Abstract
Biological tissues and biomaterials are often defined by unique spatial gradients in physical properties that impart specialized function over hierarchical scales. The structure and organization of these materials forms continuous transitional gradients and discrete local microenvironments between adjacent (or within) tissues, and across matrix-cell boundaries, which can be difficult to replicate with common scaffold systems. Here, we studied the matrix densification of collagen leading to gradients in density, mechanical properties, and fibril morphology. High-density regions formed via a fluid pore pressure and flow-driven mechanism, with increased relative fibril density (10×), mechanical properties (20×, to 94.40±18.74kPa), and maximum fibril thickness (1.9×, to >1μm) compared to low-density regions, while maintaining porosity and fluid/mass transport to support viability of encapsulated cells. Similar to the organization of the articular cartilage zonal structure, we found that high-density collagen regions induced cell and nuclear alignment of primary chondrocytes. Chondrocyte gene expression was maintained in collagen matrices, and no phenotypic changes were observed as a result of densification. Densification of collagen matrices provides a unique, tunable platform for the creation of gradient systems to study complex cell-matrix interactions. These methods are easily generalized to compression and boundary condition modalities useful to mimic a broad range of tissues.
Keywords: 3D microenvironment, physical gradient scaffold, chondrocyte phenotype and dedifferentiation, molecular crowding, plastic compression
1. Introduction
Spatial gradients abound in nature, and in biological tissues these gradients influence specific functions and local cellular responses. Gradients are found between tissues (e.g. ligament-bone insertions), within tissues (e.g. spatial patterning of cartilage, skin, and heart), and at the intercellular level (e.g. organization of the extracellular, interstitial, and pericellular matrices). Physical gradients influence the transmission of stress and strain within tissues, as well as provide transition zones between regions of specialized matrix organization or biochemical composition[1, 2]. Cells within tissues are sensitive to regional and spatially-dependent variations in mechanical properties, extracellular matrix (ECM) ultrastructure, and ligands including growth factors and ECM binding sites, among others[3, 4].
Of numerous possible materials suitable for the creation of three-dimensional (3D) matrices, polymerizable collagens are often used to investigate cell-matrix interactions due to their physiological ubiquity and biocompatibility. Collagen is the primary structural material of connective tissues, giving rise to much of the structural integrity of the tissue, and is a primary matrix component with which cells physically and chemically interact. Moreover, recent research has shown that collagen matrices can be formed into 3D matrices with near tissue-like properties [5, 6]. Integrin binding sequences in the collagen structure act as signaling mechanisms, driving gene and protein expression similar to native physiology[7]. Collagen is easily obtained, and recent collagen extraction developments have yielded collagen materials which closer represent the physicochemical structure found in the native state[8]. Further, collagen-based matrices have been extensively used with a variety of cell populations to investigate cell-matrix interactions.
However, broad application of collagen is narrowed by the low density of matrices that can be formed by conventional means. The density of collagen matrices are often limited by the solubility of the collagen, typically bounding the density to less than 20 mg/mL (2% wet weight) as compared to native tissues such as articular cartilage (200 mg/mL, 20% wet weight) or ligament (225-300 mg/mL, 22.5-30%, wet weight)[8, 9]. Recently, efforts have been made in the development of high density collagen matrices beyond the solubility limit. To date, the most successful methods in the development of high-density collagen matrices have utilized molecular crowding (100-400 mg/mL), evaporation (≤ 1000 mg/mL), and matrix compression (≤230 mg/mL)[10-12]. Of these methods, only matrix compression has been shown so far to viably encapsulate cell populations prior to formation[13]. Molecular crowding and evaporation-based methods have shown that seeded cells can survive on the surface, but do not easily infiltrate into the matrix[10, 12]. Without cellular encapsulation in the engineered environment, investigation of the 3D cellular response within the matrix is simply not possible, as cells that are seeded on the exterior and infiltrate can lead to a degraded or remodeled ultrastructure, potentially eliminating the effects of the designed microenvironment[14].
In this work, we explored densification of collagen matrices in a controlled, confined compression process, and with embedded cells, to characterize density, mechanical properties, fibril structure/morphology, and the cellular response. Although previous work has noted the development of gradients as a result of compression[5, 6, 15], the concept has not been explored further, and there remains a need to develop platforms to investigate fundamental local cell-matrix interactions in these complex structures, and model systems to gain further insight into disease etiology and regenerative medicine approaches. We addressed the following four objectives, to: (1) characterize density, ultrastructure, and mechanical properties of collagen oligomer matrices as a result of densification, (2) examine potential mechanisms that drive collagen densification in confined compression, (3) investigate the ability to densify and create viable cellularized collagen matrices, and (4) characterize chondrocyte morphology and expression of extracellular and nuclear proteins after short term culture in densified matrices, as compared to native and monolayer culture environments.
2. Results and Discussion
2.1. Densification by Confined Compression Produces Gradients in Density, Fibril Morphology, and Mechanical Properties
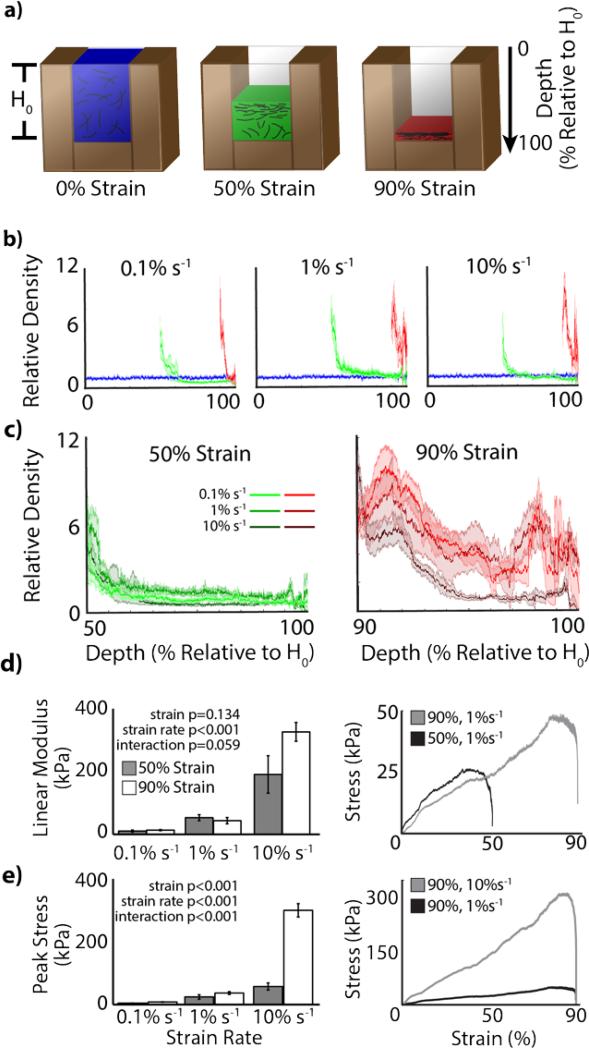
Densification of homogenous, low-concentration collagen-fibril matrices via confined compression resulted in a fibril density gradient in the direction of compression (Figure 1-2). Control samples did not exhibit any densification effects, maintaining a relatively homogenous density throughout the sample. In densified samples, high fibril concentrations were measured at the impact surface, which then decreased and approached the initial density with depth. Across strain rates, densification to 50% and 90% exhibited similar results, where maximum densification occurred near the impact surface (top), which then decreased toward control density (Figure 2b-c). At all strain rates, 50% strain samples plateaued to approximately control density at approximately 1/3 of the final height from the impact surface. Little difference was observed in densification patterns across strain rates at 50% (Figure 2c). Alternatively, 90% strain rates exhibited different densification patterns, with higher strain rates peaking at higher densifications near the impact surface. Additionally, densification often occurred throughout the depth of the matrix, resulting in fibril densities greater than initial (control) density (Figure 2c). Strain rate (p<0.001), but not strain magnitude (p=0.134) was found to significantly increase the linear modulus of the force profile required to densify the matrices (Figure 2d). Both strain rate (p<0.001) and strain magnitude (p<0.001) were found to affect the peak force required for densification (Figure 2e). With the final goal of cellular encapsulation and characterization, we chose to utilize 0.1% s−1 strain rate samples for all further analysis to minimize internal mechanical stresses.
