Abstract
To facilitate fine-scale phenotyping of whole specimens, we describe here a set of tissue fixation-embedding, detergent-clearing and staining protocols that can be used to transform excised organs and whole organisms into optically transparent samples within 1–2 weeks without compromising their cellular architecture or endogenous fluorescence. PACT (passive CLARITY technique) and PARS (perfusion-assisted agent release in situ) use tissue-hydrogel hybrids to stabilize tissue biomolecules during selective lipid extraction, resulting in enhanced clearing efficiency and sample integrity. Furthermore, the macromolecule permeability of PACT- and PARS-processed tissue hybrids supports the diffusion of immunolabels throughout intact tissue, whereas RIMS (refractive index matching solution) grants high-resolution imaging at depth by further reducing light scattering in cleared and uncleared samples alike. These methods are adaptable to difficult-to-image tissues, such as bone (PACT-deCAL), and to magnified single-cell visualization (ePACT). Together, these protocols and solutions enable phenotyping of subcellular components and tracing cellular connectivity in intact biological networks.
INTRODUCTION
Because of their intrinsic transparency, the worm Caenorhabditis elegans and the zebrafish Danio rerio provide scientists with an unobstructed, organism-wide view of tissue anatomy and cellular activity via, e.g., cell type–specific fluorescence labeling and genetically encoded calcium indicators1–3 and conventional imaging techniques4–6. In combination with their small size and genetic tractability, their whole-body transparency enables rigorous, high-throughput investigations into how environmental, cellular and genetic alterations influence biological processes from cellular signaling and apoptosis, to organism development and survival. By contrast, the comparatively large size and optical opacity of mammalian models generally has limited researchers to imaging snapshots of cellular organization on thin-sectioned tissue samples. However, it was hypothesized that if the bodies of these mammalian model organisms were to acquire the same level of optical transparency as zebrafish embryos, whole-body image data sets would theoretically become available to scientists for study (Table 1).
TABLE 1.
Current and potential biomedicaL applications of PARS and PACT.
| Application | Cleared tissue | Additional references |
|---|---|---|
| Biodistribution of chemicals or biologicals (e.g., virus infectivity, antibody and gene therapies) | Whole-body clearing (Fig. 1a–e) | 143 – 150 |
| Mapping discrete cellular niches, such as 3D genetic makeup and architecture of tumors, stem cell niches; potential for larger volume array tomography151,152 | PACT of tumor biopsies18 and whole-body PARS for rodent cancer models | 98,101, 153–157 |
| Monitoring the progression of cell death and tissue damage (i.e., in stroke, peripheral infarcts), and the corresponding neurogenesis | PARS for whole-body, targeted vasculature fixation and immunolabeling | 158,159 |
| Tract tracing complex long-range fiber bundles (e.g., vagus nerve) and whole-body vasculature (i.e., both circulatory and lymphatic systems); short- and long-range cellular 3D mapping160 (including via neuronal positioning system (NPS)133, via Brainbow161 and via array tomography151,152) | PARS with whole-body targeted IHC, PARS-CSF (spinal cord), PACT-deCAL for vertebral column, ePACT for subcellular spectral resolution of overlapping NPS vesicles, Brainbow labeling and FISH probes | 39–41,133, 162–164 |
| Following neurodevelopment (neural stem cell differentiation), neurogenesis and nerve/axon regeneration | PARS | 14,165–169 |
| Tracking myelination trajectory over lifetime and demyelination in disease states (autism, traumatic brain injury, multiple sclerosis) | PACT and PARS with IHC for myelin-associated proteins and markers of inflammation (Fig. 5d) | 165,170–173 |
| Studying the brain-gut connection, microbiome, blood-brain barrier permeability | PARS | 174,175 |
| Assessing the effects of peripheral immunoactivation on cognition and health | PARS with IHC for cytokines, inflammation and neuronal markers | 171,172,176 |
| Imaging through dense, complex tissues (e.g., bone marrow stem cells) | PACT-deCAL for through-bone labeling and imaging | 97,164, 177–182 |
| Exploring topics in microbiology, including biofilms (characterizing biofilm structure and the interaction of different cellular layers), the heterogeneity and distribution of microbes that occupy the same niche | PACT with considerations for fragile samples (e.g., PACT-hydrogel formulated with paraformaldehyde and/or bis-acrylamide) so that bacterial colonies are retained in tissue or biofilm samples during clearing | 183 – 186 |
| Diffusion tensor imaging (DTI)187 and spectral confocal reflectance microscopy (SCoRe, for label-free in vivo imaging of myelinated axons)188 | Future potential for ex vivo variation of DTI, wherein PARS-based diffusion of materials and immunolabels grants whole-organism imaging | 166,188–191 |
Several methodologies for tissue clearing have been proposed for large-scale 3D mapping of tissue macromolecular content7–21. Each of these protocols offers distinct advantages, such as preserving tissue architecture7,14,18,22,23, accommodating standard histological techniques8,15,17,18,24,25 or creating a computational workflow for acquiring and/or reconstructing thick-tissue image stacks11,15,14,26. Building on our prior CLARITY technique and concepts for generating extractable tissue-hydrogel hybrids8,27, we further developed the trio of PACT, PARS and RIMS to offer a user-friendly, rapid approach to rendering whole organs and whole organisms transparent18. These methods help to stabilize tissue architecture and preserve the macromolecular content of samples, thus enabling imaging of immunohistochemical, single-molecule RNA fluorescence in situ hybridization (smFISH), and small-molecule staining throughout thick tissues, as well as enabling long-term storage18. In this protocol, we provide detailed information about how to implement PACT, PARS and RIMS so that users can apply these methods to their own research.
Imaging of large volumes of cleared tissue can generate gigabyteto terabyte-sized data sets, which creates new challenges associated with the computational analysis of the high-resolution image stacks. Tract-tracing in particular is a difficult and laborious undertaking, whether for mapping the brain connectome or for generating a smaller-scale wiring diagram of isolated projections between specific brain regions or of peripheral nerves at target organs. Thus, in conjunction with refining methodologies to render tissues transparent, we evaluated a wide range of image analysis software packages for their ability to process data sets of cleared brain volumes. On the basis of our findings regarding the best-performing tools, we propose here sample workflows to provide users with a springboard for basic image analysis to complement and facilitate their adoption of the PACT, PARS and RIMS methods.
Advantages of tissue clearing by tissue-hydrogel hybrids
The tissue stabilization and clearing methods that we developed8,18,27 use gentle delivery of structural supportive hydrogels and removal of light-obstructing lipids through either passive clearing (PACT) or through the vasculature of intact postmortem organisms (PARS). The hydrogel mesh itself is transparent, and it secures proteins and nucleic acids into place so that we can later detect them with fluorescent labels under a microscope. There are a number of tissue clearing protocols available that combine the use of `chemical' clearing methods (i.e., the modification and/or removal of tissue components) and `optical' clearing methods (i.e., the homogenization of refractive indices (RIs) throughout the sample and sample mount, a feat that is usually accomplished through sample hyperhydration, dehydration and/or immersion in specially designed mounting solutions) in order to maximize sample transparency28. We experimented with many of these protocols alongside our initial development18 of PACT and PARS so that we could endeavor to incorporate some of their strengths and avoid major pitfalls. For example, we recognized the hydrogel-based tissue stabilization of CLARITY8,24 to be beneficial to sample integrity, the rapidity of 3DISCO14 and the decolorization of CUBIC11,21 to be highly desirable, and the risk of tissue damage via burning8,24 or unchecked swelling9 to be best avoided. Our observations are summarized in Table 2 to guide researchers in selecting a clearing protocol that best suits their clearing application.
TABLE 2.
Clearing techniques that multi-task: macromolecular extraction and refractive index matching.
| Method | Sample preparationa | Chemical clearingb | Optical clearingc | Processing timed | Size fluctuations | Fluorescence, IHC compatibilitye |
|---|---|---|---|---|---|---|
| 3DISCO14,26,192 | Graded tetrahydrofuran | Dichloromethane lipid solubilization | dibenzyl ether and/or BABBf | <1 week (1–2 d) | No tissue expansion reported; tissue shrinkage with dehydration | IHC |
| iDISCO23 | Rounds of dehydration, bleaching, rehydration | 2–3 weeks (1–2 d) | IHC | |||
| CUBIC11 | NA | Aminoalcohol-based tissue decolorization and lipid removal (15% (wt/vol) Triton X-100, 25% (wt/wt) urea, 25% (wt/wt) NNNN′ tetrakis (2-hydroxypropyl) ethylenediamine); ≥5 d per organ | Scale-based optical clearing (50% (wt/wt) sucrose, 25% (wt/wt) urea, 10% (wt/wt) nitrilotriethanol, 0.1% (wt/vol) Triton X-100); ≥2 d | 2 weeks, 10–14 d (≤ 10 d) | Transient and reversible | IHC/F |
| CUBIC decolorization21 | IHC/F | |||||
| CLARITY1,8,24,193 Advanced CLARITY15,16 | Formation of tissue-hydrogel hybrid | Electrophoretic tissue clearing: lipid removal with SDS | RI matching with Focus Clear or glycerol | 10 d | Transient, reversible tissue expansion during the process | IHC/F |
| Passive CLARITY15,193 | Weeks | IHC/F | ||||
| PARS | Formation of tissue-hydrogel hybrid | Perfusion-based or passive lipid removal with SDS | RI matching with RIMS, sRIMS or glycerol | <2 weeks (1 week) | Transient, reversible tissue expansion during the process; gradual tissue expansion in RIMS mounting medium | IHC/F |
| PACT | ≥1 month | IHC/F | ||||
| ExM94 | Formation of tissue-hydrogel hybrid | Proteinase K digestion | Clearing through water absorption; possible RI homogenization with digestion | <1 week | 4–5× linear expansion |
Sample preparation aside from standard fixation and brief postfixation (e.g., 4% PFA transcardial perfusion).
Chemically or mechanically removing tissue macromolecular components (e.g., lipids, heme) to improve light probe penetration and to reduce light scattering.
Homogenizing the refractive indices throughout heterogeneous tissues and at all material interfaces between the sample and objective lens (e.g., tissue, mounting medium, cover glass, immersion medium) to reduce light scattering during optical imaging; it often involves tissue dehydration and/or immersion in RI matching solution or solvent and/or hyperhydration, but not removal of cellular/macromolecular structures.
Processing time is the approximate time from sample collection to sample mounting for a whole adult mouse brain, not including IHC, as based on published reports; the time in parentheses is the approximate clearing time.
IHC: compatible with small-molecule and antibody-based immunohistochemistry, some restrictions in immunofluorescence (e.g., rapid signal decay16,25) and/or some reservations about harsh tissue treatments that may adversely affect tissue integrity or labeling; IHC/F: compatible with IHC and immunofluorescence labeling, validated for (>0.5 mm) depth of antibody penetration and for a wide range of fluorophore wavelengths; IHC-compatible, but only with the use of custom probes.
BABB: a mixture of benzyl-alcohol and benzyl-benzoate13.
Emerging from these different approaches to tissue clearing, PACT and PARS are notable for their versatility in preparing a variety of tissue types for high-resolution imaging at depth. The PACT hydrogel formulation and clearing process is easily optimized to render difficult-to-image tissues transparent (e.g., PACT-deCAL, for PACT delipidation and decalcification of bone), to expand tissues for better separation of compact structures (e.g., ePACT, for PACT-based expansion clearing of dense cells or projections) and to preserve tissue integrity in fragile samples through varying the degree of paraformaldehyde-tissue cross-linking. Meanwhile, PARS is positioned to tackle a variety of scientific problems that would benefit from a comprehensive, whole-body view of gene expression patterns, cellular organization and/or structural composition.
PACT- or PARS-based preparation and clearing of tissue, followed by tissue mounting in RIMS, can preserve the signal from native fluorescent proteins and improve the efficacy of postclearing immunofluorescence labeling (Fig. 1). Fluorescence signal intensity is also maintained through month-long storage periods post fixation18. Other brain-specific tissue clearing protocols (Table 2) have at least one functional drawback, such as incompatibility with endogenous fluorescent labels. Some of these limitations have been overcome by the use of automated tissue sectioning techniques29–32, which have been successfully used in tracking long-range projection axons and sparse cell populations throughout whole brains in rodents or human brain sections26,33–38, or adapted to mapping the cellular organization and innervation of peripheral whole organs in mammalian tissues14,23,26,39–41. These heavily automated imaging systems are not readily available to biology and clinical laboratories with limited resources and budgets. Conversely, traditional histology relies on the irreversible sectioning and manual reconstruction of successive slides for image analysis, which is time-consuming and potentially loses molecular information and connectivity in the process. PACT, PARS and RIMS enable deep imaging of large tissue samples without sectioning and reconstruction. Antibody expense aside, these cost-effective techniques generate detailed 3D reconstructions of intact circuits using only mainstream single-photon microscopy.
Figure 1.
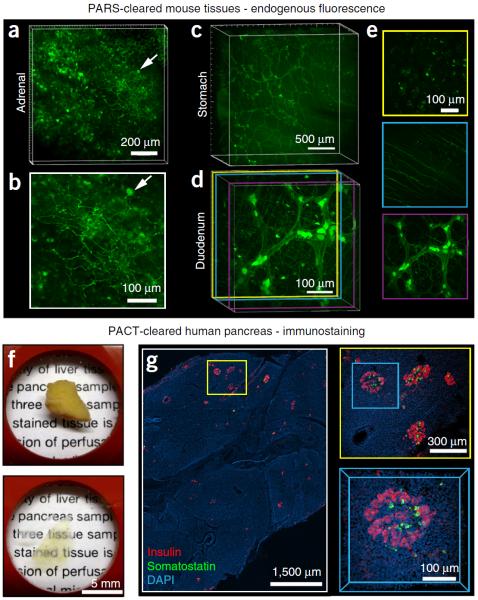
Applications of whole-organ and whole-organism clearing protocols. (a–e) PARS-based whole-body clearing for assessing cellular-level adeno-associated virus (AAV) tropism (Supplementary Methods). Three weeks after systemic injection of AAV9:CAG-GFP, mice were PARS-cleared and their organs were excised and sectioned for imaging. (a,b) Projection images show GFP+ transduced cells in the adrenal gland. Arrow highlights a GFP+ cell near the surface of the adrenal gland with neuronal morphology, which is shown in higher magnification in b. (c) Projection images show GFP+ cells in the stomach from the surface to the lumen. GFP expression is particularly high in the myenteric plexus. (d,e) AAV9 transduces cells in several layers within the intestine (duodenum). (d) Projection image of GFP fluorescence. Double colored lines correspond to the positions of 50-μm maximum projection images extracted from the data set and presented in e. (e) GFP+ cells in the intestinal crypt (top), submucosal plexus (middle) and myenteric plexus (bottom). (f,g) Islet distribution within human pancreatic tissue. (f) A 2-mm-thick section of an adult human pancreas (top) was rendered transparent (bottom) with the PACT method. Briefly, a 2-mm-thick section was cut from a 4% PFA-fixed human pancreas, incubated in 0.5% PFA and 4% (wt/vol) acrylamide at 4 °C overnight and then polymerized in fresh A4P0 hydrogel monomer with 0.25% VA-044 thermal initiator for 2 h at 37 °C. The tissue was cleared with 4% SDS-PBS (pH 7.5) for 48 h, immunostained and mounted in sRIMS (~50% (wt/vol) sorbitol in 0.02 M PB, RI of 1.44). (g) The islet distribution was visualized by immunostaining for insulin (red), somatostatin (green) and DAPI (cyan) (see Table 4 for details on antibodies and nuclear stain); panels represent an imaging stack of 70 μm. Magnified regions are designated by yellow and blue boxes. Sparsely distributed islets are easily located with only 5× magnification (left). A group of islets were identified at 10× magnification (right, top), and a 3D image of a single islet was captured with a 25× magnification (right, bottom). All images were collected on a Zeiss LSM 780 confocal microscope with the Fluar 5× 0.25 NA M27 air objective (w.d. 12.5 mm), Plan-Apochromat 10× 0.45 NA M27 air objective (w.d. 2.0 mm) and the LD LCI Plan-Apochromat 25× 0.8 NA Imm Corr DIC M27 multi-immersion objective (w.d. 0.57 mm). Experiments on vertebrates conformed to all relevant governmental and institutional regulations, and they were approved by the Institutional Animal Care and Use Committee (IACUC) and by the Office of Laboratory Animal Resources at the California Institute of Technology.
