Abstract
A popular and successful strategy in semi-rational design of protein stability is the use of evolutionary information encapsulated in homologous protein sequences. Consensus design is based on the hypothesis that at a given position, the respective consensus amino acid contributes more than average to the stability of the protein than non-conserved amino acids. Here, we review the consensus design approach, its theoretical underpinnings, successes, limitations and challenges, as well as providing a detailed guide to its application in protein engineering.
Keywords: consensus design, multiple sequence alignment, protein stability, semi-rational design, statistical sequence analysis, thermostability
Introduction
Directed evolution and informatics-based rational design are transforming the field of protein engineering (Arnold and Volkov, 1999; Jiang et al., 2008; Lutz, 2010; Brustad and Arnold, 2011; Bornscheuer et al., 2012; Joh et al., 2014; Woolfson et al., 2015). Semi-rational or knowledge-based hybrid approaches, which mix rational design with directed evolution schemes, to create small libraries of very high quality, have gained substantial momentum (Patrick and Firth, 2005; Lutz, 2010; Wijma et al., 2013, 2014; Magliery, 2015). Typically, information from protein structure, function, sequence homology and predictive computational algorithms are combined to preselect sites for focussed mutagenesis with limited amino acid diversity. This focus translates into dramatically reduced library sizes with a major increase in functional content, allowing for a more efficient sampling of sequence space.
A popular strategy in semi-rational design of stability is the use of evolutionary information encapsulated in homologous protein sequences. Multiple sequence alignments (MSAs) and phylogenetic analyses have become standard tools for exploring sequence conservation (Steipe et al., 1994) and ancestral relationships (Pauling et al., 1963; Yang et al., 1995; Thornton et al., 2003; Thornton, 2004) amongst protein homologues. Such sequences and alignments can be acquired from natural sequence databases (UniProt Consortium, 2008; NCBI Resource Coordinators, 2014), curated alignment databases (Sigrist et al., 2002, 2013; Wilson et al., 2009; Finn et al., 2016) and neutral drift experiments (Bershtein et al., 2008; Jäckel et al., 2010).
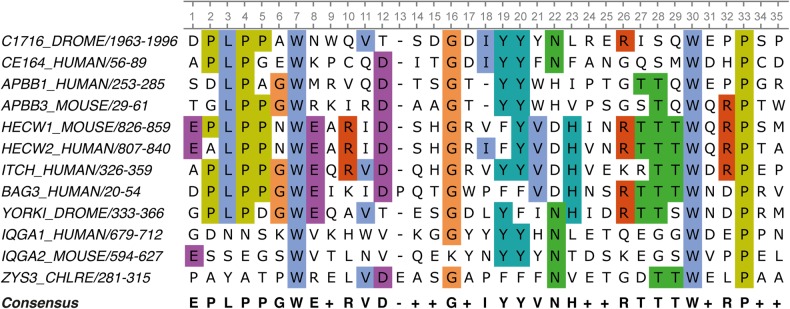
Consensus design, like ancestral sequence reconstruction, utilises evolutionary history; however, rather than inferring phylogenetic hierarchy, all sequences are aligned, and the most frequently observed amino acid is identified at each position in the alignment (Fig. 1) (Steipe et al., 1994). The consensus design approach has been widely successful in improving the stabilities of functional and non-functional proteins, for example increasing melting temperatures by 10–32°C (Wirtz and Steipe, 1999; Lehmann and Wyss, 2001; Lehmann et al., 2002; Dai et al., 2007; Lutz, 2010; Magliery et al., 2011; Magliery, 2015; Porebski et al., 2015; Paatero et al., 2016). However, only ∼50% of conserved residues are associated with improved stability, with ∼10% being stability neutral and ∼40% being destabilising, leading to challenges and trade-offs during implementation (Steipe et al., 1994; Nikolova et al., 1998; Wang et al., 1999; Lehmann et al., 2000, 2002; Lehmann and Wyss, 2001; Polizzi et al., 2006; Khersonsky et al., 2012).
Fig. 1.
Sequence alignment of 12 WW domains across several species and parent proteins. In the consensus, a ‘−’ is a gap, whilst a ‘+’ is an ambiguous position with no consensus. The most conserved residues are highlighted.
Consensus design involves the following four steps: (1) identification of a domain to be targeted (for example, boundaries within a larger sequence context), (2) acquisition and pre-processing of homologous sequences, (3) iterative assessment of several MSA regimes and removal of disruptive sequences, and (4) calculation of sequence conservation. Application of sequence conservation is typically performed in one of three ways. First, single- or multiple-point mutations of the most conserved amino acid positions can be made to a target protein (Schreiber et al., 1994; Nikolova et al., 1998; Wang et al., 1999; Polizzi et al., 2006; Ferreiro et al., 2007a), and these mutations may further be filtered or weighted by other statistical or computational methods (Socolich et al., 2005; Polizzi et al., 2006). Second, full-length sequences can be created de novo, avoiding the problem of identifying residues that are truly stabilising (Pantoliano et al., 1989; Blatt et al., 1996; Lehmann et al., 2000; Dai et al., 2007; Vazquez-Figueroa et al., 2008; Sullivan et al., 2011; Jacobs et al., 2012; Porebski et al., 2015). Third, conserved residues and positions can be spiked or targeted in directed evolution studies to increase sampling of functionally relevant sequence space (Amin et al., 2004; Socolich et al., 2005; Bershtein et al., 2008; Case and Hackel, 2016). The strategy of implementation is highly dependent on requirements and available resources; however, all approaches have seen impressive results, with an exhaustive catalogue of consensus-designed proteins shown in Supplementary Table S1.
