Abstract
Influenza A virus is a human pathogen whose genome is comprised of eight viral RNA segments that replicate in the nucleus. Two viral mRNAs are alternatively spliced. The unspliced M1 mRNA is translated into the matrix M1 protein while the ion channel M2 protein is generated after alternative splicing. These proteins are critical mediators of viral trafficking and budding. We show that influenza virus utilizes nuclear speckles to promote post-transcriptional splicing of its M1 mRNA. We assign previously unknown roles for the viral NS1 protein and cellular factors to an intranuclear trafficking pathway that targets the viral M1 mRNA to nuclear speckles, mediates splicing at these nuclear bodies, and exports the spliced M2 mRNA from the nucleus. Since nuclear speckles are storage sites for splicing factors, which leave these sites to splice cellular pre-mRNAs at transcribing genes, we reveal a functional subversion of nuclear speckles to promote viral gene expression.
Keywords: influenza virus, nuclear speckles, splicing, mRNA export
Mammalian nuclei contain ~ 20–50 speckles of varying size and they are also known as interchromatin granule clusters (IGCs). These nuclear bodies are involved in the maturation and storage of pre-mRNA splicing factors, RNA processing factors, transcription factors, and other factors that function in gene expression1–3. It has been shown that splicing factors leave nuclear speckles to function at active transcribing genes in the nucleoplasm, demonstrating the dynamic nature of this compartment4. Thus, most of gene transcription and splicing does not occur at nuclear speckles5. Although controversial, it has been reported that post-transcription splicing may be spatially mediated at nuclear speckles6. This concept is supported by accumulation of specific polyadenylated mRNAs at nuclear speckles, including mRNA originated from plasmid, which appear to be involved in splicing and/or export of these mRNAs to the cytoplasm7–10. However, most cellular pre-mRNAs do not utilize nuclear speckles, which serve as suppliers of factors with key roles in gene expression1.
Viruses, such as influenza viruses, usurp cellular factors to execute splicing of viral mRNAs11. Two influenza virus mRNAs, NS1 and M1, are alternatively spliced11. The unspliced NS1 and M1 mRNAs are translated into the non-structural 1 (NS1) and M1 proteins, respectively, while the nuclear export protein (NEP, also known as NS2) and M2 mRNAs are generated after splicing. Other alternative spliced forms of M1 mRNA have been detected and include the mRNA3, which is not known to encode a peptide12, 13, and M4 mRNA that encodes an isoform of the M2 ion channel and it is only present in certain viral strains14, 15,11. NS1, NS2, M1 and M2 proteins have critical functions during the virus life cycle; therefore, the mechanisms involved in influenza virus mRNA alternative splicing represent key steps in the viral life cycle. However, the processes involved in viral mRNA splicing have not been fully elucidated.
Results
Influenza Virus M mRNA Accumulates at Nuclear Speckles
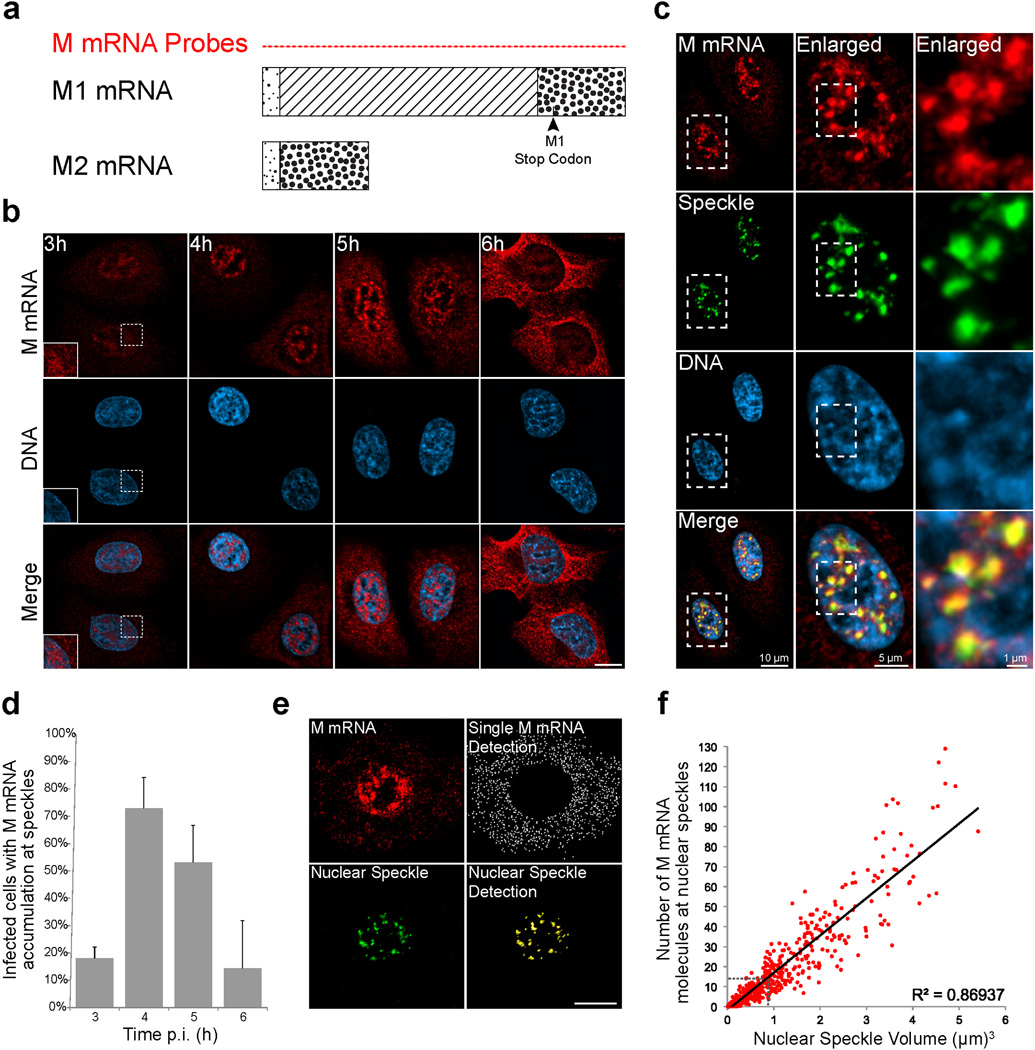
To spatially and temporally define the mechanisms involved in influenza virus M1 mRNA alternative splicing and export, we first followed M1 mRNA distribution using single molecule RNA-FISH (smFISH) in infected cells. Forty-six FISH probes covering the entire M1 mRNA of the influenza A/WSN/33 virus strain were generated to detect single M1 mRNA transcripts during influenza virus infection (Fig. 1a and Supplementary Information; M mRNA probes). While these 46 probes can recognize M1 and its alternative spliced forms, they can only detect M1 at the single molecule level due to the small size of the other spliced forms that are recognized in bulk. Hence, we label the signal detected with the 46 probes as M mRNA. We observed that at 3h after infection single M1 mRNA transcripts are detected in the nucleus and in the cytoplasm indicating efficient transcription and nuclear export of M1 mRNA (Fig. 1b). Notably, M mRNA significantly accumulates at intranuclear foci ~4–5 h post-infection (Fig. 1b). RNA-FISH and immunofluorescence staining against M mRNA and SC35 protein, respectively, showed that M mRNA is enriched at nuclear speckles (Fig. 1c and Supplementary Video 1) as SC35 is a SR protein, known splicing factor, and nuclear speckle marker1. Other influenza A virus strains, A/Texas/36/91 and A/Puerto Rico/8/34, also show M mRNA localization at nuclear speckles (Supplementary Fig. 1a,b). Approximately 6h post-infection, M mRNA enrichment at nuclear speckles is reduced and most of the M mRNA is detected in the cytoplasm, indicating efficient nuclear export (Fig. 1b,d).
Figure 1. Influenza M mRNA is localized at nuclear speckles.
(a) Schematic representation of M1 and M2 mRNAs. The diagonal lines in M1 mRNA represent an intron that is spliced out to generate M2 mRNA. Forty-six FISH probes labeled with Quasar 570 (M mRNA Probes) that cover the entire M1 and M2 mRNAs were synthesized. (b) These probes monitored the distribution of M mRNA at 3h, 4h, and 6h during A/WSN/33 infection of A549 cells at MOI 10. Inset shows enlargement of an area in the nucleus and cytoplasm. Images are representative of three independent experiments. Scale Bar = 10µm. (c) M mRNA accumulates at nuclear speckles, which were marked by SC35 immunofluorescence staining (Pearson’s Correlation Coefficient between the M mRNA channel and the Speckle channel in the nucleus = 0.72). The squared marked region was enlarged and shown in the middle panel. Further enlargement is shown in the right panel. Images are representative of three independent experiments. (d) The percentage of cells with speckle mRNA accumulation was quantified. Values are percentage mean ± s.d. of at least 99 cells that were counted in three independent experiments for each time point. p.i.- post infection. (e) M mRNA quantification at nuclear speckles. Fluorescent signal emerging from labeled M mRNA transcripts was detected in the cytoplasm (Single M mRNA Detection, white dots) while the M mRNA signal emerging from the nucleus was masked based on Hoechst staining. Nuclear speckles were also marked (Nuclear Speckle Detection, yellow punctate). Images are representative of three independent experiments. Scale Bar = 10µm. (f) Quantification of M mRNA copy number at nuclear speckles marked with SC35 antibody. M mRNA fluorescence intensity sum inside speckles was divided by the average intensity sum of individual cytoplasmic M mRNA transcripts. Cells were infected for 4 h and RNA-FISH was quantified as in e. The number of M mRNA at nuclear speckles was plotted according to the nuclear speckle volume. Dotted lines in the graph depict the average nuclear speckle volume and the corresponding M mRNA copy number. Six hundred and eighty two nuclear speckles were analyzed in 16 cells.