Figure 1.
Gradient densification of collagen matrices via confined compression. Collagen matrices were polymerized in a rectangular mold bound by cover-glass to allow for confocal microscopy analysis without matrix manipulation. Collagen matrices were compressed with a porous platen of defined porosity (15%), directing fluid flow through the porous boundary condition. Localized, high-density collagen was observed near the compression surface that decreased with depth into the matrix. High-density regions were associated with higher mechanical properties and fibril alignment/bundling. b) Collagen matrices compressed over a range of strain magnitudes (shown in absolute percent) depict the qualitative effect of local densification, where highest densities are localized to the surface which decreases over distance from the indented surface (grid spacing = 2.1mm). c) Confocal reflectance microscopy (with image intensity normalized to distinguish fibrils over all density levels) revealed high densities of collagen fibrils in (top) areas of high opacity (near the indented surface) with more distributed fibrils near the bottom, more translucent region of the matrix (bar = 100μm).
Figure 2.
Quantitative analysis of collagen density gradients. a) All quantitative analysis was depicted respective to initial height and color coded, i.e. top of control = 0%, top of 50% strain samples = 50%, and top of 90% strain samples = 90%. b) Comparison across strain magnitudes showed an agreement of quantitative and qualitative analysis, i.e. higher density regions were achieved near the indented surface with a reduction in density toward the initial (control) density with distance along the height toward the bottom (mean±standard deviation). c) Comparison across strain rates (data replotted) showed little discernable difference between similar strain magnitudes. d) During the densification process, linear modulus of the loading regime depended on strain rate, but not strain magnitude. e) Peak stress depended on both strain magnitude and rate.
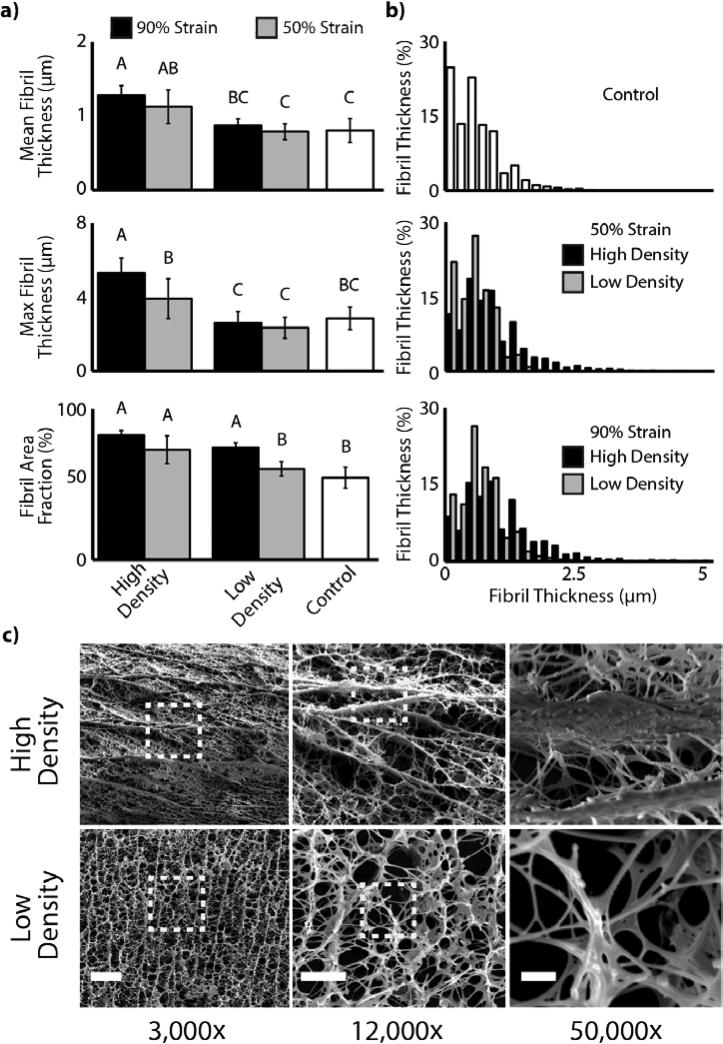
Densification of collagen oligomers resulted in fibril alignment and apparent bundling of fibrils. Control samples exhibited a collagen network of random fibril orientation and submicron fibril diameters. With densification, collagen fibrils near the impact surface were found to be highly aligned perpendicular to the axis of densification (Figure 3c). Similar to density trends, fibril alignment and bundling were seen to decrease toward the low density regions. Quantitatively, we found mean fibril thickness, maximum fibril thickness, and fibril area fraction to be dependent on compression strain magnitude (p=0.002, p=0.004, p<0.001, respectively) and density region (p<0.001, p<0.001, p=0.001, respectively) (Figure 3a-b). Each of the measured dependent variables increased with compression strain magnitude and similarly were increased in high compared to low density regions within each strain magnitude group.
Figure 3.
Collagen densification induces in local fibril alignment and bundling. a) Measurements of mean fibril thickness, maximum fibril thickness, and fibril area fraction were increased in density compared to control regions. Letters indicate statistically significant groups (p<0.004). b) Distribution of fibril thicknesses varied with strain magnitude and density region (high vs low). c) Qualitative analysis reveals fibril bundling in high-density regions (scale bar: 3000×=15μm, 12,000×=5μm, 50,000×=1μm). Dashed boxes indicate area of subsequent zoom.
