic trial interpretations and are particularly problematic when the properties possessed by one strain are confused with another. Indeed, the use of appropriate tools (such as those described here) to distinguish between closely related strains such as B. longum subsp. longum and B. longum subsp. infantis is needed to discriminate between these microorganisms with significantly different metabolic capacities that impact colonization behavior and as well as host health outcomes (20,22). Close and continued monitoring of probiotic products is recommended, and consideration should be given to those probiotic products for which validated, good manufacturing practice (GMP) production is known. Certainly federally funded clinical trials using commercial probiotics would benefit from increased strain validation solely to ensure meaningful, interpretable, data is generated.
Methods
Construction of mock communities
Type strains of various Bifidobacterium species (B. longum subsp. infantis ATCC15697, B. longum subsp. longum DJO10A, B. animalis subsp. lactis ATCC27536, B. breve ATCC15700, and B. bifidum DSM20456) were grown in de Man Rogosa Sharpe (MRS) broth supplemented with 500 mg/L of L-cysteine-HCL, and DNA extraction was performed using the Epicentre MasterPure Gram Positive DNA Purification kit (Epicentre, Madison WI). DNA concentrations were determined by nanodrop spectrophotometry and adjusted to 40 ng/ul. DNA from the various bifidobacteria was then combined in different ratios, as noted, to construct 20 mock community DNA pools. The final volume of DNA was adjusted to 15ul and diluted to a final concentration of 20ng/ul. One microliter of the resulting mix was used to perform Bif-TRFLP/BLIR as described below.
Probiotic products and DNA extraction
A search was performed both online and in local Davis, CA stores for probiotics listing bifidobacteria as an ingredient, and 16 such products were selected for evaluation. One product (#14) contained two formulations in a single package that we list as 14a and 14b. Two separate lots of each probiotic product were purchased approximately 2 years apart. Probiotics 13 and 16 had been discontinued at the time of purchase for the second lot. Genomic DNA was extracted from either two separate pills or two 100 mg aliquots of powder of each probiotic product using the Masterpure Gram Positive DNA Purification Kit (Epicentre, Madison WI) after dilution and vortexing in 9 ml of phosphate buffered saline. Label claims of bifidobacterial content of each product were recorded.
Bifidobacterial Terminal Restriction Fragment Length Polymorphism (Bif-TRFLP)
The method of Lewis et al. (33) was used to perform the Bif-TRFLP assay. Briefly, DNA from feces was amplified in triplicate by PCR using primers NBIF389 (5’-[HEX]-GCCTTCGGGTTGTAAAC) and NBIF1018 REV (GACCATGCACCACCTGTG) (Table S1). DNA was purified using the Qiagen Qiaquick PCR purification kit (Qiagen, Valencia CA) and then cut with restriction enzymes AluI and HaeIII. The resulting fragments were analyzed on an ABI 3100 Capillary Electrophoresis Genetic Analyzer at the UC Davis College of Biological Sciences Sequencing Facility and sizes were compared against the published database for species identification. The samples were analyzed with PeakScanner 2.0 software (Applied Biosystems, Carlsbad, CA). Negative controls containing DNA-free water instead of sample DNA were included with each run.
Bifidobacterium Longum-Infantis Ratio (BLIR)
A PCR-based assay, BLIR, was developed in order to determine which of the two human-associated subspecies of B. longum (B. longum subsp. longum and B. longum subsp. infantis) were present in each sample and to gain an estimate of their relative abundance to each other (20). In brief, three primers (FWD_BL_BI (5-[HEX]-AAAACGTCCATCCATCACA), REV_BL (5-ACGACCAGGTTCCACTTGAT), and REV_BI (5-CGCCTCAGTTCTTTAATGT)) targeting a conserved portion of the genome (between Blon_0424 and Blon_0425) shared by both subspecies were designed using multiple genome sequences of each subspecies. FWD_BL_BI is complementary to a sequence in both subspecies while REV_BL and REV_BI are complementary to nearby sequences in only B. longum and B. infantis, respectively. FWD_BL_BI and REV_BL amplify a fragment of the B. longum genome 145 bp in length, while FWD_BL_BI and REV_BI amplify a fragment of the B. infantis genome 114 bp in length, allowing differentiation of the amplicons.
