Abstract
Genome sequences that contain tandem repeats of guanine can form stable four-stranded structures known as G-quadruplex, or G4 DNA. While the molecular mechanisms are not fully defined, such guanine-rich loci are prone to mutagenesis and recombination. Various repair pathways function to reduce the potential for genome instability by correcting base damage and replication errors; however, it is not yet fully defined how well these processes function at G4 DNA. One frequent form of base damage occurs from cytidine deamination, resulting in deoxyuracil and UG mismatches. In duplex and single-stranded DNA, uracil bases are recognised and excised by uracil glycosylases. Here, we tested the efficiency of uracil glycosylase activity in vitro on uracil bases located directly adjacent to guanine repeats and G4 DNA. We show that uracil excision by bacterial UDG and human hUNG2 is reduced at uracils positioned directly 5′ or 3′ of a guanine tetrad. Control reactions using oligonucleotides disrupted for G4 formation or reaction conditions that do not favour G4 formation resulted in full uracil excision activity. Based on these in vitro results, we suggest that folding of guanine-rich DNA into G4 DNA results in a DNA conformation that is resistant to uracil glycosylase-initiated repair and this has the potential to increase the risk of instability at guanine repeats in the genome.
Introduction
Guanine-rich and repetitive genomic DNA has the unique ability to adopt four-stranded conformations called G4 DNA, and the impact of those structures on DNA metabolism is important for clarifying the molecular sources of genetic disease (1–3). Physiological salt and pH conditions promote G4 folding, structures with stacks of guanine ‘tetrads’, each of which is composed of four guanine bases paired to each other via Hoosteen bonds (4,5). The specific conformation and overall stability of a given G4 is influenced by the sequences, ions, and reaction conditions (6). In higher eukaryotes, G4 sequences are located at sites of programmed recombination and at a variety of unstable intergenic regions where G4 structure formation promotes site-specific instability and disease (3,7,8). Of particular note are G4-capable sequences enriched at oncogenes, where G4 structure formation may contribute to gene regulation and oncogenesis (9). G4 structures have been shown to form from guanine-repeat sequences at rearrangement sites, such as the HOX11 (10), BCL2 (11), and TCF3 (12) cancer-related genes.
When DNA becomes transiently denatured during transcription or replication, guanine repeats freed from their complement are permitted to adopt structures like G4, which can subsequently interfere with various DNA metabolic pathways (1,2,13–15). This raises the possibility that some DNA repair activities may be inhibited at G4 DNA, contributing to the apparent instability that characterises many guanine-rich loci. One major pathway, base excision repair (BER), reduces mutagenesis by correcting bases that were damaged by deamination or oxidation (16,17). BER is initiated by a lesion-specific glycosylase that removes the damaged base, followed by excision of the resulting abasic site to permit subsequent resynthesis to reconstitute the original base pairing. Emerging evidence suggests that some DNA lesions in G4 DNA may not be processed efficiently by BER. In telomeric G4, activity assays have shown that 8-oxoguanine is not excised by human glycosylases, even though further oxidation products can be processed (18,19). The APE1 enzyme, which cleaves abasic sites, was also shown to have reduced enzymatic activity at abasic sites residing within certain G4 DNA structures (19,20). These findings support the model that G4 DNA can interfere with DNA correction activities, and could therefore contribute to mutagenesis at guanine-rich loci by reducing DNA repair efficacy at those sites.
Here we asked if G4 DNA interferes with excision of a common form of DNA damage, deaminated cytidine (uracil), by uracil DNA glycosylase in vitro. Using synthetic oligonucleotide substrates, we placed a single deoxyuracil either directly 3′ or 5′ of a guanine repeat sequence or in between repeats, and then measured the activities of two uracil glycosylases, hUNG2 (human) and UDG (Escherichia coli) on those DNAs under conditions that allow G4 to fold. Activities were compared to controls that cannot adopt G4 structures. We find that the excision of uracil is reduced when the base is next to guanine repeats participating in G4 formation, but not when it is located three bases away from the tetrad. Addition of APE1 to the reactions did not increase abasic site cleavage, suggesting that the initiating steps of BER of uracil are reduced at G4 DNA. Together, our results suggest that deaminated cytidines positioned adjacent to G4 DNA structures are poor substrates for the BER pathway.
Materials and methods
Oligonucleotides
Deoxyuracil containing G4 sequences were based on a previously described G4 sequence from the TCF3 gene (12). All oligonucleotides were synthesised by Eurofins MWG Operon (Huntsville, AL, USA) and PAGE purified. Oligonucleotides were 5′ end labelled with γ-P32 ATP from MP Biomedicals (Solon, OH, USA) and with T4 PNK, New England Biolabs (NEB) (Ipswich, MA, USA). Unincorporated label was removed by spin chromatography using an Illustra Microspin G-50 column from GE Healthcare (Pittsburg, PA) using the manufacturer’s instructions.
Native PAGE analysis
G4 and GT oligonucleotides were both subjected to the same G4 folding conditions. Reactions (10 µl) contained 5 pmols of labelled oligonucleotide and 100mM KCL in TE. The DNA was first denatured in a small > 98°C water bath, which was then allowed to cool to room temperature to permit structure folding. Ficoll was added to the samples, which were then resolved by 16% native PAGE (29:1) for 600Vhrs in 0.5× TBE buffer containing 100mM KCl.