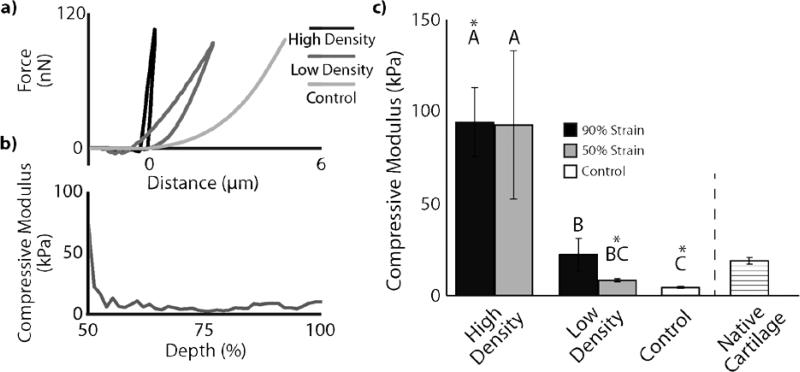
Compression of low-concentration collagen-fibril matrices and the development of gradient collagen density altered spatial mechanical properties. Densification (p<0.001) and density region (high vs. low density, p<0.001) were found to have significant effects on compressive modulus (Figure 4). Control matrices exhibited compressive modulus values of 4.49±0.53 kPa. Densification to 50% strain was found to result in a compressive modulus of 92.88±40.34 kPa in high density regions (p<0.05 compared to control) and 8.32±0.82 kPa in low density regions (p>0.05 compared to control). Densification to 90% strain was found to result in a compressive modulus of 94.40±18.74 kPa in high density regions (p<0.05 compared to control) and 22.22±8.94 kPa in low density regions (p<0.05 compared to control).
Figure 4.
AFM analysis reveals depth-dependent mechanical stiffening that corresponds with density. a) Individual AFM indentation curves showed steeper linear regions (high density>low density>control) for a 50% strain magnitude sample. b) For a single sample (50% strain magnitude) from top (left) to bottom, compressive modulus curve was qualitatively similar to density curve, confirming the correlation between local density and mechanical properties. c) Statistical analysis of strain magnitudes and density regions show up to 20-fold increases in mechanical properties over control and local mechanical properties that exceed that of native cartilage. High variability seen in high density regions is likely due to variability in porosity and fibril diameter (Figure 3c, S1). Different letters indicate statistical difference (p<0.05). *=statistically different from native cartilage (p<0.05).
Interestingly, bovine articular cartilage, measured under identical conditions, was found to have a compressive modulus of 18.88±1.80 kPa (Figure 4c). A two-sample t-test revealed that this value was not statistically different from 50% strain, high density samples (p=0.083) and 90% strain, low density samples (p=0.717). All other comparisons (90% strain high density vs. cartilage p=0.001, 50% strain low density vs. cartilage p<0.001, and 0% strain control vs. cartilage p<0.001) were found to be significantly different.
Densification via controlled, confined compression resulted in complex 3D collagen-fibril matrices with spatial gradients in density, fibril morphology, and local mechanical properties. Densified collagen matrices showed high density regions reaching over 20-fold the initial density and localizing near the impact surface, consistent with previous work[5, 15]. High mechanical properties, large collagen aggregate bundles, and directional fibril alignment were found to be correlated with regions of high density. These effects decreased with distance from the high density surface similar to the gradient decreases in relative density. Other current research in this area focuses on the development of thin collagen sheets, or gradients formed transverse to the compression direction by altering matrix dimensions prior to compression [16]. This work showed that compression does not always result in homogenous sheets, and that the formation of gradients was repeatable and results in the formation of unique environments across the different regions of the matrix.
Interestingly, we found the formation of large fibril aggregates (>1μm) in high density regions, which to our knowledge is not noted in current literature. The large range of mechanical properties seen is additionally useful, as previous studies have noted that differences in mechanical properties within this range to induce specific cell differentiation and phenotype[3, 16].
High density regions were found to have an average compressive modulus, via the AFM parameters used, that was significantly higher compared to native bovine cartilage tested under identical conditions. This was likely due to the inhomogeneous aggregation of large fibril aggregates surrounded by highly porous collagen regions, seen in densified collagen matrices (Figure 3c). As a result, compressive modulus results are highly variable, where measurements show a maximum modulus of 752.6 kPa and a minimum modulus of 5.8 kPa (similar to control) in high density regions. Conversely, native bovine cartilage is composed of smaller diameter fibrils which are more homogenously distributed (Figure S1). These effects are accounted for in the standard deviation bars seen (Figure 4c). Although the AFM results suggest that high density regions exceed the mechanical properties of native cartilage, we note that the collagen matrices can continue to be plastically deformed and resist further bulk compression in a manner similar to native cartilage.
2.2. Finite Element Analysis Reveals a Pressure Gradient Mechanism for Plastic Densification
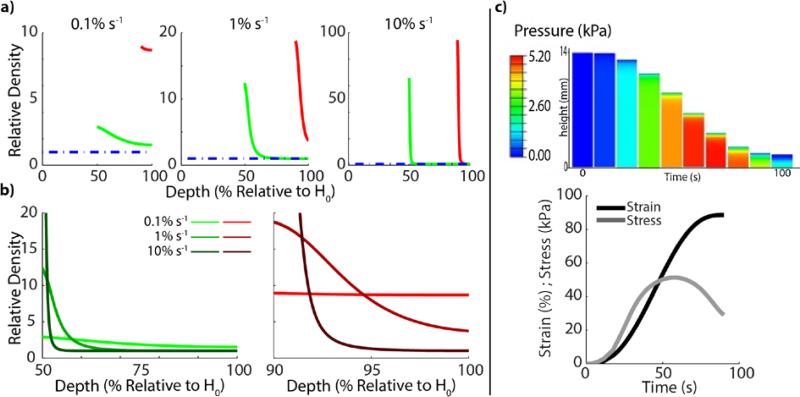
Finite Element simulations generally concur with experimental observations and reveal densification as a pressure driven mechanism. Similar to experimental density measurements (Figure 2), a steep gradient in collagen density is predicted to occur close to the porous platen (the only open boundary condition) and decrease toward the bottom. However, unlike experimental results, the simulations showed that the gradient in relative density is strain rate dependent, with the steepest gradient for the faster loading cases (Figure 5a-b). Further, the simulation shows much higher fibril densities in some samples than what is observed experimentally. Specifically in the 10% s−1 samples, local densification reaches 65- and 94-fold over the initial density for the 50% and 90% compressed samples, respectively (Figure 5a). This relative density change appears to be underestimated in the experimental data. We believe that this discrepancy between simulation and experiment occurs for several reasons. First, the majority of the density observed in the simulation above that measured experimentally is seen over the superficial 180μm depth near the platen. It is possible that entanglement of the collagen fibers in the porous platen may have led to some decompression of the matrix when the platen was removed, which was not accounted for in the simulation. Also, the SEM figures (Figure 3c) confirm extreme local densification near the outflow opening. The observed thicker fibril bundles developing in the extremely densified areas may also affect quantification of density. Such fibril aggregation is not included in the simulations.
Figure 5.
