Abstract
In the brain, CB1 cannabinoid receptors primarily mediate the effects of cannabinoids, but CB2 cannabinoid receptors (CB2Rs) have recently been discovered in the nervous system and also implicated in neuromodulatory roles. To understand the mechanisms of CB2R functions in the brain, it is essential to localize CB2Rs, but the types of cells expressing CB2Rs have been controversial. Unequivocal localization of CB2Rs in the brain has been impeded in part by the low expression levels of CB2Rs and poor specificity of detection methods. Here, we used an ultrasensitive and specific in situ hybridization method called the RNAscope to determine the spatial pattern of CB2R mRNA expression in the mouse hippocampus. CB2R mRNAs were mostly expressed in a subset of excitatory and inhibitory neurons in the CA1, CA3 and dentate gyrus areas, but rarely in microglia. CB2R knock-out mice were used as a negative control. Using the quantitative real-time polymerase chain reaction, we also found that the temporal pattern of CB2R mRNA expression was stable during postnatal development. Consistent with previous reports, the immunological detection of CB2Rs was not reliable, implying extremely low levels of the protein expression and/or insufficient specificity of the current anti-CB2R antibodies. Our findings of the expression patterns of CB2R mRNAs may help determine the cell types involved in, and hence the mechanisms of, the CB2R-mediated neuromodulation.
Keywords: cannabinoid, in situ hybridization, RNAscope, confocal microscopy, immunohistochemistry, CB2 cannabinoid receptor
INTRODUCTION
Neuropsychiatric effects of cannabinoids are primarily mediated by CB1 cannabinoid receptors (CB1Rs) in the nervous system. Although early studies have reported that CB1Rs are expressed in the brain and CB2 cannabinoid receptors (CB2Rs) are in the immune system (Devane et al., 1988; Matsuda et al., 1990; Munro et al., 1993), recent evidence has indicated that CB2Rs might also be present in the nervous system (for review, Atwood and Mackie, 2010). However, the cellular location of CB2Rs in the brain is still unclear. It is physiologically important to determine the distribution and function of CB2Rs because major endocannabinoids and Δ9-tetrahydrocannabinol (the main psychoactive ingredient of marijuana) activate both CB1Rs and CB2Rs (Mechoulam et al., 1995; Showalter et al., 1996; Sugiura et al., 2000).
CB2Rs in the brain are expressed in activated microglia whereas their expression is low or undetectable in quiescent microglia (Carlisle et al., 2002; Walter et al., 2003; Zhang et al., 2003; Maresz et al., 2005). In contrast to the microglial expression of CB2Rs, the expression of CB2Rs in neurons is still controversial. Immunohistochemical studies have shown that CB2Rs are expressed in neurons in the hippocampus (Gong et al., 2006; Onaivi, 2006; Brusco et al., 2008; Onaivi et al., 2008). However, conflicting results have been reported concerning the location of hippocampal CB2Rs; e.g., the area of the highest CB2R immunoreactivity is variable—stratum pyramidale (Gong et al., 2006) or stratum radiatum (Brusco et al., 2008)— depending on the source of antibody. Furthermore, lack of negative controls using CB2R knockout (KO) mice in these studies failed to settle the controversies surrounding the presence of CB2Rs in neurons. Contradictory results have also been reported regarding whether CB2Rs are expressed in the brain stem (Derbenev et al., 2004; Van Sickle et al., 2005; Baek et al., 2008). In fact, skepticism about the specificity of many anti-CB2R antibodies has been expressed, demonstrating the current limitation in immunological detection of CB2Rs (Ashton, 2012; Baek et al., 2013; Cecyre et al., 2014; Marchalant et al., 2014).
In situ hybridization studies have shown that CB2R mRNAs are expressed in neurons in the cerebellum (Skaper et al., 1996), globus pallidus, cerebral cortex, hippocampus (Lanciego et al., 2011; Sierra et al., 2014), ventral tegmental area (Zhang et al., 2014a), nucleus accumbens, and dorsal striatum (Zhang et al., 2015) of rodents or macaque. Negative control experiments with KO mice (Zhang et al., 2014a) or sense probes (Skaper et al., 1996; Lanciego et al., 2011; Zhang et al., 2014a) have strengthened the evidence for neuronal expression of CB2R mRNAs. However, the pattern of CB2R expression, especially in the hippocampus, remains elusive. For instance, CB2R-expressing cells are widely scattered throughout the hippocampus but the types of these cells have not been identified (Lanciego et al., 2011). Whereas most pallidal neurons express CB2R mRNAs (Lanciego et al., 2011), the proportion of hippocampal neurons that express CB2R mRNAs is unclear.
CB2Rs modulate neuronal functions, such as anxiety (Garcia-Gutierrez et al., 2012), memory (Garcia-Gutierrez et al., 2013) and pain (Anand et al., 2009). Chronic activation of CB2Rs increases excitatory synaptic transmission (Kim and Li, 2015), whereas acute activation of CB2Rs reduces inhibitory synaptic transmission (Morgan et al., 2009) and neuronal excitability (Patel et al., 2003; Sagar et al., 2005; den Boon et al., 2012; Zhang et al., 2014a). However, the cellular mechanisms of CB2R functions are unknown. For investigation of the mechanisms, it is important to determine the cellular location of CB2Rs in the brain because it may suggest the signaling pathways of CB2Rs. For example, if hippocampal CB2Rs are expressed only in interneurons as suggested (Lanciego et al., 2011), the effects of CB2Rs on pyramidal cells (Kim and Li, 2015) might occur via intercellular signaling mechanisms, but this critical matter is yet to be identified.
Here, we investigated the spatial and temporal patterns of CB2R mRNA expression in the hippocampus using an ultrasensitive in situ hybridization method called the RNAscope (Wang et al., 2012) and quantitative PCR (qPCR) with both wild type (WT) and CB2R KO mice.
EXPERIMENTAL PROCEDURES
Animals
C57BL/6J, CB2R WT and CB2R KO mice (The Jackson Laboratory) of either sex were used. CB2R KO mice (Jackson stock number 005786) were originally created by Deltagen on the background of C57BL/6J mice. We crossed homozygous KO mice (CB2R−/−) with C57BL/6J mice to obtain heterozygous CB2R+/− mice, and then bred the CB2R+/− mice with each other to generate littermates of CB2R+/+ (i.e., WT) and CB2R−/−. Animals were anesthetized with isoflurane vapor and decapitated. For RNAscope and immunohistochemistry experiments, anesthetized mice were fixed before decapitation by cardiac perfusion with 10% neutral buffered formalin (Thermo Scientific). All experiments were conducted in accordance with the animal use protocol that was approved by the Institutional Animal Care and Use Committee of Georgia Regents University.
Polymerase chain reaction (PCR)
The hippocampi were isolated from C57BL/6J mice at age 1–22 weeks. The hippocampi from 20-week-old CB2R KO mice were also used as a negative control and the spleens, which express abundant CB2Rs, from 2-week-old C57BL/6J mice were used as a positive control. Except for 1-week-old mice, the blood of anesthetized animals was removed by cardiac perfusion with ice-cold phosphate-buffered saline (PBS). After decapitation, the hippocampi were dissected out and immediately transferred to PBS. mRNAs were isolated with the RNeasy Mini kit (Qiagen) and treated with RNase-Free DNase (Qiagen), and the purity was verified with NanoDrop 2000 Spectrophotometer (Thermo Scientific). Single strand cDNA was synthesized from the extracted RNA (2.5 μg) with SuperScript III First-Strand Synthesis System (Invitrogen) and oligo-dT, and then used for PCR.
The reverse transcription PCR (RT-PCR) was performed with the synthesized cDNA (1 μ l) and three pairs of primers for Cnr2 (the gene for CB2Rs). The forward primers for the primer pairs 1, 2 and 3 were AGGACAAGGCTCCACAAGAC, GCACCCATGTGACTTGCAGA and ACAGAAGTGACCAACGGCTC, respectively. The backward primer, ATAGGTAGCGGTCAACAGCG, was common for the three pairs. The predicted sizes of products from the primer pairs 1, 2 and 3 were 494, 679 and 382 bp, respectively. Primers for Gapdh were also used as a loading control (forward, TGACCACAGTCCATGCCATC; backward, GGATAGGGCCTCTCTTGCTC; product, 539 bp). The PCR products were visualized in 1.5% agarose gels with ethidium bromide staining.
The qPCR was conducted with the cDNA and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) in triplicate using the Real-Time PCR Detection System (Bio-Rad). The primer pair for Cnr2 was GGGTCGACTCCAACGCTATC (forward) and AGGTAGGCGGGTAACACAGA (backward; product, 126 bp). Primers for Gapdh were used as an internal control (forward, CCGCATCTTCTTGTGCAGTG; backward, ATGAAGGGGTCGTTGATGGC; product, 149 bp). Each experiment included a template-free control. The PCR products were analyzed by the DNA melting curve. The relative quantities of Cnr2 PCR products were estimated with respect to the amount of Gapdh product using the ΔCt method: %GAPDH = (2Ct of Gapdh – Ct of Cnr2) × 100.
We genotyped mice using the REDExtract-N-Amp Tissue PCR Kit (Sigma-Aldrich) and the following primers: GGGGATCGATCCGTCCTGTAAGTCT, GACTAGAGCTTTGTAGGTAGGCGGG and GGAGTTCAACCCCATGAAGGAGTAC.