Circular dichroism
Circular dichroism (CD) detection of TCF3 G4 was performed as previously described (12). Briefly, we used an Aviv model 215 CD spectrometer at 37°C and a 1-cm cuvette. Oligonucleotides were prepared at 12.5 µM in 10mM Tris-HCl (pH 7.6), 1mM EDTA, and 100mM KCl. Samples were heated to 98°C in a water bath, which was then allowed to cool to room temperature. Samples were stored on ice. Molar ellipticity was taken from 200nm to 300nm in 1nm increments. The averages of three scans for each oligonucleotide were analysed.
Uracil glycosylase and APE1 cleavage assays
Purified E. coli UDG and hAPE1 were purchased from NEB (Ipswich, MA, USA) and hUNG2 purchased from Enzymax (Lexington, KY, USA). The glycosylase concentrations used in the assays were based on the unit definitions provided by the manufacturer. Each reaction contained double the amount of enzyme required for complete digestion of unstructured substrates. G4 DNA oligonucleotide structures were formed from 5 pmol of labelled DNA in 10 μl of 100mM KCl and 1× UDG reaction buffer (20mM Tris-HCl, 1mM DTT, 1mM EDTA, pH 8.0). Samples were placed in a ABI 2720 Thermal-cycler (Foster City, CA, USA) set at the following temperatures with 5-min intervals; 98°C, 90°C, 75°C, 60°C, 45°C, 30°C, 20°C. Samples were then immediately placed on ice. Cleavage assays contained indicated enzymes and incubated at 37°C for 20min (glycosylase reactions) and 60min for reactions containing both hUNG2 and APE1. Reactions were halted by adding an equal volume of 1.8% SDS and 100 μg/ml Proteinase K, Fermentas (Waltham, MA, USA). The abasic sites were cleaved by bringing the reactions to 333mM NaOH followed by heating at 55°C or 10min. DNAs were brought to 50% formamide and resolved by 16% denaturing PAGE (19:1) with 7M urea and 0.5× TBE. Electrophoresis was performed at 550V for 20–35min, depending on the oligonucleotide.
Image collection and data analysis
Images were collected using a Typhoon FLA 7000 phosphorimager and bands quantified using ImageQuant TL software (GE Healthcare). Pixel density ratios were used to score enzyme activity. Graphs show the percentage of the total DNA cleaved. Enzymatic reactions were repeated at least three times (n = 3) for each oligonucleotide and displayed with standard deviation. One representative phosphorimage was selected for each figure. A two tailed t-test for paired two sample means (95% confidence interval) was performed for each pair of G4 and GT oligonucleotide assay.
Results
G4 formation with deoxyuracil oligonucleotides
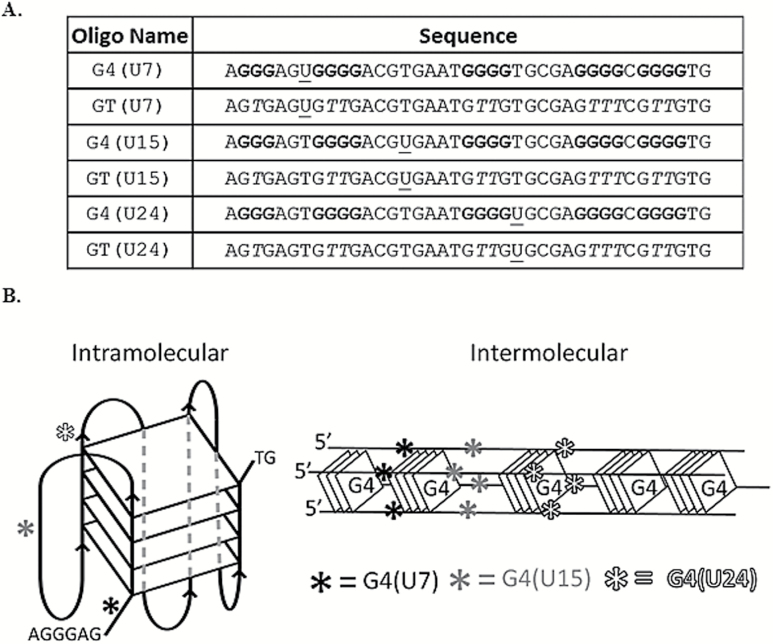
We selected a G4 DNA sequence from the TCF3 (E2A) gene as a model structure, which we previously showed folds into G4 DNA at physiological salt and pH conditions in vitro (12) (Figure 1A). The goal of this study was to simply test the efficiency of uracil excision activities on DNA containing uracil bases located directly next to (5′ or 3′) a guanine tetrad. Therefore, oligonucleotides were synthesised to contain a single uracil lesion at defined positions. The U7, U15, and U24 sequences contain a uracil at position 7, 15 and 24, respectively (Figure 1A). The uracil is placed directly 5′ (U7), 3′ (U24) or in between (U15) the guanine repeats participating in G4 formation (Figure 1A). Control DNAs were sequence paired to the uracil-containing G4 oligonucleotide, except that the guanine repeats were interrupted with thymine to eliminate stable G4 folding (Figure 1A). These oligonucleotides are distinguished from one another by the terminology ‘G4’ (with guanine repeats) or ‘GT’ (thymine substituted). One important experimental consideration regarding G4 DNA is that a sequence capable of intra-molecular G4 formation will also permit inter-molecular G4 conformations in solution (5,21). Therefore, we expected to observe a mixture of both conformations (Figure 1B). Independent of the precise G4 structure that forms, the U7 and U24 uracil bases will be adjacent to a tetrad. Structure prediction using the Mfold server (22) did not return any significant fold-back structures for the GT oligonucleotides (ΔG < −1.0 kcal/mol). For the G4-capable sequences, the QGRS G4 prediction software (23) verified that the most likely G4 conformation will involve the longest guanine repeats (using an input with a G-group of four and minimum loop of 1). That results in placement of U7 and U24 directly next to a tetrad in either intra-molecular or inter-molecular G4, and this is depicted in Figure 1B. This remains the predominant structure predicted by the program even when the input parameters are reduced to a G-group size of three. Alternate G4 conformations are theoretically possible, but if they do form they would most likely exist as a collection of various minor G4 species in addition to the two predominant inter- and intra-molecular conformations predicted by QGRS mapper (Figure 1B).