To determine the number of viral M mRNA molecules at nuclear speckles, we first quantified the fluorescence signal emerging from single mRNA molecules that could be detected in the cytoplasm (Fig. 1e). In the same cells, we used SC35 labeling to locate nuclear speckles and quantify the overall intensities of RNA-FISH labeled M mRNAs inside the nuclear speckle volume (Fig. 1e). The M mRNA fluorescent signal at individual speckles was then divided by the average intensity of a single M1 mRNA. In general, nuclear speckles vary in size ranging from one to several micrometers in diameter1. We detected and analyzed 682 SC35 labeled nuclear speckles in 16 cells (average of 42.6 nuclear speckles per nucleus) and measured the volume of these nuclear speckles. We found that the average volume of a single nuclear speckle is 0.82 µm3 (Diameter = 1.16 µm). We then calculated the accumulation of M mRNAs with respect to the nuclear speckle volume (Fig. 1f). We show that there is a linear correlation between nuclear speckle volume and the number of M mRNAs enriched at nuclear speckles (R2=0.869). Additionally, we found that inside the average size nuclear speckle, the bulk M mRNA signal is equivalent to ~ 13 M1 mRNA molecules. Thus, we estimate that hundreds of viral M mRNA transcripts enter/pass through nuclear speckles during infection.
Next we tested whether the viral NS mRNA, which is also alternatively spliced, accumulates inside nuclear speckles. For this purpose we generated an additional set of probes covering the entire NS mRNA. We then followed both NS mRNA and M1 mRNA (see below the description of specific M1 mRNA probes) distribution during infection. At 3h post-infection most of NS mRNA had already been exported to the cytoplasm while M1 mRNA was mainly in the nucleus (Supplementary Fig. 1c). In contrast to M1 mRNA, NS mRNA was not enriched at nuclear speckles (Supplementary Fig. 1c). This may indicate a major difference between M and NS mRNA biogenesis pathways and that M mRNA specifically utilizes nuclear speckles for RNA processing.
M mRNA is Post-transcriptionally Spliced at Nuclear Speckles
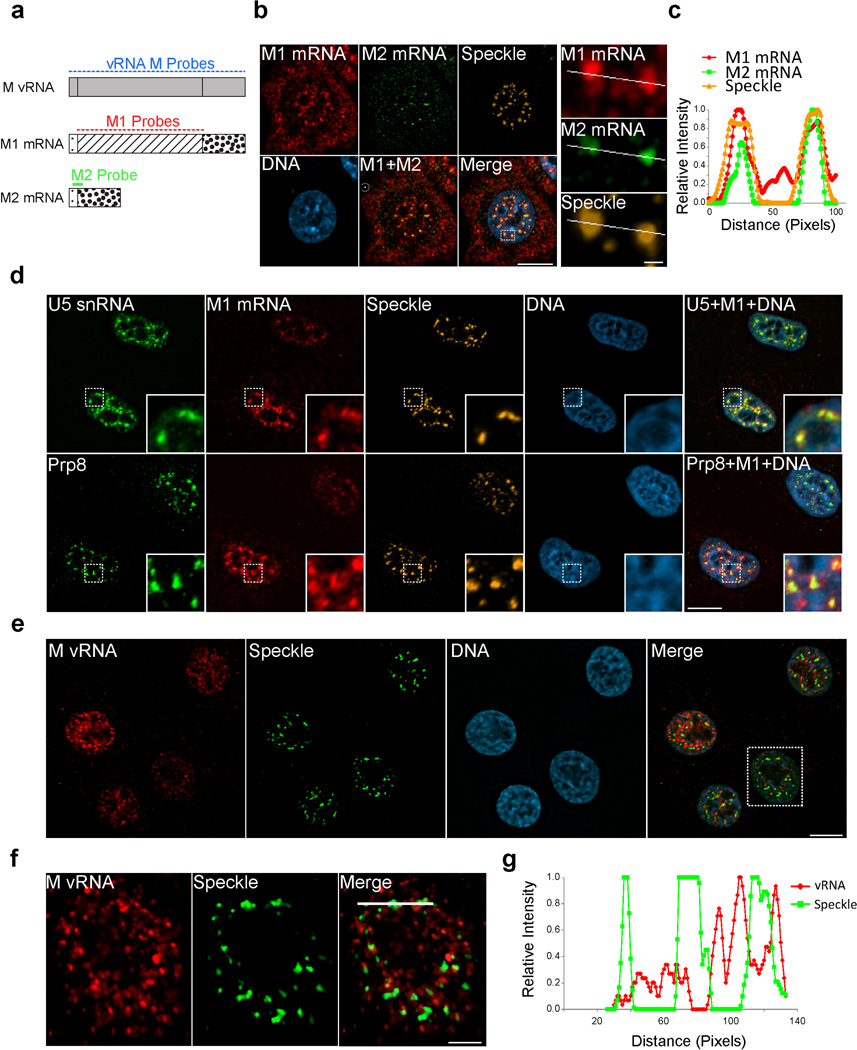
To investigate if the viral M1 mRNA is actively spliced at nuclear speckles, we generated M1 and M2 probes that specifically label the unspliced M1 mRNA and the spliced M2 mRNA, respectively (Fig. 2a andSupplementary Fig. 2a–e). We then monitored both M1 and M2 mRNAs at 4h post-infection and found that significant pools of both mRNAs were localized at nuclear speckles (Fig. 2b,c). M1 mRNA also co-localized with U5 snRNA and Prp8 at nuclear speckles, indicating accumulation at speckles that contain high concentration of key catalytic components of the splicing reaction (Fig. 2d)16. In addition, we were also able to detect mRNA3 at nuclear speckles (Supplementary Fig. 2f–h), demonstrating the presence of another spliced product of M1 mRNA at this nuclear body. Thus, the presence of both M1 mRNA and the spliced M2 mRNA and mRNA3 at nuclear speckles suggests that splicing of the M1 mRNA may occur at nuclear speckles. To reveal whether viral M1 mRNA splicing occurs co- or post-transcriptionally at nuclear speckles, we co-labeled single M vRNA segments using RNA-FISH and tracked accumulation in SC35 marked speckles. At 4 h post-infection, when M mRNAs are enriched at nuclear speckles, we did not detect single M vRNAs at nuclear speckles but they can be adjacent to these nuclear bodies (Fig. 2e–g). In fact, comparative imaging analysis of the M vRNA channel and the speckle channel shows low Pearson’s coefficient of 0.17. These findings suggest that the M1 mRNA is first generated upon M vRNA transcription in the nucleoplasm and then M1 mRNA associates with nuclear speckles for post-transcriptional splicing.
Figure 2. M1 mRNA and spliced M2 mRNA accumulate at nuclear speckles after transcription.
(a) Schematic representation of 3 sets of probes that were synthesized to specifically detect the M vRNA, and M1 and M2 mRNAs. Forty-six FISH probes labeled with Quasar 670 were synthesized to label the entire M vRNA segment. Thirty-one FISH probes labeled with Quasar 670 were synthesized to cover the M1 mRNA intron region. The M2 mRNA was detected using a single probe labeled with 2 Quasar 570 Fluorophores. This probe binds the exon junction region of M2 mRNA. (b) M1 and M2 probes were used in RNA FISH experiments performed at 4 h post-infection. Nuclear speckles were marked with SC35 antibody. A single cell was enlarged in order to demonstrate the M1 and M2 mRNA enrichment at nuclear speckles. The squared region shown in Merge was further enlarged and the M1, M2 and Speckle channels are shown on the right. (c) Relative intensity of the three channels along the white line is shown in the graph. (d) U5 snRNP components, U5 snRNA and Prp8, were co-labeled with M1 mRNA at 4 h post-infection. Nuclear speckles were marked with SC35 antibody (top) or SON antibody (bottom). (e) M vRNA probes were used in RNA FISH experiments performed at 4h post-infection. Nuclear speckles were marked with SC35 antibody. (f) The square region marked in e-Merge was enlarged and M vRNA, Speckle, and Merge are shown. (g) Relative intensity of the M vRNA and Speckle signal along the white line is shown in f-Merge. Images shown in b,d,e and f are representative of three independent experiments. Scale bar in b right panel = 1µm. Scale bar in f= 5µM. All other scale bars = 10µm.