Genomic DNA from each probiotic was amplified by PCR using 0.5 µl of 10 µM stock of each of the above primers, 12.5 µl GoTaq Green Master Mix (Promega), 1 µl of 25 mM MgCl2, 1 µl of template DNA, and 9 µl of nuclease free water. Cycling conditions were 95°C for 2 minutes, 30 cycles of 95°C for 1 minute, 54°C for 1 minute, and 72°C for 30 seconds, followed by a 72°C extension for 5 minutes. PCR products were purified from the mixture using the QIAquick PCR purification kit (Qiagen, Valencia CA) and diluted 1:10 to avoid off-scale peak sizes. Capillary electrophoresis and analysis of the amplicon sizes proceeded as with the Bif-TRFLP procedure above. A positive control of known genomic DNA was included with each PCR run to ensure potential amplification of both B. longum subsp. longum and B. longum subsp. infantis products. The percentage of peak area corresponding to each subspecies was multiplied by the percentage of B. longum species peak area from the Bif-TRFLP data to arrive at final abundances of each subspecies. Negative controls containing DNA-free water instead of sample DNA were included with each run.
Bifidobacterial Isolations
To isolate bifidobacteria, 100 mg or one pill of each product was aseptically transferred to a sterile tube, diluted tenfold with sterile phosphate buffered saline (PBS) and homogenized by vortex. Serial dilutions were prepared in PBS and inoculated on modified BSIM agar (40). Modified BSIM agar was prepared by supplementing de Man Rogosa Sharpe (MRS) media with 13 g/L agar, 500 mg/L of L-cysteine-HCL, 20 mg/L of nalidixic acid, 50 mg/ml mupirocin, 50 mg/ml kanamycin, 50 mg/ml polymixin B sulfate, 100 mg/ml Iodoacetate, 100 mg/ml 2,3,5-triphenyltetrazolium chloride. The plates were incubated for 48 hours at 37°C in an anaerobic chamber with an atmosphere containing approximately 5% carbon dioxide, 3% hydrogen, and the remainder nitrogen. Up to 10 resulting colonies from each sample with the appropriate colony appearance were streaked onto MRS-cysteine plates for purity for two passages. The resulting pure strains were grown in MRS broth supplemented with 0.05% L-cysteine and stored at −80°C in 50% glycerol.
Matrix Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) Identification of Isolates
Glycerol stocks of each isolate were streaked on MRS plates and incubated at 37°C for 48 h in anaerobic chamber. A colony from each plate was added to 300 µl nuclease free water in a 1.5 ml tube and homogenized by vortex. Next, 900 µl of 98% ethanol was added to the tube, briefly vortexed, and centrifuged for 2 min at maximum speed. The supernatant was removed and the tubes were again centrifuged for 2 minutes. All liquid was decanted and the samples were left at room temperature to allow the ethanol to evaporate. Subsequently, 25 µl formic acid was then added to each tube and homogenized by vortex, followed by the addition of 25 µl acetonitrile. Samples were then centrifuged for 2 min and one µl of extract was placed on a MALDI target plate, left to dry at room temperature, covered with an α-Cyano-4-hydroxycinnamic acid (HCCA) matrix and air dried. The MALDI target plate was then analyzed by a MALDI Biotyper (Bruker, Fremont CA) and the best database match for each isolate was recorded. The MALDI Biotyper was not found to reliably differentiate between B. longum subspecies (data not shown); accordingly BLIR was used on DNA extracted from an overnight MRS broth culture of each B. longum group isolate using the Epicentre Masterpure Gram Positive DNA Purification Kit (Epicentre, Madison WI) to identify each B. longum group isolate at the subspecies level.
Genome Sequencing
Libraries of genomic DNA from each of two isolates of probiotic 15 identified by MALDI as members of the B. longum species were created using standard Illumina library prep and sequenced on an Illumina MiSeq by the UC Davis Genome Center DNA Technologies core, using 150 bp single read settings. Reads were mapped to the B. longum subsp. infantis ATCC 15697 and B. longum subsp. longum DJO10A ggenomes and visualized using CLC Bio software (Qiagen, Boston MA) (27,29,32). An equal number of reads were mapped to both genomes.