Figure 1.
Sequences used and diagrams of G4 structures. (A) The name (left) and sequence (right) for each oligonucleotide are shown. G4 and GT indicate oligonucleotides capable or incapable of adopting G4, respectively. The nucleotide position of the uracil, U7, U15 and U24 relative to the 5′ end is indicated within each oligonucleotide name. The positions of uracil bases within each sequence are underlined and guanine repeats are bolded. Substitution of thymine for guanine interrupts G4 folding potential, shown with an italicised ‘T.’ (B) Diagram depicting intramolecular, left, or intermolecular, right, G4 conformations and the relative positions of each uracil (asterisks). All three uracil positions are depicted on a single G4 conformation for convenience. Diagrams are not scaled models. Actual G4 conformations in solution are predicted to be a mixture of intra and inter-molecular species, but U7 and U24 will be adjacent to a G-tetrad.
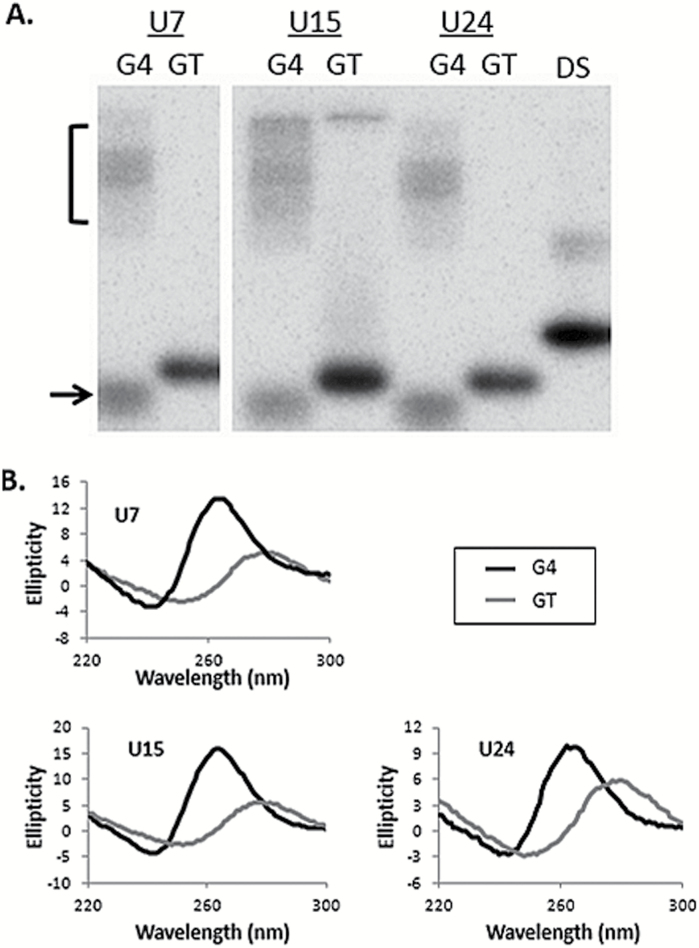
We predicted that substituting thymine for uracil in the TCF3 oligonucleotide would not disrupt G4 formation, principally because those bases differ by only a single methyl group and they are located between G-tetrads (Figure 1A and B), so we did not expect to see alterations in guanine-guanine pairing. In other G4 studies, telomeric G4 structures were shown to maintain stability when thymine was substituted for 5-hydroxymethyluracil, a major oxidation product (24,25), so we infer that changes to bases not directly participating in G4 folding will not disrupt the structure. In contrast, tandem guanine repeats are required for G4, and interruption of those sequences by thymine substitution would eliminate the possibility of G4 formation. Consistent with that, QGRS mapper returned no scores for the GT oligonucleotides used here, indicating the absence of G4 folding potential. In order to experimentally verify G4 formation we applied two different approaches, native PAGE and CD. In native PAGE, G4 DNA structures migrate faster (intramolecular) or slower (intermolecular) compared to unfolded controls of similar molecular weight (GT). In the presence of 100mM KCl, the migration patterns for G4(U7), G4(U15), and G4(U24) are entirely consistent with G4 DNA (Figure 2A). This matches previous results showing G4 formation with TCF3 sequences in KCl salt, except those oligonucleotides did not contain uracil (12). The GT controls for each G4 sequence migrate predominately as a single species (Figure 2A), as expected. The G4(U24) oligonucleotide paired with complement (double stranded) migrates predominately as a single species and slower than the single-stranded counterparts, as expected (Figure 2A).
Figure 2.
Detection of G4 formation. (A) Phosphorimages showing native PAGE resolution of 5′ 32P end labelled G4 and GT oligonucleotides in neutral pH and 100mM KCl. The name of each oligonucleotide is shown at the top. Oligonucleotides where guanine repeats are interrupted by thymine to prohibit G4 formation (GT) migrate predominately as a single species. Guanine repeat oligonucleotides (G4) migrate faster (arrow) and slower (bracket) than each control (GT). All oligonucleotides are equal nucleotide length and migration of G4(U24) duplex DNA (DS) is shown as a reference for double-stranded DNA. (B) CD of each G4 and GT uracil-containing oligonucleotide. Ellipticity is shown on the Y axis, wavelength (nm) on the X axis. Spectra for uracil-containing oligonucleotides with guanine repeats (G4) are in black, repeat interrupted (GT) oligonucleotides in grey. All oligonucleotides were assayed in 100mM KCl solution at pH 7.6.