Experimental design
The procedure comprises seven main stages: tissue preparation (Steps 1–5); formation of a tissue-hydrogel matrix (Step 6); tissue clearing (Steps 7 and 8); staining (Step 9, optional); enhancement of optical clarity using RIMS (Steps 10–13); imaging (Step 14); and image visualization and analysis (Steps 15–17). Although PACT and PARS, including their respective tissue-specific variations (PACT-deCAL, PARS-CSF18), each follow the same main stages, the decision to proceed with PACT or PARS is generally made before commencing the procedure. If the primary goal is to stabilize soft and/or amorphous samples (e.g., thymus, spleen, pancreas) for experimentation and sectioning, and not to enhance tissue transparency for imaging, users may process samples according to Steps 1–6 (PACT or PARS), and with the option to include bis-acrylamide and/or paraformaldehyde (PFA) in the PACT hydrogel formulation for increased sample rigidity.
Required expertise
Most steps of the procedure can be performed by all members of the research community. Aspects of the PARS setup (Steps 1–5) require that the scientist be approved for working with laboratory animals and/or possess the surgical dexterity to establish an intravascular route for delivery of PARS reagents. For example, to execute PARS-based clearing of whole laboratory animals (e.g., rodents, nonhuman primates) via transcardial perfusion or cannulation, the researcher should be proficient in conducting animal euthanasia via transcardial perfusion and/or basic animal surgical techniques and practices.
As whole-organ and thick-tissue imaging can generate tera-scale data sets, a computational or informatics background, although not necessary if relying on commercial software with good technical support, is very helpful in managing large data sets (file storage and handling) and in performing image analysis (Steps 15–17).
PACT or PARS?
Without the use of organic solvents, passively clearing and immunostaining larger tissue volumes (e.g., whole organs) is prohibitively slow. In terms of clearing efficiency, PACT18 (Figs. 1f,g and 2–6; Supplementary Figs. 1–4) is best suited for the quick clearing of small tissue sections (e.g., up to 1–3-mm-thick-sectioned organs (Supplementary Fig. 1 or tissue biopsies (Fig. 1f,g)). For whole-organ screening or profiling tissues throughout the entire organism, PARS greatly accelerates and simplifies the clearing process. All hydrogel monomer solutions, wash buffers, buffered detergents and phenotypic labels are driven throughout tissue vasculature via a perfusion-based pressure gradient (Fig. 7), which under whole-mouse or whole-rat PARS clearing renders most organs transparent within 4 d (Fig. 8 and Supplementary Fig. 5). To achieve these PARS clearing rates via PACT, excised organs would need to be thick-sectioned and processed individually or in batches, as for most other tissue-clearing protocols7–17,19,20. Although the PARS setup is more involved than PACT, with the PARS tubing and reagent levels requiring attentive, daily monitoring, all organs are processed simultaneously and cleared rapidly and consistently via a single perfusion line. In addition, the basic PARS system can be obtained through the repurposing of common laboratory items (Fig. 7) and standard protocols (transcardial perfusion) within biomedical research.
Figure 2.
PACT setup and procedure. To successfully hybridize tissue with hydrogel monomers via free-radical polymerization, the sample and hydrogel solution must be incubated at 37 °C in an oxygen-depleted environment. This is best accomplished within an airtight container that permits sample degassing. (a) Supplies for PACT chamber (left): 50-ml conical tube (large sample) or Vacutainer (small sample), size 7 stoppers that fit the 50-ml conical tube, PTFE tubing, needles, syringes and a razor blade or scissors to cut the syringe in half. Construct a degassing line that will allow a sample tube to be evacuated of oxygen using the house vacuum and then be placed under inert nitrogen atmosphere (a, left to right). (b) The PACT procedure for sample degassing and hydrogel polymerization is as follows (b, top row): prepare the hydrogel monomer solution, taking care to keep all reagents ice-cold; infuse the tissue sample with hydrogel monomer solution at 4 °C; insert the vacuum line needle into the stopper and place the container under house vacuum for 5–10 min; remove the vacuum line and insert both a venting needle and the hypodermic needle, which is connected to the nitrogen gas line tubing; bubble nitrogen gas through the sample and solution for 5–10 min, ensuring that the venting needle allows excess pressure to escape from the PACT container; and quickly remove both needles and place the sample and container in a 37 °C water bath for 1–3 h. (b, bottom row) Once the hydrogel has polymerized, pour off excess hydrogel, rinse the sample with 1× PBS and/or tissue off with a Kimwipe, section the sample (optional) and place the sample into a 50-ml conical tube filled with 8% (wt/vol) SDS clearing buffer. Incubate the sample at 37 °C in a shaking water bath until the sample is clear. Thoroughly wash the cleared sample, immunostain (optional) and then incubate the sample in RIMS to improve its optical clarity.
Figure 6.
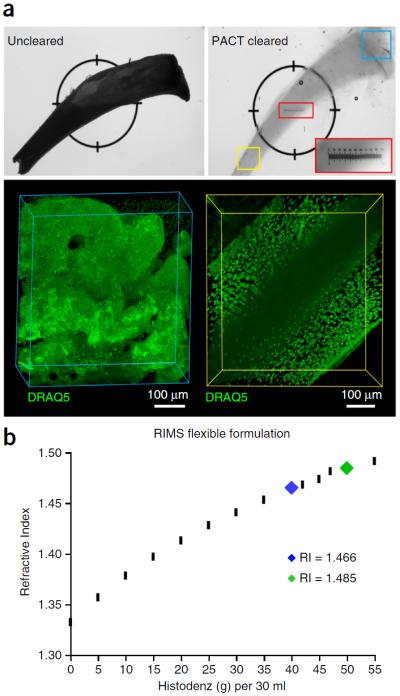
PACT-deCAL and optimized RIMS formulation for imaging decalcified bone samples. (a) After perfusion fixation with 4% PFA, the right and left tibia bones were dissected and postfixed in 4% PFA overnight. One tibia was reserved as the uncleared control (top left), whereas the other tibia bone was A4P0-embedded and cleared (top right) according to PACT-deCAL, as follows. The tibia bone was first cleared in 8% SDS-PBS (pH 8) for 24 h, and then it was transferred into 0.1 M EDTA for 2 d and finally cleared further in 8% SDS-PBS (pH 8) for 2 d at 37 °C. The cleared bone was washed in 1× PBS three times over 1 d and incubated in PBS containing 1:200 DRAQ5 for 2 d at 37 °C. The stained bone was quickly rinsed in 1× PBS and incubated in 1.49 RIMS overnight at 37 °C. The bright-field image (top right) depicts the resulting bone transparency via the placement of a ruler (small red box) underneath the tibia, wherein the tibia's outline on top of the ruler can be seen in the magnified inset of the ruler (large red box). The cleared tibia was imaged in two regions (yellow and blue boxes) on a Zeiss LSM 780 confocal microscope with the LD LCI Plan-Apochromat 25× 0.8 NA Imm Corr DIC M27 multi-immersion objective (w.d. 0.57 mm). (b) RIMS may be formulated with different concentrations of Histodenz in order to achieve an RI that aligns with the tissue density and light-scattering properties of the sample to be imaged, as well as to the optical properties of the imaging setup (objective lens with or without immersion medium). RIMS with an RI ~1.47 is well suited for most cleared soft tissues (blue tick mark), whereas cleared bones should be incubated in RIMS with RI ~1.48–1.49 (green tick mark). Rodent husbandry and euthanasia conformed to all relevant governmental and institutional regulations; animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) and by the Office of Laboratory Animal Resources at the California Institute of Technology.
Figure 7.
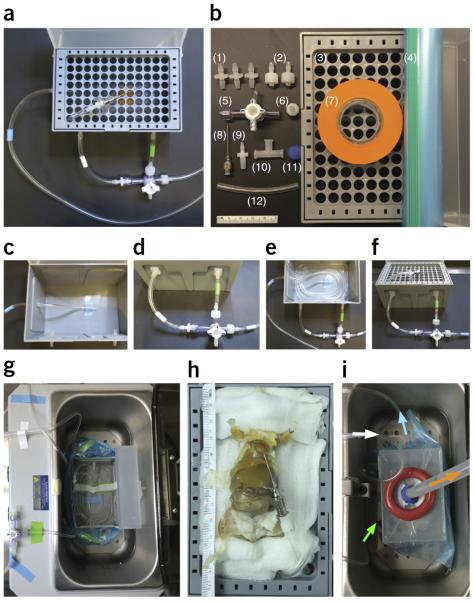
Assembling and working with the PARS chamber. (a) A completed PARS chamber used for whole-body tissue clearing. (b) Individual parts to build a PARS chamber: (1) three 1/8 × 1/8-inch barbed connectors, (2) two 3/32-inch barbed male Luers with locking nut, (3) a 1,000 μl pipette tip box, (4) a 1-gallon Ziploc freezer bag, (5) a three-way stopcock with Luer lock, (6) a 3/32-inch barbed female Luer with full tread, (7) a roll of lab tape, (8) a 22-G × 1-inch gavage needle, (9) a 1/8-inch barbed male slip Luer, (10) a female Luer tee with locks, (11) clay and (12) Tygon E-lab tubing. Ruler shown is 5 cm in length. (c) Three 1/8-inch holes are drilled into the pipette tip box: two into the box front and one into its side, all ~2 cm below the top rim of the box. The three 1/8 × 1/8-inch barbed connectors are placed into the drilled holes. To connect the outflow line (blue tape bands on outflow line tubing), a piece of Tygon tubing is connected from the bottom inside of the pipette box to the single 1/8-inch barbed connector that was inserted through the box side. (d) To continue the outflow line, a second, longer piece of blue-taped tubing is attached to the outer fitting of this same barbed connector (on the outside of the pipette tip box side), and then the other end of this tubing is threaded through the peristaltic pump, pulled back over toward the pipette box and finally connected to a three-way stopcock with a 3/32-inch barbed male Luer with locking nut (rightmost blue-banded tubing in d). To form the inflow line, a short length of tubing (green tape band) is used to connect the three-way stopcock to the front right 1/8-inch barbed connector of the pipette box. The solute flushing line and nitrogen bubbling line, which are subserved by the same tubing (white tape band), are formed by another short length of tubing that joins the third port of the stopcock to the front left 1/8-inch barbed connector. (e) The inflow line is continued inside the pipette box, with the tubing coiled several times around the base of the box so that the solute will be reheated before it passes through the feeding gavage into the subject. The solute flushing line and nitrogen bubbling line is continued inside the pipette tip box and taped to the bottom of the chamber (not shown). (f) The tip of the coiled inflow line tubing is threaded up through the tip wafer (see bird's-eye view of threaded wafer in a) and connected to a 22-G ×1-inch gavage needle with a 1/8-inch barbed male slip Luer. The gavage needle is secured with a short loop of Tygon tubing (~90 mm) threaded through two holes of the wafer. (g) During the polymerization step, the chamber is placed into a 37 °C water bath and sealed in a Ziploc bag. The tubing is attached to the chamber with three 1/8 × 1/8-inch barbed connectors punctured through the Ziploc bag. The Tygon tubing is reconnected from the outside of the bag and surrounded with clay to make an airtight seal. (h) The animal is placed onto the pipette tip box, and the 22-G × 1-inch gavage needle is used to catheterize the heart. (i) The chamber is placed into a 37 °C water bath. A female Luer tee, which is taped onto the lid of the pipette tip box, is punctured through the Ziploc bag, and this joint is sealed with clay to ensure an airtight seal. Finally, to accelerate polymerization, a vacuum line is connected to the female Luer tee to remove oxygen (orange arrow), and a nitrogen gas line (white arrow) is connected to the 1/8-inch barbed connector to deliver a steady flow of nitrogen into the bagged system. The solute is continually circulated through the animal from the outflow line (blue arrow, which also indicates the direction of flow through blue-taped tubing) and inflow line (green arrow, which also indicates the direction of flow through green-taped tubing).
Figure 8.
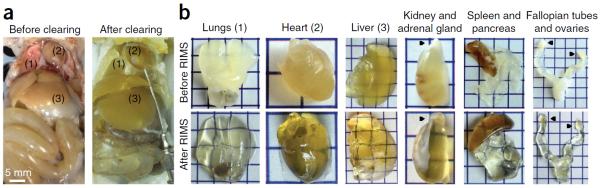
Whole-body clearing of mice with PARS. (a) A4P0-hybridized organs shown before the start of clearing (left) and after 5 d of clearing with 8% SDS-PBS (pH 8.5) and overnight washing with 1× PBS at pH 7.5 (right). Numbers correspond to the extracted organs in b. (b) Extracted organs from the cleared mouse in a, pictured before (top) and after (bottom) RIMS incubation for 3 d. Black pointers correspond to the adrenal gland on the kidney and to the ovaries on the fallopian tubes. Each square represents 0.5 cm2. Rodent husbandry and euthanasia conformed to all relevant governmental and institutional regulations; animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) and by the Office of Laboratory Animal Resources at the California Institute of Technology.