RNAscope in situ hybridization and co-immunohistochemistry
CB2R WT and KO mice at age 4 months were used. Formalin-perfused, isolated hippocampi were further stored in 10% neutral buffered formalin for 16–32 h at 23°C. In each session of RNAscope hybridization, slices from CB2R WT and KO mice were processed simultaneously. Fixed hippocampi were dehydrated in ethanol, embedded in paraffin and cut into 5-μm-thick slices. The slices on Superfrost Plus slides (Fisher Scientific) were baked in a dry oven for 1 h at 60°C and then deparaffinized with xylene (twice, 5 min each) and ethanol (twice, 1 min each). RNAscope hybridization was performed with the RNAscope 2.0 HD Detection Kit (Advanced Cell Diagnosis). The tissue sections were treated with pretreatment solutions of proprietary compositions (Advanced Cell Diagnosis): Pretreatment Solution #1 for 10 min at 23°C, Pretreatment Solution #2 for 15 min at 100–104°C, and Pretreatment Solution #3 (protease digestion) for 15 min at 40°C. The slices were incubated with a custom synthesized CB2R RNAscope probe (Advanced Cell Diagnostics) for 2 h at 40°C, followed by amplifying hybridization processes. The CB2R RNAscope probe targeted the base pairs 338–672 of the CB2R mRNA NM009924.3. The sliceswere stainedwith 4',6-diamidino-2-phenylindole (DAPI) (1 μg/ml in PBS) for 10 min at 23°C and then mounted with the EcoMount mounting medium (Biocare Medical).
A subset of slices were co-immunostained with one of the primary antibodies against neurogranin (Millipore, AB5620), GAD67 (Millipore, MAB5406) or Iba1 (Wako, 019-19741). Between the RNAscope hybridization and DAPI staining, slices were washed with PBS containing 0.1% Triton X-100 (referred to as PBST) (twice, 5 min each) and then blocked by 4% normal goat serum in PBST for 1 h at 23°C. The primary antibodies were diluted at 1:300 in PBST containing 4% normal goat serum. After incubation with a primary antibody (15–17 h at 4°C), the slices were washed with PBST and incubated with a secondary antibody—goat anti-rabbit or anti-mouse IgG conjugated with DyLight488 (Thermo Scientific)—for 2 h at 23°C. The secondary antibodies were diluted at 1:300 in PBST. After being washed with PBST, the slices were mounted with ProLong Diamond Antifade Mountant with DAPI (Life Technologies).
Western blotting and immunohistochemistry with anti-CB2R antibodies
For Western blotting, the hippocampi were isolated from C57BL/6J and CB2R KO mice aged 4 months and then transferred to an ice-cold radioimmunoprecipitation assay buffer (Thermo Scientific), which was supplemented with a protease inhibitor cocktail (Sigma–Aldrich) and a phosphatase inhibitor cocktail (Thermo Scientific). The tissue was homogenized by sonication with two 10-s pulses (one pulse every min) at 4 °C. The homogenates were centrifuged at 10,000×g for 10 min at 4 °C. The supernatants were collected and incubated in a sample buffer (200 mM Tris at pH 6.8, 5% sodium dodecyl sulfate (SDS), 0.1 mM EDTA, 8 M urea, 100 mM dithiothreitol, 0.1% bromophenol blue and 30% glycerol) for 10 min at 37 °C. Protein samples were run on a 10% polyacrylamide gel (Bio-Rad) and then transferred onto polyvinylidene difluoride membranes (Thermo Scientific). After a brief wash with tris-buffered saline containing 0.1% Tween 20 (referred to as TBST), the membranes were blocked for 1 h at 23 °C in a TBST blocking solution (TBST + 5% bovine serum albumin), and then incubated with an anti-CB2R antibody (1:1000) for 15–17 h at 4 °C. The anti-CB2R antibodies were obtained from Alpha Diagnostic International (CB22A), Thermo Scientific (PA1746A), Santa Cruz Biotechnology (sc10076 and sc25494), Abcam (ab45942 and ab3651), Alomone Labs (ACR003), Cayman Chemical (101550) and R&D Systems (MAB3655). An anti-CB1R antibody was from Cayman (101500). The membranes were incubated with horseradish peroxide-conjugated anti-IgG (1:10,000; Bio-Rad) for 1 h at 23 °C. Bands were visualized with a chemiluminescence detection solution (Thermo Scientific). The membranes were stripped in a stripping buffer (three times, 5 min each; 23 °C), which contained 50 mM glycine and 0.1% SDS (pH 2.0–2.5 with HCl), and then re-probed with an antibody against β-tubulin (1:10,000; Thermo Scientific) for 1 h at 23 °C.
For immunohistochemistry with anti-CB2R antibodies, 5-μm thick hippocampal slices were prepared as described earlier for the RNAscope method. Deparaffinized sections were treated with ethanol (100%, 95%, and then 80%) and hydrated with deionized water. Sections were boiled (95–100°C) in sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) for 20 min and then cooled for 20 min. After blocking with 4% normal serum (goat or rabbit; Thermo Scientific) in PBST for 1 h at 23°C, the sections were incubated with one of the following anti-CB2R antibodies (1:300) for 15–17 h at 4°C: Alpha Diagnostic International CB22A (host, rat), Cayman 101550 (rabbit), Abcam ab3651 (rabbit), and Santa Cruz sc10076 (goat). The following secondary antibodies (1:300) were used for 2 h at 23°C: goat anti-rat IgG, goat anti-rabbit IgG, or rabbit anti-goat IgG conjugated with DyLight488 (Thermo Scientific). The slices were mounted with ProLong Diamond Antifade Mountant with DAPI (Life Technologies).
Confocal imaging
Fluorescent signals of RNAscope hybridization and immunohistochemistry were imaged with a 40× objective lens on a laser-scanning confocal microscope (Zeiss LSM700 or LSM780). In each imaging session, we imaged both CB2R WT and KO slices that were processed simultaneously in the same experiment of hybridization or immunostaining. The microscope settings (e.g., gain, offset and resolution) were fixed in a given imaging session. We analyzed images in the ImageJ software (NIH, Bethesda, MD) using the maximum projection of all optical sections.
Hippocampal slice culture
The hippocampi were isolated from 1-week-old C57BL/6J mice and sliced at 400 μm thickness with a vibrating slicer (Leica VT1200S) in ice-cold slicing saline, which consisted of 113 mM NaCl, 2.5 mM KCl, 40 mM NaHCO3, 1 mM NaH2PO4, 0.5 mM CaCl2, 5 mM MgSO4, 10 mM glucose and 0.5 mM ascorbic acid (300 mOsm; bubbled with 95% O2 and 5% CO2). The slices were placed on culture membranes (Millipore) at the interface of the culture medium and air (with 5% CO2) at 37°C. The medium was exchanged every 2–3 d and composed of 50% Minimum Essential Media (Life Technologies, 11090-081), 25% Hank’s Balanced Salt Solution (Life Technologies, 24020-117), 25% horse serum (HyClone), 2 mM L-glutamine, 10 mM HEPES and additional 5 mM glucose. After being cultured for 20 days, slices (8 slices per mouse) were transferred to PBS for mRNA extraction.
Statistics
Comparisons among multiple groups were made with one-way ANOVA. If ANOVA revealed a significant difference (P < 0.05) among the groups, pairwise comparisons (Bonferroni t-tests) were designed to be performed with a two-tailed confidence level of P < 0.05. Comparisons between two groups were made with Student’s t-tests with a two-tailed confidence level of P < 0.05.
RESULTS
The amount of hippocampal CB2R mRNAs is stable during postnatal development
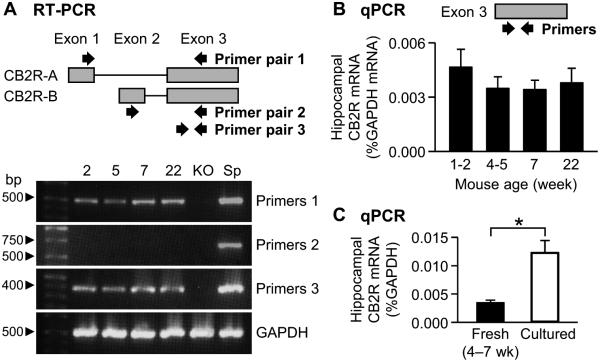
We first examined whether CB2R mRNAs are expressed in the mouse hippocampus using RT-PCR. Two pairs of primers were designed to detect two splicing variants, which have been named CB2R-A (NM_009924.3) and CB2R-B (XM_006538515.1) (Liu et al., 2009) (Fig. 1A). A third primer pair targeted the exon that is common for both transcripts (Fig. 1A). The RT-PCR results indicated that CB2R mRNAs were expressed in the hippocampus of mice at 1–22 weeks of age (Fig. 1A). The data also showed that the mRNA levels of CB2R-B in the hippocampus were almost undetectable (Fig. 1A), in accord with the results from other brain areas such as the prefrontal cortex, striatum and brain stem (Liu et al., 2009).
Figure 1.