Ethical Considerations
As no human or animal subjects were involved in this research, no ethical approval (e.g. Institutional Review Board) was sought.
Supplementary Material
Acknowledgments
We thank Steve Ho for his aid with the DNA extractions from the probiotic products.
Statement of Financial Support:
This work has been supported by the USA National Institutes of Health awards R01AT007079 and R01AT008759 and the Peter J. Shields Endowed Chair in Dairy Food Science (Davis, CA). SAF is supported by a USA National Institutes of Health Ruth Kirschstein National Research Service Award, F32AT008533. ZTL is supported by an Alfred P. Sloan Foundation Microbiology of the Built Environment Postdoctoral Fellowship (New York, NY).
Footnotes
Conflict of Interest Statement:
DAM is a co-founder and SAF an employee of Evolve Biosystems, a company focused on diet-based manipulation of the gut microbiota.
References
- 1.Fasoli S, Marzotto M, Rizzotti L, Rossi F, Dellaglio F, Torriani S. Bacterial composition of commercial probiotic products as evaluated by PCR-DGGE analysis. Int J Food Microbiol. 2003;82:59–70. doi: 10.1016/s0168-1605(02)00259-3. [DOI] [PubMed] [Google Scholar]
- 2.Canganella F, Paganini S, Ovidi M, et al. A microbiological investigation on probiotic pharmaceutical products used for human health. Microbiol Res. 1997;152:171–179. doi: 10.1016/s0944-5013(97)80009-2. [DOI] [PubMed] [Google Scholar]
- 3.Angelakis E, Million M, Henry M, Raoult D. Rapid and Accurate Bacterial Identification in Probiotics and Yoghurts by MALDI-TOF Mass Spectrometry. J Food Sci. 2011;76:568–572. doi: 10.1111/j.1750-3841.2011.02369.x. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein EJC, Citron DM, Claros MC, Tyrrell KL. Bacterial counts from five over-the-counter probiotics: Are you getting what you paid for? Anaerobe [Internet] 2014;25:1–4. doi: 10.1016/j.anaerobe.2013.10.005. Available from: http://dx.doi.org/10.1016/j.anaerobe.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Marcobal A, Underwood MA, Mills Da. Rapid determination of the bacterial composition of commercial probiotic products by terminal restriction fragment length polymorphism analysis. J Pediatr Gastroenterol Nutr [Internet] 2008;46:608–611. doi: 10.1097/MPG.0b013e3181660694. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18493222. [DOI] [PubMed] [Google Scholar]
- 6.Patro JN, Ramachandran P, Lewis JL, et al. Development and Utility of the FDA “GutProbe” DNA Microarray for Identification, Genotyping, and Metagenomic Analysis of Commercially Available Probiotics. J Appl Microbiol [Internet] 2015 doi: 10.1111/jam.12795. n/a - n/a. Available from: http://doi.wiley.com/10.1111/jam.12795. [DOI] [PubMed] [Google Scholar]
- 7.Temmerman R, Pot B, Huys G, Swings J. Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Int J Food Microbiol. 2003;81:1–10. doi: 10.1016/s0168-1605(02)00162-9. [DOI] [PubMed] [Google Scholar]
- 8.Huys G, Vancanneyt M, D’Haene K, Vankerckhoven V, Goossens H, Swings J. Accuracy of species identity of commercial bacterial cultures intended for probiotic or nutritional use. Res Microbiol. 2006;157:803–810. doi: 10.1016/j.resmic.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Madan JC, Farzan SF, Hibberd PL, Karagas MR. Normal neonatal microbiome variation in relation to environmental factors, infection and allergy. [cited 2013 Mar 5];Curr Opin Pediatr [Internet] 2012 24:753–759. doi: 10.1097/MOP.0b013e32835a1ac8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23111681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zivkovic AM, Lewis ZT, German JB, Mills DA. Establishment of a Milk-Oriented Microbiota ( MOM ) in early life?: How Babies Meet Their MOMs. Funct Food Rev. 2013;5:3–12. [Google Scholar]
- 11.La Rosa PS, Warner BB, Zhou Y, et al. Patterned progression of bacterial populations in the premature infant gut. Proc Natl Acad Sci. 2014;111:12522–12527. doi: 10.1073/pnas.1409497111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alfaleh K, Anabrees J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Evidence-Based Child Heal. 2014;9:584–671. doi: 10.1002/ebch.1976. [DOI] [PubMed] [Google Scholar]