Finite element simulations revealed a pressure and fluid flow driven mechanism of local densification. a) Simulations show similar density features to experimental data, where collagen density peaks near the impacted surface and decreases toward the control density with depth. The simulation was shown to exhibit much higher fibril density than experimental results (in higher strain rate samples), likely due to the absence of bundling in the simulation, a feature observed experimentally. b) Replotting of the data to compare across strain rates revealed the strain rate dependence of the simulation and shows similar qualitative results to the experimental data. c) Simulations of the matrix internal pressure during the compression test revealed a pressure gradient near the porous platen and the only open boundary condition. Local fluid flow as a result of the pressure gradient likely leads to matrix pore collapse and local densification as observed experimentally and in simulations.
The stress profile required to compress the samples agrees with experimental data. Only the maximum stress required to compress the 90%, 10%/s samples is higher in the experiments. Interesting concurring details include the higher stress required to compress the 50% than 90% samples around the 10-40% compression interval, and the steep decline in stress during the last 15% of the compression. With help of the model, these observations can be explained from the applied deformation rate. Rather than a constant rate, a smoother, s-shaped deformation was applied over time (Figure 5C). The toe of this curve is slightly longer for the 90% than the 50% compression curve, which means that the compression rate around the 10-40% compression interval is slightly higher for the 50% than for the 90% compressed samples. The steep decline in stress is explained by the decrease in compression rate in combination with the visco-plastic nature of the present non-cross-linked collagen gel. In such material, stress is strongly loading-rate dependent. Thus, with the decrease in loading-rate, the stress drops rapidly as well (Figure 5C). This is best illustrated in Figure 5C, which shows that the stress peaks at the time point at which the strain rate starts to decrease.
The finite element simulations further enable monitoring of the internal strain, stress, pressure, and density profiles throughout the tissue over the time course of compression, allowing insight into the densification mechanism. The simulations revealed that densification is the result of (local) fluid outflow, driven by gradients in fluid pressure (Figure 5c). Simulations revealed that throughout the sample, the pressure increases with compression rate. However, a global increase in pressure does not result in a global pressure gradient. A significant pressure gradient develops near the porous platen (outflow boundary of the system) (Figure 5c). Local fluid flow, as a result of fluid pressure gradients, occurs locally and in the regions of densification. As a result of the simulations, we believe that densification is occurring as a result of local fluid flow induced by the compressive action. Local fluid outflow occurs in the pores, causing pore collapse and leading to local densification and the aggregation of collagen fibrils as observed in the SEM analysis. It is further possible that the forced fluid flow also leads to some of the pores remaining open, seen in the porous regions between fibril aggregations in high density regions. This mechanistic examination of pressure gradient induced fluid flow for structure densification suggests that alternate boundary conditions (e.g. unconfined compression), and therefore fluid flow patterns, could lead to additional unique gradients of fibril morphology and density.
2.3. Cell Morphology Influenced by Local Ultrastructure
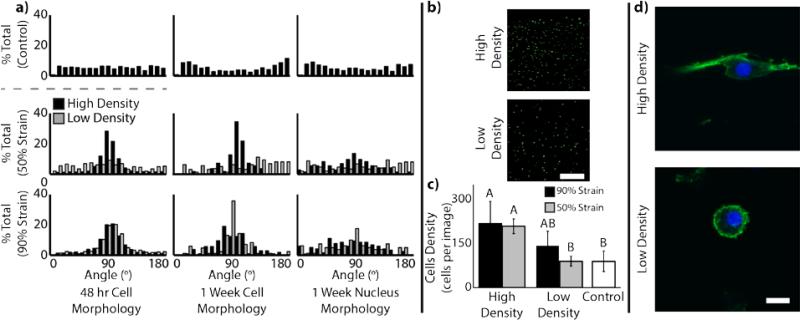
Cells were found to remain viable in control and densified matrices. After 48 hours, control, 50% strain, and 90% strain matrices were found to have 94.0±3.4%, 98.5±0.5%, and 95.5±2.9% viability, respectively. Cell morphology analysis after 48 hours showed cellular alignment in the high density regions via analysis of calcein-stained cell bodies (Figure 6a). Similarly analysis of cellular actin structure and nuclear staining shows that this preferential alignment in the high density regions persists throughout a one-week culture period (Figure 6a,d). In addition to cell morphology, cell density was also confirmed to increase in high density regions as compared to low density regions and control (Figure 6b-c). Cell density (at 48 h) was found to be dependent on both densification and density region (p<0.001 and p<0.001, respectively).
Figure 6.
Encapsulated cells respond to local microenvironment as a result of densification. a) Cellular alignment histograms revealed elongated cellular morphology at 48 h (via calcein dye) and 1 week (via F-actin and Hoescht staining) along aligned fibril bundles in high density regions. b) As a result of densification, encapsulated cells were at higher density in regions of higher collagen density (scale bar=100 μm). c) Quantitative analysis of cell density confirms higher cell density in regions of higher collagen density. Different letters indicate statistical significance between groups (p<0.05). d) At 1 week, F-actin (green) and nuclear (blue) images in cells from a 50% strain magnitude sample show continued alignment with fibril bundles in high density regions, in contrast to random orientations and circular morphology in low density regions. (scale bar=10μm)
Cells can be encapsulated in the collagen matrices prior to densification, remain viable, and respond to the morphology of the local microenvironment. During densification, it was likely that cells were subject to compressive stress from the platen as well as shear stress from the flowing fluid leaving the matrix. To minimize total stress applied to the cells, and therefore have the greatest likelihood of sustained viability, all cellularized experiments were performed in matrices compressed at a strain rate of 0.1% s−1. As a result, cells were shown to maintain high viability. It should be noted that, along with regions of high collagen density, cell density is similarly high relative to low density and control regions. Interestingly, after 48 hours, cells located in regions of fibril alignment were similarly aligned. Additionally, these cells maintained their alignment throughout the week-long cell study, suggesting minimal alteration of the surrounding microenvironment over this time period. Cellular alignment has previously been shown to induce specific cellular function and is characteristic of cellular morphology in similar physiological regions (i.e. superficial zone)[17, 18]. Previous studies in collage matrix compression have shown maintenance of cellular viability post compression[13]. However, these studies, as well as other studies investigating fibril alignment techniques, have either been unable to encapsulate cells prior to formation of the matrix, or did not characterize cellular morphology after alignment[19]. Further, other methods for the development of high density collagen matrices show an inability of cells to infiltrate the dense matrix, limiting the applicability for investigation in cell-matrix interactions[10, 13].