CD measures the absorbance of circularly polarised light by chiral molecules and G4 DNA structures produce characteristic CD spectra, with an ellipticity maximum at 264nm and a minimum at 240nm (26–29). We applied CD analysis to the uracil containing GT and G4 oligonucleotides shown in Figure 1A to measure their ability to fold into G4 DNA in the presence of 100mM KCl. Only the oligonucleotides containing guanine repeats (G4) produced spectra characteristic of G4 DNA (Figure 2B), consistent with earlier results (12). Interruption of the guanine repeats (GT) resulted in a CD spectral shift for each oligonucleotide (Figure 2B). We conclude that the TCF3 oligonucleotides containing uracil fold into G4 DNA structures and the GT control sequences do not adopt G4 DNA.
G-tetrads interfere with bacterial UDG activity
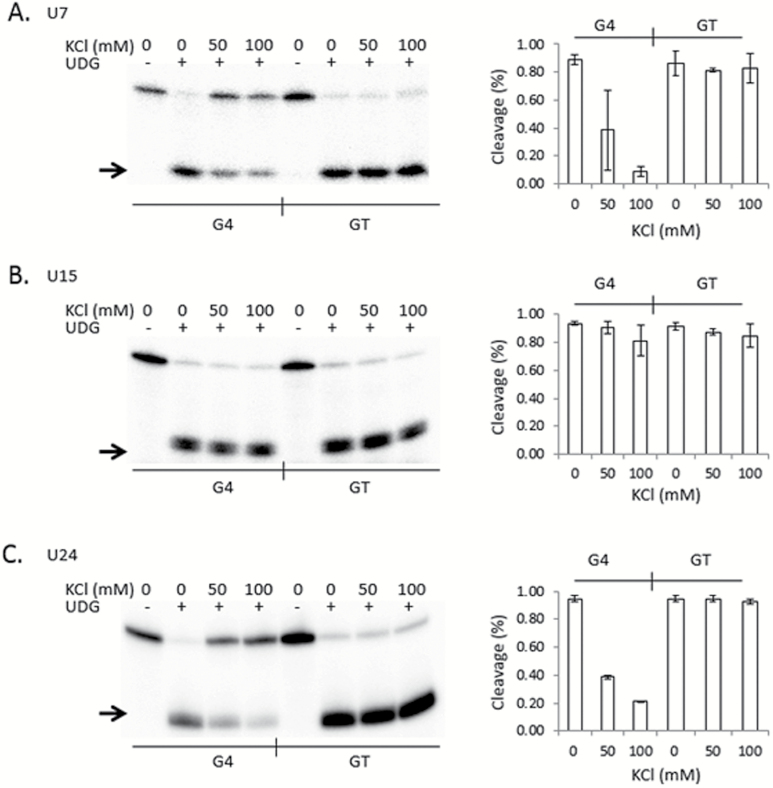
We next asked if uracil glycosylase is active at uracils residing within guanine-rich DNA and next to a G4 tetrad. We first tested E. coli UDG. Each 5′ 32P end-labelled and uracil-containing G4 and GT oligonucleotide was incubated with UDG, and after alkaline lysis of the abasic sites the cleavage products were resolved by denaturing PAGE. Generally, G4 is stabilised in solutions containing physiological concentrations of K+ and in neutral pH, and it is less stable in the absence of K+ salts or in the presence of Li+ (5,30,31). Therefore, we performed UDG reactions in either 0mM, 50mM or 100mM KCl, anticipating that 100mM KCl will result in the most stable G4 DNA. One representative phosphorimage for each PAGE-resolved cleavage assay is shown in Figure 3 (left), and to the right of each image are graphs depicting the quantitation of cleavage from at least three independent experiments. In the absence of KCl, all six oligonucleotides were cleaved with near equal efficiency, regardless of the position of the uracil (Figure 3 A–C). In contrast, UDG activity on G4(U7) and G4(U24) was sharply reduced in the presence of KCl compared to each companion GT control (Figure 3A and C; P < 0.004 and P < 0.002, respectively at 100mM KCl). KCl had only a marginal affect on the cleavage activity of UDG on G4(U15), even at 100mM (P > 0.17; Figure 3B). In contrast, the presence of KCl had a strong effect on the ability for UDG to cleave G4(U7) and G4(U24), with more than 5-fold fewer uracils removed compared to reactions that did not contain KCl (Figure 3A and C). KCl at concentrations up to 100mM did not measurably effect UDG activity on any of the GT oligonucleotides, so the reduction in UDG activity observed for G4(U7) and G4(U24) was not due to the ionic strength of the reaction. Furthermore, UDG showed equivalent activity on all oligonucleotides tested when LiCl was substituted for KCl, up to 100mM (supplementary Figure 1, available at Mutagenesis Online). Together, we concluded that uracil bases located next to guanine repeats are excised by UDG. However, UDG does not cleave the uracil as efficiently when those repeats participate in G4 formation compared single-stranded DNA of identical sequence.
Figure 3.
G-tetrads in TCF3 G4 interfere with UDG activity. (Left) Representative phosphorimages of E. coli UDG cleavage assays for each 5′ radiolabelled oligonucleotide are shown. Reactions were performed at the indicated salt conditions (0, 50 and 100mM KCl), followed by alkaline lysis and resolution of the cleavage products by denaturing PAGE. Oligonucleotides contained a single uracil at position 7 (A), 15 (B) or 24 (C) in sequences capable of forming G4 (G4) or controls where the guanine repeats were interrupted with thymine to prohibit G4 formation (GT). The presence (+) or absence (−) of UDG is indicated at the top. An arrow shows the position of the cleavage products. (Right, A–C) Quantitation of the percentage cleaved by UDG (Y axis) for each oligonucleotide at the indicated salt concentration (X axis). Values are means from three independent experiments, with standard deviation.