Quantification of CB2R mRNAs in the mouse hippocampus. A. RT-PCR with mRNAs extracted from the hippocampi of C57BL/6J and CB2R KO mice. A schematic diagram of the structure of mouse CB2R genome is illustrated on top. Approximate locations of the primers used for RT-PCR are indicated by arrows. Ages of C57BL/6J mice (2–22 weeks) were indicated above the images. KO, the hippocampus of CB2R KO mice. Sp, the spleen of C57BL/6J mice. B. qPCR with mRNAs from the hippocampus of C57BL/6J mice. The primers for qPCR targeted the exon 3 of Cnr2. The amount of CB2R mRNA was normalized to that of GAPDH mRNA. C. qPCR with mRNAs extracted from cultured hippocampal slices of C57BL/6J mice or freshly fixed hippocampus of C57BL/6J mice. The data of fresh tissue were from Figure 1B (age 4–7 weeks). *, P = 0.00003, t-test. Error bars indicate SEM.
We used qPCR to quantify the relative amount of CB2R mRNAs at various ages. The primers were designed to detect the common axon (i.e., exon 3). Levels of CB2R mRNAs in the hippocampus of 1- to 22-week-old mice varied within the range from 0.0034 ± 0.0005% (n = 3 mice at age 7 weeks) to 0.0047 ± 0.0010% (n = 10 mice at age 1–2 weeks) of the amount of GAPDH mRNA (Fig. 1B). The CB2R mRNA amounts at age 4–5 and 22 weeks were 0.0035 ± 0.0006% (n = 8) and 0.0038 ± 0.0008% (n = 3) of the GAPDH mRNA amount, respectively. There was no significant difference among the mRNA levels within 1–22 weeks of age (P = 0.73, ANOVA). CB2R mRNAs in the hippocampus of CB2R KO mice were also quantified with the same primers, but no signal was detected in the qPCR assay (data not shown). This result suggests that Cnr2 in the mouse hippocampus is expressed without significant temporal variation from age 1 week to adulthood.
Cultured or ex vivo systems have been used for functional assays of CB2Rs (Skaper et al., 1996; Zhang et al., 2014a; Kim and Li, 2015), but it is unknown whether the CB2R expression levels in these systems recapitulate those in fresh tissue. For example, culture processes activate microglia (Becher and Antel, 1996) and thus induce their CB2R expression (Carlisle et al., 2002). We examined changes in CB2R mRNA levels in slice cultures of the mouse hippocampus using qPCR. The amount of CB2R mRNAs in cultured slices was 0.012 ± 0.002% of the amount of GAPDH mRNAs (n = 3 mice; Fig. 1C). This value was 3.5 times higher than the CB2R mRNA levels in the freshly isolated hippocampus of 4- to 7-week-old adolescent mice (0.0035 ± 0.0005% of GAPDH mRNA; n = 11 from Fig. 1B; P = 0.00003, t-test). This result raises a possibility that the expression and/or function of CB2Rs could be overestimated in cultured systems.
RNAscope in situ hybridization specifically detects CB2R mRNAs in the hippocampus
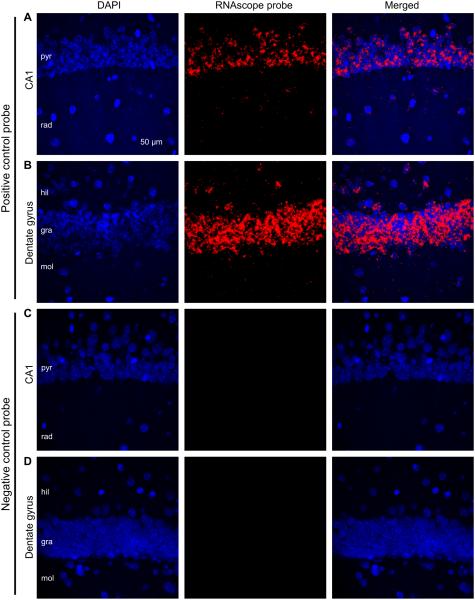
Next, we examined the spatial pattern of CB2R mRNA distribution in the mouse hippocampus. The traditional in situ hybridization method generated both negative (Munro et al., 1993; Howlett et al., 2002) and positive (Lanciego et al., 2011) results about hippocampal CB2R mRNAs. Therefore, we used an ultrasensitive in situ hybridization method called the RNAscope (Wang et al., 2012). As a first step, we needed to validate our method of RNAscope hybridization using control probes. Positive and negative control probes were designed to detect the mouse housekeeping gene Ppib and the bacterial gene DapB, respectively (Wang et al., 2012). The positive control probe produced strong fluorescent signals in the hippocampus of CB2R WT mice (Fig. 2A–B), whereas the negative control probe generated no fluorescence (Fig. 2C–D). The positive control signals overlapped or were in close proximity to DAPI staining, implying that the Ppib mRNAs are present in the nuclei and cytoplasm, which are the locations of transcription and translation, respectively. This result is consistent with the previous report (Wang et al., 2012), in which the same probes were used, and thus suggests that our hybridization was successfully carried out.
Figure 2.
Validation of the RNAscope method using a positive and a negative control probes. A–B. In situ hybridization was performed with an RNAscope probe targeting the eukaryotic housekeeping gene Ppib (i.e., positive control). The hippocampal section was made from a CB2R WT mouse and the images were taken from the CA1 region (A) and dentate gyrus (B). C– D. The RNAscope hybridization was conducted with a negative control probe targeting the bacterial gene DapB. The CA1 region (C) and dentate gyrus (D) were imaged from a hippocampal slice cut from a WT mouse. pyr, stratum pyramidale; rad, stratum radiatum; mol, stratum moleculare; gra, stratum granulosum; hil, hilus.
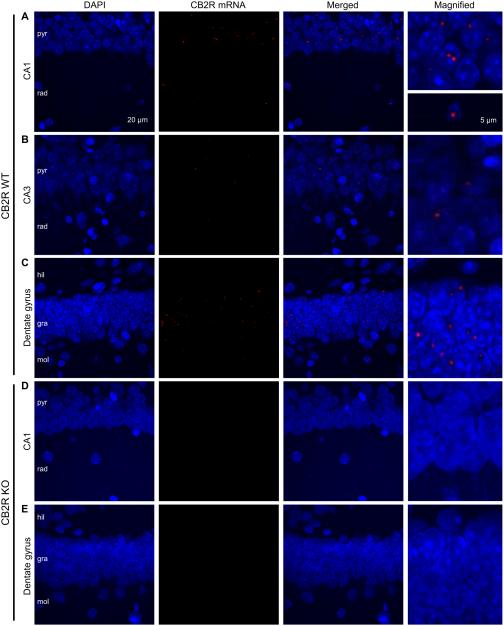
We then examined the distribution of CB2R mRNAs using a custom made RNAscope probe targeting the sequences deleted in the CB2R KO mice that we used—base numbers 122– 456 in the common axon 3. CB2R mRNAs were observed in strata pyramidale and radiatum in the CA1 and CA3 regions, and strata granulosum and moleculare and hilus in the dentate gyrus (Fig. 3A–C). Similar hybridization patterns were observed in a total of 11 WT slices. Because the expression pattern of CB2R mRNAs in the CA3 area (Fig. 3B) resembled that in the CA1 region, further experiments were focused on the CA1 area and dentate gyrus. The punctate pattern of fluorescence in the RNAscope hybridization is typically observed for mRNAs of low expression levels (Wang et al., 2012), and therefore, our data suggest that the expression level of CB2R mRNAs might be low. The fluorescent puncta of CB2R mRNAs were localized near or together with DAPI staining, implying that most CB2R mRNAs were located in the somata, i.e., at the location of either transcription or translation, as observed with typical mRNAs in other in situ hybridization experiments including the RNAscope (Wang et al., 2012; McIsaac et al., 2013). It appeared that CB2R mRNAs were expressed only in a subset of cells because only part of DAPI signals were associated with CB2R mRNAs (Fig. 3A–C). In a total of 11 slices from CB2R KO mice, the CB2R RNAscope probe generated little fluorescent signal (Fig. 3D–E), suggesting that our detection method was specific for CB2R mRNAs.
Figure 3.
RNAscope hybridization demonstrates the expression of CB2R mRNAs in a subset of hippocampal cells. A–C. RNAscope hybridization was performed with a probe that targets the deleted sequence in CB2R KO mice. Hippocampal slices were made from CB2R WT mice. Images were taken from the CA1 (A), CA3 (B) and dentate gyrus (C) regions. Right column, magnified views of the arrow heads shown in the merged images. D–E. RNAscope hybridization in hippocampal sections made from CB2R KO mice. Images from the CA1 region (D) and dentate gyrus (E) show no signal of CB2R mRNAs. The areas indicated by arrow heads are magnified in the right column. pyr, stratum pyramidale; rad, stratum radiatum; mol, stratum moleculare; gra, stratum granulosum; hil, hilus.