NS1 Protein Promotes M mRNA Splicing and Nuclear Export Via Nuclear Speckles
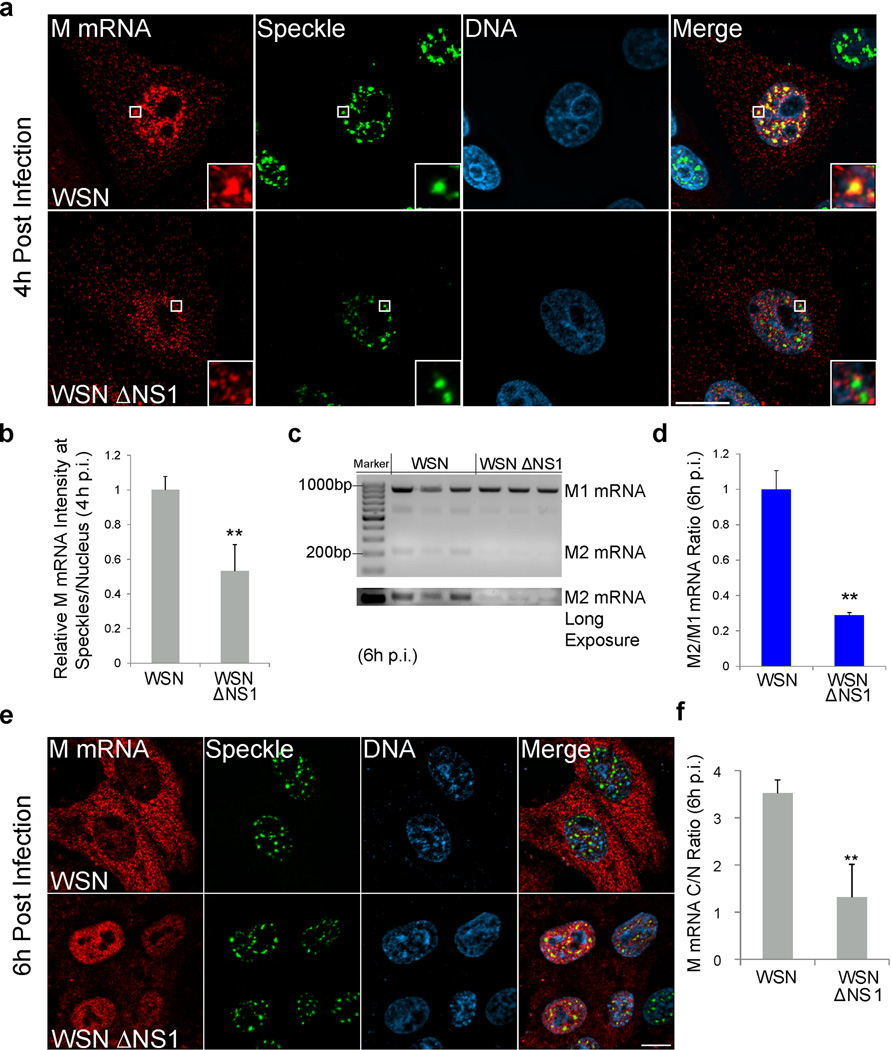
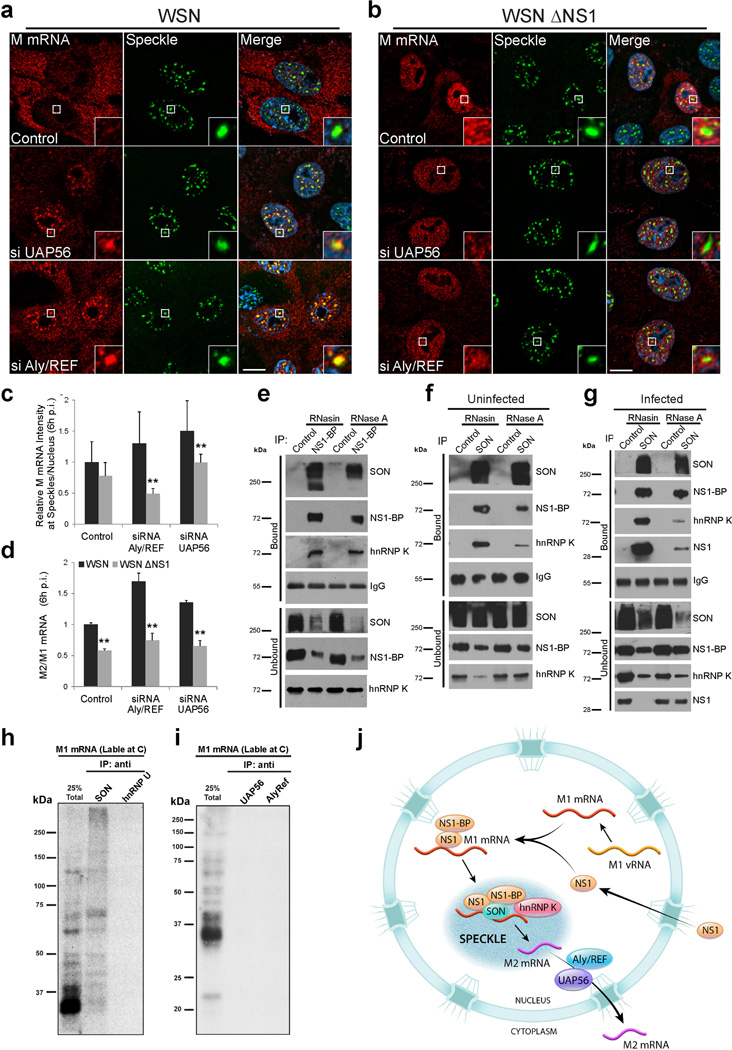
M1 mRNA splicing has been linked to the viral NS1 protein17 and M1 mRNA can interact with NS117–19. Additionally, NS1 has been previously shown to alter the SC35 punctate pattern, suggesting that NS1 may induce changes in nuclear speckle structure and function20. Hence, we tested whether NS1 is involved in M mRNA localization and splicing at nuclear speckles using an influenza virus strain that lacks NS1 (WSN ΔNS1)21. These infection studies were performed both in VERO cells, which are interferon-deficient and therefore allow replication of this mutant virus, and in A549 cells, which are immune-competent. We found that the lack of NS1 protein significantly reduced accumulation of M mRNA at nuclear speckles (Fig. 3a,b and Supplementary Fig. 3a–c) and diminished M1 mRNA splicing (Fig. 3c,d and Supplementary Fig. 3d) in both cell types. Furthermore, after 6h post-infection, we observed a decrease in M mRNA nuclear export in cells infected with WSN ΔNS1 (Fig. 3e,f and Supplementary Fig. 3e,f). Consequently, these effects contribute to the decrease in M1 and M2 protein levels in cells infected with virus lacking NS1 as compared to cells infected with wild-type virus (Supplementary Fig. 3g–i). These results point to a specific and previously unknown function of NS1 in promoting M1 mRNA localization at nuclear speckles, which lead to M1 to M2 splicing. This function of NS1 seems to be independent from its inhibitory role of interferon expression since M mRNA mislocalization and splicing-export defects observed upon WSN ΔNS1 infection occurred in both A549 and VERO cells.
Figure 3. Viral NS1 protein promotes M1 mRNA splicing at nuclear speckles and nuclear export.
(a) VERO cells were infected with WSN virus or mutated virus that lack NS1 (WSN ΔNS1) at MOI 10. After 4h, cells were fixed and M mRNA was labeled by RNA-FISH. Speckles were marked by immunofuorescence with SON antibody. Insets are enlargements of the marked white squares showing nuclear speckle areas. Scale Bar = 10µm. (b) Quantification performed as in Supplementary Fig. 3a for measuring M mRNA intensity at speckles after WSN or WSN ΔNS1 infection. Data represents three independent experiments. M mRNA intensity sum at speckles was normalized to the intensity sum in the nucleus. Normalized intensities at speckles of WT virus infected cells were set to 1 and the relative fold change in WSN ΔNS1 virus infected cells is shown. Values are mean ± s.d. measured in 25 WSN and 29 WSN ΔNS1 infected cells. p.i.- post infection. (c) VERO cells were infected with WSN virus or WSN ΔNS1 at MOI 1 for 6 h. Purified RNA was subjected to RT-PCR and the products were run on agarose gel. Data represents three independent experiments. (d) RT-qPCR quantification of M2/M1 mRNA ratio in WSN or WSN ΔNS1 VERO infected cells. The average M2/M1 mRNA ratio from cells infected with WSN (M2/M1 mRNA = 0.62) was set to 1 and the relative average M2/M1 mRNA ratio in WSN ΔNS1 infected cells is shown. Values are mean ± s.d. ratios from three independent experiments. (e) VERO cells were infected with WSN virus or WSN ΔNS1. After 6h, cells were fixed and M mRNA was labeled by RNA-FISH. Speckles were marked by immunofluorescence with SON antibody. Scale Bar = 10µm. (f) Quantification was performed as shown in Supplementary Fig. 3a and M mRNA cytoplasmic to nuclear ratio (C/N) was determined after 6h of WSN or WSN ΔNS1 infection. Values are mean ± s.d. measured in 41 WSN and 50 WSN ΔNS1 infected cells. Scale Bar = 10µm. Images shown in a and e are representative of three independent experiments.** T test p value < 0.01.