Tissue stabilization by the formation of a tissue-hydrogel matrix
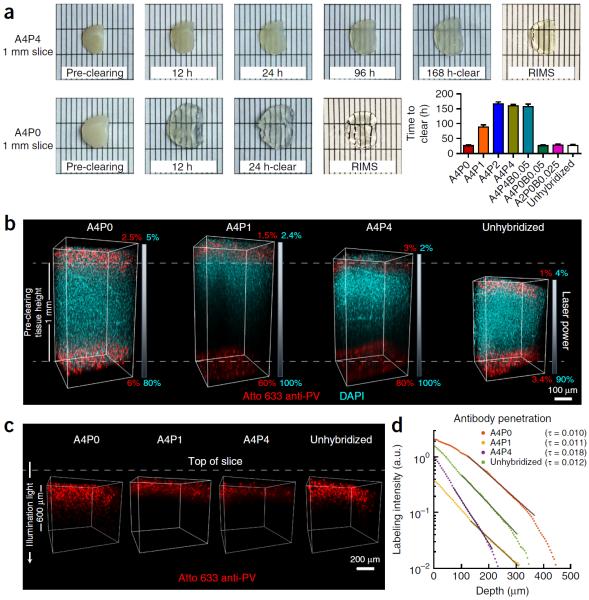
In unstabilized tissue, the prolonged incubation in detergent at 37 °C required for PACT and the perfusive force used in PARS would be detrimental to tissue integrity. Thus, the hybridization of amine-containing and PFA-cross-linked biomolecules to a hydrogel scaffold serves to stabilize tissue architecture and nonlipid content throughout all aspects of PACT and PARS tissue processing. During PARS, the rodent's intact connective tissue and inflexible skeleton provide an additional degree of structural support. To support rapid delipidation in the absence of potentially tissue damaging electrophoretic clearing (ETC)24, the composition of the PARS/PACT18 hydrogel monomer solution bares a few major changes from our originally proposed CLARITY hydrogel8, which consisted of 4% (wt/vol) acrylamide, 4% (wt/vol) PFA and 0.05% (wt/vol) bis-acrylamide (A4P4B0.05). First, the cross-linker bis-acrylamide must be excluded from the PARS hydrogel formulation to prevent hydrogel blockages in vasculature and perfusion lines. Its exclusion from the PACT hydrogel as well, and the reduced exposure of tissues to PFA in both protocols, accelerates clearing and immunolabeling steps. With a final composition of 4% (wt/vol) acrylamide and 0% PFA (A4P0), the resulting minimal polymeric scaffold of the PARS and PACT tissue-hydrogel matrices suffices not only to retain tissue proteins (Fig. 3a and Supplementary Fig. 2) and stabilize tissue macrostructure18 during clearing, but it also allows SDS micelles to diffuse more freely through tissue for efficient clearing (Figs. 4a and 8; Supplementary Fig. 2c). Similarly, a lower cross-link density ensures that antibodies can better access tissue epitopes during immunolabeling (Fig. 4b–d and Supplementary Fig. 3e).
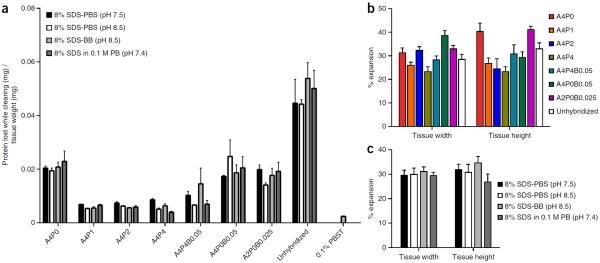
Figure 3.
PACT protein loss and tissue expansion for different hydrogel and clearing conditions. A detailed comparison of the protein loss and tissue expansion for eight different hydrogel matrix compositions: A4P0, A4P1, A4P2, A4P4, A4P0B0.05, A4P4B0.05, A2P0B0.025 and unhybridized, and four different clearing buffers: 8% SDS-PBS (pH 7.5), 8% SDS-PBS (pH 8.5), 8% SDS-BB (pH 8.5) and 8% SDS in 0.1 M PB (pH 7.4). Perfused and fixed mouse brains were sliced into 1-mm-thick coronal slices, and combinations of all the different hydrogel and clearing conditions were performed on slices from comparable locations. Slices were monitored and imaged every 12 h, and the clearing buffer was collected for protein loss measurements and replaced. (a) Total protein content within each sample of clearing buffer collected throughout the clearing process was measured by the bicinchoninic acid assay by extrapolating the concentration of protein from a standard curve of BSA concentration in each clearing buffer (Supplementary Fig. 2a). Protein amounts from each time point were summed until each slice was completely clear, resulting in a measure for the total amount of protein lost while clearing for each slice. This total protein loss was then compared with the initial weight of each slice (n = 3). A comparison was also made with the protein loss of 100-μm-thick slices that were not cleared, but were permeabilized with PBST overnight (n = 9). (b) Comparison between total width and height tissue expansion between hydrogel compositions (n = 4). (c) Tissue expansion comparisons with different clearing conditions (n = 8). (a–c) Data are presented as mean ± s.e.m. Experiments on vertebrates conformed to all relevant governmental and institutional regulations, and they were approved by the Institutional Animal Care and Use Committee (IACUC) and by the Office of Laboratory Animal Resources at the California Institute of Technology.
Figure 4.
Clearing time course and antibody penetration of PACT-processed samples. Quantitative comparison of the effect of different hydrogel-embedding conditions and clearing buffers on time to clear and antibody penetration during immunostaining. 1-mm-thick mouse coronal slices were hybridized and cleared with the array of previously used PACT conditions (Fig. 3). Slices were monitored for the time they took to become transparent. Once cleared, slices were washed and then immunostained. (a) Representative images of two 1-mm-thick coronal brain slices (~1.0–0.0 mm anterior to bregma142) through the time course for PACT clearing and a comparison of time to clear (mean ± s.e.m.) for each PACT hydrogel composition. For the representative images, slices were cleared with 8% SDS-PBS (pH 8.5) and incubated in RIMS for 24 h. (b) Imaging of antibody penetration through different PACT tissue preparations. Previously cleared and washed 1-mm-thick slices were immunostained for parvalbumin (red) and nuclei stained with DAPI (cyan), using 2-d incubations with the primary and Fab format secondary antibodies (for immunostaining reagents, see Table 4), transferred to RIMS for 5 h and then RIMS-mounted. Samples from the cortex, traversing the depth of the slice, were imaged on a Zeiss LSM 780 confocal microscope with a Plan-Apochromat 10× 0.45 NA M27 air objective (w.d. 2.0 mm). To ensure even illumination throughout the depth of the slice for fair antibody detection, we applied laser power z-correction (Zen software, Zeiss): power was changed linearly for each slice, shown as a gradient next to each image; starting power values at the top were chosen to match the level of fluorescence at the surface across slices, whereas the range of powers varied for different PACT conditions. Shown are images of staining through A4P0, A4P1 and A4P4 hydrogel-embedded samples, as well as unhybridized tissue, cleared with 8% SDS-PBS (pH 7.5). As antibody and small-molecule dye diffused through both the top and bottom surfaces of the slice simultaneously, the images show that within 2 d DAPI has fully penetrated in all of the conditions, whereas antibody labeling has progressed to varying extents, depending on the PACT condition. As slices cleared with the different conditions also swell to different extents during the process (indicated by their difference in height relative to the pre-clearing height of 1 mm, as indicated by the white dotted lines in b), penetration of antibody through a more swollen sample will either require longer diffusion time or faster diffusion rate to reach an equivalent anatomical depth as in a less swollen sample. Incomplete detection of the DAPI signal in A4P1 and A4P4 slices is due to the difficulty of achieving similar light penetration in highly cross-linked slices. (c) Depiction of parvalbumin staining through same slices as in b. DAPI signal has been removed to better show the variable penetration of the antibody over the course of a 2-d period. (d) Quantification of antibody penetration through PACT conditions depicted in b and c. Antibody fluorescence signal was scaled by the average DAPI intensity for each z-section inside the volume and the average scaled fluorescence along a line perpendicular to the tissue surface produced a final estimate of labeling intensity (in arbitrary units, a.u.) as a function of tissue depth (Supplementary Methods). Antibody diffusion was fit to an exponential model [f(x) = a × exp (−tau × x) + b], with the exponent tau being inversely proportional to the square root of the diffusivity, wherein a larger tau indicates slower diffusion. Labeling intensities for A4P0, A4P1, A4P4 and unhybridized samples cleared with 8% SDS-PBS (pH 7.5), as a representative sample of all the different buffers, are plotted on a logarithmic scale. The amount of PFA contained in the hydrogel-tissue matrix is inversely proportional to immunohistochemical staining efficiency. Experiments on vertebrates conformed to all relevant governmental and institutional regulations, and they were approved by the Institutional Animal Care and Use Committee (IACUC) and by the Office of Laboratory Animal Resources at the California Institute of Technology.
Tissue clearing
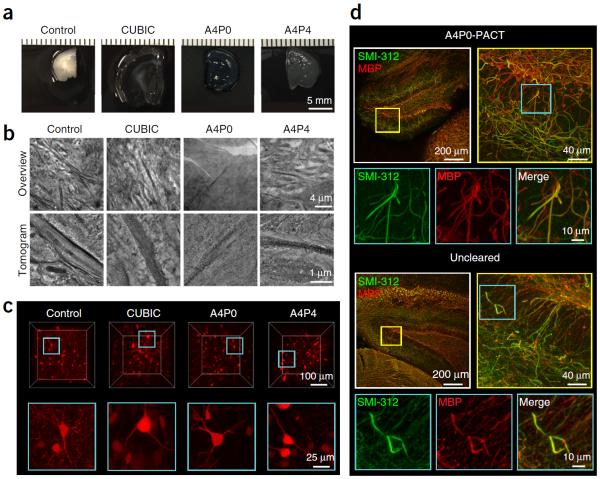
Traditionally, tissue clearing protocols have aimed to render samples transparent via homogenizing the RIs of the various tissue components, and matching their RIs with the lens and mounting setup (e.g., glass coverslip interfaces). This has often been accomplished via exchanging the aqueous fraction of tissue (RI ~1.33) with a mounting medium of higher RI, which includes organic solvents such as BABB (RI ~1.53–1.57)13,14,42, dibenzyl ether (RI ~1.56)12,23, methyl salicylate (RI ~1.52–1.54)43 and 2,2′thiodiethanol (RI ~1.52)44; polyol and saturated sugar solutions such as glycerol (RI ~1.43–1.47)8, sucrose and fructose (RI ~1.49–1.50)7,22; and amides such as formamide (RI ~1.44)10 and urea (RI ~1.38)9,11. Aside from passive CLARITY15 and PACT18, few passive clearing protocols endeavor to alter the chemical composition of tissue, by removing major tissue components from samples so that they become less light-scattering (see `Chemical clearing' in Table 2). One notable example is CUBIC11,21, which also combines the use of passive delipidation and RI matching to achieve transparency. Thus, we sought to compare the level of delipidation that was achieved with PACT-based clearing (A4P0 and A4P4 hydrogels) and CUBIC-based clearing. To examine the efficacy of tissue delipidation, we used transmission electron microscopy (TEM; Fig. 5b and Supplementary Methods). Indeed, as illustrated by membrane permeabilization and extraction, lipid removal was noticed in all conditions, and it was highest in A4P0, in which a high degree of fine structure loss is evident. In contrast, A4P4 tissue, although extracted, still retains enough contrast for identifying fine structural detail, such as membrane-bound organelles and small neurites. With respect to structural preservation, the CUBIC samples are between the two PACT conditions, showing nearly complete lipid extraction but with some cytoskeletal elements in the axon preserved. Although samples embedded in A4P0 hydrogel showed adequate protein and nucleic acid retention for imaging endogenous fluorescence (Fig. 5c) and detecting myelin-binding proteins (Fig. 5d), if an enhanced level of tissue preservation is desired it is best to embed samples in a hydrogel with a higher order of tissue cross-linking by including PFA (for example, by adding 1–4% (wt/vol) PFA to the 4% acrylamide hydrogel solution, termed A4P1-4). Alternatively, samples can be processed in parallel, and adjacent areas can be directed either to TEM or to hydrogel-embedding and clearing to obtain both ultrastructural and volume information, respectively.
Figure 5.
Preservation of tissue architecture during delipidation. The differential effects of individual clearing conditions on cellular architecture and endogenous and stained fluorescence imaging. (a–c) Mice that received bilateral intracranial injections in the lateral septum of AAV expressing the tdTomato transgene were perfusion-fixed with 4% PFA, and a subset of 1-mm-thick unhybridized coronal brain sections were prepared for microscopy without clearing (control, first column), or they were first rendered transparent via the CUBIC method11,21 (second column). The second subset of 1-mm-thick sections underwent PACT-processing: A4P0 embedding (third column) or A4P4 embedding (fourth column) and clearing with 8% SDS-PBS (pH 7.5), followed by preparation for ultrastructural study or RIMS mounting. (a) Brain sections were photographed after fixation (control) or immediately after clearing (CUBIC, A4P0 and A4P4) to illustrate the degree of tissue swelling that occurred for each condition. (b) Control (unhybridized, uncleared), CUBIC-cleared and PACT-cleared (A4P0, A4P4) tissues were then processed identically for ultrastructural examination using electron microscopy and tomography (Supplementary Methods). Overviews (top row) from each of the four samples illustrate the relative amount of lipid loss attributable to the different clearing methods, in terms of contrast between structures. Tomographic reconstruction (bottom row) of subregions of the overviews, each showing a portion of an axon and surrounding cellular structures, indicates the extent of change at the fine-structural level. (c) Control, PACT- and CUBIC-cleared brain sections were mounted in RIMS or CUBIC reagent-2 (refs. 11,21), respectively, and the endogenous expression of tdTomato was imaged on a Zeiss LSM 780 confocal microscope with the LD LCI Plan-Apochromat 25× 0.8 NA Imm Corr DIC M27 multi-immersion objective (w.d. 0.57 mm). Volume renderings (top: x,y,z = 300 μm for PACT- and CUBIC-cleared samples and x,y,z = 300, 300, 140 μm for control) and maximum intensity projections (bottom: x,y,z = 100,100,50 μm) are shown. In all images except the uncleared control, cells are visualized throughout the volume imaged. In the control image, light is unable to penetrate through the sample to image at depth. (d) Preservation of myelin proteins. 200-μm-thick A4P0-PACT-cleared mouse brain sections and 50-μm-thick uncleared sections were immunostained for SMI-312 and for myelin basic protein (MBP), using Atto 488–conjugated and Atto 647N–conjugated Fab format secondaries (see Table 4 for details). After a 2-h RIMS incubation, the transparent sections were mounted and imaged on a Zeiss LSM 780 confocal microscope with the Plan-Apochromat 10× 0.45 NA M27 air objective (w.d. 2.0 mm) and the LD LCI Plan-Apochromat 25× 0.8 NA Imm Corr DIC M27 multi-immersion objective (w.d. 0.57 mm). The images correspond to a 50-μm-thick maximum intensity projection over the dentate gyrus; Top: A4P0-PACT cleared, Bottom: uncleared smaller panels are high-magnification images of the boxed areas showing myelinated axons. Experiments on vertebrates conformed to all relevant governmental and institutional regulations, and they were approved by the Institutional Animal Care and Use Committee (IACUC) and by the Office of Laboratory Animal Resources at the California Institute of Technology.
The denaturing anionic detergent sodium dodecyl sulfate (SDS), used for lipid removal in PACT/PARS, is also very effective in dissociating DNA from proteins (e.g., for cell nuclei removal) and in disrupting extracellular matrices to facilitate protein removal (e.g., ionic interactions of SDS with membrane proteins allow for their removal and purification). For example, retrograde perfusion of a cadaveric rat heart with 1% (wt/vol) SDS for 12 h results in its complete decellularization45. By contrast, SDS solubilization of lipid bilayers via a micellar mechanism is a slower process. Thus, to guard against the extraction of peptide and nucleic acid content during SDS clearing, it is important that nonlipid tissue components have been hybridized to a hydrogel scaffold.