CB2R mRNAs are expressed in glutamatergic neurons
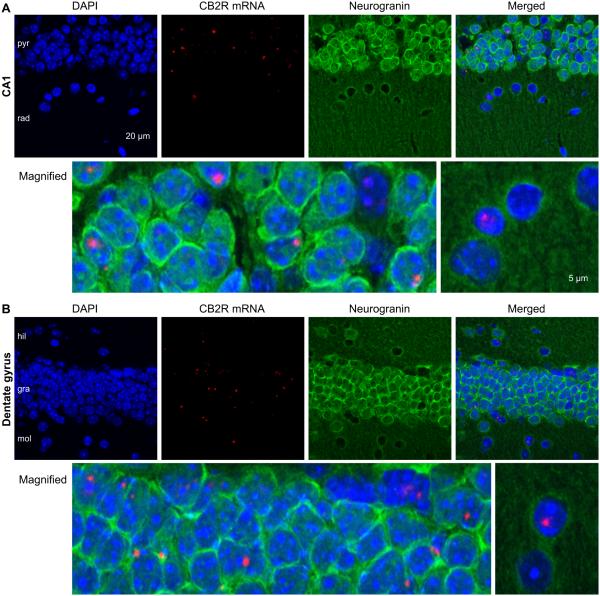
Because CB2R mRNAs were located in multiple layers of the hippocampus (Fig. 3), the types of cells expressing CB2R mRNAs needed to be determined by co-immunohistochemistry with an antibody against a cell marker. We first used an anti-neurogranin antibody to label glutamatergic neurons along with the RNAscope in situ hybridization. In CA1 stratum pyramidale, most cell bodies were strongly labeled by an anti-neurogranin antibody and CB2R mRNAs were localized in neurogranin-expressing cells (Fig. 4A), suggesting that CB2R mRNAs were expressed in pyramidal neurons. In CA1 stratum radiatum, CB2R mRNAs were also observed in neurogranin-negative cells (Fig. 4A), indicating that non-pyramidal cells, perhaps interneurons and/or glia, might also express CB2R mRNAs. In the dentate gyrus, CB2R mRNAs were expressed in both neurogranin-positive granule cells and neurogranin-negative cells in hilus and stratum moleculare (Fig. 4B).
Figure 4.
Expression of CB2R mRNAs in a subset of neurogranin-positive, excitatory neurons. A. A hippocampal section made from a CB2R WT mouse was hybridized with the CB2R RNAscope probe and co-immunostained against neurogranin, a marker for excitatory neurons. Bottom row, magnified views of the areas indicated by arrow heads in the merged image. B. Images of the dentate gyrus in a section made from a CB2R WT mouse. The experiments were similar to those in A.
It was not possible to distinguish individual cells only with DAPI staining (Fig. 3) because the staining did not reveal clear margins of nuclei in strata pyramidale and granulosum. However, the immunostaining against neurogranin enabled us to discern and thus count individual cells (Fig. 4). We counted the cells in strata pyramidale and granulosum based on the DAPI signal surrounded by neurogranin and also the cells in other layers based on the DAPI signal alone. In the CA1 area, 16% of neurogranin-positive cells (52/319 cells in 6 slices) and 19% of neurogranin-negative cells (27/144 cells in 6 slices) expressed CB2R mRNAs. In the dentate gyrus, 15% of neurogranin-positive cells (89/579 cells in 6 slices) and 23% of neurogranin-negative cells (45/195 cells in 6 slices) expressed CB2R mRNAs. These results demonstrate that approximately 20% of cells—either glutamatergic or non-glutamatergic—in the CA1 area and dentate gyrus expressed CB2R mRNAs.
CB2R mRNAs are also expressed in interneurons
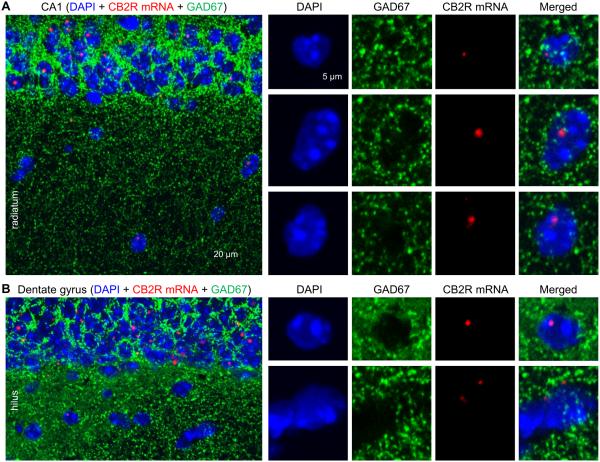
Glutamatergic neurons, GABAergic neurons and microglia are the cell types known to express CB2Rs in the brain (for reviews, Stella, 2004; Atwood and Mackie, 2010). First, to determine whether CB2R-positive cells in CA1 stratum radiatum are interneurons, we immunostained hippocampal slices against GAD67 and also hybridized with the RNAscope probe. GAD67 was detected in the strata pyramidale and radiatum (Fig. 5A), as expected from its known localization in somata and axon terminals (Pinal and Tobin, 1998; Chen et al., 2011). In CA1 stratum radiatum, the DAPI signals that were positive for or close to (≤5 μm) CB2R mRNAs overlapped or were surrounded by GAD67 immunoreactivity (Fig. 5A). In the stratum moleculare and hilus in the dentate gyrus, the expression pattern was also similar; the DAPI signals associated with CB2R mRNAs were colocalized with GAD67 (Fig. 5B). These results, consistent in a total of 7 slices, suggest that the CB2R-expressing non-glutamatergic cells in the CA1 area and dentate gyrus are likely to be GABAergic interneurons.
Figure 5.
Co-localization of CB2R mRNAs with GAD67. A. A hippocampal slice made from a CB2R WT mouse was hybridized with the CB2R RNAscope probe and co-immunostained against GAD67, a marker for inhibitory neurons. The cells indicated by arrow heads in stratum radiatum were magnified in the right four columns. B. Experiments were performed with a CB2R WT section as done in A, but the images were taken from the dentate gyrus.
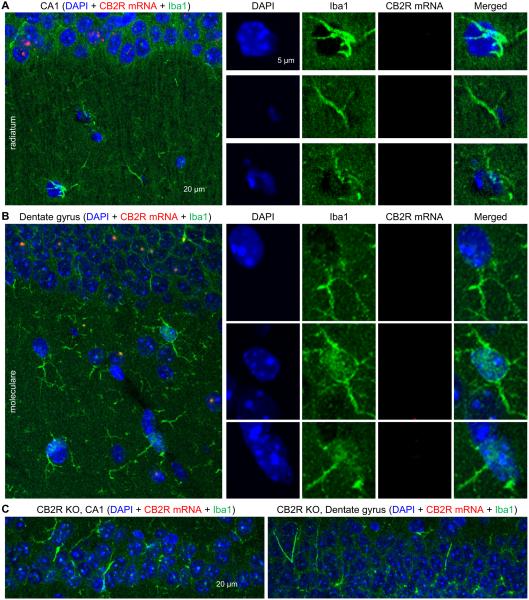
We examined whether CB2R mRNAs were also expressed in microglia by co-immunostaining against Iba1. Immunoreactivity of Iba1 was observed in the stratum radiatum in the CA1 area and stratum moleculare and hilus in the dentate gyrus, but these Iba1-expressing cells lacked CB2R mRNAs when determined by the RNAscope (Fig. 6A–B). The similar patterns of staining were observed in 8 slices. This result indicates that CB2R mRNAs might not be expressed in microglia in the hippocampus or, if expressed, their levels could be extremely low and thus below the detection threshold of our assay. In CB2R KO mice, CB2R mRNAs were not detected when hippocampal slices were co-immunostained against Iba1 (Fig. 6C), indicating that the specificity of the RNAscope hybridization was still maintained when the sections were processed for both hybridization and immunohistochemistry. It should be emphasized here that we deliberately made hippocampal sections from the normal (i.e., healthy) brain because the expression of microglial CB2Rs can be induced under pathological conditions but is very low in resting tissues (Carlisle et al., 2002; Walter et al., 2003; Zhang et al., 2003; Maresz et al., 2005).
Figure 6.
Lack of CB2R mRNAs in Iba1-expressing microglia. A–B. Hippocampal slices made from CB2R WT mice were hybridized with the CB2R RNAscope probe and co-immunostained against Iba1, a marker for microglia. The cells indicated by arrow heads in stratum radiatum (A) or stratum moleculare (B) were magnified in the right four columns. C. Hippocampal slices made from CB2R KO mice were hybridized with the CB2R RNAscope probe and co-immunostained against Iba1. Strata pyramidale (CA1) and granulosum (dentate gyrus) are shown.