Factors to consider during consensus design
Acquisition of homologous sequences
The most efficient way to acquire sequences is via sequence alignment databases such as Pfam (Sonnhammer et al., 1998; Finn et al., 2016), Prosite (Sigrist et al., 2013), SMART (Letunic et al., 2015) and Superfamily (Wilson et al., 2009). These databases contain small, manually curated seed alignments for the development of hidden Markov model (HMM) profiles (Finn et al., 2011) or motif-specific rules and patterns (Sigrist et al., 2002), which can then be applied to larger collections such as the UniProtKB/Swiss-Prot (UniProt Consortium, 2008), Protein Data Bank (PDB) and NCBI (NCBI Resource Coordinators, 2014) sequence databases.
If a protein target is not well represented in existing alignment databases, the best approach is to query the UniProtKB or NCBI sequence databases for a small number (<20) of the most homologous sequences, which can be used to curate a representative alignment that can be subjected to HMM profiling with the HMMER suite (Finn et al., 2011), and subsequently align to more distant homologues. In the unfortunate instance that the target protein is minimally or not represented in the UniProtKB or NCBI sequence databases, the next option is to generate diversity through the use of neutral drift studies (Bershtein et al., 2008; Jäckel et al., 2010). In neutral drift experiments, a target protein is subjected to rounds of random mutagenesis and selected purely on whether it is folded and/or functional, and is then sequenced. This approach was used as a means of generating unbiased sequence diversity in the consensus design of a chorismate mutase, showing the method to be successful with fewer than 30 selected sequences (Jäckel et al., 2010).
Homology
The effect of sequence homology on consensus design is poorly understood and highly likely to be a function of the target protein's biophysical properties, evolutionary history and the taxonomic representation in sequencing databases. Theoretically, inclusion of evolutionarily distant or diverse sequences should improve the probability of identifying more conserved features, as increased distance may imply increased sampling of sequence space. Although there are reports that too little (Jacobs et al., 2012) and too much diversity of the input MSA is problematic (Parmeggiani et al., 2008; Jäckel et al., 2010; Sullivan et al., 2011), this area has not been thoroughly explored.
Determining the right amount of diversity is challenging. Sullivan et al. noted this in the consensus design of a triosephosphate isomerase (TIM) using comparisons of two Pfam alignments from database versions 18 and 22 (Sullivan et al., 2011). The input sequences of version 18 were a roughly even mixture of bacterial and eukaryotic sequences, resulting in a weakly active and poorly folded consensus protein. However, the version 22 alignment was composed of predominantly bacterial sequences and resulted in a well-folded and fully active protein (Sullivan et al., 2011). To further complicate matters, the version 22 sequences were filtered to be roughly the same length, and duplicate entries were removed, which may have had other effects and therefore reduced the general applicability of this approach. Highly successful designs such as FN3con (Porebski et al., 2015) and cLRRTM2 (Paatero et al., 2016) used sequences that were predominantly or exclusively from higher-order eukaryotes without the need to filter based on sequence length, suggesting that spanning the MSA over taxonomic domains or kingdoms may negatively affect results. Parmeggiani also observed a similar problem as a result of too broad protein family selections of armadillo repeat proteins (Parmeggiani et al., 2008). However, rather than filtering or removing sequences, they subclassified their MSA into closer taxonomic groups and combined conserved residues from each subclassification, resulting in a well-expressed and stable protein.
Extending sequence homology too far may result in poor conservation, which can prevent accurate alignment and lead to design failure. For example, sequence conservation within the β-defensin family is <33%, even though structural similarity is very high (Bauer et al., 2001). Here, alignment within a specific species is challenging (Rost, 1999), and consensus design would likely be impossible using natural sequences—leaving neutral drift studies as the only solution for generating homology and a chance of successful alignment (Jäckel et al., 2010). Managing homology is therefore a balance between sequence similarity, which is good for computing a MSA, and sequence diversity, which provides a greater coverage of sequence space that can be sampled during design.
Bias
In contrast to diversity, the weighting or skew of the MSA may bias consensus design towards a predominant clade, such as a taxon, species or protein classification (Jäckel et al., 2010). This is typically the result of preferences from genome sequencing projects, which tends to over-represent particular species or proteins in sequence databases. Bias is more likely to be an issue for domains, motifs or repeat proteins that are found within larger proteins. In some instances, bias may be intentional, as to preserve functional networks of a protein family from a single species or subclassification. In the interest of purely identifying robustness and stability, it is reasonable to assume that bias and over-representation should generally be avoided; these traits may mask conserved and possibly stabilising features from other less represented evolutionary lineages. Bias reduction of natural sequences can be performed with relative ease using the sequence clustering software CD-HIT (Huang et al., 2010) or by using likelihood-based methods to account for phylogeny (Bloom and Glassman, 2009).