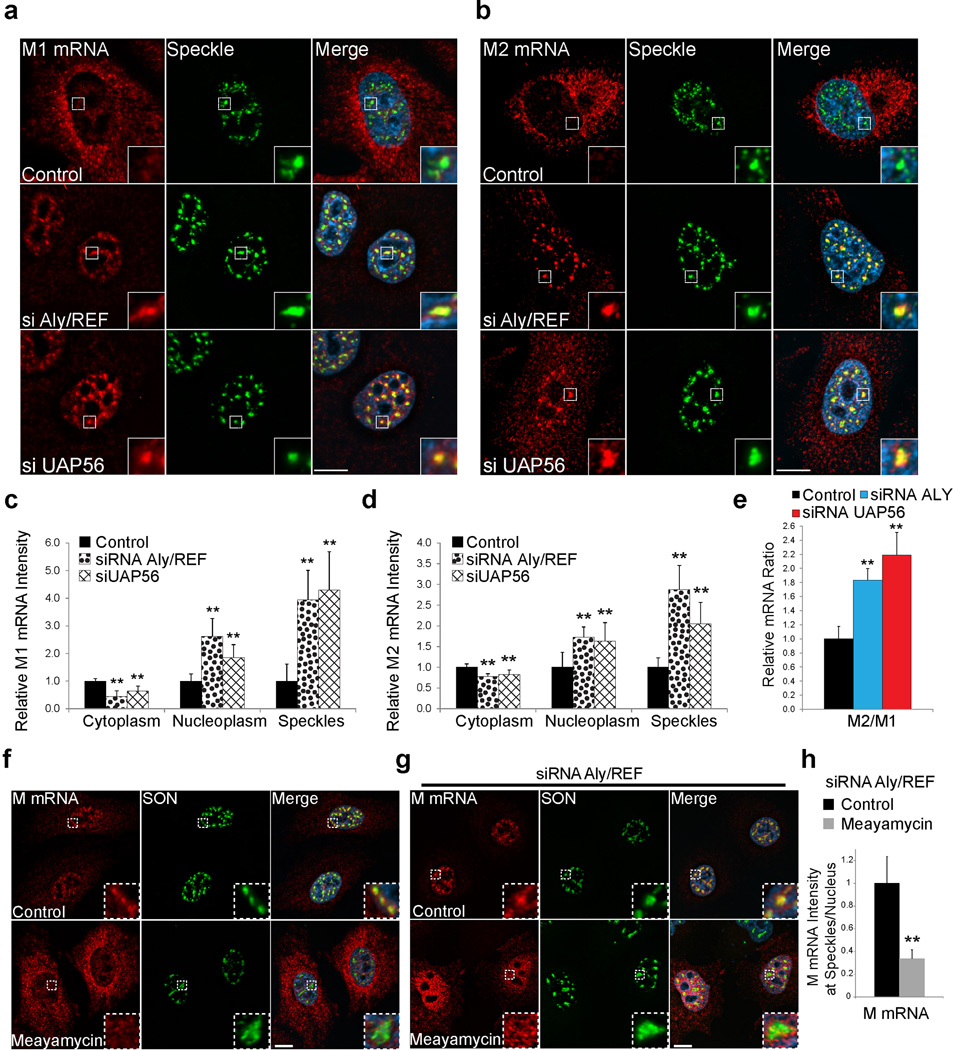
Cellular NS1-BP, hnRNP K, and SON Proteins Mediate M1 mRNA Processing at Nuclear Speckles
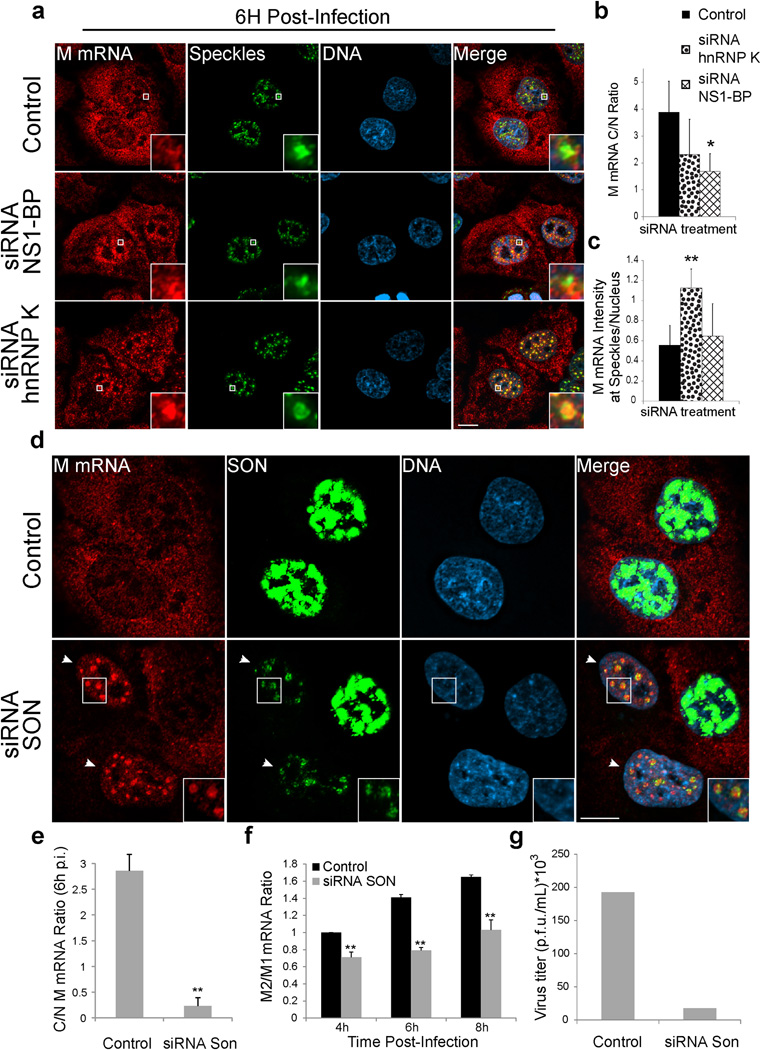
We have previously shown that the cellular protein NS1-BP, which interacts with the viral NS1 protein22, forms a complex with the cellular hnRNP K protein and they are required for proper M1 mRNA splicing into M2 mRNA23. NS1-BP and hnRNP K are RNA binding proteins involved in viral RNA splicing and the latter has also been linked to cellular pre-mRNA splicing24–27. Thus, we tested whether NS1-BP and hnRNP K have a role on M mRNA localization at nuclear speckles. First, we show that a pool of both NS1-BP and hnRNP K is found at nuclear speckles (Supplementary Fig. 4a). NS1-BP has been previously found at nuclear speckles22. In NS1-BP depleted cells, M mRNA accumulation at nuclear speckles was reduced after 4h post-infection but the cytoplasmic to nuclear M mRNA ratio C/N was not altered (Supplementary Fig. 4b,d,e,f). At 6h post-infection, we observed an inhibition of M mRNA nuclear export in cells depleted of NS1-BP (Fig. 4a,b). The observed M mRNA mislocalization shown here and the splicing defects we reported upon NS1-BP knockdown23 are similar to what we showed in cells infected with WSN ΔNS1(Fig. 3 and Supplementary Fig. 3a–f). These data suggest that NS1 and NS1-BP may function together to recruit M1 mRNA to nuclear speckles for splicing and support the concept that M1 mRNA that is generated at 4–6 h post-infection is not efficiently exported without being processed at nuclear speckles.
Figure 4. M mRNA splicing at nuclear speckles is mediated by the host factors NS1-BP, hnRNP K and SON.
(a) A549 cells were transfected with control siRNA, hnRNP K siRNA, or NS1-BP siRNA. After 72h, cells were infected with WSN at MOI 10 for 6 h. Cells were then fixed for M mRNA labeling by RNA-FISH. Nuclear speckles were marked with SON antibody. Insets show an enlarged nuclear speckle region. Scale Bar = 10µm. (b–c) Distribution of M mRNA in the nuclear and cytoplasmic compartments (b) and at speckles (c) was quantified as shown in Supplementary Figure 3a. Values are mean ± s.d. of at least 20 cells that were analyzed for each treatment. (d) A549 cells were transfected with control siRNA or SON siRNA. After 48h, cells were infected with WSN at MOI 10 for 6h. Cells were subjected to RNA-FISH. Nuclear speckles were marked with SON antibody. SON signal was intensified to show the abnormal speckle structure that form in SON depleted cells (marked by white arrows). Insets from cells depleted of SON show an enlarged area with abnormally shaped nuclear speckles. Scale Bar = 10µm. (e) Quantification of M mRNA cytoplasmic to nuclear ratios (C/N ratios). Values are mean ± s.d. measured in 61 cells transfected with control siRNA and in 27 cells transfected with SON siRNA and infected with WSN. p.i.- post infection. (f) RT-qPCR quantification of M2/M1 mRNA ratios from cells transfected with control siRNA or SON siRNA and infected for 4h, 6h, and 8h. The average M2/M1 mRNA ratio from cells treated with control siRNA at 4h post-infection (M2/M1 mRNA = 0.59) was set to 1 and the relative average M2/M1 mRNA ratios at the depicted time points are shown. Values are mean ± s.d. from three independent experiments. (g) WSN replication was determined after 24h in control cells or in SON depleted cells. Virus titer was determined by plaque assay. Values are mean from two independent experiments. Images shown in a and d are representative of three independent experiments.* T test p value < 0.05, ** T test p value < 0.01.