2.4. Densified Collagen Maintains Chondrocyte Gene Expression
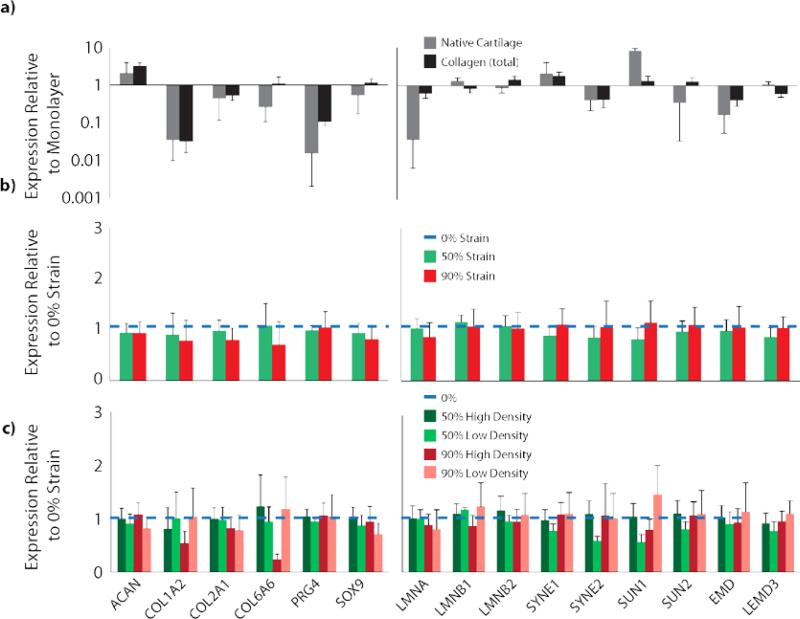
qPCR analysis revealed differences in gene expression between the plated chondrocytes, collagen encapsulated chondrocytes, and chondrocytes cultured in their native microenvironment. Significant differences were found in post-hoc Tukey analysis between plate, cartilage, and collagen matrix groups for COL1A2, PRG4, LMNA, LMNB1, SYNE2, SUN1, SUN2, EMD, and LEMD3 (p<0.001, p<0.001, p<0.001, p=0.0526, p=0.0523, p<0.0001, p=0.041, p<0.001, p=0.003, respectively), where no significant differences were found in ACAN, COL2A1, COL6A1, SOX9, SYNE1, and LMNB2 (p=0.053, p=0.066, p=0.496, p=0.235, and p=0.196, respectively) (Figure 7a). Post-hoc comparisons showed collagen matrices exhibited non-significant gene expression as compared to native cartilage for COL1A2, PRG4, SYNE2, and EMD. However, chondrocytes cultured in collagen matrices were not statistically different from monolayer cultures for LMNB1, SUN1 and SUN2 (Table S1). Within collagen matrices, no influence of strain magnitude or density region was found on the expression of any of the measured genes (p>0.05 for all, Figure 7b-c).
Figure 7.
Chondrocyte gene expression was dependent on encapsulation in collagen matrices. a) Chondrocytes encapsulated in collagen matrices showed different gene expression from both native cartilage cells and monolayer-cultured chondrocytes (p<0.05, log-scale, full statistical analysis Table S1). Genes analyzed include extracellular matrix proteins and chondrogenic markers (left), and nuclear structural proteins (right), to measure the cellular response to their microenvironment normalized to the negative control monolayer group (total gene and associated proteins listed in Table S2). b) Comparison across strain magnitudes (linear scale) and c) density regions (linear scale) showed trends but no statistical difference (p>0.05) in expression of any gene measured at 1 week of culture compared to the nondensified collagen matrices. All data presented as mean±SEM.
Encapsulated chondrocytes were found to exhibit specific gene expression based on the microenvironment in which they are cultured. Our results indicate that plated chondrocytes exhibited no significant difference in aggrecan expression and a significant increase in type I collagen production as compared to native cartilage, which is not inconsistent with existing literature[20]. It is important to note that our results with respect to type II collagen, PRG4, and SOX9 deviate from previous literature. Type II collagen, PRG4, and SOX9 are all typically found to be downregulated after several monolayer passages[21], where our data showed significant upregulation in type II collagen and PRG4 and no significant difference in SOX9 expression, as compared to native cartilage. We believe these inconsistencies are due to the low passage number and relatively short time period of our study, which is notably different from studies investigating dedifferentiation (2+ passages) of monolayer cultured chondrocytes[20, 21].
We did not find any genes that exhibited differences between strain magnitude or density region within the strain magnitude groups, although trends suggest downregulation of types I and VI collagen in high compared to low density collagen. Trends for nuclear structural proteins (e.g. nesprins) indicate down regulation in some, but not all, high versus low density collagen. This suggests that, over the term of the study, chondrocyte gene expression was not profoundly sensitive to the different mechanical and morphological environments in high and low density regions. Gene expression can be altered across a range of substrate stiffnesses[22], but it is possible that the stiffness experienced by the terminally differentiated chondrocytes in our study was not within the sensitivity range to cause altered gene expression, or are influenced also by other changing factors, such as fibril alignment or diameter. Additionally, the duration of our study is a possible limitation on the degree of altered gene expression observed. Our results are in further contrast to the cell morphological changes that were exhibited, where densified regions were found to induce cellular and nuclear alignment that was apparent for the week long study. At this point it is unclear whether gene expression of the collagen matrices is a result of 3D culture or the interaction of 3D culture with signaling from type I collagen. This issue merits further investigation as previous studies have shown chondrogenic gene expression utilizing 3D culture in a non-collagen based matrix[23], although other studies note the influence of collagen-based materials in chondrogenic differentiation[24].
Gene expression analysis of nuclear structural proteins provide further insight into the morphological changes and the cellular response to an altered mechanical microenvironment. Significant up- and down-regulation of nearly all LINC complex genes examined (LMNA, LMNB1, SYNE1, SYNE2, SUN1, SUN2, EMD, and LEMD3) suggest cellular response to a changing mechanical environment. Upregulation of SYNE1 and LMNA (coding for nesprin-1 and lamin-A/C, respectively) is likely indicative of increased cytoskeletal linkage and nuclear stiffening. However, the response between collagen encapsulated and monolayer cultured cells are similar, suggesting a similar response to a new environment, with little distinction between the two environments despite the large difference in mechanical properties.