Sequence count
One of the key advantages of consensus design over other sequence-based methods is its ability to identify stability enhancing mutations from a MSA with as few as four members. Examples include Subtilisin BPN’ (from 4 members) (Pantoliano et al., 1989) and FN3 repeats (15 members) (Jacobs et al., 2012). In the latter study, the top 10 most stable sequences were less successful in promoting thermodynamic stability than all 15 members (Jacobs et al., 2012), demonstrating that even the less stable sequences contribute to the overall stability of the resulting consensus design. In this case, more sequences provide greater diversity, thus improving the signal-to-noise ratio, and therefore the detection of conserved residues in weakly conserved regions. This effect is exemplified by recent consensus designs using very large alignments, such as FN3con (2123 sequences, ΔTm of >27°C) (Porebski et al., 2015) and cLRRTM2 (6271 sequences, ΔTm of 32°C) (Paatero et al., 2016).
Quality of the sequence alignment
Difficulties arise with MSAs containing sequences of varying length, or when there are clusters of sequences that are locally, but not globally, homologous (Rost, 1999; Pearson, 2013). Large insertions and deletions between members can affect the identification of weakly conserved positions, for example resulting in the design of a weakly active and poorly folded protein (Sullivan et al., 2011). In this specific case, filtering the homologous sequences to be roughly the same length and removing duplicate entries, the design was greatly improved, resulting in a well-folded and fully active protein. Interestingly, sequence differences between the ‘raw’ versus the filtered design were in predominantly non-conserved stretches of the protein. By sequence assessment alone, there was no obvious reason for why these differences resulted in vastly different biophysical properties. It is therefore possible that filtering sequences to those that are more homologous improved the alignment, which allows for better identification of weakly conserved residues (Sullivan et al., 2011).
Generating a ‘good’ MSA can be difficult and may actually be considered more art than science (Morrison, 2015). Unfortunately, MSA methods tend to vary significantly, and there is currently no quantitative measure for the quality of alignment (Nuin et al., 2006; Kemena and Notredame, 2009; Pearson, 2013). This is further compounded by homology, bias and sequence count and its convoluted interplay with the particular evolutionary history of a target protein and its family. Therefore, it is highly recommended to carefully examine resulting alignments prior to consensus design, possibly with an overlay of secondary structure to gauge conservation boundaries and gaps (Durani and Magliery, 2013). Iterative rounds of phylogenetic assessment and sequence pruning can improve alignment quality, which should be inspected for aligned columns that correspond with structural motifs or secondary-structure elements that have few insertions, deletions and gaps.
Statistical enhancements to consensus design
It is intriguing that consensus design is successful despite its assumption of amino acid independence, ignoring the known importance of cooperativity and coupling of amino acids (Horovitz and Fersht, 1992; Matthews, 1993). Furthermore, successes rival and often exceed those of rational design and directed evolution, which is impressive given the relative ease in which consensus design can be performed. Coupling manifests as simple pairwise interactions, through to dense and complex inter-atomic networks (LiCata and Ackers, 1995; Chen and Stites, 2001; Luque et al., 2002). For consensus design to work, coupling must be encoded into the evolutionary history and represented by amino acid conservation to some extent, which might explain why ∼40% of reported consensus mutations are destabilising (Steipe et al., 1994; Nikolova et al., 1998; Wang et al., 1999; Lehmann et al., 2000, 2002; Lehmann and Wyss, 2001; Polizzi et al., 2006; Khersonsky et al., 2012).
Attempts to improve consensus design have typically utilised additional statistical analysis that identifies coupling or covariation (Göbel et al., 1994; Lockless and Ranganathan, 1999; Atchley et al., 2000; Socolich et al., 2005; Talavera et al., 2015) and have generally been very successful in the engineering of stability (Magliery and Regan, 2004; Ozer and Ray, 2006; Sullivan et al., 2011, 2012; Durani and Magliery, 2013). The inclusion of both conserved and coupled mutations was necessary for statistical coupling analysis (SCA) in the design of a WW domain as consensus design alone was insufficient in creating a protein that folded correctly (Socolich et al., 2005). However, two previous studies had no difficulty in generating folded and stable WW domains (Macias et al., 2000; Jiang et al., 2001), suggesting that failure of consensus design may have been a result of the MSA composition rather than a limitation of the WW domain itself. Another approach used the mutual information method to calculate the pairwise statistical interactions between positions in the MSA and chose to avoid making mutations to those positions, thereby improving the accuracy of identifying stabilising mutations from ∼50 to 90% (Sullivan et al., 2012). However, this approach may not always be necessary as the pairwise covariation within and between ankyrin repeat motifs was found to be well represented by consensus design alone (Mosavi et al., 2002).