Antibody-based detection of CB2Rs is currently unreliable
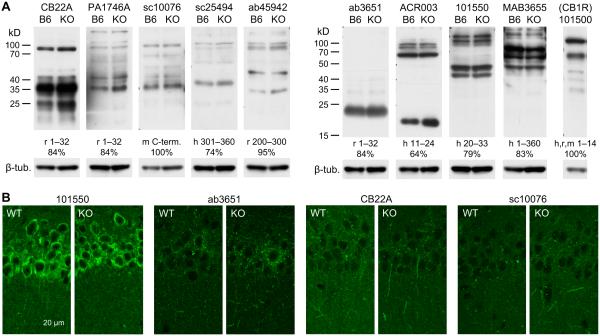
An important issue to be determined is the expression pattern of CB2R proteins in the hippocampus. This characterization requires a specific anti-CB2R antibody. Although anti-CB2R antibodies have been successfully (i.e., with KO controls) used to detect CB2Rs in some studies (Van Sickle et al., 2005; Zhang et al., 2014a), other reports have demonstrated that the specificity of many anti-CB2R antibodies is not reliable at least in some types of tissues (Ashton, 2012; Baek et al., 2013; Cecyre et al., 2014; Marchalant et al., 2014). Thus, we first tested the specificity of anti-CB2R antibodies obtained from major commercial vendors using hippocampal samples. In Western blots of proteins extracted from the mouse hippocampus, 5 out of 9 antibodies showed a band or bands near 38 kD, which was estimated from the amino acid sequence of the mouse CB2R, but these bands also appeared in the protein samples from CB2R KO mice (Fig. 7A). The blots with 4 other antibodies also showed close similarities between C57BL/6J and CB2R KO mice (Fig. 7A). This result implies that the antibodies we used might not be very specific for CB2Rs. Because G protein-coupled receptors (GPCRs) may form homodimers with each other (Wager-Miller et al., 2002) and CB2Rs can dimerize with GPR55 (38 kD) (Balenga et al., 2014), it is possible that dimerized CB2Rs in immunoblots might be detected near 76 kD. However, the bands of this size were also similarly visible in both C57BL/6J and CB2R KO samples (Fig. 7A), suggesting again the poor specificity of these antibodies for CB2Rs. We verified whether we could effectively isolate membrane-bound GPCRs using CB1Rs as a control for GPCR isolation. An immunoblot with an anti-CB1R antibody displayed strong bands at ~60 and 120 kD (Fig. 7A), which corresponded to monomers and homodimers of CB1Rs, respectively (Wager-Miller et al., 2002), implying that GPCRs might be successfully isolated under our conditions. Our data are in agreement with the previous reports of the low specificity of anti-CB2R antibodies currently available from major vendors (Ashton, 2012; Baek et al., 2013; Cecyre et al., 2014; Marchalant et al., 2014).
Figure 7.
Tests for the specificity of anti-CB2R antibodies. A. Western blotting tests for anti-CB2R antibodies. Nine antibodies against CB2R were tested with proteins extracted from the hippocampi of C57BL/6J (B6 lanes) and CB2R KO (KO lanes) mice. The vendors’ product numbers of antibodies are indicated above the blots. An anti-CB1R antibody (Cayman 101500) was used for the hippocampus of C57BL/6J mice as a control for GPCR extraction. Indicated below each blot are the species and amino acid sequence of the epitope for each antibody, as well as percent homology of the epitope with the corresponding mouse sequences (h, human; r, rat; m, mouse). The same membranes were used after stripping for β-tubulin detection. B. Fluorescent immunohistochemistry with anti-CB2R antibodies and hippocampal slices cut from CB2R WT and KO mice. The product numbers of antibodies are indicated above each pair of images of strata pyramidale and radiatum of the CA1 area.
Because of the differences in protein denaturation, non-specific epitopes in Western blots might be masked in tissue sections used for immunohistochemistry (Ashton et al., 2014), as observed in the retina (Cecyre et al., 2014). We performed immunohistochemistry to test whether anti-CB2R antibodies could specifically detect CB2Rs in tissue sections. Four anti-CB2R antibodies were selected based on the reported success in immunohistochemistry (Cayman 101550 and Alpha Diagnostic International CB22A) (Van Sickle et al., 2005; Gong et al., 2006; Cecyre et al., 2014), the species of epitope (mouse; Santa Cruz sc10076), and the small number of bands in Western blots (Abcam ab3651). However, none of these antibodies showed distinct differences in staining patterns between WT and CB2R KO tissues (Fig. 7B). This result suggests that our immunohistochemical method has a limitation in specific detection of CB2Rs, in accord with other immunohistochemical studies that also used KO controls (Ashton, 2012; Baek et al., 2013).
DISCUSSION
We demonstrate that, in the mouse hippocampus, CB2R mRNAs are expressed in glutamatergic and GABAergic neurons, and the levels of CB2R mRNAs remain stable from the age of 1 week to adulthood. CB2R mRNAs were not detected in microglia, which can express CB2Rs after injury, perhaps because healthy animals were used in our experiments. Although CB2Rs have been found in the brain, controversies surrounding what types of cells express CB2Rs have not been settled in part because 1) negative control data from CB2R KO tissues were not included in most CB2R detection studies except for limited cases (e.g., Van Sickle et al., 2005; Zhang et al., 2014a), and 2) the levels of CB2R expression in the healthy brain are low. We determined the location of CB2R mRNAs by using a very sensitive in situ hybridization method called the RNAscope with a probe that targeted the deleted region of Cnr2 in CB2R KO mice. Validation with KO controls ensured the specificity of our assay.
The RNAscope method has been used to probe CB2R mRNAs in the ventral tegmental area, but did not show any difference between WT and KO samples in contrast to a successful result from the traditional in situ hybridization method in the same study (Zhang et al., 2014a). This discrepancy might be because the RNAscope assay was not optimized and/or fragmented CB2R mRNAs were still expressed in the CB2R KO mouse strain used in the previous study. In our experience, the RNAscope method is a very delicate procedure and requires rigorous optimization. For instance, background fluorescence and false positive signals could be successfully reduced by adjusting various conditions such as the type of fixative, reagents of embedding and de-embedding, thickness of slices, and duration of each pre-treatment. To use CB2R KO mice as a negative control, we derived the sequence of our RNAscope probe from the deleted sequence in CB2R KO mice. Our design of the probe enabled us to ensure the detection specificity, and might have caused the difference between the previously reported RNAscope result (Zhang et al., 2014a) and ours. The puncta of RNAscope signal in our data were in close proximity to or colocalized with DAPI staining. This staining pattern is consistent with the idea that mRNAs are present at the site of transcription (i.e., nucleus) or exported to the cytoplasm for translation, and also in accord with other in situ hybridization results (Wang et al., 2012; McIsaac et al., 2013). Although there is a concern that the RNAscope probe might have detected genomic DNA instead of mRNA, such a possibility appears to be low because our hybridization protocol was not likely to denature double-stranded chromosomal DNA. Indeed, modifications of the hybridization protocol are required for detection of chromosomal DNA using an RNAscope probe (Wang et al., 2012), but we did not employ the DNA denaturation protocol. In addition, if our RNAscope probe had bound to DNA, most cells would have been positive for the RNAscope signal and the fluorescent puncta would have appeared in pairs in each nucleus (Wang et al., 2012). Because our results showed that only ~20% of the cells were positive for the RNAscope signal and most RNAscope-positive cells had a single fluorescent dot per cell, the RNAscope signal is unlikely to be from DNA.
Whether the low levels of CB2R mRNAs are translated into functional CB2R proteins in the brain remains unanswered, but genetic and pharmacological studies implicate CB2Rs in neurophysiological effects. The deletion of CB2Rs in mice induces schizophrenia-like symptoms, such as impairment in sensory-motor gating, an increase in depressive behavior, and a deficit in long-term, passive avoidance memory (Ortega-Alvaro et al., 2011; Garcia-Gutierrez et al., 2013). In humans, the polymorphism of CNR2 is related to schizophrenia (Ishiguro et al., 2010; Tong et al., 2013), depression (Onaivi et al., 2008) and bipolar disorder (Minocci et al., 2011). Activation of CB2Rs reduces pain (for review, Anand et al., 2009) (but also see Brownjohn and Ashton, 2012), impulsive behaviors (Navarrete et al., 2012) and locomotor activity (Valenzano et al., 2005; Onaivi et al., 2008; Xi et al., 2011) of rodents, and also vomiting of ferrets (Van Sickle et al., 2005). Chronic activation or blockade of CB2Rs in rodents increases or decreases, respectively, anxiety (Garcia-Gutierrez et al., 2012). Activation of CB2Rs decreases the excitability of cortical pyramidal neurons (den Boon et al., 2012) and dopaminergic neurons in the ventral tegmental area (Zhang et al., 2014a). CB2Rs also modulate excitatory and inhibitory synaptic transmission (Morgan et al., 2009; Ando et al., 2012; Kim and Li, 2015). Although global knockout of CB2Rs in mice could not determine the role of CB2Rs located specifically in the brain, pharmacological studies indicate that activation of CB2Rs in certain brain areas in vivo (Xi et al., 2011) or in vitro (Morgan et al., 2009; Ando et al., 2012; den Boon et al., 2012; Zhang et al., 2014a; Kim and Li, 2015) modulates neuronal activity, raising the possibility that functional CB2Rs might be present in the brain. This possibility can be further tested in future studies by the development of conditional KO mice, in which CB2Rs are deleted only in a specific types of cells, e.g., excitatory or inhibitory neurons, in a certain area of the brain.
Long-term activation of CB2Rs in the hippocampus increases spine density and excitatory synaptic transmission in CA1 pyramidal cells (Kim and Li, 2015), but the mechanisms of these effects are unclear. If stimulation of CB2Rs increases spine density in a cell-autonomous manner, only the cells that express CB2Rs should display an increase in synaptic density. However, it remains puzzling that only 20% of CB2R-positive neurons (as estimated from Fig. 4) contributed to such significant increases in spine density and glutamatergic transmission. Another possibility is that the activation of CB2Rs in a subset of neurons might induce broader changes in neuronal activity, and then this change in network activity might result in the enhancement of spine density in a large population of neurons. In this network activity scenario, delayed induction of homeostatic plasticity could play a role in altering synaptic transmission. If so, the time course of CB2R effects needs to be characterized in more detail for both excitatory and inhibitory synaptic transmission. Regarding the cell types and hippocampal sub-regions, there are many important questions unanswered. For example, it needs to be studied whether CB2Rs in both glutamatergic and GABAergic neurons are all involved in increases in excitatory transmission; whether CB2R-expressing interneurons represent a certain type of interneurons; and whether CB2Rs in the dentate gyrus play similar roles to those in the CA1 region.