- 13.Bokulich NA, Mills DA, Underwood MA. Surface microbes in the neonatal intensive care unit: changes with routine cleaning and over time. [cited 2014 Jan 25];J Clin Microbiol [Internet] 2013 51:2617–2624. doi: 10.1128/JCM.00898-13. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23740726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooks B, Firek BA, Miller CS, et al. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome [Internet] 2014;2:1. doi: 10.1186/2049-2618-2-1. Available from: http://www.microbiomejournal.com/content/2/1/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohishi A, Takahashi S, Ito Y, et al. Bifidobacterium Septicemia Associated with Postoperative Probiotic Therapy in a Neonate with Omphalocele. J Pediatr [Internet] 2010;156:679–681. doi: 10.1016/j.jpeds.2009.11.041. Available from: http://dx.doi.org/10.1016/j.jpeds.2009.11.041. [DOI] [PubMed] [Google Scholar]
- 16.Jenke A, Ruf EM, Hoppe T, Heldmann M, Wirth S. Bifidobacterium septicaemia in an extremely low-birthweight infant under probiotic therapy. Arch Dis Child - Fetal Neonatal Ed. 2012;97:F217–F218. doi: 10.1136/archdischild-2011-300838. [DOI] [PubMed] [Google Scholar]
- 17.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. [cited 2012 May 9];Nature [Internet] 2012 486:222–227. doi: 10.1038/nature11053. Available from: http://www.nature.com/doifinder/10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turroni F, Peano C, Pass Da, et al. Diversity of Bifidobacteria within the Infant Gut Microbiota. [cited 2012 May 14];PLoS One [Internet] 2012 7:e36957. doi: 10.1371/journal.pone.0036957. Available from: http://dx.plos.org/10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuda S, Toh H, Hase K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. [cited 2011 Jul 12];Nature [Internet] 2011 469:543–547. doi: 10.1038/nature09646. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21270894. [DOI] [PubMed] [Google Scholar]
- 20.Huda MN, Lewis ZT, Kalanetra KM, et al. Stool Microbiota and Vaccine Responses of Infants. [cited 2014 Jul 13];Pediatrics [Internet] 2014 134:e362–e372. doi: 10.1542/peds.2013-3937. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25002669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romond M-B, Colavizza M, Mullié C, et al. Does the intestinal bifidobacterial colonisation affect bacterial translocation? [cited 2014 Apr 16];Anaerobe [Internet] 2008 14:43–48. doi: 10.1016/j.anaerobe.2007.09.003. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17988900. [DOI] [PubMed] [Google Scholar]
- 22.Chichlowski M, De Lartigue G, German JB, Raybould HE, Mills Da. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. [cited 2013 Feb 21];J Pediatr Gastroenterol Nutr [Internet] 2012 55:321–327. doi: 10.1097/MPG.0b013e31824fb899. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22383026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheil B, MacSharry J, O’Callaghan L, et al. Role of interleukin (IL-10) in probiotic-mediated immune modulation: an assessment in wild-type and IL-10 knock-out mice. [cited 2013 Feb 21];Clin Exp Immunol [Internet] 2006 144:273–280. doi: 10.1111/j.1365-2249.2006.03051.x. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1809667&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanabe S, Kinuta Y, Saito Y. Bifidobacterium infantis suppresses proinflammatory interleukin-17 production in murine splenocytes and dextran sodium sulfate-induced intestinal inflammation. Int J Mol Med. 2008;22:181–185. [PubMed] [Google Scholar]