We then depleted hnRNP K, which interacts with NS1-BP23. In contrast to NS1-BP depletion and similar to control cells, hnRNP K depleted cells efficiently accumulated M mRNA at nuclear speckles4h post-infection (Supplementary Fig. 4c,d,e). However, at 6h post-infection we found a significant enrichment of M mRNA at nuclear speckles in hnRNP K depleted cells (Fig. 4a,c) whereas most of M mRNA is not present at nuclear speckles during this time in the control infected cells, as it has already been exported to the cytoplasm (Fig. 4a–c). Since M mRNA enters and accumulates at speckles for a prolonged time in hnRNP K knockdown cells, but does not efficiently splice23, hnRNP K appears to function on M1 to M2 mRNA splicing at nuclear speckles. Although M mRNA accumulation at nuclear speckles upon hnRNP K knockdown is prolonged, the nucleoplasmic M mRNA pool was not enriched, so the C/N ratio of M mRNA was not significantly altered relative to control cells (Fig. 4a,b). InhnRNP K and NS1-BP knockdown cells, the mislocalization phenotypes may be specific to the viral M mRNA, as cellular GAPDH mRNA localization remained unchanged after NS1-BP and hnRNP K depletion (Supplementary Fig. 4g).
To further determine the importance of nuclear speckles for M1 to M2 splicing and export, we perturbed nuclear speckle structure by knocking down the SON protein. SON was shown to be a scaffold/structural nuclear speckle factor and its depletion leads to dispersion of splicing factors from nuclear speckles to the nucleoplasm28, 29. SON was also shown to regulate cell cycle progression, development, and disease28, 29. In control cells where SON is not depleted, we found that most of the M mRNA is in the cytoplasm at 6h post-infection (Fig. 4d,e). In contrast, in SON depleted cells the M mRNA is accumulated at nuclear speckles that contain low levels of SON (Fig. 4d), while the distribution of GAPDH mRNA remains unchanged (Supplementary Fig. 4h). Although some M mRNA was detected in the cytoplasm of SON depleted cells, the C/N ratio of M mRNA was significantly reduced (Fig. 4e), indicating defect in M mRNA nuclear export in these cells. SON knockdown also reduced the M2/M1 mRNA ratio during infection (Fig. 4f). As a result, SON depletion reduced virus replication as compared to control cells (Fig. 4g). This effect is consistent with a previous screen that identified SON as an essential cellular protein for influenza virus replication30. Together, these findings reveal a novel role for the SON protein in the influenza virus life cycle, supporting the concept of M1 mRNA splicing at nuclear speckles.
TREX Components Couple Viral M mRNA Nuclear Export to Splicing via Nuclear Speckles
There is some evidence that the TREX components UAP56 and Aly/REF proteins are involved in the release of certain cellular mRNAs from nuclear speckles7. In the model of mRNA nuclear export, UAP56 is recruited to the nascent transcript and in turn recruits Aly/REF. UAP56 is then displaced by the NXF1-NXT1 mRNA export receptors and Aly/REF enhances NXF1-NXT1 binding to the RNA31. NXF1-NXT1 then mediates the mRNA translocation through the nuclear pore complex. We show that Aly/REF is present at nuclear speckles (Supplementary Figure 5a) and UAP56 has been previously localized at these nuclear bodies32. Additionally, Aly/REF and UAP56 have been shown to be involved in nuclear export of M1 and M2 mRNAs33. Thus, we tested whether depletion of these factors would lead to a change in M mRNA localization or splicing. By monitoring M1 and M2 mRNA, we found that in both Aly/REF and UAP56 siRNA treated cells M1 and M2 mRNA accumulated at nuclear speckles and in the nucleoplasm after 6h, in contrast to control in both A549 and VERO cells (Fig. 5a–5d; Supplementary Fig. 5b,c; Figure 6a; and Supplementary Fig. 6). Surprisingly, RT-qPCR analysis of the M2/M1 mRNA ratio revealed that M1 mRNA splicing was significantly enhanced by the Aly/REF and UAP56 knockdown (Fig. 5e). Despite of this splicing enhancement, both M1 and M2 mRNAs were significantly blocked in the nucleus upon depletion of Aly/REF and UAP56, which resulted in low levels of M1 and M2 proteins (Supplementary Fig. 5d). Thus, M1 mRNA nuclear export block seems to prolong the accumulation of M1 mRNA at nuclear speckles, which likely enhanced M1 mRNA splicing.
Figure 5. Aly/REF and UAP56 depletion inhibits M1 and M2 mRNA nuclear export and mediates speckle dependent M1 to M2 splicing enhancement.
(a and b) A549 cells were transfected with control siRNA, Aly/REF siRNA, or UAP56 siRNA. After 48h, cells were infected with WSN at MOI 10 for 6h. M1 (a) and M2 (b) mRNA labeling was performed by RNA-FISH. Nuclear speckles were marked with SON antibody. (c and d) Distribution of M1 (c) and M2 (d) mRNAs in the cytoplasm, nucleoplasm, and nuclear speckles was quantified as in Supplementary Figure 3a. Intensity sum in each compartment was normalized to the general fluorescence intensity in the cell. Values obtained from cells transfected with control siRNA were set to 1 and the relative fold changes in Aly/REF and UAP56 siRNA transfected cells are shown. Values are mean ± s.d. of at least 9 cells that were analyzed for each condition. (e) RT-qPCR quantification of M2/M1 mRNA ratio in cells transfected with control siRNA, Aly/REF siRNA, or UAP56 siRNA and infected for 6h. Values are mean of ratios ± s.d. from 3 independent experiments. (f) A549 cells were treated with 100 nM meayamycin or 0.1% DMSO (control) for 2h and infected with WSN at MOI 10 for 1h. Virus was removed and 100 nM meayamycin or 0.1% DMSO was added for 3h. Cells were subjected to RNA-FISH. Nuclear speckles were marked with SON antibody. (g) A549 cells were transfected with control siRNA or Aly/REF siRNA. After 48h, cells were processed as in f. (h) M mRNA intensities at nuclear speckles in cells treated as in g were quantified. M mRNA signal at speckles was normalized to the average speckle volume measured within each cell. Values obtained from cells transfected with Aly/REF siRNA (control)were set to 1 and the relative fold changes in meayamycin treated cells depleted of Aly/REF are shown. Values are mean ± s.d. of at least 9 cells that were analyzed for each condition. Images shown in a,b,f and g are representative of three independent experiments. All insets are enlargement of nuclear speckle areas. All scale bars = 10µm.
Figure 6. Nuclear Speckle Assembly Factor SON Interacts with M1 mRNA and Mediators of M1 mRNA Splicing.
(a) VERO cells were transfected with control siRNA, Aly/REF siRNA, or UAP56 siRNA. After 48h, cells were infected with WSN (a) or WSN ΔNS1(b) at MOI 10 for 6h. Cells were then fixed for M mRNA labeling by RNA-FISH. Nuclear speckles were marked with SON antibody. Insets are enlargement of areas showing nuclear speckles. Images shown in a and b are representative of three independent experiments. Scale bar = 10µm. (c) Accumulation of M mRNA at nuclear speckles was quantified and compared between WSN and WSN ΔNS1 infected cells transfected with siRNA as in a and b. Values are mean ± s.d. of at least 11 cells that were analyzed in each treatment. p.i.- post infection. (d) RT-qPCR quantification of M2/M1 mRNA ratio in VERO cells transfected with control siRNA, Aly/REF siRNA, or UAP56 siRNA and infected for 6h with WSN or WSN ΔNS1 at MOI 1. The average M2/M1 mRNA ratio from control siRNA transfected cells infected with WSN (M2/M1 mRNA = 0.495) was set to 1 and the relative average M2/M1 ratio at the depicted conditions are shown. Values are mean ± s.d. from three independent experiments. ** T test p value < 0.01. (e–g) Cell extracts from uninfected or infected A549 cells treated with RNase inhibitor (RNasin) or with RNase A were subjected to immunoprecipitations with antibodies specific to NS1-BP or SON. Immunoprecipitates (Bound) and unbound fractions were subjected to western blot using antibodies specific to the depicted proteins. (h–i) SON binds directly to M1 mRNA. The entire M1 mRNA was radiolabeled uniformly at C residues, incubated with JSL1 nuclear extract under splicing conditions, crosslinked with 254 nm light and digested with RNase. The reaction was then either resolved by SDS-PAGE (25% Total) or incubated in separate reactions with the antibodies indicated (IP: anti-). Total reaction or immunoprecipitated proteins were resolved by SDS-PAGE. (j) Model for influenza virus M1 mRNA trafficking and splicing. Data shown in e–i are representative of three independent experiments.