In the initial Nature paper describing CLARITY8, the dense tissue-hydrogel cross-linking conferred by A4P4B0.05 tissue embedding prohibited rapid passive clearing of large tissue blocks24. The advanced CLARITY protocol15 suggests decreasing acrylamide concentrations to as low as 0.5% (A0.5P4B0.0125) when clearing is performed passively rather than with ETC-based rate enhancement. After the initial, thorough perfusion-fixation step with 4% PFA, PACT and PARS tissues are infused with A4P0 monomer18. Although bis-acrylamide may be included in the hydrogel formulation to stabilize fragile samples, we have not found the addition of bis-acrylamide to be beneficial in preventing protein loss (Fig. 3a) in either A4P0-hybridized (A4P0B0.05) or A4P4-hybridized (A4P4B0.05) tissues. Furthermore, although protein retention is similar for all A4P0-4 formulations (Fig. 3a), higher concentrations of PFA, which anchors tissue to the hydrogel mesh and increases tissue cross-linking, result enhanced fine structure preservation (Fig. 5b) and limits anisotropic tissue-hydrogel expansion (Fig. 3b,c). The resulting less-porous tissue-hydrogel matrix curtails protein solubilization by SDS (Fig. 3a and Supplementary Fig. 2b); clearing speed (Fig. 4a and Supplementary Fig. 2c), overall tissue transparency (Fig. 4a) and the efficiency of antibody labeling (Fig. 4b–d) are all reduced. Thus, PFA-containing hydrogel formulations are only recommended for samples that will be used for in-depth profiling of fine structures, in which protein and nucleic acid retention is of maximum importance.
The importance of pH and temperature in clearing
Here we describe two modes of detergent-based tissue clearing: passive lipid removal (PACT: Step 6A for hydrogel permeation and embedding, Step 7A for PACT clearing) and active delipidation (PARS: Step 6B for hydrogel perfusion and embedding, Step 7C for PARS clearing). Several factors, including the chemical properties of the detergent solution, the pH of the detergent solution46 and the tissue components to be extracted (i.e., peptide, lipid and nucleic acid), affect micelle formation and composition, and hence the clearing efficiency. The role of pH is heightened in scenarios, such as tissue clearing, in which relatively high SDS concentrations (4–8% (wt/vol) SDS) are used. A slightly basic clearing solution will help to counteract proton buildup at the negatively charged surface of SDS micelles47. Conversely, a clearing solution that becomes too acidic has the potential to impair lipid extraction by disrupting the structure of the ionic micelles, as well as to encourage protein extraction via their denaturation and release from membranes. For these reasons, and to avoid damage to tissue and to endogenous fluorescent proteins48, it is important to maintain a physiological to slightly basic pH during tissue clearing. Among the alkaline buffers that are best suited for PACT and PARS are 0.2 M boric acid (pH = 8.5) and 0.01 M PBS (pH = 7.5 (ref. 18) and 8.5), with the more basic 8% (wt/vol) SDS solutions offering a slight rate enhancement to delipidation (Supplementary Fig. 2c).
Temperature represents a second important factor that influences the solubilization process, and, in particular, the micellular composition49. For SDS in aqueous medium, the average micelle volume decreases, but the total number of micelles increases as the temperature rises50. It is hypothesized that smaller micelles may more readily diffuse through the tissue-hydrogel matrix, and thus increasing the temperature of the clearing bath will accelerate lipid extraction. Higher temperatures (~50 °C), which may enhance clearing efficiency7,15, will promote protein denaturation, which has the potential to damage relevant protein epitopes or to incur fluorescent protein signal loss. Thus, both PACT and PARS clearing steps are performed at 37 °C. To accelerate lipid extraction, the concentration of SDS is raised from 4% to 8% (wt/vol) SDS relative to CLARITY, which has a similar effect as raising the clearing temperature.
Labeling
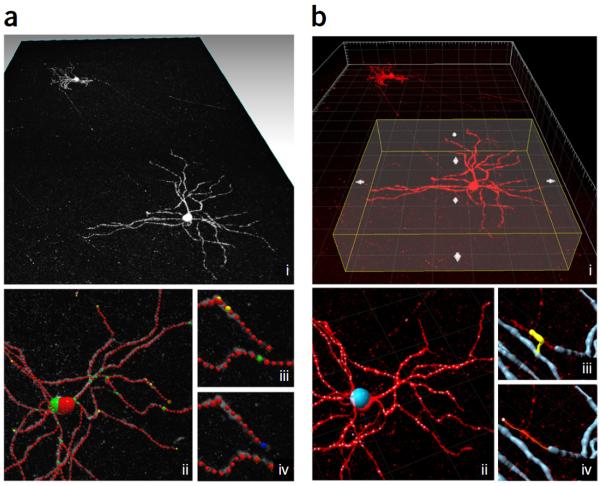
PACT- and PARS-prepared tissues are amenable to most standard histological techniques, including those that use immunohistochemical, small-molecule and fluorescent protein–based labels, as well as bright-field stains. Small-molecule dyes such as nuclear stains rapidly distribute throughout thick tissue sections, such that hour-long to overnight incubations are sufficient for most samples. The slow diffusion of full-format antibodies (150 kDa) through thick samples, and their tendency to denature and degrade over time, necessitates the use of, on average, tenfold more concentrated antibody dilutions in primary and secondary incubations of thick sections than in 40-μm-thin sections. The use of smaller antibody formats (fragment antigen-binding (Fab): 55 kDa, Fab dimer (F(ab′)2): 110 kDa,) for secondary antibody labeling is suggested, particularly given their commercial availability. Herein, we can achieve adequate labeling of 1-mm-thick sections by Fab format antibodies within 48 h. Even smaller formats, most notably camelid nanobodies (15 kDa)51,52, are ideally suited for labeling thick tissue (Supplementary Fig. 6), as at 10× smaller than full IgGs they penetrate tissue rapidly and thoroughly. In addition, their stability (e.g., over a wide pH range, at high concentrations, and at temperatures of up to 90 °C) and protease resistance allows them to remain intact throughout long incubations conducted at room temperature (RT, 18–25 °C) (Supplementary Fig. 6b).
Enhancement of optical clarity
Infusing and mounting cleared tissues in RIMS helps to minimize the mismatch between the RIs of the sample and the microscope objective. This so-called optical clearing, which is detailed in Steps 10–13, greatly enhances the optical clarity of cleared samples (Figs. 1a–e, 2b, 4, 5a,c,d, 6a and 8; Supplementary Figs. 3, 5 and 6b). One could substitute a different mounting solution for RIMS (e.g., sRIMS (Fig. 1f,g), cRIMS, glycerol dilution, FocusClear24, Cargille Labs optical liquids15 and 2,2′-thiodiethanol44). Optionally, one could measure the RI of the chosen mounting medium using a refractometer, dilute glycerol with ddH2O to the same RI, and then use this glycerol dilution as the immersion medium for dipping objectives.
Imaging
To use tissue clearing to its best advantage, the microscope setup must be capable of acquiring high-resolution image stacks through thick, cleared samples. Of utmost importance are the detection optics. A high numerical aperture (NA ~1.0) and long-working distance (w.d. = 5–10 mm) objective will provide high resolving power even when viewing deep tissue structures. In addition, as objectives are designed according to the optical properties of a target sample and sample mount, an objective that has been optimized to the RI range of the RIMS-mounted tissue and immersion medium (RI ~1.46–1.49) will minimize spherical aberrations, maximize lateral and axial resolution and help preserve fluorescent signal intensity while imaging through thick, cleared tissues. To this end, numerous manufacturers have developed specialized multi-immersion and air objectives that are well suited to imaging PACT- and PARS-cleared fluorescent samples at depth: e.g., Olympus 10× 0.6 NA UIS2-XLPLN10XSVMP and 25× 1.0 NA UIS2-XLSLPLN25XGMP objectives (w.d. 8.0 mm) for samples with RI ~1.33–1.52 and RI ~1.41–1.52; Leica HC FLUOTAR L 25× 1.0 NA IMM motCORR VISI (w.d. 6.0 mm) for samples with RI = 1.457; Zeiss Scale-optimized 20× 1.0 NA objective (w.d. 5.6 mm) for samples with RI = 1.38; and Zeiss CLARITY/CUBIC-optimized EC Plan-NEOFLUAR 5× 0.16 NA objective and LSFM clearing 20× 1.0 NA objective (w.d. 5.6 mm) for samples with RI = 1.45.
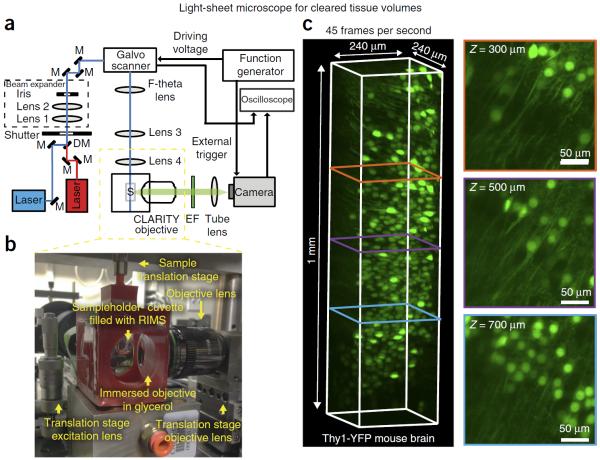
Imaging cleared tissues via two-photon or confocal microscopy can generate extremely high-resolution data sets. However, these imaging modalities are time-consuming, particularly when scanning a large field of view at depth. Light-sheet fluorescence microscopy (LSFM) permits rapid scanning through comparatively large sample volumes, which alleviates the imaging bottleneck that can occur with the high-throughput preparation of cleared samples. In addition, because image acquisition requires only brief plane illumination, LSFM minimizes sample photobleaching, a major drawback in using point-scanning confocal systems to image large fluorescently labeled samples. Given the widespread availability of confocal microscopes but the obvious benefits of LSFM, we provide imaging guidelines for each system, as well as design schematics for a cost-efficient LSFM system (Fig. 9).
Figure 9.
Light-sheet microscopy enables fast and high-resolution imaging of cleared samples. (a) A schematic diagram of the light-sheet microscope; M, mirror; DM, dichroic mirror; S, sample; EF, emission filter. The scientific CMOS camera (Zyla 4.2 sCMOS, Andor) is running in a light-sheet mode, in which the readout direction of the camera is unidirectional and synchronized with the scanning direction and speed of the light source. In this configuration, only the pixels that are illuminated will be recorded, thus improving the signal to noise ratio of the image. For ease of synchronization, the function generator, the camera and the oscilloscope are controlled using a custom MATLAB program. (b) An image of the 3D-printed immersion chamber (see design in Supplementary Data 1), in which the CLARITY objective (Olympus 25× 1.0 NA multi-immersion objective, w.d. 8.0 mm) is immersed in glycerol, whereas the sample is within a quartz cuvette filled with RIMS. (c) A volume rendering (Imaris, Bitplane) and cross-sections at different depths of a cleared Thy1-YFP mouse brain section (1 mm thick), taken with the light-sheet microscope. The intensity of the layers was normalized as per Imaris image processing function, i.e., the mean and s.d. of each layer were equalized to the mean and s.d. of the entire stack using linear transformation. The images were acquired at 45 frames per second (voxel size: 0.117 μm × 0.117 μm × 0.25 μm, bit depth: 12). The cross-sections at different depths, which are perpendicular to the scan direction, are maximum intensity projections (Imaris) across a 5-μm volume. A parts list for this setup is available in Supplementary Table 1.
Data analysis
Following on the heels of the `OME' focus of the past few decades53 and spurred by the efforts of the BRAIN Initiative (http://www.braininitiative.nih.gov/index.htm), the Human Brain Project (https://www.humanbrainproject.eu) and the Allen Brain Institute, the quest to map the human connectome has recently taken center stage. It is unlikely that the connectome project can be tackled by a select few taking a top-down approach, as was possible in the elucidation of the human genome54,55. Instead, by tasking multiple groups with mapping discrete neural circuits, one can envision the draft of a connectome gradually emerging through stitching together these individual wiring diagrams (e.g., http://www.openconnectomeproject.org)56–58. When combined with long-working-depth objectives and high-throughput imaging (LSFM, e.g., CLARITY optimized light-sheet microscopy15; and the custom-made, economical system presented here in Fig. 9, Supplementary Table 1, and Supplementary Data 1 and 2), PACT and PARS provide a means for efficiently acquiring information on the spatial position of neurons within large tissue volumes at high resolution. For this information to be applied to mapping the connectome59, however, these gigabytes or even terabytes of raw image data (e.g., for a whole mouse brain at 25× magnification) must be converted into a complex network of neuron projection pathways and neural contacts, a feat that poses substantial demands on both storage hardware and image analysis software. Many available software tools and image file formats were not designed with tera-scale data sets in mind and assume that entire image volumes fit in computer RAM.
To this end, we have evaluated a range of software packages for processing, visualization and analysis of cleared brain volumes, including both general image processing platforms and more specialized tools focused on stitching or filament tracing. Table 3 includes a summary list of those tools that we found to be stable, functional, user-friendly and well supported. For general image analysis, we recommend Fiji60 (a distribution of ImageJ61) and Vaa3D62,63, which are open-source, extensible platforms for image analysis and visualization that have a rich collection of plug-ins for carrying out specific tasks including stitching and fiber tracing64–66. We recommend neuTube67 for semiautomated tracing of neurites (Fig. 10a and Supplementary Fig. 7a; see Supplementary Data 3 for the raw 3D image stack from which the neuTube traces were generated). Commercial software packages Imaris (Bitplane) and Neurolucida68 provide similar functionality, currently offer better support for very large image files and can be more stable and user-friendly (Fig. 10b and Supplementary Fig. 7b; see Supplementary Data 3 for the raw 3D image stack from which the Imaris traces were generated)—making them a good starting point for laboratories with less image-processing expertise.
TABLE 3.
Image analysis and visualization tools.
| Name | Platform | Stitching | Out-of-core visualization | Semiautomated tracing | Notes |
|---|---|---|---|---|---|
| Imaris (Bitplane) | Commercial Win/Mac | No | Yes | Yes Filament Tracer plug-in |
Best loading and volumetric rendering of large (out-of-core) images |
| Vaa3D62,63,66 | Open Source Win/Mac/Linux | Yes iStitch70 or TeraStitcher69 plug-in |
Yes TeraFly plug-in66 |
Yes Vaa3D-Neuron2 plug-in66 |
|
| Fiji60,61 | Open Source Win/Mac/Linux | Yes Stitching plug-in71 |
Partial Virtual Stacks, Data Browser plug-ina, or BigDataViewerb (ref. 80) |
Yes Simple Neurite tracer64,81 |
|
| neuTube67 | Open Source Win/Mac/Linux | No | No | Yes | Fastest semiautomated tracing interface |
A list of image analysis tools appropriate for processing data from cleared tissue volumes including functionality for stitching, visualization and tracing. This list is far from exhaustive (consult refs. 87,88 for a broad survey), but it includes those that we tested and found most effective on our data sets.
Data Browser ImageJ plug-in (LOCI), see http://loci.wisc.edu/software/data-browser.
BigDataViewer, see http://fiji.sc/BigDataViewer.
Figure 10.