2.5. Chondrogenic ECM Protein Expression in Collagen Encapsulated Cells
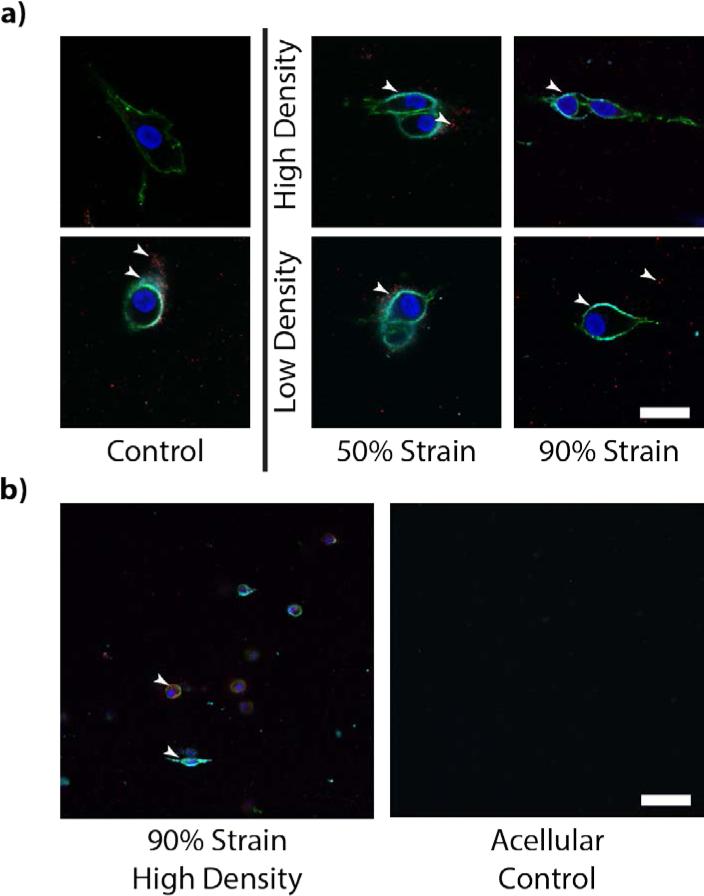
Immunofluorescent analysis reveals protein expression in the collagen matrices, confirming the results in the qPCR analysis for the proteins visualized and providing qualitative insight into ECM protein excretion (Figure 8). Throughout all collagen matrices, expression of type VI collagen and aggrecan were visually confirmed by microscopy. Aggrecan expression appears locally near cells, but does not appear to be present in the interstitium between chondrocytes. Type VI collagen expression appears to form a shell around the cell membrane, similar to that seen in native cartilage[25] and potentially indicative of the formation of a pericellular matrix and the formation of chondrons.
Figure 8.
Cells in all samples were shown to express aggrecan and type VI collagen. a) Protein-level expression of type VI collagen and aggrecan was observed immediately surrounding to the cell body as imaged via immunofluorescence (scale bar=10μm). Arrows pointing to fluoresced type VI collagen (cyan) and aggrecan (red). Indications of minimal non-specific staining suggest localizations of aggrecan in the interstitial space are excreted by the cells rather than background fluorescence. b) Comparison of cellularized samples against acellular control indicated minimal nonspecific binding (scale bar=100μm).
3. Conclusion
To the authors’ knowledge, this work represents the first report of cellular response in a native material matrix across a 3D gradient feature with similar properties to a physiological tissue. This work demonstrates the utility of controlled compression modalities for the creation of unique, cellularized microenvironments of gradient density, fibril morphology, and mechanical properties. Additionally, this work adds to the current literature of compressed collagen matrices, examining the unique gradient features induced by the compression modality. This work shows similar effects of collagen matrix compression utilizing controlled strain rates [5]. Induced gradient matrices are mechanically robust and their ability to encapsulate cells allows for unique investigation of cellular response to specific and controlled microenvironments of various properties. Further, these matrices exhibited fibril ultrastructure similar to tissue structures, e.g. the superficial and middle zone of articular cartilage, further showing the potential for use in studies of cell response to pseudo-native microenvironments. Densification was shown via finite element analysis to be dependent on fluid flow during compression and dependent on the boundary conditions, allowing for further study of collagen compression across a range of geometry and porous/solid boundary conditions. Further, the ability to encapsulate cells and investigate their morphology and gene expression throughout a culture period shows the utility of this collagen platform in the investigation of cell response to complex microenvironments. Interestingly, we showed chondrocytes morphologically respond to the different microenvironments contained in the gradient, and trends in gene expression over the short term may have been linked to property differences contained in high and low density regions. The range of properties seen in this platform will allow for continued investigation of cell response to high and low density collagen environments across a gradient and a range of cell types.
4. Experimental Section
Matrix Polymerization and Densification
Oligomeric collagen was derived and sterilized as previously described[26]. This formulation was standardized based upon purity and polymerization potential as described in the ASTM standard guidance document F3089-14[27]. Collagen was neutralized and polymerized at 5 mg/mL in specific molds (10×5×14mm, w×t×h) allowing for densification, confocal microscopy visualization, and AFM analysis. Additionally, the format of the molds facilitated confined compression, which was our chosen mode of densification. Collagen matrices were densified using a porous polyethylene platen (porosity=15%, pore size=35 μm, Small Parts Inc., Miami Lakes, FL) on a controlled displacement, servoelectric mechanical testing system (TestResources, Shakopee MN) over a range of strains (0%, 50%, 90%) and strain rates (0.1% s−1, 1% s−1, 10% s−1) to investigate the viscoelastic effect on the densification process. Linear modulus and peak stress were measured during compression (n=8).
Density Analysis
Densification was assessed via confocal reflectance microscopy (488 nm) intensity analysis (n=8, 40× water immersion objective, N.A.=0.8, Fluoview FV1000, Olympus Corp., Tokyo, Japan). Images were acquired 100 μm into the matrix (to eliminate edge effects) along the entire height of the sample. Microscope settings were established such that no saturation would occur in the highest density regions while retaining signal in the lowest density regions. Spatial density was assessed via line summation intensity throughout the depth of each sample. Relative density was analyzed and normalized to a standard curve between 0-5 mg/mL control polymerizations, where 5 mg/mL approaches the upper limit of homogenous collagen oligomer polymerization. All densification measurements were plotted as mean ± standard deviation. Considering that little difference in fibril density was seen as a result of altered strain rate, all subsequent analyses were assessed using 0.1% s−1 strain rate for all strain levels.
Ultrastructural Analysis
The effect of densification on ultrastructure was assessed via cryo-SEM (Nova NanoSEM, FEI, Hillsboro OR) using an Everhart-Thornley (<10,000× magnification) or immersion lens (>10,000× magnification) detector. Densification samples (n=3) were slush frozen in liquid nitrogen, fractured to the imaging plane, and sublimated for 13 minutes to reveal collagen ultrastructure. Ultrastructure was imaged along the density gradient similar to confocal microscopy analysis. Images were acquired at magnifications between 2000× and 50,000× to visualize gross structure and unique fibril bundles seen in high-density regions. Images at 2000× were obtained throughout the height of the samples to examine changes from high to low density. Mean fibril thickness, maximum fibril thickness, and fibril area fraction were quantified and analyzed (n=3, N=9) using the Thickness and Volume Fraction functions in BoneJ, developed to measure these properties in trabecular bone, as previously described[28]. SEM images were thresholded and binarized prior to analysis. Prior to analysis, BoneJ software was validated through the development of a custom MATLAB script, outputting artificial fibril networks of specified mean size and distribution.
AFM Measurement of Regional Mechanics
AFM (Series 5500, Agilent, Santa Clara CA) analysis was utilized to measure the regional mechanical properties caused via densification (n=5). Hertzian contact measurements were performed by colloidal probe microscopy using triangular silicon nitride AFM cantilevers with borosilicate microspheres (spring constant = 0.77 N/m; sphere diameter = 10 μm; Novascan Technologies, Ames IA). Indentations were performed along the height of the matrix (analogous to the relative density measurement) with a force trigger of 100 nN and an indentation velocity of 15 μm/s, as described previously[29]. Compressive modulus was calculated as previously described[29]. Modulus measurements were averaged in high- and low-density regions (with at least 10 data points per region per sample) for densified samples and compared to controls. Additionally, measurements were made on explanted bovine articular cartilage with identical parameters for physiological comparison [30].