G-tetrads interfere with hUNG2 activity
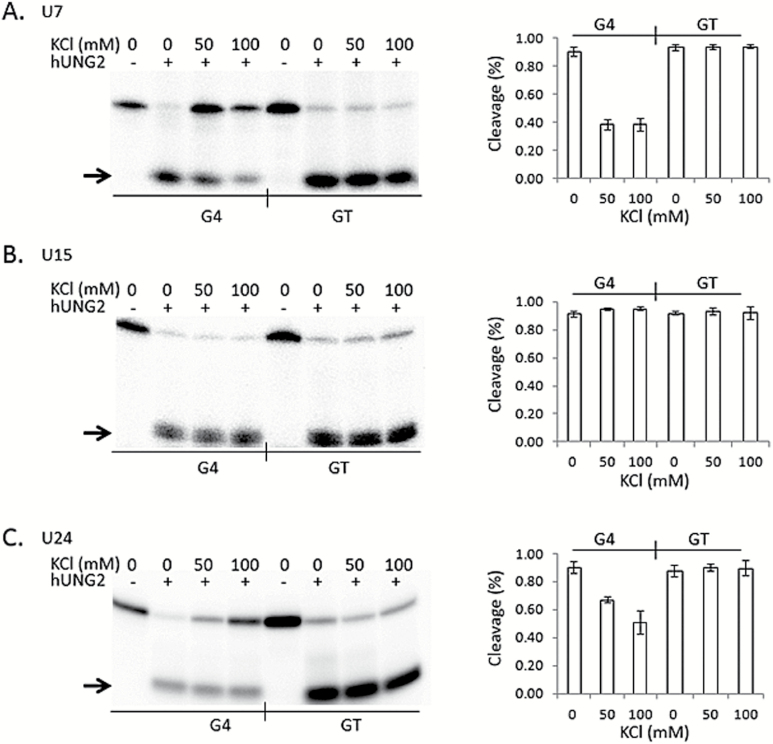
Even though uracil DNA glycosylase is a highly conserved enzyme (32), we next asked if the human UNG2 enzyme shows the same reluctance to process uracil near G4 DNA structures. We performed an identical set of cleavage assays, except that hUNG2 was substituted for E. coli UDG (Figure 4). The activities observed for hUNG2 on each oligonucleotide paralleled results obtained for E. coli UDG (Figure 3). Compared to the GT oligonucleotide, hUNG2 showed reduced activity on G4(U7) and G4(U24) oligonucleotides, but only when KCl and guanine repeats are present (Figure 4A and C), whereas activity on G4(U15) was similar to GT(U15) (Figure 4B). At 100mM KCl, the reduction in cleavage activity on G4(U7) and G4(U24) compared to their GT companions was highly significant (P < 0.001 and P < 0.008, respectively), and not significant for G4(U15) and GT(U15) (P > 0.42). The reduced activity is most likely due to the formation of G4 structures because it was only observed when K+ ions were included in the reaction. Identical reactions performed in the presence of LiCl instead of KCl resulted in full hUNG2 activity (supplementary Figure 2, available at Mutagenesis Online). We conclude that hUNG2 activity is inhibited at uracils positioned adjacent to the G4-tetrads, at least for G4 DNA structures formed by the TCF3 oligonucleotide. Further, it appears that weak cleavage activity at G4 DNA is likely a conserved property of the human hUNG2 and E. coli UDG enzymes.
Figure 4.
G-tetrads in TCF3 G4 interfere with hUNG2 activity. (Left) Representative phosphorimages of hUNG2 cleavage assays for each 5′ radiolabelled oligonucleotide are shown. Reactions were performed at the indicated salt conditions (0, 50 and 100mM KCl), followed by alkaline lysis and resolution of the cleavage products by denaturing PAGE. Oligonucleotides contained a single uracil at position 7 (A), 15 (B) or 24 (C) in sequences capable of forming G4 (G4) or controls where the guanine repeats were interrupted with thymine to prohibit G4 formation (GT). The presence (+) or absence (−) of hUNG2 is indicated at the top. An arrow shows the position of the cleavage products. (Right, A–C) Quantitation of the percentage cleaved by hUNG2 (Y axis) for each oligonucleotide at the indicated salt concentration (X axis). Values are means from three independent experiments, with standard deviation.