We next inhibited splicing of M mRNA to determine whether this perturbation would affect its nuclear speckle localization. We used the splicing inhibitor meayamycin that bindsSF3b and therefore prevents the formation of protein-RNA complexes essential for early spliceosome assembly34. M mRNA localization was assessed by RNA-FISH in the absence or presence of this chemical probe. In agreement with previous findings, meayamycin, like its analogue spliceostatin A, increased nuclear speckle sizes (Figure 5f–g and Supplementary Figure 5f)35. Additionally, we found that meayamycin treatment promotes premature nuclear export of M mRNA (Fig. 5f) in addition to inhibiting M1 to M2 splicing (Supplementary Fig. 2d,e). To prevent this premature nuclear export effect and still inhibit M mRNA splicing, we took advantage of the Aly/REF phenotype, which inhibits M mRNA nuclear export and increases both accumulation of M mRNA at nuclear speckles and splicing (Fig. 5a–e and Fig. 5g control), and treated cells with both Aly/REF siRNA and meayamycin (Fig. 5g and Supplementary Figure 5e). We found that meayamycin treatment prevents M mRNA nuclear speckle accumulation and splicing even in Aly/REF depleted cells that contain high levels of nuclear M1 mRNAs (Fig, 5g,h and Supplementary Figure 5e). When meayamycin was removed from the medium, M2 mRNA could again be detected after an additional 4 hours (Supplementary Figure 5e). Examination of both M1 and M2 mRNA distribution by RNA-FISH revealed that M1 and M2 mRNA speckle localization was restored upon meayamycin removal (Supplementary Fig. 5f). These results indicate that the accumulation of M1 mRNA at nuclear speckles is not a random process but rather a mechanism to induce M1 mRNA splicing.
Next we examined the effect of Aly/REF and UAP56 depletion during WSN ∆NS1 virus infection. As expected, both WSN and WSN ∆NS1 infected cells depleted from Aly/REF and UAP56 exhibited a significant M mRNA nuclear export defect (Fig. 6a,b). Importantly, the nuclear speckle accumulation and splicing enhancement seen after Aly/REF and UAP56 knockdown in WSN ∆NS1 infected cells were reduced as compared to WSN infected cells (Fig. 6a–d and Supplementary Fig. 6). These results indicate that the splicing enhancement in Aly/REF and UAP56-deficient cells is dependent on nuclear speckle accumulation and reinforces the new role of NS1 described here as a driver of M mRNA to speckles to undergo splicing.
SON Interacts with M1 mRNA and with Factors Required for M1 mRNA Processing
We then investigated whether NS1 and the cellular proteins involved in M1 mRNA localization and splicing at nuclear speckles physically interact with SON, a major player in nuclear speckle assembly and alternative splicing28, 29. The major form of the SON protein is ~263 kD and runs as a broad band in SDS- PAGE as it is likely post-translationally modified. Lower molecular weight forms of SON are predicted to be alternative spliced forms. We show that NS1-BP and hnRNP K interact with SON in non-infected or infected cells (Fig. 6e–g). In addition, NS1 was pulled-down by SON antibody (Fig. 6g). To this end, NS1 protein from influenza B virus has also been shown to interact with nuclear speckle domains36, further indicating the importance of this intranuclear body to the different strains of influenza virus. The NS1-BP and hnRNP K interactions with SON were detected by immunoprecipitations performed with either anti-NS1-BP antibody or anti-SON antibody in the presence or absence of RNA (Fig. 6e–g). In uninfected cells, the interactions of SON with NS1-BP (Fig. 6e) or SON with hnRNP K (Fig. 6f) were partially dependent on RNA. During infection, interactions of SON with hnRNP K and with NS1showed partial RNA dependence as well (Fig. 6g). Then, we tested whether SON directly interacted with M1 mRNA. M1 mRNA was capped, 32P-labeled, and incubated with nuclear extracts. This was followed by UV treatment to crosslink RNA-protein interactions. RNA was digested and immunoprecipitations were performed with antibodies specific to SON, hnRNP U, Aly/REF and UAP56. hnRNP U was used as negative control based on our previous work, which demonstrated direct interaction of hnRNP K with M1 mRNA and lack of interaction with hnRNP U23. As shown in Figure 6h, SON (~263 kD broad band) and its putative alternative spliced forms directly bound M1 mRNA whereas hnRNP U, Aly/REF and UAP56 did not interact with M1 mRNA (Fig. 6h,i). Lack of direct interaction between Aly/REF or UAP56 with M1 mRNA is not surprising as they likely require adaptor proteins to bind RNA. Thus, SON interaction with NS1, NS1-BP and hnRNP K in addition to directly bind M1 mRNA support the functional data that indicates M1 mRNA splicing at nuclear speckles.
Discussion
Taken together, our data indicates that M1 mRNA trafficking to nuclear speckles is dependent on the viral NS1 protein and the cellular NS1-BP protein. These findings support a model in which NS1 interacts with NS1-BP and directs the M1 mRNA to nuclear speckles during infection for hnRNP K-dependent splicing into M2 mRNA. It has been previously reported that NS1 can bind M1 mRNA and regulate M1 mRNA splicing17. Here, we revealed a previously unknown function of NS1 as a mediator of M1 mRNA targeting to nuclear speckles and found that this promotes M1 to M2 mRNA splicing (Fig. 6j). This may explain the unique M1 mRNA splicing kinetics in which M1 production predominates early in infection when NS1 synthesis is low, and then M1 to M2 mRNA splicing is likely enhanced by the high levels of NS1 at late stages of infection37. M2 protein has been shown to be cytotoxic to cells when expressed alone38, which suggests that M2 levels need to be tightly regulated during infection. Furthermore, M2 is critical for viral budding39, indicating that timing of expression is likely important for production of infectious viral particles, as premature synthesis could lead to defective particles.
The localization of M mRNA into speckles and M1 to M2 mRNA splicing also appears to be coupled to nuclear export. When M1 mRNA localization into intact speckles was impaired by infection with NS1-deficient virus or by depletion of NS1-BP or SON, we observed an accumulation of unspliced M1 mRNA in the nucleus. This effect may be related to previous reports suggesting that mRNA splicing can enhance mRNA nuclear export40–43. While our findings indicate that NS1 promotes viral M1 mRNA targeting to nuclear speckles for splicing into M2 mRNA that is efficiently exported to the cytoplasm, NS1 is known to inhibit cellular mRNA export to prevent proper host gene expression and antiviral response44–47, which happens later than the infection times used here. Together, these results indicate a differential role for NS1 in the nuclear export of viral versus host mRNAs, which may occur via different RNA-protein interactions. Another possibility is that the actions of NS1 on intranuclear trafficking of M mRNA are indirect, as NS1 is known to be a multifunctional protein involved in regulation of gene expression at the RNA and protein levels48.
Regarding M mRNA nuclear export, we showed that the cellular mRNA export factors Aly/REF and UAP56 are required for M1 and M2 mRNA export to the cytoplasm. In addition, knockdown of Aly/REF and UAP56 support the compartmentalized M1 mRNA splicing at nuclear speckles. Depletion of these mRNA export factors enhanced the accumulation ofM1 and M2 mRNAs at nuclear speckles and increased M1 to M2 mRNA splicing. Since M1 mRNA was blocked in the nucleus, the pool of M1 mRNA available for splicing is high and therefore may explain the increased levels of M1 mRNA splicing. Upon meayamycin treatment of Aly/REF depleted cells, where nuclei were highly enriched with M1 mRNA, nuclear speckle accumulation of M1 mRNA was significantly reduced and a diffusive pattern was observed. This suggests that initial spliceosome assembly and specifically U2 snRNP deposition on M1 mRNA is required for M1 mRNA nuclear speckle accumulation and splicing as meayamycin analogues were shown to bind the SF3b complex35. Importantly, enhancement of M1 mRNA splicing by Aly/REF and UAP56 knockdown was found to be dependent on entry into nuclear speckles, which was mediated by the viral NS1 protein. This finding underscores the importance of NS1 protein as a mediator of M1 mRNA targeting and splicing at nuclear speckles.