The role of covarying residues is even less understood than those of consensus mutations, although it appears that in some instances conserved residues encode most, if not all cooperativity. Therefore, consensus design and its enhancement by filtering correlated residues are dependent on how well the cooperativity is encoded into conserved residues, and whether other such correlations are statistically discernible from the alignment. Consensus design also appears to suffer when there are incompatible conserved residues and couplings as a result of divergent evolution, although this can be corrected by covariation methods (Magliery and Regan, 2004; Socolich et al., 2005; Talavera et al., 2015). However, covariation methods may not work in all scenarios; they typically require large MSAs to discern mutual amino acid dependencies (Socolich et al., 2005; Talavera et al., 2015) and are not applicable in situations where neutral drift studies are required, due to the rare event of coevolution. Interestingly, covaried residues in many cases actually have no physiospatial interactions with one another, recently sparking debate over what these methods are actually measuring (Talavera et al., 2015). Covarying substitutions are often found on different branches of the phylogenetic tree and are perhaps independent events that may or may not be attributable to molecular coevolution (Talavera et al., 2015). In the case of consensus design, highly conserved residues tend to be found within the protein core, evolve slowly and are therefore unlikely to be detected by covariation analysis even in very large alignments (Zvelebil et al., 1987; Bartlett et al., 2002; Talavera et al., 2015). Regardless, covariation methods overall do seem to have utility and appear to generally identify favourable pairs of residues that can be used on their own and in conjunction with consensus design.
Engineering thermodynamic stability
The origin of consensus mutant stabilisation is currently described as that at a given position in a MSA of homologous proteins, the respective consensus amino acid contributes more than average to the stability of the protein than non-consensus amino acids (Fig. 1) (Steipe et al., 1994; Nikolova et al., 1998; Wang et al., 1999; Lehmann et al., 2000, 2002; Lehmann and Wyss, 2001; Polizzi et al., 2006; Khersonsky et al., 2012). That is, a conserved residue is more likely to be stabilising than a random mutation at that same position (Polizzi et al., 2006; Tokuriki et al., 2007; Tokuriki and Tawfik, 2009a). However, this does not explain why conserved residues are likely to be more stabilising. A possible explanation is that as proteins evolved from a non-specialised but stable common ancestor, evolutionary drift allowed for the sampling of different stabilising mutations needed for adequate stability. Through the evolution of specialist function, many proteins now exist on a knife-edge of stability and function (Shoichet et al., 1995; Tokuriki et al., 2008; Tokuriki and Tawfik, 2009a, 2009b); for this reason, stabilising residues tend to be conserved. Consensus design is therefore able to leverage on millions of years of evolution and identify stabilising features from numerous protein homologues—amalgamating mostly additive mutations that no single protein has needed to amass.
Much of the discussion about consensus design focusses primarily on the general trend of improving thermostability (Lehmann and Wyss, 2001; Lutz, 2010; Magliery et al., 2011; Magliery, 2015). Indeed, consensus design reports a wide range of improvements to melting temperature from the modest increase of the marginally stable antibody VH domain (Tm of 36.4°C) by 6.1°C (Wirtz and Steipe, 1999), the modest increase of the highly stable Azami green fluorescent protein (Tm of ∼90°C) by 5.5°C (Dai et al., 2007), through to the large increase of the moderately stable Mouse Leucine Rich Repeat Transmembrane Neuronal 2 (LRRTM2) (Tm of ∼50°C) by 32°C (Paatero et al., 2016). However, improvements to thermodynamic stability are not necessarily the only observed effects of consensus design.
Protein evolvability
Proteins are often mutationally robust, with more than half of random single-point mutants retaining native function (Bloom et al., 2005; Tokuriki et al., 2007; Tokuriki and Tawfik, 2009a). However, extra thermodynamic stability is known to increase the robustness of the native structure to random mutations by increasing the fraction of variants that continue to possess the minimal stability required to fold (Nikolova et al., 2000; Bloom et al., 2005, 2006). The mechanism by which this occurs is not fully understood, although is thought to involve a combination of raw stability and ‘global suppressor’ residues that buffer the effect of deleterious mutations (Poteete et al., 1997; Steipe, 1999; Nikolova et al., 2000; Tokuriki et al., 2008). In the context of consensus design, raw stability is definitely observed (Supplementary Table S1); however, without extensive mutagenesis studies, it is unclear whether conserved residues infer global suppressor like properties. Given that global suppressor residues appear to be transferrable across protein homologues, such as the case in TEM β-lactamases (Brown et al., 2010), it is reasonable to suggest that conserved residues which happen to be global suppressors will induce similar effects when made in consensus design. Consensus design has been used to enhance the evolvability of a computationally designed Kemp eliminase (KE59) (Khersonsky et al., 2012). Optimisation of activity by directed evolution was initially desired; however, the stability of KE59 was insufficient to tolerate mutations, rapidly producing unfolded proteins, thereby trapping the evolutionary trajectory in a local minimum. To boost KE59's evolvability, conserved residues were spiked into the directed evolution library, thereby improving protein stability and allowing for fresh downhill evolution of function. A similar result was also reported for the directed evolution of a consensus-designed ankyrin repeat protein (DARPin) for binding to HER2 (Zahnd et al., 2007). These studies therefore demonstrate the capacity for consensus design to provide stabilising features for downstream engineering studies.