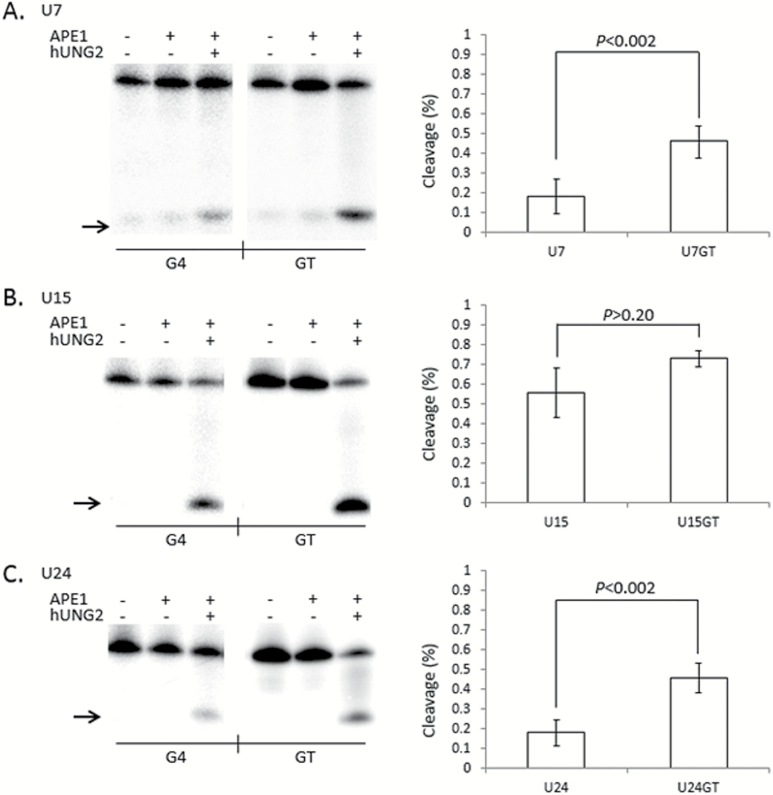
In humans, abasic DNA is cleaved by the AP-endonuclease (APE1) enzyme and it was recently demonstrated that APE1 activity is inhibited at G4 DNA (20), suggesting that BER may be compromised at guanine-rich loci. We further tested the efficiency of uracil repair by co-incubating hUNG2 and APE1 with U7, U15 and U24 G4 and GT oligonucleotides all under G4 folding conditions. As expected, uracil was poorly cleaved from the G4(U7) and G4(U24) oligonucleotides by hUNG2 and APE1 compared to GT(U7) and GT(U24) (Figure 5A and C). The cleavage difference between G4 and GT was highly significant (P < 0.002). The G4(U15) oligonucleotide was processed at essentially the same efficiency as GT(U15), although there was a modest, but not significant (P > 0.20), reduction in abasic site cleavage for the G4(U15) oligonucleotide (Figure 5B). This compares well to alkaline cleavage results (Figure 4), with the exception that APE1 displayed a lower overall ability to cleave even the unstructured substrates (Figure 5). This suggests that not all abasic sites were cut by APE1 because treatment with NaOH resulted in ~90% of the GT oligonucleotides cleaved in every uracil glycosylase assay (Figures 3 and 4, supplementary Figures 1 and 2, available at Mutagenesis Online). We conclude that uracils located next to a G-tetrad are poorly excised by hUNG2, independent of the mechanism of abasic site cleavage.
Figure 5.
G-tetrads in TCF3 G4 interfere with the cleavage step of Base Excision Repair. (Left) Representative phosphorimages showing the activity of both hUNG2 and APE1 on single uracil bases placed at positions 7 (A), 15 (B) or 24 (C) in G4 DNA folded from the TCF3 oligonucleotide (G4) or control oligonucleotides that cannot fold into G4 structures (GT). Reactions contained 100mM KCl and the presence (+) or absence (−) of hUNG2 or APE1. Cleavage products (arrow) were resolved by denaturing PAGE. (Right) Quantitation of a least three independent experiments are displayed on the right of each representative phosphorimage. The percentage cleaved by APE1 activity (Y axis) is shown for each G4 (X axis left) or GT (X axis right), with standard deviation and P values (two-tailed t-test). Both G4(U7) (P < 0.002) and G4(U24) (P < 0.002) display significant inhibition compared to GT, but G4(U15) (P > 0.20) does not.
Discussion
G4 DNA structures that form in the genome promote genetic instability and human disease (3,8), yet the reasons why are not fully defined. In this work we asked if G4 DNA structures, or more precisely the presence of G-tetrads, are an impediment to uracil repair in vitro. Both UDG and hUNG2 have been well characterised for their activity on uracils residing in either single or double stranded DNA substrates. We found that UDG and hUNG2 efficiently cleaved uracil from the unstructured single-stranded TCF3 DNA and from the repeat disrupted (GT) oligonucleotides (Figures 3 and 4), showing equal activity for the repeat containing and control DNAs. This was in contrast to the lower activity observed on the G4 oligonucleotides U7 and U24 under G4 folding conditions (Figures 3A and C, and 4A and C). The addition of salt to the reactions did not reduce excision activity on the G4(U15) oligonucleotide (Figures 3B and 4B), arguing that it is the position of the uracil relative to G4 and not the ionic strength of the reaction that affected cleavage efficiency. Furthermore, reactions that replaced KCl with a salt that does not support TCF3 G4, LiCl (12), returned full cleavage activity on the G4(U7) and G4(U24) (supplementary Figures 1 and 2, available at Mutagenesis Online). We suggest that the G4 structure formation within G4(U7) and G4(U24) place the uracil in a molecular geometry that is less accessible to either UDG or hUNG2 activity. Indeed, moving the uracil three nucleotides from a tetrad G4(U15) results in nearly full glycosylase activity compared to single-stranded conformations (Figures 3B and 4B). While these experiments do not resolve whether G-tetrads are an impediment to substrate recognition or cleavage by UDG and hUNG2, both enzymes do appear to share the property of having relatively weak activity next to G4 DNA structures.