Active spliceosomes are present in nuclear speckles and appear to be involved in post-transcription splicing6. Here we found that M vRNAs that serve as templates for the generation of M1 mRNA were not present at nuclear speckles. This indicates that M1 mRNA is not generated at nuclear speckles but is post-transcriptionally spliced at this nuclear body. At steady state, active spliceosomes residing at nuclear speckles represent ~9–25% of the entire active splicesome population6 as most cellular pre-mRNAs are spliced at transcribing genes5, 49 and not inside nuclear speckles1. Thus, influenza virus subverts nuclear speckles to likely usurp splicing factors present in high concentration to promote efficient splicing, and possibly inhibit host gene expression and antiviral response. Alternatively, the virus may utilize specific factors that are active at nuclear speckles for its M1 mRNA processing. The localization of M1 mRNA at nuclear speckles is consistent with a previous study showing differential intranuclear fractionation and splicing of M1 mRNA into M2 mRNA and mRNA3 when the M segment was expressed using a SV40 recombinant virus as opposed to M1 mRNA splicing during influenza virus infection50. Thus, transcription of M1 mRNA by the specific influenza virus polymerase machinery and other viral factors may dictateM1 mRNA splicing at nuclear speckles. Finally, inhibition of this specific viral mRNA pathway through nuclear speckles may provide a strategy for identification of antivirals.
Methods
Cell Culture
Human lung adenocarcinoma epithelial cells (A549), VERO cells and MDCK cells were cultured in high-glucose DMEM (Gibco), 10% FBS (Atlas), 100 units/ml Pen/Strep antibiotics. All cells were maintained at 37°C with 5% CO2. Cells were tested negative for mycoplasma. Cell lines were obtained and authenticated by ATCC.
Viruses
Influenza virus (A/WSN/33, A/Texas/36/91, A/Puerto Rico/8/34) was propagated in MDCK cells. Virus titer was determined by plaque assay23. Influenza virus WSNΔNS1 was generated as previously described21.
RNA interference and transfections
For imaging experiments, cells were grown on glass coverslips (Fisherbrand, Fisher Scientific) coated with 1µl 0.1% gelatin (Sigma-Aldrich). The following day, siRNA oligos were transfected (see Supplementary Information for siRNA oligos specifications) using RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Several siRNA oligos were tested for each mRNA target to exclude off target effects. Plasmids (See Supplementary Information) were transfected with Lipofectamine 2000 Transfection Reagent (Life technologies) according to the manufacturer’s protocol. After the indicated incubation time and/or infection, cells were either fixed for imaging or harvested for RNA or protein purification.
Immunofluorescence
Cells grown on coverslips were fixed for 15 min in 4% Paraformaldehyde (PFA) (Electron Microscopy Sciences) and then permeabilized for 5 min in 0.5% Triton X-100. For detection of Aly/REF (see Supplementary Information for buffers, Plasmids, antibodies and probes specifications)at speckles, cells were permeabilized on ice with 0.05% Triton X-100 for 5 min before fixation. After blocking (20 min, 5% BSA), cells were immunostained for 1h with a primary antibody (see Supplementary Information for buffers, antibodies, and probes specifications). Primary antibody was washed 3 times with PBS and then labeled with secondary antibody for 1h. Coverslips were then washed twice with PBS, stained with 1µg/ml Hoechst 33258 (Molecular Probes-Life technologies) for 10min, and briefly washed with PBS. Coverslips were mounted in ProLong Gold antifade reagent (Life technologies).
Fluorescence in situ hybridization and Immunofluorescence
Cells were fixed for 15 min in 4% PFA, incubated in ethanol for 12 h at 4°C, and then permeabilized for 5 min in 0.5% Triton X-100. For detection of Prp8 at speckles, cells were permeabilized on ice with 0.05% Triton X-100 for 5 min before fixation. Next, cells were briefly washed with PBS and immunostained for 1h with a primary antibody diluted in Inhibition Buffer (see Supplementary Information for buffers, antibodies and probes specifications). Coverslips were briefly washed with 0.2% Triton X-100 and PBS. Wash Buffer (WB) was added for 5 min. WB was removed and FISH probe in Hybridization Buffer (HB) was added. Hybridization was carried at 37°C for 4h. Probe was removed and cells were washed with WB for 30 min at 37°C and briefly washed with PBS. Cells were labeled with secondary antibody for 1h. Coverslips were washed twice for 5 min in PBS, stained with 1 µg/ml Hoechst 33258 (Molecular Probes-Life Technologies) for 10 min, and then briefly washed with PBS. Coverslips were mounted in ProLong Gold antifade reagent (Life Technologies). In situ hybridization alone was performed as described above without triton or antibody treatment.
Fluorescence microscopy and data analysis
Images were obtained with a Zeiss Axiovert 200 M automated microscope controlled by the AxioVision software. Images were taken with the Zeiss 60× Plan-APOCHROMAT lens, 1.4 numerical aperture, and captured by the Zeiss AxioCam MRm camera. Multiple z plans were captured for collecting signal from both the cytoplasm and nuclear compartments. Z stacks images were deconvolved with the AutoQuant software. Pearson’s Correlation Coefficients were analyzed by Imaris (Bitplane) using the ImarisColoc tool. Pearson’s Correlation Coefficients were measured in the nucleus using Hoechst channel for masking. Additionally, deconvolved images were analyzed by Imaris (Bitplane) using the Spots tool for single RNA detection and quantification, and the Surfaces tool for segmentation and signal analysis within the cytoplasm, nucleus, and nuclear speckles. The M mRNA cytoplasmic fluorescent signal presented in Figure 1e–1f was calculated based on volume analysis. Eleven, 0.3µm z-sections were taken for each 3d image analyzed. Signal was then deconvolved using the AutoQuant software (3d deconvolution, 10 iterations). Then, deconvolved images were analyzed using the Imaris spots tool, which detected individual spots (isolated spots could be resolved in the cytoplasm) and quantified the intensity sum of voxels within the detected spot volume. The distributions of single mRNA florescent intensities detected are unimodal and the average intensity was calculated for further analysis of the bulk M mRNA signal at nuclear speckles.
RNA purification and RT-qPCR
Total RNA was isolated from A549 cells with TRIzol Reagent (Ambion) according to the manufacturer’s instructions. RNA was reverse transcribed into cDNA by SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s protocol. cDNA samples were amplified using the SYBR green I (Roche) and specific primers (see Supplementary Information for primers specifications) in a LightCycler 480 quantitative real-time PCR (qPCR) system (Roche). In the case of RT-PCR, cDNA samples were amplified using Advantage 2 Polymerase Mix (Clontech) in a T100 Thermo Cycler (Bio-Rad), and products were analyzed by agarose gel electrophoresis.
Meayamycin treatment
Cells were treated with either 100 nM meayamycin (in DMEM containing 0.1% DMSO) or 0.1% DMSO in DMEM (control) for 2h and then infected with WSN at MOI 10. After 1h, virus was removed and 100 nM meayamycin or 0.1% DMSO was added in Infection Media for either 4h or 6h. Cells were then either fixed for imaging or harvested for RNA purification. In Supplementary Figure 5, cells were infected at MOI 10. After 1h, virus was removed and 50 nM meayamycin or 0.1% DMSO was added to the Infection Media for 3h. Media was then removed and Infection Media containing either 50 nM meayamycin or 0.1% DMSO was added for an additional 4h. Cells were then either fixed for imaging or harvested for RNA purification.
Immunoprecipitation
Immunoprecipitation was performed as previously described23.
UV crosslinking and RNA-protein interaction
The full length M1 mRNA was transcribed from a linearized plasmid-based template by T7 polymerase using 32P-CTP to 32P- label the RNA throughout its length. Labeled M1 mRNA (10 nM) was incubated with 40% JSL1 nuclear extract in a total volume of 10.2 µl under splicing condition, which contains (final concentration): 12 mM Tris-HCl, pH 7.5, 3.2 mM MgCl2, 1 mM ATP, 20 mM CP, 0.12 mM EDTA, 60 mM KCl, 1.3% PVA, 250 ng yeast tRNA, 200 ng BSA, and 12% glycerol. Reactions were incubated at 30°C for 20 min, crosslinked using UV light (254 nm) for 20 min on ice, and digested with RNaseT1 and RNase A for 20 min at 37°C. Reactions were resuspended in 2× SDS loading buffer, denatured for 5 min at 95°C, analyzed under denaturing conditions on an SDS-PAGE gel (Acrylamide/Bis 37.5:1, BioRad), and detected by autoradiography. Immunoprecipitation after crosslinking was carried out with the antibodies against SON (GTX129778 GeneTex), UAP56 (SAB1307254 Sigma), AlyRef (A9979 Sigma), and hnRNP U (ab10297 abcam).
Statistic Analysis
Statistical analyses were performed using the Two-Sample, Two-tailed, T-Test Assuming Equal Variance. For statistical analysis of M mRNA (M mRNA, M1 mRNA, or M2 mRNA) imaging studies, the average intensity sum at nuclear speckles (~20–50 speckles/cell) within each cell −9 to 29 cells were analyzed depending on the experimental condition and is described in the figure legend. Therefore, a minimum of 200 speckles were analyzed for each condition. For all imaging studies, a one-sample Kolmogorov–Smirnov test was conducted. A normal distribution can be assumed for all populations (P > 0.05). For the statistical analysis of M2/M1 mRNA ratios, 3 independent experiments were performed for each condition and Two-Sample, Two-tailed, T-Test was performed assuming equal variance.