Engineering the energy landscape
Protein folding and the kinetic stability is an often overlooked property in protein design projects due to many proteins exhibiting irreversible folding on denaturation and the associated complexities of studying multistate folding pathways (Sanchez-Ruiz, 2010). However, thermodynamic stability alone does not guarantee that the protein will fold or remain folded in the native state for extended periods of time under biological or arduous industrial conditions. In vivo, the biological function of many proteins requires a rugged energy landscape, which puts them at risk of misfolding and aggregation (Dinner et al., 2000; Dobson, 2003; Ferreiro et al., 2007b; Sanchez-Ruiz, 2010; Gershenson et al., 2014; Gianni et al., 2014). The delicate balance between function and misfolding is exemplified by members of the serine protease inhibitor or serpin superfamily (Gettins, 2002; Lomas and Carrell, 2002; Law et al., 2006; Krishnan and Gierasch, 2011). Inhibitory members fold to a metastable native state that undergoes a major conformational change in order to inhibit target proteases (Huntington et al., 2000). As such, serpins have evolved a relatively complicated folding mechanism required for their function, with sequence and structural diversity within the superfamily reflecting specialised functional and regulatory requirements. We recently used consensus design to create a synthetic serpin, based on the hypothesis that a serpin molecule reflecting optimal sequence conservation may offer insight into the serpin folding function trade-off (Porebski et al., unpublished). Remarkably, the consensus serpin uniquely exhibits reversible two-state folding, is functional, thermostable and resistant to polymerisation. Structural and biophysical analysis suggests that consensus design remodelled its folding landscape, thereby reducing the lifetime of aggregation-prone intermediates (Porebski et al., unpublished). We also observed similar, though less dramatic effects with FN3con (Porebski et al., 2015), where consensus design led to a large increase in the folding rate and decrease in the unfolding rate (high kinetic stability), suggesting a more smooth and funnel-like energy landscape.
Although consensus design nearly always modifies energy landscapes, improvements to folding and kinetic stability are unlikely to be universal (for example see Main et al., 2003; Sullivan et al., 2011; Parker et al., 2014). Success likely depends on the specific functional requirements and evolutionary history of the MSA as these will dictate the consensus energy landscape. In the case of serpins, off-pathway folding and polymerization is probably the result of independent evolutionary fine-tuning of the energy landscape as a means for conformational control of function. As these independent pathways are not highly conserved in a sequence alignment, they are removed by consensus design, thus remodelling the energy landscape to be smoother and funnel-like, whilst still retaining conserved conformational properties necessary for function (Fig. 2). It is tempting to speculate that consensus design may prove to be a fruitful avenue for investigating and engineering the risky energy landscapes of functional proteins (Barrick et al., 2008; Gershenson et al., 2014).
Fig. 2.
Smoothing of five hypothetical energy landscapes by consensus design. Five protein homologues exhibit differences in their energy landscapes, with three containing kinetic traps that present a propensity for misfolding. As the kinetic traps are not conserved across all five homologues, consensus design is capable of smoothing out the energy landscape to eliminate non-conserved features.
Function
The function of consensus-designed proteins can be preserved and is influenced by the implementation of design (Supplementary Table S1). In general, consensus mutations, especially those that are distal from the catalytic site, give the highest odds of completely preserving function (Polizzi et al., 2006; Risso et al., 2014), whilst full sequence (de novo) designs are likely to reduce catalytic rates and specificity, as can be seen in Supplementary Table S1. Although full sequence designs often reduce catalytic rates and specificity, they tend to retain function at elevated temperatures and wider ranges of pH (Lehmann et al., 2000; Sullivan et al., 2011; Stevens et al., 2016). Full sequence designs likely yield these results in a similar manner to the proposed energy landscape smoothing, with the finer features of catalytic activity being less conserved across all homologues, and are therefore removed during design.
Immunogenicity
The application of consensus design to the reduction of immunogenicity, an important factor in the design of protein therapeutics (Chirino et al., 2004; De Groot and Scott, 2007; Jawa et al., 2013), remains largely unexplored. Interestingly, what appears to be the first consensus-designed protein, alfacon-1 (Infergen), is less immunogenic and significantly more active than recombinant interferon-α (IFN-α) (Alton et al., 1983; Blatt et al., 1996). Created using a sequence alignment of 25 IFN-α subtypes, alfacon-1 is the only known consensus-designed protein that has been marketed as a therapeutic drug. Although alfacon-1 is the only reported experimental study of immunogenicity for a consensus-designed protein, computational predictions using FN3 domains suggest that Tencon is also of low immunogenic potential (Jacobs et al., 2012). However, it is of course possible that the absence of reports may reflect a general failure in immunogenicity reduction by consensus design. Regardless, the possibility that consensus design can reduce immunogenicity warrants further investigation.
Concluding remarks
Consensus design is a proven and highly effective sequence-based method that is typically overlooked in protein engineering in favour of directed evolution and rational design methodologies. Given the challenges in computational modelling of entropy and non-native states, consensus design provides an additional tool for the protein engineer to not only stabilise the native state but also modify the folding landscape.
Author contributions
B.T.P. and A.M.B. wrote the paper.
Supplementary data
Funding
B.T.P. is a Medical Research Council Career Development Fellow. A.M.B. is a National Health and Medical Research Council Senior Research Fellow (1022688). Funding to pay the Open Access publication charges for this article was provided by Monash University.