Our study is limited to the analysis of a single uracil placed at three different positions relative to guanine repeats in a synthetic oligonucleotide that can participate in G4 formation. The model G4 sequence selected for this study is from a portion of the transcription factor 3 (TCF3 or E2A) gene (12). The TCF3 gene encodes a key transcriptional regulator in immune cell differentiation (33,34) and is mutated or altered in both B and T cell lymphoma (35,36). Based on our results, it seems likely that a uracil located directly adjacent to any guanine tetrad will be poorly repaired by UNG-initiated BER, and that our findings extend beyond the model (TCF3) G4 sequence used here. Since we have used a single sequence here (albeit more than one structural conformation) we cannot exclude the possibility that uracil excision activity may be more efficient at G4s folded from other genomic sequences or other G4 conformations. It is possible that G-tetrads are a general impediment to uracil excision activity by UNG enzymes, but cytidine deaminations within sequence contexts that support alternate G4 conformations, such as anti-parallel or Na+ stabilised G4s, could feasibly be better substrates. Some deaminated cytidines in telomeric G4, or at positions located a single base away from a tetrad may also be substrates for repair. Furthermore, considering the apparent abundance and complexity of unstable G4 loci in the human genome (3,7,8), it will be important to further characterise the limits of uracil excision activity on a wider range of G4 structural conformations.
During transcription or replication, when DNA is transiently denatured, cytosine is exposed to hydrolytic deamination at a rate orders of magnitude higher than that of duplex DNA (37). Presumably, unpaired cytidines present within G4 DNA structures will also be prone to deamination, and our results suggest hUNG2 may not efficiently remove these particular base lesions. Even so, there are three other glycosylases that are capable of removing uracil from DNA with varying efficiencies (16,17,38), and some or all could feasibly substitute for hUNG2 at G4 DNA. Using telomeric G4 substrates and glycosylases specific to oxidised DNA damage, it was shown that 8-oxyguanine was not processed, but other oxidation products were excised by the NEIL glycosylases (18) suggesting that G4 DNA does not inhibit every glycosylase. Furthermore, some repair pathways appear to not be affected by G4 DNA, O(6)-alkylguanine in telomeric G4 was shown to be repaired by alkylguanine alkyltransferase at levels comparable to duplex DNA (39).
One prediction from our results is that repeat sequences that support G4, like the telomeres (30,31), would not contain cytidines adjacent to G4. This is because unrepaired cytidine deaminations caused by reduced BER at those sites would promote C to T mutagenesis. There is some evidence to support that model. A compilation of known G-rich telomere repeat sequences from animals, plants and fungi species shows that cytosines are rare and, if they are present in the repeat, they do not reside directly next to tandem guanines (40) and would therefore not be directly adjacent to a G4 tetrad. Even C-type variant repeats found enriched in telomeres of ALT cells contain cytidine one nucleotide away from the guanine triplet (41). It is therefore interesting to speculate that the frequency of G4 formation in the cell may be a reflection of the sequence context and size of the looped regions. It is possible that G4-capable sequences containing cytosine-rich loops infrequently participate in stable or persistent G4 formation. Indeed, it was recently shown in yeast that both minisatellite instability and the thermal stability of G4 structures increases for sequence variants that contain shortened pyrimidine loops, which happen to be rare G4 sequences in the yeast genome (42). If selection acts to favour genetic stability at a given G4 locus, our results provide a possible molecular mechanism for that evolutionary force and an explanation for the paucity of short pyrimidine loops between G-tetrads. Cytidine damage may be poorly repaired in the context of G4 DNA, and if damage occurs within stable G4 structures this could result in unresolved DNA repair intermediates, or mutagenesis, at those sites.
In conclusion, we have used a model G4 forming sequence from the TCF3 gene to ask if G4 DNA structures have the potential to interfere with uracil excision activities. Both bacterial UDG and hUNG2 shared a reluctance to cleave uracils next to G-tetrads, but showed full activity on control oligonucleotides that cannot adopt DNA structure, and full activity on guanine repeat oligonucleotides in conditions that do not support G4 formation. The addition of APE1 to the reaction did not improve excision activity, supporting the notion that the BER pathway is not fully activated at uracil bases in G4 DNA. Guanine-rich loci are known to be genetically unstable, and our results suggest that defects in BER of uracil at G4 DNA can contribute to mutagenesis.
Supplementary data
Supplementary Figures 1 and 2 are available at Mutagenesis Online.
Funding
This work was supported by the School of Biological Sciences, College of Arts and Sciences at Illinois State University and the National Institutes of Health, R15CA182978 to E.D.L.
Supplementary Material
Acknowledgements
The authors would like to thank Jonathan D. Williams for assistance in the CD analysis and the entire Larson lab for insightful discussion throughout the project.
Conflict of interest statement: None declared.