The limitation of our study is that the distribution of CB2R proteins could not be determined. Therefore, the present data show which types of cells express CB2R mRNAs, but do not provide information about the relative levels of CB2R expression among cell types or subcellular compartments. Immunological electronmicrographs indicate that CB2Rs are expressed in both cell bodies and dendrites of hippocampal neurons (Brusco et al., 2008), but the relative levels of expression in these compartments are still vague; the expression appears to be the highest in the stratum radiatum (i.e., dendrites) in one report (Brusco et al., 2008), but in the stratum pyramidale (i.e., pyramidal cell bodies or basket cell terminals) in another (Gong et al., 2006). Given that these results have not been validated with KO controls, one of the reasons for this discrepancy might be attributed to the low specificity of anti-CB2R antibodies. Unequivocal determination of the subcellular location of CB2R proteins would be possible with a new, specific detection method. Most of the CB2R mRNA signals in our data were colocalized with or near DAPI staining, but a small proportion (<10%) of CB2R mRNA signals were observed without adjacent DAPI signals (i.e., >5 μm from DAPI) in the strata radiatum and moleculare. Examples of such CB2R mRNAs are found in Figs. 5A (left image) and 6A (left image). This result raises the possibility that a small population of CB2R mRNAs could be located outside the somata. It needs to be further determined whether CB2R mRNAs are translocated to the dendrites for local translation into CB2R proteins.
We estimated that about 20% of hippocampal cells—excitatory and inhibitory neurons— express CB2R mRNAs (Fig. 4). In the dorsal motor nucleus of the rat vagus, a majority of neurons are positive for CB2R immunostaining (Van Sickle et al., 2005). In situ hybridization studies indicate that most cells express CB2R mRNAs in the ventral tegmental area, prefrontal cortex, nucleus accumbens, and dorsal striatum in rodents (Zhang et al., 2014a; Zhang et al., 2015) and the cerebral cortex and globus pallidus in macaque (Lanciego et al., 2011). Such widespread expression of CB2Rs in these regions clearly contrasts our observation of the sparse expression in the hippocampus. Expression of CB2R mRNAs in a limited number of cells throughout the macaque hippocampus has also been reported (Lanciego et al., 2011), but because cell types were not determined, it has been uncertain whether most interneurons express CB2Rs or only part of excitatory and inhibitory neurons do. Our study reveals that the latter is the case. The RNAscope method is sensitive enough to detect a single mRNA molecule (Wang et al., 2012). However, it cannot be ruled out that the sparse expression of CB2R mRNAs in our study might have resulted, in part, from underestimation of the proportion of CB2R-positive cells, some of which might express CB2R mRNAs at extremely low levels, below the detection threshold of our method.
The components of the endocannabinoid system—cannabinoid receptors, endocannabinoids, endocannabinoid degradation enzymes and endocannabinoid synthases—may undergo developmental changes in their expression and/or function (Fernandez-Ruiz et al., 2000). Postnatal expression of CB1Rs, for example, increases in the cerebellum and decreases in the cerebral cortex and hippocampus with age (Fernandez-Ruiz et al., 2000). In contrast, the expression of Cnr2 in the mouse hippocampus does not significantly vary with age until adulthood in our study (Fig. 1B). This result from mice is in accord with the stable developmental pattern of CNR2 expression in the postmortem prefrontal cortex of humans (Choi et al., 2012). However, it still needs to be determined whether the expression of CB2Rs in individual cell types is also constant during postnatal development. Because the mRNAs in our qPCR experiment were pooled from the entire hippocampus, it cannot be ruled out that CB2R expression in certain populations of cells might change during postnatal development while changes in CB2Rs in other types of cells counterbalance it. RNA sequencing data from 7-day old mice indicate that, in the cerebral cortex, CB2R mRNAs are expressed mostly in microglia but very little in neurons and astrocytes (Zhang et al., 2014b). In contrast, in situ hybridization experiments with the cerebral cortex from older animals demonstrate widespread expression of CB2R mRNAs including strong expression in neurons (Lanciego et al., 2011; Zhang et al., 2015). Whether the age of animals contributed to this difference remains to be resolved.
Our data also show that the expression of CB2R mRNAs is increased in cultured slices (Fig. 1C). Given that the expression of CB2Rs in microglia can be induced by injury (Carlisle et al., 2002; Walter et al., 2003; Zhang et al., 2003; Maresz et al., 2005), it is possible that culture procedures may insult cells and thus stimulate CB2R expression in slice cultures. A culture-induced increase in CB2R expression might have contributed, in part, to the more pronounced effects of a CB2R agonist on the excitability of cultured dopaminergic neurons than those in vivo (Zhang et al., 2014a). In spite of such a quantitative difference, cultured and non-cultured neurons respond to CB2R activation in a qualitatively similar way (Zhang et al., 2014a; Kim and Li, 2015). These similarities might be because 1) the culture-induced increases in CB2R expression is only mild (e.g., about a 3.5 fold; Fig. 1C) and/or 2) the CB2R-mediated neuromodulation might not involve microglial CB2Rs. Our data suggest that CB2R-related data from cultures or insulted preparation such as synaptosomes (Ando et al., 2012) should be interpreted with caution to avoid potential exaggeration of CB2R functions. It will be an important task to determine whether the increase in CB2R mRNA expression in cultured slices occurred in microglia or neurons.
As reported (Baek et al., 2013; Cecyre et al., 2014; Marchalant et al., 2014), our validation of anti-CB2R antibodies also failed. Most antibodies we used displayed multiple bands in Western blots and all the bands at any molecular weight similarly appeared in both C57BL/6J and CB2R KO mouse samples. Eight out of nine antibodies were produced using rat or human epitopes that displayed 64–95% sequence homology with the mouse sequences and one was produced using a mouse epitope (Fig. 7A). However, the antibody specificity, judged from the KO control, was not dependent on the degree of sequence homology. In the CB2R KO mice that we used, 112 N-terminal amino acids (from 26th to 137th) were deleted, but the location of epitope does not appear to affect the antibody specificity because the epitopes of our antibodies were from various locations, e.g., N- and C-termini, as well as the whole sequence.
Major endocannabinoids, e.g., 2-arachidonoylglycerol and anandamide, can activate both CB1Rs and CB2Rs with a 3–4 fold higher affinity for CB1R than for CB2R (Han et al., 2013). Δ9-tetrahydrocannabinol can also bind to CB1R and CB2R with the same affinity (Han et al., 2013). Therefore, it is conceivable that changes in endocannabinoid levels or consumption of marijuana might induce CB2R-mediated responses. Although the investigation on cannabinoid effects has been focused on CB1Rs, the roles of cannabinoids can be more completely understood with further characterization of the function, cellular and subcellular location, and developmental profiles of CB2Rs. Our present study may provide insights into these future studies.
CONCLUSIONS
The effects of cannabinoids in the brain are mostly mediated by CB1Rs, but the recent discoveries of CB2Rs in the nervous system also implicate CB2Rs in neuropsychological effects of cannabinoids. To understand the cellular mechanisms of CB2R functions in the brain, it is essential to determine the types of cells expressing CB2Rs. Our data indicate that, in the mouse hippocampus, CB2R mRNAs are primarily expressed in a subset of excitatory and inhibitory neurons. The amount of CB2R mRNAs in the mouse hippocampus is stable during postnatal development, from age 1 week to adulthood. The expression of CB2R mRNAs is elevated in cultured hippocampal slices compared with the freshly isolated hippocampus, raising a possibility of potential overestimation of CB2R expression and/or function in a cultured system. Localization of CB2R expression might help determine the mechanisms of CB2R action, such as intra- and/or interneuronal signaling cascades.
HIGHLIGHTS.
Hippocampal excitatory and inhibitory neurons in mice express CB2 receptor mRNAs.
Only a subset of hippocampal neurons express CB2 cannabinoid receptor mRNAs.
Levels of hippocampal CB2 receptor mRNAs are stable during postnatal development.
The expression of CB2 cannabinoid receptor mRNAs is augmented in cultured tissue.
ACKNOWLEDGEMENTS
This work was partially supported by the National Institute on Aging Grant R01AG036794. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors thank the Alomone Labs (Jerusalem, Israel) for the generous gift of antibody.