Supplementary Material
References
- Alton K., Stabinsky Y., Richards R., Ferguson B., Goldstein L., Altrock B., Miller L., Stebbing N. (1983) The Biology of Interferon System. 2nd ed. Amsterdam: Elsevier Science Publishers, pp. 119–128. [Google Scholar]
- Amin N., Liu A.D., Ramer S., Aehle W., Meijer D., Metin M., Wong S., Gualfetti P., Schellenberger V. (2004) Protein Eng., Des. Sel., 17, 787–793. [DOI] [PubMed] [Google Scholar]
- Arnold F.H., Volkov A.A. (1999) Curr. Opin. Chem. Biol., 3, 54–59. [DOI] [PubMed] [Google Scholar]
- Atchley W.R., Wollenberg K.R., Fitch W.M., Terhalle W., Dress A.W. (2000) Mol. Biol. Evol., 17, 164–178. [DOI] [PubMed] [Google Scholar]
- Barrick D., Ferreiro D.U., Komives E.A. (2008) Curr. Opin. Struct. Biol., 18, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett G.J., Porter C.T., Borkakoti N., Thornton J.M. (2002) J. Mol. Biol., 324, 105–121. [DOI] [PubMed] [Google Scholar]
- Bauer F., Schweimer K., Klüver E., Conejo-Garcia J.R., Forssmann W.G., Rösch P., Adermann K., Sticht H. (2001) Protein Sci., 10, 2470–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S., Goldin K., Tawfik D.S. (2008) J. Mol. Biol., 379, 1029–1044. [DOI] [PubMed] [Google Scholar]
- Blatt L.M., Davis J.M., Klein S.B., Taylor M.W. (1996) J. Interferon Cytokine Res., 16, 489–499. [DOI] [PubMed] [Google Scholar]
- Bloom J.D., Glassman M.J. (2009) PLoS Comput. Biol., 5, e1000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J.D., Silberg J.J., Wilke C.O., Drummond D.A., Adami C., Arnold F.H. (2005) Proc. Natl. Acad. Sci., 102, 606–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J.D., Labthavikul S.T., Otey C.R., Arnold F.H. (2006) Proc. Natl. Acad. Sci., 103, 5869–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornscheuer U.T., Huisman G.W., Kazlauskas R.J., Lutz S., Moore J.C., Robins K. (2012) Nature, 485, 185–194. [DOI] [PubMed] [Google Scholar]
- Brown N.G., Pennington J.M., Huang W., Ayvaz T., Palzkill T. (2010) J. Mol. Biol., 404, 832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustad E.M., Arnold F.H. (2011) Curr. Opin. Chem. Biol., 15, 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case B.A., Hackel B.J. (2016) Biotechnol. Bioeng., doi:10.1002/bit.25931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Stites W.E. (2001) Biochemistry, 40, 14012–14019. [DOI] [PubMed] [Google Scholar]
- Chirino A.J., Ary M.L., Marshall S.A. (2004) Drug Discov. Today, 9, 82–90. [DOI] [PubMed] [Google Scholar]
- Dai M., Fisher H.E., Temirov J., Kiss C., Phipps M.E., Pavlik P., Werner J.H., Bradbury A.R.M. (2007) Protein Eng. Des. Sel., 20, 69–79. [DOI] [PubMed] [Google Scholar]
- De Groot A.S., Scott D.W. (2007) Trends Immunol., 28, 482–490. [DOI] [PubMed] [Google Scholar]
- Dinner A.R., Sali A., Smith L.J., Dobson C.M., Karplus M. (2000) Trends Biochem. Sci., 25, 331–339. [DOI] [PubMed] [Google Scholar]
- Dobson C.M. (2003) Nature, 426, 884–890. [DOI] [PubMed] [Google Scholar]
- Durani V., Magliery T.J. (2013) Methods Enzymol., 523, 237–256. [DOI] [PubMed] [Google Scholar]
- Ferreiro D.U., Cervantes C.F., Truhlar S.M.E., Cho S.S., Wolynes P.G., Komives E.A. (2007a) J. Mol. Biol., 365, 1201–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreiro D.U., Hegler J.A., Komives E.A., Wolynes P.G. (2007b) Proc. Natl. Acad. Sci. USA, 104, 19819–19824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Clements J., Eddy S.R. (2011) Nucleic Acids Res., 39, W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Coggill P., Eberhardt R.Y. et al. (2016) Nucleic Acids Res., 44, D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenson A., Gierasch L.M., Pastore A., Radford S.E. (2014) Nat. Chem. Biol., 10, 884–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettins P.G.W. (2002) Chem. Rev., 102, 4751–4804. [DOI] [PubMed] [Google Scholar]
- Gianni S., Camilloni C., Giri R., Toto A., Bonetti D., Morrone A., Sormanni P., Brunori M., Vendruscolo M. (2014) Proc. Natl. Acad. Sci. USA, 111, 14141–14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göbel U., Sander C., Schneider R., Valencia A. (1994) Proteins, 18, 309–317. [DOI] [PubMed] [Google Scholar]
- Horovitz A., Fersht A.R. (1992) J. Mol. Biol., 224, 733–740. [DOI] [PubMed] [Google Scholar]
- Huang Y., Niu B., Gao Y., Fu L., Li W. (2010) Bioinformatics, 26, 680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington J.A., Read R.J., Carrell R.W. (2000) Nature, 407, 923–926. [DOI] [PubMed] [Google Scholar]