Two different workflows for cell tracing in neuTube and Imaris. (a) Tracing using neuTube67. (b) Tracing using Imaris 7.1 (Bitplane). Results shown here took 25 min for a novice user with ~5 h of total experience using each tracing tool. Total tracing time to achieve similar results was generally comparable, but we found neuTube to be more efficient for quickly tracing isolated neurites. (a) neuTube 3D visualization (i), neuTube semiautomated tracing result (ii), tracing error (iii), and manual correction (iv). (b) Imaris ROI selection (i), Imaris `Autopath' seeding (ii), manual correction of tracing error (iii), and trace extension using `Autopath' (iv). The original test mage on which semiautomated tracing was performed is provided in Supplementary Data 3.
Image stitching
Confocal and light-sheet microscopes equipped with motorized stages usually support tiled acquisition, which is essential for imaging large volumes at cellular resolution. These tiles can then be aligned to pixel accuracy and blended together using microscope acquisition software such as Leica Application Suite (Leica Microsystems), Zen (Zeiss), cellSense (Olympus) and NIS Elements (Nikon Instruments), or manipulated off-line using open-source tools such as the TeraStitcher69, the Vaa3D iStitch plug-in70, the ImageJ stitching plug-in71, and XuvTools72.
When stitching together multiple tiles, systematic variations in brightness across the image field caused by nonuniform illumination, vignetting or imprecise optical alignment often result in significant variations in image brightness that can make downstream visualization and processing difficult. One solution is to capture smaller tiles from the central field of view where illumination tends to be more uniform. However, this increases capture time, as, for example, reducing the field of view to the center one-third requires capturing and stitching nine times as many tiles. An alternate approach is to directly measure the illumination profile using a uniform calibration slide (e.g., see protocols73 and `How flat is your confocal illumination profile? Want to find out?' at http://www.spectral.ca/Downloads?f=2745809748.pdf) or CIDRE74, and then to apply the estimated correction to each acquired image tile. This so-called `flat field' or shading correction from a reference image is often supported by acquisition software such as the μManager MultiChannelShading plug-in (see http://nic.ucsf.edu/dokuwiki/doku.php?id=flatfieldimageacquisition and ref. 75), shading reference in NIS Elements (Nikon Instruments), Leica Application Suite (Leica Microsystems), Zen (Zeiss) and cellSense (Olympus), or it can be carried out using an off-line workflow such as with the ImageJ ImageCalculator; see `How to correct background illumination in bright-field microscopy' by G. Landini at http://imagejdocu.tudor.lu/doku.php?id=howto:working:how_to_correct_background_illumination_in_bright-field_microscopy.
Visualization
Image stacks can be visualized using commercial software such as Imaris (Bitplane), Amira (FEI), MetaMorph (Molecular Devices) and others (Zen (Zeiss), Leica Application Suite (Leica Microsystems), NIS Elements (Nikon Instruments), cellSense (Olympus), MetaMorph (Molecular Devices), Volocity (PerkinElmer), Huygens (SVI), Arivis Vision4D (Arivis), or using free or open-source tools such as Fiji 3D Viewer76, Vaa3D62, Icy77, BioImageXD78, VolView (Kitware, see http://www.kitware.com/opensource/volview.html) or Bioview3D79. These tools all support 3D volumetric rendering of image data that can be interactively rotated and zoomed by the user, as well as functionality for selecting subvolumes, virtual 2D sectioning, image contrast and other colormap adjustments and manual annotation. Here we provide example workflows and estimated processing times based on tests with a large tiled image, 144 fields of view taken on an LSM 780 at 5× magnification, stitched in ZEN (Zeiss) to produce a single channel, 8-bit, 30-GB image stack of size 3.3 × 1010 voxels (16,384 × 9,216 × 220) covering ~1.165 mm3 (2.72 × 1.53 × 0.28 mm) of tissue.
Stitching very large acquisition volumes can easily produce image files that are too slow to load and display directly on machines with limited memory. Table 3 indicates which software tools support `out of core' visualization, using read on demand, caching and multiresolution representations to process and visualize data sets that are too large to fit in memory while preserving interactivity. In particular, TeraFly69, Imaris (Bitplane) and BigDataViewer80 (Fiji, see http://fiji.sc/BigDataViewer) use custom multiresolution, tiled file formats for storing image data on disk. This aids interactive visualization (even on machines with substantial amounts of RAM), as low-resolution views can be displayed quickly with higher-resolution detail filled in as soon as it can be read from disk.
Morphometric analysis
Tracing of neurites can be carried out using plug-ins provided in general processing tools such as Imaris Filament Tracer (BitPlane), Amira Skeletonization plug-in (FEI), Metamorph NX Neurite Tracing (Molecular Devices), Fiji Simple Neurite Tracer64,81 and Vaa3D-Neuron266. Alternatively, one can use special-purpose software such as Neurolucida68, neuTube67, Neural Circuit Tracer82, flNeuronTool83, Farsight trace editor84, Neuron Studio84 and Neuromantic85. Several of these tools provide automated or semiautomated workflows that allow a user to trace neurites by clicking on a few points along a given neurite, which can greatly accelerate initial tracing of long-range projections. The 3D Image Analysis workflow below gives estimates of time required to produce a rough-draft trace for a test image (Supplementary Data 3). Semiautomated tracing tools are computationally intensive and currently have slow performance on volumes larger than a few gigabytes. Efficient use of these tools thus requires manual selection or cropping of regions of interest (ROIs) during annotation, and the resulting traces need to be merged in a post-processing step.
Morphology of traced neurites can be saved in SWC (a standard file format developed by the Southampton Neurosciences Group; see the SWC file format specifications on the Computational Neurobiology and Imaging Center of the Mount Sinai School of Medicine website: http://research.mssm.edu/cnic/swc.html), NeuroML86 or NEURON .hoc file formats, which in turn can be used with a wide range of downstream neuroinformatics tools87,88 including statistical morphometry (e.g., Scorcioni et al.89), assembly and simulation of biophysical models (e.g., Gleeson et al.90) and deposition in online searchable databases (e.g., http://www.neuromorpho.org/ (ref. 91)).
Applications of the method
PACT, PARS and RIMS clear a variety of tissues, from laboratory mice and rats (organs and adult whole bodies) to primates (Fig. 1f,g, tumor biopsy18), and they are compatible with endogenous fluorescence, immunohistochemistry, long-term sample storage18, smFISH and microscopy with cellular and subcellular resolution18. Furthermore, the potential exists to apply PARS to the clearing and staining of large, isolated whole organs when the vasculature is preserved during organ excision92,93. Akin to paraffin embedding, the increased rigidity of hydrogel-embedded, uncleared samples may allow unstructured soft tissues (e.g., pancreas, thymus) and amorphous biological samples (e.g., sputum) to be stabilized for manual sectioning, as well as for automated slicing and imaging systems, such as serial two-photon tomography29,37. When these tissue-hydrogel hybrids are cleared with PACT or PARS rather than thin-sectioned for imaging, whole organs and thick tissue blocks become amenable to visualization with modern microscopy methods such as LSFM (which rapidly scans large sample volumes, thereby minimizing photobleaching but maximizing the phenotypic content within the image stack) and super-resolution microscopy29,37. Bridging these microscale and nanoscale imaging modalities, the recent method of expansion microscopy (ExM)94 recruits a by-product of CLARITY8 and PACT18 hydrogel-embedding, namely the capacity to absorb water, to great advantage. By deliberately swelling tissue-hydrogel hybrids, isotropically expanded tissues can be mined for qualitative information of subcellular structures at synaptic resolution using only a conventional confocal microscope.
Accelerating biomedical discovery with tissue clearing
The current and potential biomedical applications of PARS and PACT are summarized in Table 1. PARS and PACT enable detailed structural information from peripheral tissue and organ samples to be obtained, aiding in the study of distinct cellular populations or environments within their unsevered tissue milieu. For example, stem cell niches embedded within relevant tissue environments, such as small intestinal crypts95,96 and the bone marrow97, can be studied. Tumor architecture and morphology can be mapped98, including tumor margins, tumor vascularization, cellular heterogeneity and metastatic foci across the entire organism, for both research and diagnostic purposes. Whole-body optical clearing by PARS and imaging could facilitate obtaining better peripheral nerve maps39,99, which can then facilitate an understanding of the neural processing that accompanies peripheral nerve/organ function and dysfunction41.
Importantly, PARS may also facilitate whole-body screening of therapeutics for off-target and on-target binding, and for imaging the biodistribution of administered agents as a method for the qualitative determination of their pharmacokinetic–pharmacodynamic (PK/PD) properties. Similarly, PARS can be used to expedite the slow, labor-intensive process of screening novel viral vector variants for specific tropism characteristics. Typically, researchers perform conventional tissue slicing and histology on numerous tissues across multiple samples, which is an exceedingly laborious process. Whole-body screening through PARS can improve throughput and reduce the risk of sampling errors.
The described protocols for tissue stabilization and lipid removal allow for rapid phenotyping of whole organs and whole organisms, and therefore they could advance biomedical research with respect to the study of changing tissue pathology during aging or during disease progression. One obstacle to studying the progression of cell death that occurs during neurodegeneration (e.g., in Parkinson's disease, Alzheimer's disease, epilepsy, stroke) is the inability to visualize cells that have already died and have been removed by macrophages before the tissue was dissected for histological analysis. A similar cellular mapping confound exists in ablation experiments, wherein toxins are used to damage cells for studies that aim to causally link the function of a defined neuronal population (compact or sparsely distributed) to brain activity and behavior. The postquantification is rarely accurate, as it relies on inferring the exact distribution of ablated cells on the basis of their representative distribution in placebo-treated brains. By combining PARS with TEMPEST27—a precursor to CLARITY—the in vivo expression of long-lasting keratin filaments (that outlive the cells themselves while keeping a loyal blueprint of the morphology) within populations of interest can facilitate accurate postmortem quantification and brain-wide mapping of long-degenerated cells.
Size fluctuations in tissue-hydrogel hybrids: challenge and opportunities
Most protocols that render tissues transparent cause notable sample volume fluctuations. In general, clearing protocols that entail dehydration steps for clearing with organic solvents or some concentrated RI-matching solutions cause tissue shrinkage, whereas protocols that involve prolonged incubations in aqueous detergent-based solutions tend to cause gradual tissue expansion (Table 2)7,9,22,100. In part a consequence of the water-absorbing properties of polyacrylamide, a nitrogen-containing derivative of the super-absorber polyacrylic acid, tissue-hydrogel expansion has previously been reported with CLARITY and PACT-processing8,18 (Fig. 3b,c), and indeed it has been used to great advantage in ExM94. Several factors have been shown to influence the swelling properties of water-absorbing hydrogels. The most notable are pH; the dissolved ion content of the aqueous swelling medium (i.e., clearing buffer) and the tissue-hydrogel microstructure, including the ordering of monomeric units within a polymerized hydrogel; the degree of cross-linking; and the mechanical rigidity of the embedded tissue. With respect to tissue clearing, as detergent gradually solubilizes and extracts tissue biomacromolecules, not only can water migrate into this additional space in the tissue-hydrogel matrix but also there is less mechanical resistance from tissue components to polymer swelling as water continues to diffuse in.
For neuron tracing and brain mapping purposes (i.e., connectomics101, expansion-contraction cycles should be minimized so as not to sever fine processes, distort the spatial arrangement of cells within local niches or alter cellular connectivity7. Similarly, gross size changes, particularly when anisotropic, complicate image registration with existing atlases such as the Allen Brain Atlas102 We have previously proposed a few modifications to passive CLARITY-based protocols in order to counteract tissue expansion that occurs during clearing and to minimize the occurrence of morphological artifacts that could be introduced with fluctuating tissue size7,9,100 They include using in-skull clearing protocols (e.g., PARS-CSF18 and PARS, see Figs. 7 and 8; Supplementary Fig. 5); extending the postfixation step for perfused, excised organs, including the brain, before the start of any clearing protocol; and/or performing PACT with a hydrogel monomer formulation that contains increasing amounts of PFA (e.g., a hydrogel solution of 4% (wt/vol) acrylamide and 1–4% (wt/vol) PFA, A4P1–4)). With respect to the latter, the inclusion of PFA in hydrogel monomer compositions not only combats hydrogel swelling but also the expansion becomes increasingly isotropic (Fig. 3b). Thus, for improved tissue preservation, it is advisable to supplement the A4P0 hydrogel recipe with PFA (1–4% PFA in the monomer solution).
The inclusion of PFA in monomer solutions also curtails tissue size changes in mounting medium. Upon their initial immersion in RIMS, tissue samples contract during the first hour (~20% for A4P0-embedded coronal rodent brain sections), followed by a gradual rebound back to their pre-RIMS size. Imaging during this time window should be avoided, as these slight size fluctuations could introduce apparent tissue deformities or sample drift issues during image acquisition. With adequate equilibration in RIMS (e.g., hours to days, depending on sample size, tissue permeability and so on), sample size and transparency will reach a steady state for high-resolution, deep imaging18.
It follows that a motivating factor behind the development of PARS was to neutralize this potential risk of tissue expansion during clearing. Although the tissue becomes more permissive to hydrogel swelling as lipid membranes are permeabilized and extracted, the skin encasing, bone structure and connective tissue will continue to restrict water absorption by the hydrogel and thus minimize size changes of the internal organs. Consequently, the addition of PFA to the PARS monomer solution is not necessary.
However, swelling—if isotropic—can be advantageous. By expanding hydrogel-embedded tissue uniformly, dense cell populations can be distributed spatially for cell counting or for analyzing local cell contacts (Supplementary Fig. 4 and Supplementary Methods); similarly, dense cell and/or fiber tracts, such as the corpus callosum, the spinal cord and individual muscles, may be expanded for easier anatomical study. For example, ExM recently recruited the swelling properties of CLARITY tissue-hydrogel hybrids8,18 to visualize samples at nanometer resolution94 Through altering the monomer components and concentration, scientists may quickly adjust the overall volume occupied by the hydrogel-embedded tissues, shrinking tissues to fit within the working distance of an objective or swelling tissues for facile high-resolution imaging of diffraction-limited spots103 Such measurements require separate validation with well-established super-resolution microscopy methods104–108 until one can confirm that specimen preparation does not introduce anisotropic distortions.
Limitations of the method
As discussed in the previous section, some tissue deformation is expected with all tissue clearing protocols (see examples in Table 2), wherein the tendency for tissue to expand and/or shrink moderately during sample clearing and/or mounting is frequently noted7,8,10. Whether these volume changes cause structural damage that would confound the interpretation of sample images is widely debated. Although we have observed some tissue swelling during PACT and PARS clearing, tissues subsequently contract to approximately their original size in RIMS medium. Although it is difficult to test exhaustively by individual efforts, the net impact of these changes on overall cellular architecture appears to be minimal, as demonstrated by the preservation of fine cellular morphology across a range of tissue types18. However, such changes in tissue volume could complicate image registration. To compare an image stack of an experimental sample with a representative data set or with a reference atlas, it will be necessary to use structural landmarks or tissue stains rather than the sample size to align images. Validated tissue stains that can help with registration include the following: Nissl or Golgi stain for the brain; membrane and organelle stains, including the use of lectins to label vascular networks, as well as H&E stain to simultaneously visualize hematoxylin-stained nucleic acid and eosin-stained red blood cells, cytoplasmic material, cell membranes and extracellular structures and protein; and fuchsin to label collagen, smooth muscle or mitochondria. Finally, unstained structural components (e.g., distinctive neuron arborization109, large tissue structures110 and cortical surfaces110–112 can be imaged at lower resolution to create reference images to aid in registering high-resolution, small volumes113) and artificial landmarks (e.g., gold-seeding or quantum dot deposition114, electron microscopy (EM) platform grids for serial section mounting and imaging115) may be used as fiducial markers for registering sample data sets collected during light and electron microscopy, as well as for cross-referencing in vivo imaging scans with subsequent ex vivo data sets.