ABBREVIATIONS
- CB1R
CB1 cannabinoid receptor
- CB2R
CB2 cannabinoid receptor
- DAPI
4',6-diamidino-2-phenylindole
- EDTA
ethylenediaminetetraacetic acid
- GABA
γ-aminobutyric acid
- GPCR
G protein-coupled receptor
- KO
knock-out
- PBS
phosphate-buffered saline
- PBST
PBS with triton X-100
- PCR
polymerase chain reaction
- qPCR
quantitative PCR
- RT-PCR
reverse transcription PCR
- SDS
sodium dodecyl sulfate
- TBST
tris-buffered saline with tween 20
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anand P, Whiteside G, Fowler CJ, Hohmann AG. Targeting CB2 receptors and the endocannabinoid system for the treatment of pain. Brain Res Rev. 2009;60:255–266. doi: 10.1016/j.brainresrev.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando RD, Biro J, Csolle C, Ledent C, Sperlagh B. The inhibitory action of exo- and endocannabinoids on [3H]GABA release are mediated by both CB1 and CB2 receptors in the mouse hippocampus. Neurochem Int. 2012;60:145–152. doi: 10.1016/j.neuint.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Ashton JC. The use of knockout mice to test the specificity of antibodies for cannabinoid receptors. Hippocampus. 2012;22:643–644. doi: 10.1002/hipo.20946. [DOI] [PubMed] [Google Scholar]
- Ashton JC, Zheng Y, Darlington C, Baek JH, Smith PF. Cannabinoid CB2 receptor immunolabelling in the healthy brain—still a live possibility. Naunyn Schmiedebergs Arch Pharmacol. 2014;387:301. doi: 10.1007/s00210-013-0948-y. [DOI] [PubMed] [Google Scholar]
- Atwood BK, Mackie K. CB2: a cannabinoid receptor with an identity crisis. Br J Pharmacol. 2010;160:467–479. doi: 10.1111/j.1476-5381.2010.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek JH, Darlington CL, Smith PF, Ashton JC. Antibody testing for brain immunohistochemistry: brain immunolabeling for the cannabinoid CB2 receptor. J Neurosci Methods. 2013;216:87–95. doi: 10.1016/j.jneumeth.2013.03.021. [DOI] [PubMed] [Google Scholar]
- Baek JH, Zheng Y, Darlington CL, Smith PF. Cannabinoid CB2 receptor expression in the rat brainstem cochlear and vestibular nuclei. Acta Otolaryngol. 2008;128:961–967. doi: 10.1080/00016480701796944. [DOI] [PubMed] [Google Scholar]
- Balenga NA, Martinez-Pinilla E, Kargl J, Schroder R, Peinhaupt M, Platzer W, Balint Z, Zamarbide M, Dopeso-Reyes IG, Ricobaraza A, Perez-Ortiz JM, Kostenis E, Waldhoer M, Heinemann A, Franco R. Heteromerization of GPR55 and cannabinoid CB2 receptors modulates signalling. Br J Pharmacol. 2014;171:5387–5406. doi: 10.1111/bph.12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher B, Antel JP. Comparison of phenotypic and functional properties of immediately ex vivo and cultured human adult microglia. Glia. 1996;18:1–10. doi: 10.1002/(SICI)1098-1136(199609)18:1<1::AID-GLIA1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Brownjohn PW, Ashton JC. Spinal cannabinoid CB2 receptors as a target for neuropathic pain: an investigation using chronic constriction injury. Neuroscience. 2012;203:180–193. doi: 10.1016/j.neuroscience.2011.12.028. [DOI] [PubMed] [Google Scholar]
- Brusco A, Tagliaferro P, Saez T, Onaivi ES. Postsynaptic localization of CB2 cannabinoid receptors in the rat hippocampus. Synapse. 2008;62:944–949. doi: 10.1002/syn.20569. [DOI] [PubMed] [Google Scholar]
- Carlisle SJ, Marciano-Cabral F, Staab A, Ludwick C, Cabral GA. Differential expression of the CB2 cannabinoid receptor by rodent macrophages and macrophage-like cells in relation to cell activation. Int Immunopharmacol. 2002;2:69–82. doi: 10.1016/s1567-5769(01)00147-3. [DOI] [PubMed] [Google Scholar]
- Cecyre B, Thomas S, Ptito M, Casanova C, Bouchard JF. Evaluation of the specificity of antibodies raised against cannabinoid receptor type 2 in the mouse retina. Naunyn Schmiedebergs Arch Pharmacol. 2014;387:175–184. doi: 10.1007/s00210-013-0930-8. [DOI] [PubMed] [Google Scholar]
- Chen Y, Dong E, Grayson DR. Analysis of the GAD1 promoter: trans-acting factors and DNA methylation converge on the 5' untranslated region. Neuropharmacology. 2011;60:1075–1087. doi: 10.1016/j.neuropharm.2010.09.017. [DOI] [PubMed] [Google Scholar]
- Choi K, Le T, McGuire J, Xing G, Zhang L, Li H, Parker CC, Johnson LR, Ursano RJ. Expression pattern of the cannabinoid receptor genes in the frontal cortex of mood disorder patients and mice selectively bred for high and low fear. J Psychiatr Res. 2012;46:882–889. doi: 10.1016/j.jpsychires.2012.03.021. [DOI] [PubMed] [Google Scholar]
- den Boon FS, Chameau P, Schaafsma-Zhao Q, van Aken W, Bari M, Oddi S, Kruse CG, Maccarrone M, Wadman WJ, Werkman TR. Excitability of prefrontal cortical pyramidal neurons is modulated by activation of intracellular type-2 cannabinoid receptors. Proc Natl Acad Sci U S A. 2012;109:3534–3539. doi: 10.1073/pnas.1118167109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbenev AV, Stuart TC, Smith BN. Cannabinoids suppress synaptic input to neurones of the rat dorsal motor nucleus of the vagus nerve. J Physiol. 2004;559:923–938. doi: 10.1113/jphysiol.2004.067470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devane WA, Dysarz FA, 3rd, Johnson MR, Melvin LS, Howlett AC. Determination and characterization of a cannabinoid receptor in rat brain. Mol Pharmacol. 1988;34:605–613. [PubMed] [Google Scholar]
- Fernandez-Ruiz J, Berrendero F, Hernandez ML, Ramos JA. The endogenous cannabinoid system and brain development. Trends Neurosci. 2000;23:14–20. doi: 10.1016/s0166-2236(99)01491-5. [DOI] [PubMed] [Google Scholar]
- Garcia-Gutierrez MS, Garcia-Bueno B, Zoppi S, Leza JC, Manzanares J. Chronic blockade of cannabinoid CB2 receptors induces anxiolytic-like actions associated with alterations in GABAA receptors. Br J Pharmacol. 2012;165:951–964. doi: 10.1111/j.1476-5381.2011.01625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gutierrez MS, Ortega-Alvaro A, Busquets-Garcia A, Perez-Ortiz JM, Caltana L, Ricatti MJ, Brusco A, Maldonado R, Manzanares J. Synaptic plasticity alterations associated with memory impairment induced by deletion of CB2 cannabinoid receptors. Neuropharmacology. 2013;73:388–396. doi: 10.1016/j.neuropharm.2013.05.034. [DOI] [PubMed] [Google Scholar]
- Gong JP, Onaivi ES, Ishiguro H, Liu QR, Tagliaferro PA, Brusco A, Uhl GR. Cannabinoid CB2 receptors: immunohistochemical localization in rat brain. Brain Res. 2006;1071:10–23. doi: 10.1016/j.brainres.2005.11.035. [DOI] [PubMed] [Google Scholar]
- Han S, Thatte J, Buzard DJ, Jones RM. Therapeutic utility of cannabinoid receptor type 2 (CB2) selective agonists. J Med Chem. 2013;56:8224–8256. doi: 10.1021/jm4005626. [DOI] [PubMed] [Google Scholar]
- Howlett AC, Barth F, Bonner TI, Cabral G, Casellas P, Devane WA, Felder CC, Herkenham M, Mackie K, Martin BR, Mechoulam R, Pertwee RG. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol Rev. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Horiuchi Y, Ishikawa M, Koga M, Imai K, Suzuki Y, Morikawa M, Inada T, Watanabe Y, Takahashi M, Someya T, Ujike H, Iwata N, Ozaki N, Onaivi ES, Kunugi H, Sasaki T, Itokawa M, Arai M, Niizato K, Iritani S, Naka I, Ohashi J, Kakita A, Takahashi H, Nawa H, Arinami T. Brain cannabinoid CB2 receptor in schizophrenia. Biol Psychiatry. 2010;67:974–982. doi: 10.1016/j.biopsych.2009.09.024. [DOI] [PubMed] [Google Scholar]
- Kim J, Li Y. Chronic activation of CB2 cannabinoid receptors in the hippocampus increases excitatory synaptic transmission. J Physiol. 2015;593:871–886. doi: 10.1113/jphysiol.2014.286633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciego JL, Barroso-Chinea P, Rico AJ, Conte-Perales L, Callen L, Roda E, Gomez-Bautista V, Lopez IP, Lluis C, Labandeira-Garcia JL, Franco R. Expression of the mRNA coding the cannabinoid receptor 2 in the pallidal complex of Macaca fascicularis. J Psychopharmacol. 2011;25:97–104. doi: 10.1177/0269881110367732. [DOI] [PubMed] [Google Scholar]