References
- 1. Tarsounas M. and Tijsterman M (2013) Genomes and G-quadruplexes: for better or for worse. J. Mol. Biol., 425, 4782–4789. [DOI] [PubMed] [Google Scholar]
- 2. Bochman M. L. Paeschke K. and Zakian V. A (2012) DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet., 13, 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maizels N. (2015) G4-associated human diseases. EMBO Rep., 16, 910–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sen D. and Gilbert W (1988) Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature, 334, 364–366. [DOI] [PubMed] [Google Scholar]
- 5. Sen D. and Gilbert W (1990) A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature, 344, 410–414. [DOI] [PubMed] [Google Scholar]
- 6. Burge S. Parkinson G. N. Hazel P. Todd A. K. and Neidle S (2006) Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res., 34, 5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davis L. and Maizels N (2011) G4 DNA: at risk in the genome. EMBO J., 30, 3878–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cea V. Cipolla L. and Sabbioneda S (2015) Replication of Structured DNA and its implication in epigenetic stability. Front. Genet., 6, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eddy J. and Maizels N (2006) Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res., 34, 3887–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nambiar M. Srivastava M. Gopalakrishnan V. Sankaran S. K. and Raghavan S. C (2013) G-quadruplex structures formed at the HOX11 breakpoint region contribute to its fragility during t(10;14) translocation in T-cell leukemia. Mol. Cell. Biol., 33, 4266–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nambiar M. Goldsmith G. Moorthy B. T. Lieber M. R. Joshi M. V. Choudhary B. Hosur R. V. and Raghavan S. C (2011) Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res., 39, 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Williams J. D. Fleetwood S. Berroyer A. Kim N. and Larson E. D (2015) Sites of instability in the human TCF3 (E2A) gene adopt G-quadruplex DNA structures in vitro. Front. Genet., 6, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Kregten M. and Tijsterman M (2014) The repair of G-quadruplex-induced DNA damage. Exp. Cell Res., 329, 178–183. [DOI] [PubMed] [Google Scholar]
- 14. Wu Y. and Brosh R. M. Jr (2010) G-quadruplex nucleic acids and human disease. FEBS J., 277, 3470–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brooks T. A. Kendrick S. and Hurley L (2010) Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J., 277, 3459–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krokan H. E. Sætrom P. Aas P. A. Pettersen H. S. Kavli B. and Slupphaug G (2014) Error-free versus mutagenic processing of genomic uracil–relevance to cancer. DNA Repair (Amst)., 19, 38–47. [DOI] [PubMed] [Google Scholar]
- 17. Krokan H. E. and Bjørås M (2013) Base excision repair. Cold Spring Harb. Perspect. Biol., 5, a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou J. Liu M. Fleming A. M. Burrows C. J. and Wallace S. S (2013) Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J. Biol. Chem., 288, 27263–27272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou J. Fleming A. M. Averill A. M. Burrows C. J. and Wallace S. S (2015) The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res., 43, 4039–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Broxson C. Hayner J. N. Beckett J. Bloom L. B. and Tornaletti S (2014) Human AP endonuclease inefficiently removes abasic sites within G4 structures compared to duplex DNA. Nucleic Acids Res., 42, 7708–7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sen D. and Gilbert W (1992) Guanine quartet structures. Methods Enzymol., 211, 191–199. [DOI] [PubMed] [Google Scholar]
- 22. Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res., 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kikin O. D’Antonio L. and Bagga P. S (2006) QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res., 34, W676–W682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Konvalinová H. Dvořáková Z. Renčiuk D. Bednářová K. Kejnovská I. Trantírek L. Vorlíčková M. and Sagi J (2015) Diverse effects of naturally occurring base lesions on the structure and stability of the human telomere DNA quadruplex. Biochimie, 118, 15–25. [DOI] [PubMed] [Google Scholar]
- 25. Virgilio A. Esposito V. Mayol L. Giancola C. Petraccone L. and Galeone A (2015) The oxidative damage to the human telomere: effects of 5-hydroxymethyl-2’-deoxyuridine on telomeric G-quadruplex structures. Org. Biomol. Chem., 13, 7421–7429. [DOI] [PubMed] [Google Scholar]
- 26. Balagurumoorthy P. Brahmachari S. K. Mohanty D. Bansal M. and Sasisekharan V (1992) Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res., 20, 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kypr J. Kejnovská I. Renciuk D. and Vorlícková M (2009) Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res., 37, 1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dapić V. Abdomerović V. Marrington R. Peberdy J. Rodger A. Trent J. O. and Bates P. J (2003) Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res., 31, 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vorlíčková M. Kejnovská I. Sagi J. Renčiuk D. Bednářová K. Motlová J. and Kypr J (2012) Circular dichroism and guanine quadruplexes. Methods, 57, 64–75. [DOI] [PubMed] [Google Scholar]
- 30. Sundquist W. I. and Klug A (1989) Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature, 342, 825–829. [DOI] [PubMed] [Google Scholar]
- 31. Williamson J. R. Raghuraman M. K. and Cech T. R (1989) Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell, 59, 871–880. [DOI] [PubMed] [Google Scholar]
- 32. Olsen L. C. Aasland R. Wittwer C. U. Krokan H. E. and Helland D. E (1989) Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J., 8, 3121–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kee B. L. Quong M. W. and Murre C (2000) E2A proteins: essential regulators at multiple stages of B-cell development. Immunol. Rev., 175, 138–149. [PubMed] [Google Scholar]
- 34. Miyazaki K. Miyazaki M. and Murre C (2014) The establishment of B versus T cell identity. Trends Immunol., 35, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Steininger A., Möbs M., Ullmann R., et al. (2011) Genomic loss of the putative tumor suppressor gene E2A in human lymphoma. J. Exp. Med., 208, 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schmitz R., Young R.M., Ceribelli M., et al. (2012) Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature, 490, 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barnes D. E. and Lindahl T (2004) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet., 38, 445–476. [DOI] [PubMed] [Google Scholar]
- 38. Krokan H. E. Drabløs F. and Slupphaug G (2002) Uracil in DNA–occurrence, consequences and repair. Oncogene, 21, 8935–8948. [DOI] [PubMed] [Google Scholar]
- 39. Hellman L. M. Spear T. J. Koontz C. J. Melikishvili M. and Fried M. G (2014) Repair of O6-methylguanine adducts in human telomeric G-quadruplex DNA by O6-alkylguanine-DNA alkyltransferase. Nucleic Acids Res., 42, 9781–9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Podlevsky J. D. Bley C. J. Omana R. V. Qi X. and Chen J. J (2008) The telomerase database. Nucleic Acids Res., 36, D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Conomos D. Stutz M. D. Hills M. Neumann A. A. Bryan T. M. Reddel R. R. and Pickett H. A (2012) Variant repeats are interspersed throughout the telomeres and recruit nuclear receptors in ALT cells. J. Cell Biol., 199, 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Piazza A., Adrian M., Samazan F., et al. (2015) Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J., 34, 1718–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.