- Jäckel C., Bloom J.D., Kast P., Arnold F.H., Hilvert D. (2010) J. Mol. Biol., 399, 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S.A., Diem M.D., Luo J., Teplyakov A., Obmolova G., Malia T., Gilliland G.L., O'Neil K.T. (2012) Protein Eng. Des. Sel., 25, 107–117. [DOI] [PubMed] [Google Scholar]
- Jawa V., Cousens L.P., Awwad M., Wakshull E., Kropshofer H., De Groot A.S. (2013) Clin. Immunol., 149, 534–555. [DOI] [PubMed] [Google Scholar]
- Jiang X., Kowalski J., Kelly J.W. (2001) Protein Sci., 10, 1454–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Althoff E.A., Clemente F.R. et al. (2008) Science, 319, 1387–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joh N.H., Wang T., Bhate M.P., Acharya R., Wu Y., Grabe M., Hong M., Grigoryan G., DeGrado W.F. (2014) Science, 346, 1520–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemena C., Notredame C. (2009) Bioinformatics, 25, 2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky O., Kiss G., Röthlisberger D., Dym O., Albeck S., Houk K.N., Baker D., Tawfik D.S. (2012) Proc. Natl. Acad. Sci. USA, 109, 10358–10363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan B., Gierasch L.M. (2011) Nat. Struct. Mol. Biol., 18, 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law R.H.P., Zhang Q., McGowan S. et al. (2006) Genome Biol., 7, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M., Kostrewa D., Wyss M., Brugger R., D'Arcy A., Pasamontes L., van Loon A.P. (2000) Protein Eng., 13, 49–57. [DOI] [PubMed] [Google Scholar]
- Lehmann M., Loch C., Middendorf A., Studer D., Lassen S.F., Pasamontes L., van Loon A.P.G.M., Wyss M. (2002) Protein Eng., 15, 403–411. [DOI] [PubMed] [Google Scholar]
- Lehmann M., Wyss M. (2001) Curr. Opin. Biotechnol., 12, 371–375. [DOI] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2015) Nucleic Acids Res., 43, D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LiCata V.J., Ackers G.K. (1995) Biochemistry, 34, 3133–3139. [DOI] [PubMed] [Google Scholar]
- Lockless S.W., Ranganathan R. (1999) Science, 286, 295–299. [DOI] [PubMed] [Google Scholar]
- Lomas D.A., Carrell R.W. (2002) Nat. Rev. Genet., 3, 759–768. [DOI] [PubMed] [Google Scholar]
- Luque I., Leavitt S.A., Freire E. (2002) Annu. Rev. Biophys. Biomol. Struct., 31, 235–256. [DOI] [PubMed] [Google Scholar]
- Lutz S. (2010) Curr. Opin. Biotechnol., 21, 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias M.J., Gervais V., Civera C., Oschkinat H. (2000) Nat. Struct. Biol., 7, 375–379. [DOI] [PubMed] [Google Scholar]
- Magliery T.J. (2015) Curr. Opin. Struct. Biol., 33, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliery T.J., Lavinder J.J., Sullivan B.J. (2011) Curr. Opin. Chem. Biol., 15, 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliery T.J., Regan L. (2004) J. Mol. Biol., 343, 731–745. [DOI] [PubMed] [Google Scholar]
- Main E.R.G., Xiong Y., Cocco M.J., D'Andrea L., Regan L. (2003) Structure, 11, 497–508. [DOI] [PubMed] [Google Scholar]
- Matthews B.W. (1993) Annu. Rev. Biochem., 62, 139–160. [DOI] [PubMed] [Google Scholar]
- Morrison D.A. (2015) Syst. Bot., 40, 14–26. [Google Scholar]
- Mosavi L.K., Minor D.L., Peng Z.Y. (2002) Proc. Natl. Acad. Sci., 99, 16029–16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators (2014) Nucleic Acids Res., 42, D7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova P.V., Henckel J., Lane D.P., Fersht A.R. (1998) Proc. Natl. Acad. Sci. USA, 95, 14675–14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova P.V., Wong K.B., DeDecker B., Henckel J., Fersht A.R. (2000) Embo J., 19, 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuin P.A.S., Wang Z., Tillier E.R.M. (2006) BMC Bioinf., 7, 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer H.G., Ray W.C. (2006) Nucleic Acids Res., 34, W133–W136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paatero A., Rosti K., Shkumatov A.V. et al. (2016) Biochemistry, 55, 914–926. [DOI] [PubMed] [Google Scholar]
- Pantoliano M.W., Whitlow M., Wood J.F., Dodd S.W., Hardman K.D., Rollence M.L., Bryan P.N. (1989) Biochemistry, 28, 7205–7213. [DOI] [PubMed] [Google Scholar]
- Parker R., Mercedes-Camacho A., Grove T.Z. (2014) Protein Sci., 23, 790–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmeggiani F., Pellarin R., Larsen A.P., Varadamsetty G., Stumpp M.T., Zerbe O., Caflisch A., Plückthun A. (2008) J. Mol. Biol., 376, 1282–1304. [DOI] [PubMed] [Google Scholar]
- Patrick W.M., Firth A.E. (2005) Biomol. Eng., 22, 105–112. [DOI] [PubMed] [Google Scholar]
- Pauling L., Zuckerkandl E., Henriksen T., Lövstad R. (1963) Acta Chem. Scand., 17(suppl.), 9–16. [Google Scholar]