Supplementary Material
Acknowledgments
Funding was provided by NIH R01 GM113874-01, AI079110, R01 AI089539, CPRIT RP121003-RP120718-P2 to B.F; NIH T32CA124334 to A.M; CRIP (Center for Research in Influenza Pathogenesis), NIAID funded Center of Excellence for Influenza Reserch and Surveillance (CEIRS, contract # HHSN272201400008C) to A-GS. We thank Richard Cadagan and Katherine Phelps (Live Cell Imaging Core Facility) for assistance.
Footnotes
Author Contributions
A.M, A.W., K.Z., M.T. - designed and conducted experiments
R. M-M. - conducted experiments and generated important reagents
M.E. – conducted experiments
K.K. – generated important reagent
K.W.L. – designed experiments
A.M., A. G-S., and B.M.A.F. – designed experiments and wrote the paper
COMPETING FINANCIAL INTEREST: The authors declare no competing financial interests.
References
- 1.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harbor perspectives in biology. 2011;3 doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saitoh N, et al. Proteomic analysis of interchromatin granule clusters. Molecular biology of the cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 5.Tilgner H, et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Girard C, et al. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nature communications. 2012;3:994. doi: 10.1038/ncomms1998. [DOI] [PubMed] [Google Scholar]
- 7.Dias AP, Dufu K, Lei H, Reed R. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nature communications. 2010;1:97. doi: 10.1038/ncomms1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akef A, Zhang H, Masuda S, Palazzo AF. Trafficking of mRNAs containing ALREX-promoting elements through nuclear speckles. Nucleus. 2013;4:326–340. doi: 10.4161/nucl.26052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishihama Y, Tadakuma H, Tani T, Funatsu T. The dynamics of pre-mRNAs and poly(A)+ RNA at speckles in living cells revealed by iFRAP studies. Experimental cell research. 2008;314:748–762. doi: 10.1016/j.yexcr.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 10.Melcak I, Melcakova S, Kopsky V, Vecerova J, Raska I. Prespliceosomal assembly on microinjected precursor mRNA takes place in nuclear speckles. Molecular biology of the cell. 2001;12:393–406. doi: 10.1091/mbc.12.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubois J, Terrier O, Rosa-Calatrava M. Influenza viruses and mRNA splicing: doing more with less. mBio. 2014;5:e00070–e00014. doi: 10.1128/mBio.00070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson D, Lamb RA. The influenza A virus spliced messenger RNA M mRNA3 is not required for viral replication in tissue culture. The Journal of general virology. 2008;89:3097–3101. doi: 10.1099/vir.0.2008/004739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb RA, Lai CJ, Choppin PW. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:4170–4174. doi: 10.1073/pnas.78.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shih SR, Suen PC, Chen YS, Chang SC. A novel spliced transcript of influenza A/WSN/33 virus. Virus Genes. 1998;17:179–183. doi: 10.1023/a:1008024909222. [DOI] [PubMed] [Google Scholar]
- 15.Wise HM, et al. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS pathogens. 2012;8:e1002998. doi: 10.1371/journal.ppat.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robb NC, Fodor E. The accumulation of influenza A virus segment 7 spliced mRNAs is regulated by the NS1 protein. J Gen Virol. 2012;93:113–118. doi: 10.1099/vir.0.035485-0. [DOI] [PubMed] [Google Scholar]
- 18.Bier K, York A, Fodor E. Cellular cap-binding proteins associate with influenza virus mRNAs. J Gen Virol. 2011;92:1627–1634. doi: 10.1099/vir.0.029231-0. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, et al. Imaging and characterizing influenza A virus mRNA transport in living cells. Nucleic acids research. 2008;36:4913–4928. doi: 10.1093/nar/gkn475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fortes P, Lamond AI, Ortin J. Influenza virus NS1 protein alters the subnuclear localization of cellular splicing components. J Gen Virol. 1995;76(Pt 4):1001–1007. doi: 10.1099/0022-1317-76-4-1001. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, et al. Inhibition of pyrimidine synthesis reverses viral virulence factor-mediated block of mRNA nuclear export. The Journal of cell biology. 2012;196:315–326. doi: 10.1083/jcb.201107058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolff T, O'Neill RE, Palese P. NS1-binding protein (NS1-BP): A novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nucleus of infected cells. J. Virol. 1998;72:7170–7180. doi: 10.1128/jvi.72.9.7170-7180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai PL, et al. Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza A virus RNA splicing. PLoS Pathog. 2013;9:e1003460. doi: 10.1371/journal.ppat.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao W, Razanau A, Feng D, Lobo VG, Xie J. Control of alternative splicing by forskolin through hnRNP K during neuronal differentiation. Nucleic acids research. 2012;40:8059–8071. doi: 10.1093/nar/gks504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchand V, et al. Identification of protein partners of the human immunodeficiency virus 1 tat/rev exon 3 leads to the discovery of a new HIV-1 splicing regulator, protein hnRNP K. RNA biology. 2011;8:325–342. doi: 10.4161/rna.8.2.13984. [DOI] [PubMed] [Google Scholar]
- 26.Motta-Mena LB, et al. A disease-associated polymorphism alters splicing of the human CD45 phosphatase gene by disrupting combinatorial repression by heterogeneous nuclear ribonucleoproteins (hnRNPs) J Biol Chem. 2011;286:20043–20053. doi: 10.1074/jbc.M111.218727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venables JP, et al. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol. 2008;28:6033–6043. doi: 10.1128/MCB.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma A, Takata H, Shibahara K, Bubulya A, Bubulya PA. Son is essential for nuclear speckle organization and cell cycle progression. Molecular biology of the cell. 2010;21:650–663. doi: 10.1091/mbc.E09-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu X, Ng HH, Bubulya PA. The role of SON in splicing, development, and disease. Wiley interdisciplinary reviews. RNA. 2014;5:637–646. doi: 10.1002/wrna.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karlas A, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 31.Hautbergue GM, Hung ML, Golovanov AP, Lian LY, Wilson SA. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc Natl Acad Sci U S A. 2008;105:5154–5159. doi: 10.1073/pnas.0709167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gatfield D, et al. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol. 2001;11:1716–1721. doi: 10.1016/s0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- 33.Read EK, Digard P. Individual influenza A virus mRNAs show differential dependence on cellular NXF1/TAP for their nuclear export. J Gen Virol. 2010;91:1290–1301. doi: 10.1099/vir.0.018564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albert BJ, et al. Meayamycin inhibits pre-messenger RNA splicing and exhibits picomolar activity against multidrug-resistant cells. Molecular cancer therapeutics. 2009;8:2308–2318. doi: 10.1158/1535-7163.MCT-09-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaida D, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nature chemical biology. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 36.Schneider J, Dauber B, Melen K, Julkunen I, Wolff T. Analysis of influenza B Virus NS1 protein trafficking reveals a novel interaction with nuclear speckle domains. J Virol. 2009;83:701–711. doi: 10.1128/JVI.01858-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valcarcel J, Portela A, Ortin J. Regulated M1 mRNA splicing in influenza virus-infected cells. J Gen Virol. 1991;72(Pt 6):1301–1308. doi: 10.1099/0022-1317-72-6-1301. [DOI] [PubMed] [Google Scholar]
- 38.Ilyinskii PO, Gabai VL, Sunyaev SR, Thoidis G, Shneider AM. Toxicity of influenza A virus matrix protein 2 for mammalian cells is associated with its intrinsic proton-channeling activity. Cell Cycle. 2007;6:2043–2047. doi: 10.4161/cc.6.16.4564. [DOI] [PubMed] [Google Scholar]
- 39.Nayak DP, Balogun RA, Yamada H, Zhou ZH, Barman S. Influenza virus morphogenesis and budding. Virus Res. 2009;143:147–161. doi: 10.1016/j.virusres.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mor A, et al. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nature cell biology. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- 41.Cheng H, et al. Human mRNA export machinery recruited to the 5' end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 42.Luo MJ, Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci U S A. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valencia P, Dias AP, Reed R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:3386–3391. doi: 10.1073/pnas.0800250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3' end formation of cellular pre-mRNAs. Mol. Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 45.Qiu Y, Krug RM. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A) J Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Satterly N, et al. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci U S A. 2007;104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, et al. Inhibition of Pyrimidine Synthesis Reverses Viral Virulence Factor-Mediated Block of mRNA Nuclear Export. J. Cell Biol. 2012;196:315–326. doi: 10.1083/jcb.201107058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ayllon J, Garcia-Sastre A. The NS1 Protein: A Multitasking Virulence Factor. Current topics in microbiology and immunology. 2015;386:73–107. doi: 10.1007/82_2014_400. [DOI] [PubMed] [Google Scholar]
- 49.Kornblihtt AR, et al. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14:153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 50.Valcarcel J, Fortes P, Ortin J. Splicing of influenza virus matrix protein mRNA expressed from a simian virus 40 recombinant. J Gen Virol. 1993;74(Pt 7):1317–1326. doi: 10.1099/0022-1317-74-7-1317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.