- 25.Preising J, Philippe D, Gleinser M, et al. Selection of bifidobacteria based on adhesion and anti-inflammatory capacity in vitro for amelioration of murine colitis. [cited 2013 Feb 21];Appl Environ Microbiol [Internet] 2010 76:3048–3051. doi: 10.1128/AEM.03127-09. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2863435&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Underwood MA, Kalanetra KM, Bokulich NA, et al. A Comparison of Two Probiotic Strains of Bifidobacteria in Premature Infants. [cited 2013 Sep 19];J Pediatr [Internet] 2013 163:1585–1591. doi: 10.1016/j.jpeds.2013.07.017. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23993139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sela DA, Chapman J, Adeuya A, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A [Internet] 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2596198&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LoCascio RG, Ninonuevo MR, Freeman SL, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem [Internet] 2007;55:8914–8919. doi: 10.1021/jf0710480. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17915960. [DOI] [PubMed] [Google Scholar]
- 29.LoCascio RG, Desai P, Sela DA, Weimer B, Mills DA. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. [cited 2011 Aug 6];Appl Environ Microbiol [Internet] 2010 76:7373–7381. doi: 10.1128/AEM.00675-10. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2976205&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrido D, Kim JH, German JB, Raybould HE, Mills DA. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. [cited 2011 Aug 29];PLoS One [Internet] 2011 6:e17315. doi: 10.1371/journal.pone.0017315. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3057974&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganguli K, Meng D, Rautava S, Lu L, Walker WA, Nanthakumar N. Probiotics prevent necrotizing enterocolitis by modulating enterocyte genes that regulate innate immune-mediated inflammation. Am J Physiol Gastrointest Liver Physiol [Internet] 2013;304:G132–G141. doi: 10.1152/ajpgi.00142.2012. Available from: http://ajpgi.physiology.org/content/304/2/G132.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schell Ma, Karmirantzou M, Snel B, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A [Internet] 2002;99:14422–14427. doi: 10.1073/pnas.212527599. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=137899&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis ZT, Bokulich NA, Kalanetra KM, Ruiz-Moyano S, Underwood MA, Mills DA. Use of bifidobacterial specific terminal restriction fragment length polymorphisms to complement next generation sequence profiling of infant gut communities. [cited 2013 Feb 4];Anaerobe [Internet] 2013 19:62–69. doi: 10.1016/j.anaerobe.2012.12.005. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23261904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ewaschuk JB, Diaz H, Meddings L, et al. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am J Physiol Gastrointest Liver Physiol. 2008;295:G1025–G1034. doi: 10.1152/ajpgi.90227.2008. [DOI] [PubMed] [Google Scholar]
- 35.Lewis ZT, Totten SM, Smilowitz JT, et al. Maternal Fucosyltransferase 2 Status Affects the Gut Bifidobacterial Communities of Breastfed Infants. Microbiome. 2015;3:1–21. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J-H, Karamychev VN, Kozyavkin Sa, et al. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics. 2008;9:247. doi: 10.1186/1471-2164-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ofek Shlomai N, Deshpande G, Rao S, Patole S. Probiotics for preterm neonates: what will it take to change clinical practice? [cited 2014 Jan 30];Neonatology [Internet] 2014 105:64–70. doi: 10.1159/000354891. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24281648. [DOI] [PubMed] [Google Scholar]
- 38.Janvier A, Malo J, Barrington KJ. Cohort Study of Probiotics in a North American Neonatal Intensive Care Unit. [cited 2014 Jan 20];J Pediatr [Internet] 2014 :1–6. doi: 10.1016/j.jpeds.2013.11.025. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24411521. [DOI] [PubMed] [Google Scholar]
- 39.Donovan SM. Promoting bifidobacteria in the human infant intestine: why, how, and which one? [cited 2011 Jul 22];J Pediatr Gastroenterol Nutr [Internet] 2011 52:648–649. doi: 10.1097/MPG.0b013e31821e2799. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21505364. [DOI] [PubMed] [Google Scholar]
- 40.Ruiz-Moyano S, Totten SM, Garrido Da, et al. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of bifidobacterium breve. Appl Environ Microbiol. 2013;79:6040–6049. doi: 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.