To image whole organs or thick tissue sections, the image data file sizes will be tera-scale; thus, it is important to use a computational workstation with substantial RAM (this will be highly dependent on the individual software requirements, user-specific variables such as the average file size and the desired image analysis capabilities). Our experience showed that as much as 64–256 GB might be needed, depending on data and analysis type, multicore CPUs and an excellent graphics card (e.g., Windows platform: AMD Radeon R9 290X 4.0 GB; Mac platform: AMD FirePro D700 6 GB).
MATERIALS
REAGENTS
Sample to be imaged. This protocol describes imaging of brain and body samples prepared from wild-type mice (C57BL/6N and FVB/N, both male and female), Thy1-YFP mice (line H) and TH-cre rats ▲ CRITICAL Experiments on vertebrates must conform to all relevant governmental and institutional regulations. Animal husbandry and all experimental procedures involving mice and rats were approved by the Institutional Animal Care and Use Committee (IACUC) and by the Office of Laboratory Animal Resources at the California Institute of Technology.
Euthasol (Virbac, cat. no. 710101)
Perfusion solutions
Paraformaldehyde (PFA; 16% and/or 32% (wt/vol) PFA in aqueous solution; Electron Microscopy Sciences, cat. no. 15710-S)
1× PBS
Sodium nitrite (Sigma-Aldrich, cat. no. 237213-500G) ▲ CRITICAL As a vasodilator, 0.5% (wt/vol) sodium nitrite is added to the heparinized saline perfusion buffer to facilitate thorough blood removal from vasculature and perfusion ease. Alternatively, nitroglycerin may be substituted for sodium nitrite.
Heparin sodium salt from porcine intestinal mucosa (Sigma-Aldrich, cat. no. H3149) ! CAUTION PFA is toxic. Perform all procedures in a fume hood.
Hydrogel monomer solution
Acrylamide solution (40% (wt/vol); Bio-Rad, cat. no. 161-0140) ! CAUTION Acrylamide monomers are toxic. Perform all procedures in a fume hood.
Bis-acrylamide (2% (wt/vol); Bio-Rad, cat. no. 161-0142) ! CAUTION Bis-acrylamide monomers are toxic. Perform all procedures in a fume hood.
Polymerization thermal initiator VA044: 2,2′-Azobis[2-(2-imidazolin-2-yl) propane]dihydrochloride (Wako, cat. no. VA-044)
10× PBS
Clearing solutions
Boric acid (Sigma-Aldrich, cat. no. B7901 or B6768)
Sodium hydroxide pellets (EMD, cat. no. SX0590-3)
SDS (Sigma-Aldrich, cat. no. L3771) or 20% (wt/vol) SDS solution in water (Sigma-Aldrich, cat. no. 05030)
10× PBS ▲ CRITICAL We have successfully used either `homemade' PBS or PBS from a variety of suppliers.
0.5 M EDTA solution (Lonza AccuGENE, cat. no. 51234; or Sigma-Aldrich, cat. no. 03690)
Refractive index matching solution (RIMS)
Histodenz (Sigma-Aldrich, cat. no. D2158)
Phosphate buffer, 0.02 M
Sodium azide (Fisher Scientific, cat. no. 71448-16) ! CAUTION To prevent microbial growth, sodium azide should be added to all mounting media (RIMS and sRIMS), as well as to all immunostaining dilutions and wash buffers that are used in extended incubations. As a stock solution, 5% (wt/vol) sodium azide in water is highly toxic. Keep sodium azide solid and 5% stock solutions away from ignition sources. Store it in a cool, dry, well-ventilated area away from incompatible substances. Do not store it in metal containers, and keep the storage containers tightly closed. Contact with acids liberates very toxic and explosive hydrazoic acid vapor, and some hydrazoic acid may form in aqueous solutions prepared with sodium azide. Hydrazoic acid volatilizes readily at 99 °F; thus, do not autoclave any solutions that are prepared with sodium azide. Sodium azide is an eye and skin irritant, it may be highly toxic if inhaled or ingested and it is metal-reactive; wear gloves and eye protection when preparing stock solutions that contain sodium azide, and do not weigh out solid sodium azide using a metal spatula (use plastic instead). ! CAUTION With respect to long-term storage, sodium azide is chemically stable. However, it will decompose upon heating. Sodium azide waste disposal should be conducted according to federal, state and local regulations.
Sorbitol-based RIMS (sRIMS)
Sorbitol, 70% (wt/vol) (Sigma-Aldrich, cat. no. 309532)
Refractive index matching solution for cold storage (cRIMS)
Histodenz (Sigma-Aldrich, cat. no. D2158)
0.005 M phosphate buffer
Sodium azide (Fisher Scientific, cat. no. 71448-16)
Immersion medium and alternative mounting medium
Glycerol (87% (vol/vol)): prepare 80–90% (vol/vol) glycerol (Sigma-Aldrich, cat. no. G5516) in dH2O.
Immunostaining reagents
1× PBS with Triton X-100 (0.1% (vol/vol))
Primary and secondary antibodies (see Table 4 for examples of antibodies used in this and related work)
Normal donkey serum (Jackson ImmunoResearch Laboratories, cat. no. 017-000-121)
Agarose, low-melt temperature (Research Products International Corp., cat. no. 9012-36-6)
TABLE 4.
Antibodies and small-molecule stains validated for cell-phenotyping in PACT- and PARS-processed tissues.
| Label | Supplier and/or formulation |
|---|---|
| Nucleic acid and molecular labels, cell-type and tissue-type small-molecule stains | |
| DAPI | Life Technologies (no. D-1306); 0.1–10 μg/ml |
| Hoechst | Cell Signaling |
| NeuroTrace 530/615 Red Fluorescent Nissl Stain | Life Technologies (no. N-21482); 1:50 dilution |
| SYTO 24 | Life Technologies (no. S-7559); 1:200 dilution |
| DRAQ5 | Cell Signaling (no. 4084); 1:200 dilution |
| Acridine Orange | Life Technologies (no. A-1301); 100 μg/ml dilution |
| Lectin, DyLight 488 conjugate | Vector Laboratories (no. L-1174); 1:100 dilution |
| Methylene blue | Sigma-Aldrich (no. 66720); 1 μg/ml |
| Atto-565-conjugated phalloidin | Sigma-Aldrich (no. 94072); 1:100 dilution |
| Alexa Fluor 647–phalloidin | Life Technologies (no. A-22287); 0.5 μM in PBS |
| Primary antibodiesa | |
| Mouse anti-pan-cytokeratin (AE1/AE3) antibody, Alexa Fluor 488–conjugated | EBiosciences (no. 53–9003); 1:100 dilution |
| Synthetic anti-GFAP nanobody, Atto 488–conjugated | GFAP nanobody producing according to published methods194,195 1:100 dilution (Purified GFAP nanobodies were first conjugated to Atto 488, Sigma-Aldrich, diluted in dH2O to 1 mg/ml stock and then diluted 1:100 for tissue labeling) |
| Chicken anti-tyrosine hydroxylase (TH) IgY | Aves Labs (no. TYH); 1:400 dilution |
| Chicken anti-glial fibrillary acidic protein (GFAP) IgY | Aves Labs (no. GFAP); 1:400 dilution |
| Rabbit anti-ionized calcium-binding adapter molecule 1 (Iba1) IgG | Biocare Medical (no. CP 290A); 1:200 dilution |
| Rabbit anti–integrin-β4 and anti-integrin β5 IgGs | Santa Cruz Biotechnology (β4: sc-9090, β5: sc-14010); 1:200 dilution |
| Rabbit anti–β-tubulin IgG | Santa Cruz Biotechnology (no. sc-9104); 1:200 dilution |
| Rabbit anti-parvalbumin antibody | Abcam (ab11427); 1:200–1:400 dilution |
| Mouse anti–β-spectrin II antibody | BD Biosciences (612563); 1:200 dilution |
| Rabbit anti–α-adducin antibody | Abcam (ab51130); 1:200 dilution |
| Goat anti–myelin basic protein antibody | Santa Cruz Biotechnology (C-16: sc-13914); 1:200 dilution |
| Mouse anti–pan-axonal neurofilament SMI-312 antibody | BioLegend (SMI-312); 1:500 dilution |
| Guinea pig anti-insulin antibody | DAKO (A0564); 1:500 dilution |
| Goat anti-somatostatin antibody | Santa Cruz Biotechnology (sc7819); 1:500 dilution |
| Secondary antibodiesb | |
| Donkey anti-chicken IgY, available as Cy2c, Cy3d, Cy5e, Alexa Fluor 594f, Alexa Fluor 647g conjugates | Jackson ImmunoResearch (703-###-155), where ### = fluorophore code; 1:200 dilution |
| Donkey anti-goat IgG, available as Cy2c, Cy3d, Cy5e, Alexa Fluor 594f, Alexa Fluor 647g conjugates | Jackson ImmunoResearch (703-###-155), where ### = fluorophore code; 1:200 dilution |
| Donkey anti-guinea pig IgG, available as Cy2c, Cy3d, Cy5e, Alexa Fluor 594f, Alexa Fluor 647g conjugates | Jackson ImmunoResearch (703-###-155), where ### = fluorophore code; 1:200 dilution |
| Donkey anti-mouse IgG, available as Cy2c, Cy3d, Cy5e, Alexa Fluor 594f, Alexa Fluor 647g conjugates | Jackson ImmunoResearch (703-###-155), where ### = fluorophore code; 1:200 dilution |
| Donkey anti-rabbit IgG, available as Cy2c, Cy3d, Cy5e, Alexa Fluor 594f, Alexa Fluor 647g conjugates | Jackson ImmunoResearch (703-###-155), where ### = fluorophore code; 1:200 dilution |
| Reagentsh for FISH labeling18 | |
| 20-mer oligo probes toward β-actin | 1 nM each of 24 Alexa Fluor 594–labeled 20-mer oligo probes toward β-actin prepared in hybridization buffer |
| Slowfade Gold + DAPI | Life Technologies (no. S-36938); mounting medium for smFISH samples |
Atto fluorescent dyes that possess an NHS ester moiety (available from Sigma-Aldrich) may be conjugated to the primary antibody; this eliminates the need to perform a secondary antibody incubation when imaging tissues via fluorescence microscopy.
Cyanine dyes are traditionally better able to withstand dehydration and embedding in nonpolar, plastic media, whereas DyLight and Alexa Fluor dyes are perceived as brighter than Cyanine dyes in aqueous medium. Both seem to work well in labeling thick, cleared tissue samples.
Cy2 code: 225.
Cy3 code: 165.
Cy5 code: 175.
Alexa Fluor 594: 585.
Alexa Fluor 647N: 605.
See Supplementary Methods for protocol.
FISH reagents
FISH reagents18-are optional (see Supplementary Methods and published methods116–119)
10× PBS, pH 7.4, RNase-free (Life Technologies, cat. no. AM9625)
Ethanol, absolute (J.T. Baker, cat. no. 8025) ! CAUTION Ethanol is flammable.
RNase-free sterile H2O (Life Technologies, cat. no. 10977-015)
Dextran sulfate, 10% (wt/vol)-(Sigma-Aldrich, cat. no. D8906)
Formamide, deionized, nuclease-free (EMD Millipore, cat. no. 344206; or Life Technologies, cat. no. AM9342) ! CAUTION Formamide is a toxic chemical and a teratogen; handle it inside a fume hood with appropriate protective gear (gloves, goggles, lab coat).
Saline sodium citrate buffer, 20×, RNase-free (Life Technologies, cat. no. AM9763)
Slowfade Gold + DAPI (Life Technologies, cat. no. S-36938)
Aminosilane-treated coverslips ((3-aminopropyl) triethoxysilane; Sigma-Aldrich, cat. no. 440140)
Sodium borohydride (Sigma-Aldrich, cat. no. 213462; or Santa Cruz biotechnology, cat. no. CAS 16940-66-2) ! CAUTION Sodium borohydride is highly flammable when in contact with moisture, and it is very toxic to the skin. Do not leave the flask uncapped. Prepare dilutions fresh, on ice, in a fume hood or in a chemical hood. Close tightly after weighing, seal with Parafilm and return the reagent-to its containment canister (if applicable to institutional laboratory practices).