Mixture Theory Analysis of Densification
To further investigate the mechanism of confined compression densification, a biphasic (i.e., fluid/solid phase) model was implemented under the various applied testing conditions. A mesh representing the 14 mm high sample was created in ABAQUS (v11, Pawtucket, RI) consisting of 716 two-dimensional plane strain pore pressure elements of decreasing size towards the outflow opening. The matrix was mechanically defined as a linear hyperelastic compressible material using measured parameters for Young's modulus (1 kPa) and Poisson's ratio (0.001). Permeability of the material was strain-dependent, decreasing linearly with collagen density from 1×10−10 m4/Ns at the initial collagen density of 0.5% (v/v) to 1×10−11 m4/Ns when collagen density reached 99.99%. Confined compression was simulated by fixing the bottom nodes in all directions, allowing frictionless displacement of the other nodes only in the vertical direction, and allowing free fluid outflow from the top surface under the indenter. Deformation was prescribed in agreement with the experimental test conditions, to 50% or 10% of the original sample height, following a smooth s-shaped curve which averaged to 0.1, 1 or 10%/s strain rate (Figure 5c, bottom).
Cell Viability
Short-term viability studies were performed to assess the effects of densification on cell survival. Primary bovine chondrocytes were obtained[31] and encapsulated in collagen matrices prior to polymerization at a concentration of 1×106 cells/mL. Matrices were densified immediately (within 10 min) post-polymerization and subsequently transferred to media for culture. Viability was assessed after 48 hours via Live/Dead (calcein/propidium iodide) staining protocol (n=5). All cell studies were performed in F-12 Dulbecco's Modified Eagle Medium (Invitrogen, Carlsbad CA) supplemented with 10% fetal bovine serum, 100 units/mL penicillin, 100 μg/mL streptomycin, 50 μg/mL ascorbate-2-phosphate and 0.1% bovine serum albumin (cell culture grade). All cells for viability analyses were utilized before the fourth passage.
Gene Expression
To investigate the ability of densified collagen microenvironments to maintain chondrocyte gene expression and morphology, a one-week culture study was performed. This time period was selected to measure equilibrium response of the chondrocytes to the microenvironment, with a duration that was short enough such that significant remodeling of the matrix was unlikely to have occurred. Identical to cell viability studies, cells were encapsulated in collagen matrices prior to polymerization, matrices were polymerized and subsequently densified, and the constructs were placed in culture (n=5 per group). After one week of culture, samples were removed from culture and cut in half in the direction of the density gradient, where one half was fixed in 4% paraformaldehyde for 30 min (for immunofluorescence studies) and the other half was immediately analyzed by quantitative polymerase chain reaction (qPCR). As a positive control for chondrogenic gene expression, bovine cartilage discs were harvested and placed in identical culture medium for 1 week, then analyzed via qPCR (n=5). As a negative control gene expression, chondrocytes were plated on plastic petri dishes in identical culture medium for one week, and then analyzed via qPCR (n=5). All cells utilized for short-term culture studies were passage 0, directly harvested from bovine articular cartilage and utilized without intermediate plating or culture to maintain native initial phenotype.
Total RNA isolation was performed using the Aurum Total RNA Mini Kit (Bio-Rad, Hercules CA, USA). Collagen matrices were homogenized (TissueRuptor, QIAzol Lysis Reagent, Qiagen, The Netherlands) and followed by a protein precipitation. Bovine cartilage plugs were homogenized in liquid nitrogen and the resulting powder was subject to similar protein precipitation as collagen matrices. Total RNA was determined by spectrometry (NanoDrop ND-1000, Thermo Fischer Scientific, Waltham MA, USA) and equal amounts were reverse transcripted into cDNA (iScript Reverse Transcription Supermix, Bio-Rad) using a thermocycler (CFX96 Touch, Bio-Rad). Quantitative Real-Time PCR was performed using a final concentration of 500nM for each primer and 3ng cDNA template per 20μl reaction. The cycling protocol was as follows: 95°C for 30s, 40 cycles of 95°C for 10s and 60°C for 20s. Melting curves were analyzed to verify primer specificity.
Housekeeping genes were found using the Genevestigator gene expression databank tool (https://www.genevestigator.com/gv/) by searching for stably expressed genes between chondrocytes and fibroblasts, as with ongoing passages cells seem to recapitulate a fibroblastic phenotype. From selected potential candidates (CSNK1A1, HPRT1 and RPL10A), HPRT1 showed the most stable expression. All primers were specific for all known isotypes and are separated by at least one intron or span an exon-exon junction, if splicing information was available (Table S2).
Immunofluorescence
As a result of the PCR analysis, we chose to confirm the presence and investigate the localization of extracellular matrix protein production indicative of mature chondrocytes. For preparation, samples were fixed in 4% paraformaldehyde, permeabilized via incubation in 0.1% Triton-X, and blocked with goat serum for 48 h at 4°C followed by rinsing in 1× PBS 3×30 min. Samples were then stained with primary antibodies for 48 h at 4°C with gentle rocking, rinsed with PBS 3×30 min at room temperature, then stained with fluorescently-labeled secondary probes for 48 h at 4°C with gentle rocking, rinsed again with PBS 3×30 min at room temperature and imaged immediately afterward. Primary labels were aggrecan (1:100, mouse monoclonal; Abcam ab3778; Cambridge, UK) and type VI collagen (1:200, rabbit; Millipore AB7821; Darmstadt, Germany) and were counterstained with Hoechst (1:500; to label DNA), phalloidin (1:200, to label f-actin), goat anti-mouse AF546 (to label aggrecan; Invitrogen A-11003; Grand Island, NY), and goat anti-rabbit AF633 (to stain type VI collagen, Invitrogen A-21070; Grand Island, NY). All imaging was performed on a confocal microscope (Nikon A1Rsi; Melville, NY) using 20× and 40× water immersion objectives.
Statistical Analysis
Compressive modulus, cell density, mean fibril diameter, max fibril diameter, and fibril area fraction were analyzed using a general linear model analysis of variance (ANOVA) with strain magnitude (0%, 50%, and 90%) and region (high vs low density) as main effects, where region was considered as a nested variable within strain magnitude. Compressive modulus data was logarithmically transformed to meet normality assumptions for ANOVA. Cell viability was analyzed via one-way ANOVA with strain magnitude as the main effect. Comparisons of PCR data between monolayer culture, native cartilage culture, and expression in chondrocytes encapsulated in densified collagen matrices was analyzed via one-way ANOVA. For the purposes of this bulk comparison, all data from collagen samples (all strain magnitudes and density regions) were aggregated for comparison against monolayer and native cartilage samples. Further analysis of differences within collagen matrix samples was performed via General Linear ANOVA with strain magnitude and region as main effects, with similar nesting as described previously. All measures reported as mean±standard deviation except gene expression data, which is reported as mean±standard error.