- Liu QR, Pan CH, Hishimoto A, Li CY, Xi ZX, Llorente-Berzal A, Viveros MP, Ishiguro H, Arinami T, Onaivi ES, Uhl GR. Species differences in cannabinoid receptor 2 (CNR2 gene): identification of novel human and rodent CB2 isoforms, differential tissue expression and regulation by cannabinoid receptor ligands. Genes Brain Behav. 2009;8:519–530. doi: 10.1111/j.1601-183X.2009.00498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchalant Y, Brownjohn PW, Bonnet A, Kleffmann T, Ashton JC. Validating Antibodies to the Cannabinoid CB2 Receptor: Antibody Sensitivity Is Not Evidence of Antibody Specificity. J Histochem Cytochem. 2014;62:395–404. doi: 10.1369/0022155414530995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresz K, Carrier EJ, Ponomarev ED, Hillard CJ, Dittel BN. Modulation of the cannabinoid CB2 receptor in microglial cells in response to inflammatory stimuli. J Neurochem. 2005;95:437–445. doi: 10.1111/j.1471-4159.2005.03380.x. [DOI] [PubMed] [Google Scholar]
- Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- McIsaac RS, Silverman SJ, Parsons L, Xu P, Briehof R, McClean MN, Botstein D. Visualization and analysis of mRNA molecules using fluorescence in situ hybridization in Saccharomyces cerevisiae. J Vis Exp. 2013;76:e50382. doi: 10.3791/50382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechoulam R, Ben-Shabat S, Hanus L, Ligumsky M, Kaminski NE, Schatz AR, Gopher A, Almog S, Martin BR, Compton DR, Pertwee RG, Griffin G, Bayewitch M, Barg J, Vogel Z. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem Pharmacol. 1995;50:83–90. doi: 10.1016/0006-2952(95)00109-d. [DOI] [PubMed] [Google Scholar]
- Minocci D, Massei J, Martino A, Milianti M, Piz L, Di Bello D, Sbrana A, Martinotti E, Rossi AM, Nieri P. Genetic association between bipolar disorder and 524A>C (Leu133Ile) polymorphism of CNR2 gene, encoding for CB2 cannabinoid receptor. J Affect Disord. 2011;134:427–430. doi: 10.1016/j.jad.2011.05.023. [DOI] [PubMed] [Google Scholar]
- Morgan NH, Stanford IM, Woodhall GL. Functional CB2 type cannabinoid receptors at CNS synapses. Neuropharmacology. 2009;57:356–368. doi: 10.1016/j.neuropharm.2009.07.017. [DOI] [PubMed] [Google Scholar]
- Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- Navarrete F, Perez-Ortiz JM, Manzanares J. Cannabinoid CB2 receptor-mediated regulation of impulsive-like behaviour in DBA/2 mice. Br J Pharmacol. 2012;165:260–273. doi: 10.1111/j.1476-5381.2011.01542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaivi ES. Neuropsychobiological evidence for the functional presence and expression of cannabinoid CB2 receptors in the brain. Neuropsychobiology. 2006;54:231–246. doi: 10.1159/000100778. [DOI] [PubMed] [Google Scholar]
- Onaivi ES, Ishiguro H, Gong JP, Patel S, Meozzi PA, Myers L, Perchuk A, Mora Z, Tagliaferro PA, Gardner E, Brusco A, Akinshola BE, Hope B, Lujilde J, Inada T, Iwasaki S, Macharia D, Teasenfitz L, Arinami T, Uhl GR. Brain neuronal CB2 cannabinoid receptors in drug abuse and depression: from mice to human subjects. PLoS One. 2008;3:e1640. doi: 10.1371/journal.pone.0001640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Alvaro A, Aracil-Fernandez A, Garcia-Gutierrez MS, Navarrete F, Manzanares J. Deletion of CB2 cannabinoid receptor induces schizophrenia-related behaviors in mice. Neuropsychopharmacology. 2011;36:1489–1504. doi: 10.1038/npp.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel HJ, Birrell MA, Crispino N, Hele DJ, Venkatesan P, Barnes PJ, Yacoub MH, Belvisi MG. Inhibition of guinea-pig and human sensory nerve activity and the cough reflex in guinea-pigs by cannabinoid (CB2) receptor activation. Br J Pharmacol. 2003;140:261–268. doi: 10.1038/sj.bjp.0705435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinal CS, Tobin AJ. Uniqueness and redundancy in GABA production. Perspect Dev Neurobiol. 1998;5:109–118. [PubMed] [Google Scholar]
- Sagar DR, Kelly S, Millns PJ, O'Shaughnessey CT, Kendall DA, Chapman V. Inhibitory effects of CB1 and CB2 receptor agonists on responses of DRG neurons and dorsal horn neurons in neuropathic rats. Eur J Neurosci. 2005;22:371–379. doi: 10.1111/j.1460-9568.2005.04206.x. [DOI] [PubMed] [Google Scholar]
- Showalter VM, Compton DR, Martin BR, Abood ME. Evaluation of binding in a transfected cell line expressing a peripheral cannabinoid receptor (CB2): identification of cannabinoid receptor subtype selective ligands. J Pharmacol Exp Ther. 1996;278:989–999. [PubMed] [Google Scholar]
- Sierra S, Luquin N, Rico AJ, Gomez-Bautista V, Roda E, Dopeso-Reyes IG, Vazquez A, Martinez-Pinilla E, Labandeira-Garcia JL, Franco R, Lanciego JL. Detection of cannabinoid receptors CB1 and CB2 within basal ganglia output neurons in macaques: changes following experimental parkinsonism. Brain Struct Funct. 2014;220:2721–2738. doi: 10.1007/s00429-014-0823-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaper SD, Buriani A, Dal Toso R, Petrelli L, Romanello S, Facci L, Leon A. The ALIAmide palmitoylethanolamide and cannabinoids, but not anandamide, are protective in a delayed postglutamate paradigm of excitotoxic death in cerebellar granule neurons. Proc Natl Acad Sci U S A. 1996;93:3984–3989. doi: 10.1073/pnas.93.9.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella N. Cannabinoid signaling in glial cells. Glia. 2004;48:267–277. doi: 10.1002/glia.20084. [DOI] [PubMed] [Google Scholar]
- Sugiura T, Kondo S, Kishimoto S, Miyashita T, Nakane S, Kodaka T, Suhara Y, Takayama H, Waku K. Evidence that 2-arachidonoylglycerol but not N-palmitoylethanolamine or anandamide is the physiological ligand for the cannabinoid CB2 receptor. Comparison of the agonistic activities of various cannabinoid receptor ligands in HL-60 cells. J Biol Chem. 2000;275:605–612. doi: 10.1074/jbc.275.1.605. [DOI] [PubMed] [Google Scholar]
- Tong D, He S, Wang L, Jin L, Si P, Cheng X. Association of single-nucleotide polymorphisms in the cannabinoid receptor 2 gene with schizophrenia in the Han Chinese population. J Mol Neurosci. 2013;51:454–460. doi: 10.1007/s12031-013-0062-0. [DOI] [PubMed] [Google Scholar]
- Valenzano KJ, Tafesse L, Lee G, Harrison JE, Boulet JM, Gottshall SL, Mark L, Pearson MS, Miller W, Shan S, Rabadi L, Rotshteyn Y, Chaffer SM, Turchin PI, Elsemore DA, Toth M, Koetzner L, Whiteside GT. Pharmacological and pharmacokinetic characterization of the cannabinoid receptor 2 agonist, GW405833, utilizing rodent models of acute and chronic pain, anxiety, ataxia and catalepsy. Neuropharmacology. 2005;48:658–672. doi: 10.1016/j.neuropharm.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Van Sickle MD, Duncan M, Kingsley PJ, Mouihate A, Urbani P, Mackie K, Stella N, Makriyannis A, Piomelli D, Davison JS, Marnett LJ, Di Marzo V, Pittman QJ, Patel KD, Sharkey KA. Identification and functional characterization of brainstem cannabinoid CB2 receptors. Science. 2005;310:329–332. doi: 10.1126/science.1115740. [DOI] [PubMed] [Google Scholar]
- Wager-Miller J, Westenbroek R, Mackie K. Dimerization of G protein-coupled receptors: CB1 cannabinoid receptors as an example. Chem Phys Lipids. 2002;121:83–89. doi: 10.1016/s0009-3084(02)00151-2. [DOI] [PubMed] [Google Scholar]
- Walter L, Franklin A, Witting A, Wade C, Xie Y, Kunos G, Mackie K, Stella N. Nonpsychotropic cannabinoid receptors regulate microglial cell migration. J Neurosci. 2003;23:1398–1405. doi: 10.1523/JNEUROSCI.23-04-01398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi ZX, Peng XQ, Li X, Song R, Zhang HY, Liu QR, Yang HJ, Bi GH, Li J, Gardner EL. Brain cannabinoid CB2 receptors modulate cocaine's actions in mice. Nat Neurosci. 2011;14:1160–1166. doi: 10.1038/nn.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HY, Bi GH, Li X, Li J, Qu H, Zhang SJ, Li CY, Onaivi ES, Gardner EL, Xi ZX, Liu QR. Species differences in cannabinoid receptor 2 and receptor responses to cocaine self-administration in mice and rats. Neuropsychopharmacology. 2015;40:1037–1051. doi: 10.1038/npp.2014.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HY, Gao M, Liu QR, Bi GH, Li X, Yang HJ, Gardner EL, Wu J, Xi ZX. Cannabinoid CB2 receptors modulate midbrain dopamine neuronal activity and dopamine-related behavior in mice. Proc Natl Acad Sci U S A. 2014a;111:E5007–5015. doi: 10.1073/pnas.1413210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hoffert C, Vu HK, Groblewski T, Ahmad S, O'Donnell D. Induction of CB2 receptor expression in the rat spinal cord of neuropathic but not inflammatory chronic pain models. Eur J Neurosci. 2003;17:2750–2754. doi: 10.1046/j.1460-9568.2003.02704.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014b;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]