EQUIPMENT
Hydrogel polymerization
House vacuum line or vacuum pump
Nitrogen gas supply (any)
Polytetrafluoroethylene (PTFE) tubing (McMaster-Carr) and/or Masterflex L/S 14 tubing (Cole-Parmer), for connection to vacuum line and inert gas supply
PACT equipment
Sample vials, either commercially available Vacutainers (10-ml Vacutainer serum blood collection tubes; BD Biosciences, cat. no. 366430) or 50-ml conical tubes with commercially available rubber stoppers for hydrogel-embedding step
Commercially available stoppers for 50-ml conical tubes: folding skirt rubber stopper, 30.7 mm diameter (Cole-Parmer, cat. no. EW-62995-87) or Saint Gobain folding skirts rubber stoppers, 31.4 mm diameter (Spectrum Chemical Mfg. Corp, cat. no. 142-55179) or Twistit rubber stopper size 6 (Fisher Scientific, cat. no. 14-131D; Sigma-Aldrich, cat. no. Z164364; eBay, various)
Air-Tite Vet premium hypodermic needles, 22-gauge (22-G) × 4 inches, (Air-Tite Products Co., Lot: 14-11563, SKU N224)
1- to 1.5-inch needles for venting sample containers during hydrogel-embedding (16–22-G)
3- to 5-ml syringes (BD syringes)
PARS equipment
Masterflex Tygon E-Lab tubing (Cole Palmer, cat. no. EW-06460-48), or Tygon S3 laboratory tubing E-3603 (VWR, 0.125 inch inner diameter (i.d.): cat. no. 89403-854; 0.09375 inch i.d.: 89404-000)
Three-way stopcock with Luer lock (World Precision Instruments, cat. no. 14035-10)
Luer-to-tubing coupler kit (World Precision Instruments, cat. no. 500895)
Barbed fitting assortment kit (World Precision Instruments, cat. no. 500890)
22-G × 1-inch gavage needle (e.g., 22-G, 1.25-mm-tip-diameter straight feeding needle; Fine Science Tools, cat. no 18061-22; Braintree Scientific, cat. no. N-PK 002)
Pipette tip boxes; we use empty 1,000-μl racked filter tip boxes (USA Scientific)
Optional: 20-G blunt needle (BD Biosciences, cat. no. 305183) and tubing (PlasticsOne) for PARS-CSF18
C & B Metabond (Parkell, cat. no. S380)
Tape (any)
Modeling clay (e.g., Sargent Art, cat. no. 22-4400)
Peristaltic pump or circulator (e.g., Cole Palmer Masterflex L/S, cat. no. 77800-60; or Cole-Palmer Masterflex L/S Easy Load II head and pump drive, cat. nos. 77200-62 and 7557-12)
Freezer bags, 1 gallon (Ziploc, or equivalent reusable and re-sealable airtight freezer bags made of durable plastic with a zipper closure)
General equipment and supplies
Silicon aquarium sealant (e.g., 3 M marine grade silicone sealant, clear, cat. no. PN08019)
Platform shaker (VWR, rocking platform model 200) and/or nutating mixer (VWR, cat. no. 82007-202)
Bath incubator (Fisher Scientific, Isotemp model 2223) or 37 °C warm room
Shaking water bath (Thermo Forma, cat. no. 003-8830)
Razor blades and/or scissors (any)
mColorpHast pH test strips (EMD Millipore, cat. nos. 1.09543.0001 and 1.09584.0001)
Sample mounting and imaging for confocal microscopy
Spacers, 7.0 or 3.0 mm (iSpacer, SunJin Lab Co.), or 0.5 mm or 2.5 mm spacers (Silicone Isolator, Electron Microscopy Sciences; or GRACE Bio-Labs), or silicone rubber sheet (any)
Putty (e.g., Bostik Blu-Tack adhesive putty)
Clear nail polish or Entellan (Electron Microscopy Sciences, cat. no. 14800)
Microscope slides (Thermo Scientific, cat. no. 10143352; VWR, cat. no. 48382-173; Brain Research Laboratories, cat. no. 5075-plus)
Coverslips (VWR, cat. no. 48404-452, 16004-344, 16004-322; Brain Research Laboratories, cat. no. 4860-11/2)
Vacuum grease (Sigma-Aldrich, cat. no. Z273554)
Optional: Refractometer (Reichert AR200 digital handheld refractometer, cat. no. 13950000)
Confocal or light-sheet microscope – any, as available; data here were obtained with a Zeiss LSM 780 single-photon microscope or a custom-made LSFM system (see supplies listed in Supplementary Table 1, immersion chamber and sample holder in Supplementary Data 1 and 2, respectively)
Microscope objectives for thick-section imaging, such as the CLARITY-optimized objectives now produced by major microscopy companies, including Leica and Olympus. Images presented here and in Yang et al.18 were obtained using the following Zeiss objectives: Fluar 5×/0.25 M27 objective (w.d. 12.5 mm), Plan-Apochromat 10×/0.45 M27 objective (w.d. 2.0 mm), LD SC Plan-Apochromat 20×/1.0 Corr M32 85mm scale-immersion objective (w.d. 5.6 mm), LD LCI Plan-Apochromat 25×/0.8 Imm Corr DIC M27 multi-immersion objective (w.d. 0.57 mm) and Olympus 25× 1.0 NA multi-immersion objective (w.d. 8.0 mm)
Image handling software, such as Imaris (Bitplane)
REAGENT SETUP
0.1 M phosphate buffer (PB)
Add 3.1 g of NaH2PO4 (monohydrate) and 10.9 g of Na2HPO4 (anhydrous) in dH2O to a total volume of 1 liter at pH 7.4; filter-sterilize the solution and store it at RT or at 4 °C for up to several months. For RIMS, dilute the buffer fivefold to 0.02 M phosphate buffer, and adjust the final RIMS pH to 7.5.
0.01 M PBS (1× PBS)
Combine 8 g of NaCl, 0.2 g of KCl, 1.42 g of Na2HPO4 and 0.245 g of KH2PO4 in dH2O to a total volume of 1 liter; adjust the pH to 7.4, filter-sterilize or autoclave the solution and store it at RT or at 4 °C for up to several months. Alternatively, purchase 1× PBS mix (Sigma-Aldrich, cat. no. P5368) or pre-made solution (Lonza, cat. no. 04-409R) from a commercial supplier; adjust the final pH when necessary. Use 1× PBS at pH 7.4 unless otherwise noted (e.g., in clearing buffers).
10× PBS stock
For 10 liters of the 10× stock, dissolve 800 g of NaCl, 20 g of KCl, 144 g of Na2HPO4 dihydrate and 24 g of KH2PO4 in 8 liters of dH2O. Add additional water to a total volume of 10 liters; filter-sterilize or autoclave the solution. Upon dilution to 1× PBS, the pH should approach 7.4. The pH may be adjusted with hydrochloric acid or sodium hydroxide, as needed. The resulting 1× PBS should have a final concentration of 10 mM PO4, 137 mM NaCl and 2.7 mM KCl. Alternatively, purchase 10× PBS premade solution (e.g., Lonza, cat. no. 17-517Q) from a commercial supplier.
Heparinized PBS (hPBS)
For flushing vasculature of blood at the start of perfusion, prepare 1× PBS with 0.5% (wt/vol) sodium nitrite and 10 units per ml heparin, pH 7.4. Place it on ice until use or refrigerate it for up to a few weeks.
4% PFA (for perfusion fixation)
To prepare 40 ml of 4% PFA (wt/vol, final concentration), combine 4 ml of 10× PBS, 5 ml of 32% (wt/vol) PFA solution and 31 ml of ice-cold water. Adjust the pH to 7.4 and keep it on ice or refrigerate it until use (same day).
1× PBS containing 0.1% (vol/vol) Triton X-100 (PBST)
Add 1 ml of Triton X-100 to 1× PBS for a total volume of 1 liter; adjust the pH to 7.4. PBST may be stored at RT for a few months when it is filter-sterilized; vortex or stir the solution on a stir plate for several minutes before use.
Boric acid buffer (BB)
Prepare a 1 M boric acid buffer stock solution by stirring 61.83 g of boric acid and 10 g of NaOH in 900 ml of water with gentle heating. Once sodium hydroxide pellets and boric acid are fully dissolved, adjust the pH to 8.5 with NaOH and add water to a total volume of 1 liter; store it at RT for up to a few months. To prepare fresh borate-buffered clearing solutions, such as 8% (wt/vol) SDS in 0.2 M BB at pH 8.5 (8% SDS-BB) for PACT and PARS, dilute 400 ml of 20% (wt/vol) SDS and 200 ml of 1 M boric acid buffer stock to 1 liter with distilled and deionized water (ddH2O); adjust the pH to 8.5, if necessary. To make a boric acid wash buffer (BBT, 0.2 M boric acid buffer with 0.1% (vol/vol) Triton X-100, pH 8.5), dilute the 1 M boric acid stock to 0.2 M boric acid in ddH2O, adding 1 ml of Triton X-100 per liter of BBT and stirring on a stir plate for 10 min. BBT may be stored at RT for several weeks, barring contamination; vortex the solution or stir it on a stir plate for several minutes before use.
PACT monomer solution
For rapid preparation of samples that are amenable to both standard immunohistochemistry and fluorescence imaging, as well as smFISH, prepare an A4P0 hydrogel: 4% (wt/vol) acrylamide (0% PFA) in 1× PBS. For 200 ml of hydrogel monomer solution, add 20 ml of 40% (wt/vol) acrylamide and 20 ml of 10× PBS to 160 ml of ice-cold dH2O. Stir 500 mg of thermoinitiator 2,2′-azobis[2-(2-imidazolin-2-yl)propane]dihydrochloride into ice-cold monomer solution (0.25% (wt/vol) final concentration). Hydrogel monomer solutions must remain cold before use to prevent premature polymerization; we generally prepare solutions freshly on ice, but they may be stored short-term (several hours) at 4 °C or on ice, or long-term (several months) at −20 °C, protected from light. ▲ CRITICAL We have tested various hydrogel monomer formulations: including combinations of 2% or 4% (wt/vol) acrylamide with 0% or 4% PFA and/or 0.05–0.25% (wt/vol) bis-acrylamide. We found that A4P0 without bis-acrylamide granted rapid clearing and good antibody penetration during immunohistochemistry (IHC) without compromising the macromolecular content and cellular structure of tissue samples. In comparison with CLARITY, we eliminated 4% PFA from the hydrogel monomer solution; however, we ensure the thorough PFA-mediated cross-linking of tissue proteins before hydrogel monomer incubation via 4% PFA transcardial perfusion and 4% PFA postfixation steps. ▲ CRITICAL To enlarge the hydrogel pores for faster sample clearing and immunolabeling, we excluded bis-acrylamide and PFA from the hydrogel recipe proposed for CLARITY8. Although tissue proteins and overall tissue architecture was preserved during PACT and PARS processing18, specific native and non-native biomolecules (e.g., non-membrane-associated proteins, cytoplasmic signaling molecules, commensal and pathogenic micro-organisms) may be more sensitive to the clearing process. Herein, either or both of these hydrogel components may be re-introduced into the hydrogel formulation to increase cross-linking density and thus better stabilize sparse epitopes. However, any increase in the net concentrations of hydrogel monomers will result in slower diffusion of SDS micelles and of antibody-based labels during clearing and immunostaining, respectively.
A4P1, A4P2 and A4P4 monomer solutions
To preserve a sensitive sample's structural integrity during clearing, include 1, 2, or 4% PFA (wt/vol, final concentration) in the hydrogel formulations (i.e., A4P1, A4P2, and A4P4, respectively). For example, for 200 ml of A4P4 monomer solution, add 20 ml of 40% (wt/vol) acrylamide, 25 ml of 32% (wt/vol) PFA and 20 ml of 10× PBS to 135 ml of ice-cold dH2O. Stir 500 mg of thermoinitiator 2,2′-azobis[2-imidazolin-2-yl)propane] dihydrochloride into ice-cold monomer solution (0.25% (wt/vol) final concentration). Hydrogel monomer solutions must remain cold before use to prevent premature polymerization; we generally prepare solutions freshly on ice, but they may be stored short-term at 4 °C or long-term at −20 °C, protected from light.
PACT-deCAL
Combine 10 ml of 0.5 M EDTA and 40 ml of 1× PBS; adjust the pH to 8 and store it at RT for up to a year, barring contamination.
Detergent for tissue clearing
PACT and PARS tissue clearing is accomplished via exposing the tissue to an 8% (wt/vol) SDS detergent solution or, in special cases (PACT-deCAL, ePACT), to a 10% (wt/vol) SDS detergent solution. All initial validation of PACT and PARS was performed using a range of SDS concentrations (4–16% (wt/vol) SDS), prepared in a range of buffers (1× PBS at pH 7.5, 1× PBS at pH 8.0 (for PACT-deCAL), 1× PBS at pH 8.5 and in 0.2 M sodium borate buffer at pH 8.5). Aside from a slight clearing rate enhancement at more alkaline pH (i.e., 8% SDS-BB and 8% SDS-PBS at pH 8.5), there was no apparent trade-off in the quality or characteristics of cleared soft tissue. Thus, PARS and PACT tissue clearing in 1× PBS at pH 7.5 (abbreviated 8% SDS-PBS (pH 7.5)) may hold added convenience for many users. We suggest periodically replacing the clearing solution if it begins to acidify (i.e., monitor the clearing solution pH with pH indicator strips every 72 h). For PACT-deCAL, we strongly recommend increasing the SDS concentration and clearing solution pH, as both will favor more rigorous and efficient calcium removal; the results presented in Figure 6 were obtained using a clearing solution of 10% (wt/vol) SDS in 1× PBS, with a final pH of 8.0 (abbreviated 10% SDS-PBS (pH 8)). ▲CRITICAL It is worthwhile to note that sodium borate buffer possesses antimicrobial and antifungal characteristics that make it an ideal buffer for extended tissue incubations. When 1× PBS instead of sodium borate is used, an appropriate antimicrobial agent should be added to the buffer (e.g., a final concentration of 0.01% (wt/vol) sodium azide in buffer solutions). We prepare clearing solutions fresh for each round of tissue clearing, with RT storage (up to several weeks) of excess clearing solution for buffer exchanges. ▲CRITICAL Although a clearing solution of 8% (wt/vol) SDS is proposed, users may wish to vary the SDS concentration according to their needs. As a starting point, lower SDS concentrations should be used for larger samples, as this prevents the detergent-exposed outer layers from overclearing while the sample center remains opaque. Thinner tissue sections (e.g., 250-μm brain slices from electrophysiology) may be cleared rapidly with 10–15% (wt/vol) SDS; however, overclearing and loss of biomolecules are a greater risk.
Antibody incubation buffer (IHC buffer)
The dilution of antibodies used in PACT and PARS will be highly dependent on, among other things, the quality of the antibody, the size and tissue type of the sample to be labeled, and the cellular location and concentration (i.e., expression level) of the target biomolecule. We recommend a starting dilution of ~1:200–400 and/or staining reagents in 1× PBS containing 2% (vol/vol) normal donkey serum, 0.1% (vol/vol) Triton X-100 and 0.01% (wt/vol) sodium azide; however, the exact antibody concentrations will need to be validated on a case-by-case basis. Freshly prepare IHC buffer.
RIMS
For a mounting medium with an RI = 1.47 (RIMS-1.47), which is used for all samples presented here unless otherwise noted, dissolve 40 g of Histodenz in 30 ml of sterile-filtered 0.02 M phosphate buffer. This is most easily accomplished by adding Histodenz, phosphate buffer and a magnetic stir bar to the final storage container (e.g., a 125-ml glass jar with a lid); sealing the container to minimize evaporation and contamination; and stirring the solution on a stir plate for ~10 min, vigorously shaking the closed jar by hand a few times during the stirring process. Once the Histodenz has dissolved, add sodium azide to a total concentration of 0.01% (wt/vol) and adjust the pH to 7.5 with NaOH. RIMS may be prepared with a lower or higher RI by varying the final concentration (wt/vol) of dissolved Histodenz (Fig. 6b). RIMS may be stored at RT for several months; discard it if microbial contamination occurs. Do not autoclave any solutions containing sodium azide. ▲CRITICAL There are numerous commercial and home-made RIMS alternatives, including FocusClear24, Cargille Labs optical liquids15, 2,2′-thiodiethanol44 and diluted glycerol. We have only verified the compatibility of FocusClear and glycerol with our PACT and PARS prepared samples, and thus we provide a glycerol-based mounting medium recipe here.
sRIMS
Prepare a 70% (wt/vol) sorbitol solution in 0.02 M phosphate buffer with 0.01% (wt/vol) sodium azide (pH adjusted to 7.5 with NaOH); store sRIMS at RT for up to several months, barring microbial contamination. This sorbitol-based mounting medium outperforms 80–90% (vol/vol) glycerol as RIMS for rodent brain samples. At a net cost of ~$0.2 per ml, sRIMS offers the greatest cost advantage over commercial RI matching solutions that we have tested, such as FocusClear8, and without a sacrifice in performance.
cRIMS
Prepare a stock buffer solution of sterile-filtered 0.005 M phosphate buffer. For a mounting medium with RI = 1.47, dissolve 40 g of Histodenz in 30 ml of this stock buffer solution; this is most easily accomplished on a stir plate (see instructions for RIMS). Once the Histodenz has dissolved, add sodium azide to 0.01% (wt/vol) and adjust the pH to 7.5 with NaOH. cRIMS may be stored at 4 °C for several months, barring microbial contamination. Samples that require short-term storage at 4 °C may be mounted in cRIMS, whereas RIMS-mounted tissue will become cloudy or turbid placed at 4 °C—the lower salt concentration of cRIMS reduces the appearance of salt precipitate at colder temperatures. Do not autoclave any solutions that contain sodium azide.