Supplementary Material
Acknowledgements
The authors would like to acknowledge funding from NIH R01 AR063712, NSF CAREER 1349735, OVPR Incentive Grant, and NSF GRFP DGE-0833366 (TN). The authors would like to thank technical assistance from Aaron Taylor, Ph.D., in the Purdue Life Science Microscopy Facilities, and Sarah Calve, Ph.D., in the Weldon School of Biomedical Engineering at Purdue.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Wu X, Jiang P, Chen L, Yuan F, Zhu YT. PNAS. 2014;111:7197. doi: 10.1073/pnas.1324069111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomopoulos S, Williams GR, Gimbel JA, Favata M, Soslowsky LJ. J Orthop Res. 2003;21:413. doi: 10.1016/S0736-0266(03)00057-3. [DOI] [PubMed] [Google Scholar]
- 3.Tse JR, Engler AJ. PLoS One. 2011;6:e15978. doi: 10.1371/journal.pone.0015978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reilly GC, Engler AJ. J Biomech. 2010;43:55. doi: 10.1016/j.jbiomech.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Cheema U, Brown RA. Adv Wound Care. 2013;2:176. doi: 10.1089/wound.2012.0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown RA, Wiseman M, Chuo CB, Cheema U, Nazhat SN. Adv Funct Mater. 2005;15:1762. [Google Scholar]
- 7.Hwang NS, Varghese S, Li H, Elisseeff J. Cell Tissue Res. 2011;344:499. doi: 10.1007/s00441-011-1153-2. [DOI] [PubMed] [Google Scholar]
- 8.Bailey JL, Critser PJ, Whittington C, Kuske JL, Yoder MC, Voytik-Harbin SL. Biopolymers. 2011;95:77. doi: 10.1002/bip.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amiel D, Kuiper SD, Wallace CD, Harwood FL, VandeBerg JS. J Gerontol. 1991;46:B159. doi: 10.1093/geronj/46.4.b159. [DOI] [PubMed] [Google Scholar]
- 10.Saeidi N, Karmelek KP, Paten JA, Zareian R, DiMasi E, Ruberti JW. Biomaterials. 2012;33:7366. doi: 10.1016/j.biomaterials.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosser G, Anglo A, Helary C, Bouligand Y, Giraud-Guille MM. Matrix Biol. 2006;25:3. doi: 10.1016/j.matbio.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Mi S, Khutoryanskiy VV, Jones RR, Zhu X, Hamley IW, Connon CJ. J Biomed Mater Res A. 2011;99:1. doi: 10.1002/jbm.a.33152. [DOI] [PubMed] [Google Scholar]
- 13.Bitar M, Salih V, Brown RA, Nazhat SN. J Mater Sci Mater Med. 2007;18:237. doi: 10.1007/s10856-006-0685-1. [DOI] [PubMed] [Google Scholar]
- 14.Yow SZ, Quek CH, Yim EK, Lim CT, Leong KW. Biomaterials. 2009;30:1133. doi: 10.1016/j.biomaterials.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alekseeva T, Hadjipanayi E, Abou Neel EA, Brown RA. Eur Cell Mater. 2012;23:28. doi: 10.22203/ecm.v023a03. [DOI] [PubMed] [Google Scholar]
- 16.Hadjipanayi E, Mudera V, Brown RA. Cell Motil Cytoskeleton. 2009;66:121. doi: 10.1002/cm.20331. [DOI] [PubMed] [Google Scholar]
- 17.Dubey N, Letourneau PC, Tranquillo RT. Biomaterials. 2001;22:1065. doi: 10.1016/s0142-9612(00)00341-0. [DOI] [PubMed] [Google Scholar]
- 18.Martel-Pelletier J, Boileau C, Pelletier JP, Roughley PJ. Best Pract Res Clin Rheumatol. 2008;22:351. doi: 10.1016/j.berh.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Shields KJ, Beckman MJ, Bowlin GL, Wayne JS. Tissue Eng. 2004;10:1510. doi: 10.1089/ten.2004.10.1510. [DOI] [PubMed] [Google Scholar]; Caliari SR, Harley BA. Biomaterials. 2011;32:5330. doi: 10.1016/j.biomaterials.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Z, Fitzgerald JB, Xu J, Willers C, Wood D, Grodzinsky AJ, Zheng MH. J Orthop Res. 2008;26:1230. doi: 10.1002/jor.20523. [DOI] [PubMed] [Google Scholar]
- 21.Cheng T, Maddox NC, Wong AW, Rahnama R, Kuo AC. J Orthop Res. 2012;30:234. doi: 10.1002/jor.21503. [DOI] [PubMed] [Google Scholar]
- 22.Schuh E, Kramer J, Rohwedel J, Notbohm H, Müller R, Gutsmann T, Rotter N. Tissue Eng Part A. 2010;16:1281. doi: 10.1089/ten.TEA.2009.0614. [DOI] [PubMed] [Google Scholar]
- 23.Caron MM, Emans PJ, Coolsen MM, Voss L, Surtel DA, Cremers A, van Rhijn LW, Welting TJ. Osteoarthritis Cartilage. 2012;20:1170. doi: 10.1016/j.joca.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Chang CF, Lee MW, Kuo PY, Wang YJ, Tu YH, Hung SC. J Biomed Mater Res A. 2007;80:466. doi: 10.1002/jbm.a.30963. [DOI] [PubMed] [Google Scholar]
- 25.Neu CP, Novak T, Gilliland KF, Marshall P, Calve S. Osteoarthritis Cartilage. 2014 doi: 10.1016/j.joca.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey JL, Critser PJ, Whittington C, Kuske JL, Yoder MC, Voytik-Harbin SL. Biopolymers. 2011;95:77. doi: 10.1002/bip.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.American Society for Testing and Materials Standard Guide for Characterization of Type I Collagen as Starting Material for Surgical Implants and Substrates for Tissue Engineered Medical Products. ASTM Standard # F3089-14. :2008. [Google Scholar]
- 28.Doube M, Kłosowski MM, Arganda-Carreras I, Cordelières FP, Dougherty RP, Jackson JS, Schmid B, Hutchinson JR, Shefelbine SJ. Bone. 2010;47:1076. doi: 10.1016/j.bone.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLeod MA, Wilusz RE, Guilak F. J Biomech. 2013;46:586. doi: 10.1016/j.jbiomech.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neu CP, Khalafi A, Komvopoulos K, Schmid TM, Reddi AH. Arthritis Rheum. 2007;56:3706. doi: 10.1002/art.23024. [DOI] [PubMed] [Google Scholar]
- 31.Novak T, Voytik-Harbin SL, Neu CP. Acta Biomater. 2015;11:274. doi: 10.1016/j.actbio.2014.09.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.