- Pearson W.R. (2013) Curr Protoc Bioinformatics, 42, 3.1:3.1.1–3.1.8. [Google Scholar]
- Polizzi K.M., Chaparro-Riggers J.F., Vazquez-Figueroa E., Bommarius A.S. (2006) Biotechnol. J., 1, 531–536. [DOI] [PubMed] [Google Scholar]
- Porebski B.T., Nickson A.A., Hoke D.E., Hunter M.R., Zhu L., McGowan S., Webb G.I., Buckle A.M. (2015) Protein Eng. Des. Sel., 28, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete A.R., Rennell D., Bouvier S.E., Hardy L.W. (1997) Protein Sci., 6, 2418–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso V.A., Gavira J.A., Gaucher E.A., Sanchez-Ruiz J.M. (2014) Proteins, 82, 887–896. [DOI] [PubMed] [Google Scholar]
- Rost B. (1999) Protein Eng., 12, 85–94. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ruiz J.M. (2010) Biophys. Chem., 148, 1–15. [DOI] [PubMed] [Google Scholar]
- Schreiber G., Buckle A.M., Fersht A.R. (1994) Structure, 2, 945–951. [DOI] [PubMed] [Google Scholar]
- Shoichet B.K., Baase W.A., Kuroki R., Matthews B.W. (1995) Proc. Natl. Acad. Sci., 92, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C.J.A., Cerutti L., Hulo N., Gattiker A., Falquet L., Pagni M., Bairoch A., Bucher P. (2002) Briefings Bioinf., 3, 265–274. [DOI] [PubMed] [Google Scholar]
- Sigrist C.J.A., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., Bougueleret L., Xenarios I. (2013) Nucleic Acids Res., 41, D344–D347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolich M., Lockless S.W., Russ W.P., Lee H., Gardner K.H., Ranganathan R. (2005) Nature, 437, 512–518. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L., Eddy S.R., Birney E., Bateman A., Durbin R. (1998) Nucleic Acids Res., 26, 320–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steipe B. (1999) Curr. Top. Microbiol. Immunol., 243, 55–86. [DOI] [PubMed] [Google Scholar]
- Steipe B., Schiller B., Plückthun A., Steinbacher S. (1994) J. Mol. Biol., 240, 188–192. [DOI] [PubMed] [Google Scholar]
- Stevens A.J., Brown Z.Z., Shah N.H., Sekar G., Cowburn D., Muir T.W. (2016) J. Am. Chem. Soc., 138, 2162–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.J., Durani V., Magliery T.J. (2011) J. Mol. Biol., 413, 195–208. [DOI] [PubMed] [Google Scholar]
- Sullivan B.J., Nguyen T., Durani V., Mathur D., Rojas S., Thomas M., Syu T., Magliery T.J. (2012) J. Mol. Biol., 420, 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera D., Lovell S.C., Whelan S. (2015) Mol. Biol. Evol., 32, 2456–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton J.W. (2004) Nat. Rev. Genet., 5, 366–375. [DOI] [PubMed] [Google Scholar]
- Thornton J.W., Need E., Crews D. (2003) Science, 301, 1714–1717. [DOI] [PubMed] [Google Scholar]
- Tokuriki N., Stricher F., Schymkowitz J., Serrano L., Tawfik D.S. (2007) J. Mol. Biol., 369, 1318–1332. [DOI] [PubMed] [Google Scholar]
- Tokuriki N., Stricher F., Serrano L., Tawfik D.S. (2008) PLoS Comput. Biol., 4, e1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N., Tawfik D.S. (2009a) Curr. Opin. Struct. Biol., 19, 596–604. [DOI] [PubMed] [Google Scholar]
- Tokuriki N., Tawfik D.S. (2009b) Science, 324, 203–207. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium (2008) Nucleic Acids Res., 36, D190–D195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Figueroa E., Yeh V., Broering J.M., Chaparro-Riggers J.F., Bommarius A.S. (2008) Protein Eng., Des. Sel., 21, 673–680. [DOI] [PubMed] [Google Scholar]
- Wang Q., Buckle A.M., Foster N.W., Johnson C.M., Fersht A.R. (1999) Protein Sci., 8, 2186–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijma H.J., Floor R.J., Janssen D.B. (2013) Curr. Opin. Struct. Biol., 23, 588–594. [DOI] [PubMed] [Google Scholar]
- Wijma H.J., Floor R.J., Jekel P.A., Baker D., Marrink S.J., Janssen D.B. (2014) Protein Eng. Des. Sel., 27, 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D., Pethica R., Zhou Y., Talbot C., Vogel C., Madera M., Chothia C., Gough J. (2009) Nucleic Acids Res., 37, D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz P., Steipe B. (1999) Protein Sci., 8, 2245–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfson D.N., Bartlett G.J., Burton A.J., Heal J.W., Niitsu A., Thomson A.R., Wood C.W. (2015) Curr. Opin. Struct. Biol., 33, 16–26. [DOI] [PubMed] [Google Scholar]
- Yang Z., Kumar S., Nei M. (1995) Genetics, 141, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahnd C., Wyler E., Schwenk J.M. et al. (2007) J. Mol. Biol., 369, 1015–1028. [DOI] [PubMed] [Google Scholar]
- Zvelebil M.J., Barton G.J., Taylor W.R., Sternberg M.J. (1987) J. Mol. Biol., 195, 957–961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.