EQUIPMENT SETUP
Degassing the container for hydrogel polymerization
Glass Vacutainers work well for degassing and hydrogel-embedding small rodent organs and tissue samples. However, for rat whole brains and larger tissue samples, a larger container is sometimes useful. One solution is to purchase rubber stoppers that are compatible with 50-ml conical tubes and replace the conical screw-cap with an air-tight rubber stopper during degassing and hydrogel polymerization steps (Fig. 2).
PARS chamber
To perfuse PARS reagents through vasculature in a contained environment, we construct a PARS chamber using components that are readily found in most of the biological research laboratories (Fig. 7). The necessary components of a PARS setup are as follows: a feeding needle catheter to deliver PARS reagents to vasculature; a perfusate catch-basin (pipette box) in which recirculating PARS reagents may pool once they exit the vasculature;-Tygon tubing threaded through a peristaltic pump so that pooling reagents may be collected from the catch-basin and recirculated back into a subject's vasculature; Luer-to-tubing couplers; and a Ziploc bag to contain the entire PARS chamber setup.
To construct the PARS chamber, drill two 1/8-inch holes into the front and one 1/8-inch hole into the left side wall of an empty 1,000-μl pipette tip box. The holes are drilled just below the tip wafer (in Fig. 7, the holes are ~2 cm below the top rim). Next, snap 1/8 × 1/8-inch barbed connectors into each of the drilled holes. The outflow line will circulate solvents from the pipette box chamber to the 3-way stopcock. To join the outflow line to a 3-way stopcock, use a 10-cm piece of Tygon tubing and connect one end to the inner left side 1/8 × 1/8-inch barbed connector and tape the other end to the inside bottom of the pipette tip box. Continue this line through a peristaltic pump by using a new piece of Tygon tubing, and connect one end to the outer left side 1/8 × 1/8-inch barbed connector and then thread the tubing through a peristaltic pump. Next, join the free end to a three-way stopcock with a 3/32-inch barbed male Luer with locking nut. The inflow line circulates solvents from the three-way stopcock to the vasculature. To link the inflow line to the pipette tip box, use a piece of 15-cm Tygon tubing and connect one end to the outer right front 1/8 × 1/8-inch barbed connector and the other end to the three-way stopcock with a 3/32-inch barbed male Luer with locking nut. To finish the inflow line, connect a piece of 75-cm Tygon tubing to the inner right front 1/8 × 1/8-inch barbed connector. Next, coil the inflow line to the bottom of the pipette tip box. This will equilibrate inflowing solvent to the desired temperature before it enters the subject's vasculature. Tape the coiled tubing to the pipette tip box. To quickly circulate bubbles formed during the changing of solutions without disconnecting the inflow line and for use of bubbling nitrogen gas into the solution during the polymerization step, a line linking the outflow line back to the pipette tip box is connected by joining a piece of 15-cm Tygon tubing to the three-way stopcock with a full-thread 3/32-inch barbed female Luer to the outer left front 1/8 × 1/8-inch barbed connector. This line is continued inside the pipette tip box and taped to the bottom. To finish the chamber, thread the inflow line through the top-left corner of the tip wafer and connect it to a feeding tube with a 1/8-inch barbed male slip Luer. As a forewarning, SDS and salt precipitate will begin to accumulate within these narrow lines over time. It is important to flush the lines (e.g., with ddH2O) between subjects, and to replace occluded lines with new Tygon tubing (e.g., after every few subjects).
During hydrogel polymerization, the chamber must be enclosed inside a Ziploc freezer bag. To do this, disconnect the outer Tygon tubing that connects to the barbed connectors of the pipette tip box, and puncture three holes into the Ziploc bag to accommodate the 1/8 × 1/8-inch barbed connectors. Reconnect the Tygon tubing to their original 1/8 × 1/8-inch barbed connector. To connect a vacuum line to this bagged PARS box for withdrawing oxygen, tape a female Luer tee onto the lid of the pipette box and puncture one hole through the Ziploc. Finally, make the Ziploc airtight by placing clay around the punctured regions in the Ziploc.
As a final note, a 1,000-μl tip box has a volume of ~750 ml. Thus, during hydrogel polymerization and during clearing, 200–300 ml of solution may be placed in the pipette box for recirculation without risk of the pipette box overflowing or the solution splashing out during its transport. Similarly, to conserve reagents during PARS clearing and immunostaining of smaller samples, a 200-μl tip box may be used to construct the PARS chamber; only 100 ml of reagent is necessary to fill such a chamber about one-third full (Fig. 7).
Light-sheet microscope
The light-sheet microscope that we use was built based on the laser-scanning single-side illumination method120. Key to the design are objectives that offer a long w.d. while maintaining high NA (e.g., CLARITY objectives with 8 mm w.d. and NA of 1.0). The system described below provides a cost-effective and relatively easy-to-replicate alternative to CLARITY optimized light-sheet microscopy(COLM)15, a recently introduced light-sheet microscope for CLARITY. We used cost-effective optical components, especially when creating, shaping and projecting the illumination light sheet. The immersion chamber and sample holder are custom-made with a 3D printer. The list of components and the .stl design files can be found in the Supplementary Table 1 and in the Supplementary Data 1 and 2 (.stl format), for the immersion chamber and sample holder, respectively.
The microscope is built onto a 4 × 6-foot optical table (Fig. 9b). The various lasers are combined with dichroic filters to one beam, which is then expanded using a Galilean telescope and shaped with an iris to match the Galvanometer scanner mirror size (6 mm in diameter). The Galvanometer scanner, coupled with an f-theta lens, is then used to generate the scanning light sheet, which is projected to the sample holder using two achromatic doublet lenses. The resulting light sheet has a full-width-half-maximum of 5–7 μm, depending on the wavelength of illumination (473–632 nm).
The detection objective lens (25×, 1.0 NA CLARITY objective, Olympus) is inserted into the immersion chamber. To prevent leakage of the medium from the chamber, we sealed the gap between the chamber and the objective with an O-ring and a flexible latex film, in which only the tip of the detection objective is immersed. This setting allows the objective to move uninterruptedly during data acquisition while maintaining a sealed connection. The immersion chamber is created with a 3D printer (ABS plastic) and it is filled with glycerol to prevent evaporation-induced aberrations in RIMS medium.
Adjacent to the detection objective, we use tube lenses with different focal lengths to change the magnification of the light-sheet microscope and, consequently, its field of view. Higher magnification is used to digitally sample the acquired images in line with the high NA of the detection lens. We typically use magnification values between 25× and 55×, with the corresponding field of view of 0.28–0.06 mm2. To acquire the images, a camera with a light-sheet mode feature is used (Zyla 4.2 sCMOS, Andor), in which the scanning light sheet and the camera pixel readout are synchronized to improve the signal-to-noise ratio121.
To rapidly scan large volumes, the sample is constantly translated using a xyz-theta stage, whereas the light sheet remains stationary. The xyz-theta stage is mounted on heavy-duty stainless steel bars to prevent sample vibration during data acquisition. To connect the sample holder to the xyz-theta stage, we first place the sample in a quartz cuvette filled with RIMS solution. The cuvette is then attached to a 3D printed cap that has a Luer lock female connector mounted on top, and laboratory Parafilm is used to seal the connector-cuvette interface. The sample holder is then attached to a xyz-theta stage via the Luer lock male connector.
To automatically scan large volumes using the microscope, we wrote a MATLAB program, which runs μManager75 and serial communication, both controlling and synchronizing the various mechanical components. This program finds the synchronization parameters to run the camera in a light-sheet mode, performs autofocus for the detection objective and optimizes the lateral position of the illumination light sheet. To this end, both the illumination lens and the detection objective are mounted on computer-controlled linear stages.
Computer for visualization workflow
We perform visualization workflows for a sample single channel, 8-bit, 30-GB image on a 64-bit Windows 8 machine with Intel i7-3770 CPU and 16 GB of RAM.
PROCEDURE
Tissue preparation for PACT and PARS ● TIMING 2 h–1 d
-
1|
Prepare the perfusion and hydrogel monomer solutions, including 1× PBS containing 0.5% (wt/vol) NaNO2 (optional, for vasodilation) and 10 U/ml heparin (optional, for anticoagulation; hPBS), 4% PFA in 1× PBS and the A4P0 hydrogel solution.
▲ CRITICAL STEP 4% PFA should be freshly prepared. A4P0 may be freshly prepared or stored at −20 °C until use. For the latter, thaw A4P0 on ice before use. Perfusion solutions should be ice-cold. Discard PFA and hydrogel stock solutions if a precipitate is observed.
-
2|
Anesthetize the animal with an intraperitoneal (i.p.) injection of Euthasol (100 mg/kg of body mass for mice and rats), or according to institutional guidelines for rodent euthanasia (e.g., carbon dioxide inhalation until loss of consciousness, injection of pentobarbital or similar).
! CAUTION Follow appropriate institutional and governmental guidelines and regulations for husbandry, handling and euthanasia of laboratory animals.
-
3|
Transcardially perfuse122 the subject with ice-cold hPBS until the perfusate drains clear from the right atrium (~20 ml at 10 ml/min for mice, ~100 ml at 50 ml/min for rats).
▲ CRITICAL STEP The perfusion pressure (flow rate) during transcardial perfusion and during PARS (unless otherwise noted) should approximate the physiological pressure of the subject's circulatory system; at night, mice and rats have systolic and diastolic pressures (mm Hg) of ~125/90 (mouse) and 121/84 (rat), respectively. When using a peristaltic pump123 or alternative pressurized system122, we suggest a rate of 10 ml/min for mice and ~100 ml/min for rats given that their cardiac output is reported to be ~10–35 ml/min (mouse124–126) and ~50–120 ml/min (rat127), respectively, depending on sex, strain, age and so on124–127. If the perfusate is observed to leak out of the subject's nostrils, the cerebral vasculature was probably compromised by too high a flow rate; it is not advisable to proceed with PARS-based clearing of this subject, as PARS reagents may not reach all tissues; instead, organs of interest can be excised and cleared by PACT. Decrease the perfusion flow rate for all subsequent transcardial perfusions and PARS-based clearing steps. For the initial perfusion fixation (Steps 3–4), gravity alone may be used to draw hPBS and PFA through rodent vasculature.
? TROUBLESHOOTING
-
4|
Without introducing air to the perfusion tubing, continue to perfuse the animal with ice-cold PFA (~50–70 ml at 10 ml/min for mice, ~100 ml at 50 ml/min for rats).
▲ CRITICAL STEP Although there are several alternative fixatives to 4% PFA, many of them carry consequences that are particularly detrimental to the hydrogel-embedding process and/or subsequent imaging of thick cleared samples. Mechanistically, formaldehyde augments the conjugation of tissue components to the acrylamide scaffold via its formation of methylene bridges between peptide amines and acrylamide, and thus fixatives that lack this cross-linking ability will result in limited tissue-hydrogel hybridization. Although glutaraldehyde, which is commonly used for EM sample preparation, can penetrate and cross-link tissue more efficiently than formaldehyde, it also generates high autofluorescence that is more difficult to counteract in thicker tissues via standard aldehyde blocking measures.
-
5|
For PACT-based clearing of excised tissue samples, including bones (PACT-deCAL variation), prepare tissue as described in option A. For PARS-based whole-body or whole-organ clearing using continuous perfusion through intact vasculature, proceed to option B. PARS-CSF allows within-skull clearing through the use of an indwelling guide cannula, which was either previously inserted for neurobiological or pharmacological studies or positioned specifically for PARS-CSF clearing. For PARS-CSF whole-brain or whole-spinal cord clearing using continuous perfusion through an intracranial cannula, proceed to option C.
▲ CRITICAL STEP After perfusion fixation, all tissue samples that contain endogenous fluorophores must be protected from light. Minimize unnecessary light exposure during long incubations (>1 h) by, for example, wrapping the sample containers in aluminum foil.
-
(A)Tissue preparation for PACT
-
(i)Carefully excise whole organs and tissues to be processed with PACT.
-
(ii)If appropriate, slice whole organs into 0.5- to 3.0-mm-thick sections. Alternatively, pliable or fragile organs may be easier to thick-section, if required, immediately after hydrogel-embedding, which greatly increases their firmness, before proceeding to clearing (Step 7).
-
(iii)Postfix the samples in 4% PFA for 1–2 h at RT with gentle agitation on a rocking platform shaker.■ PAUSE POINT The samples may be postfixed overnight at 4 °C. Fixing samples for longer periods of time, especially smaller, thinly sectioned tissues, may result in overfixation and antigen masking.
-
(i)
-
(B)Tissue preparation for PARS
-
(i)Set up the PARS chamber, tubing and pump. Prefill the PARS tubing with PFA so that no air bubbles are introduced into vasculature or-tissue. Fill the pipette box with PFA until it is about one-third to one-half full.
-
(ii)Transfer the perfused subject to the PARS chamber, laying the subject on top of the pipette wafer.
-
(iii)PARS reagents will be delivered through the same feeding needle catheter used during transcardial perfusion. Thus, after transferring the rodent to the PARS chamber, check the placement of the feeding needle catheter. The catheter should enter the left ventricle. If it sits stably in the ventricle, leave it as is. Otherwise, advance the catheter through the left ventricle and into the aorta, just before the level of the aortic arch. Connect the PARS tubing to the feeding needle catheter.▲ CRITICAL STEP Be careful not to tear rodent vasculature.? TROUBLESHOOTING
-
(iv)Set the peristaltic pump to a flow rate of 1 ml/min and postfix the subject for 1–2 h at RT. As 4% PFA is pumped through the feeding needle, PFA perfusate should exit the right atrium and drain into the pipette box. This perfusate is then drawn up through tubing and recirculated through the subject.▲ CRITICAL STEP If necessary, add additional 4% PFA to the pipette box so that there is always enough PFA pooled in the pipette box to be recirculated through the tubing and subject vasculature. The amount of solution required for continuous recirculation will depend on the individual setup (size of the pipette box, liquid volume to fill tubing, evaporation from PARS chamber, species of subject and so on).
-
(v)To prevent PFA from cross-linking acrylamide within vasculature during subsequent steps, perfuse 1× PBS for 45 min at RT.
-
(i)
-
(C)Tissue preparation for PARS-CSF
-
(i)If an indwelling guide cannula is not available, insert an intracranial brain shunt (e.g., 20-G blunt needle) into the cisterna magna for spinal cord clearing, or lower the cannula through the skull (by drilling a hole in the area of interest and using tweezers to create an opening in the dura), to the level of the subarachnoid space, directly above the dorsal inferior colliculus18. Ensure that the cannula, whether newly inserted or existing, is firmly attached to the skull using dental acrylic (C&B-Metabond, Parkell), and that it is free from blockages.
-
(ii)Set up the PARS chamber, tubing and pump; prefill the PARS tubing with 4% (wt/vol) acrylamide monomer solution (A4P0) so that no air bubbles are introduced into vasculature or tissue. Partially fill the pipette box with cold A4P0.
-
(iii)Transcardially perfuse with 4% PFA, and then briefly perfuse the subject with 1× PBS (≤30–60 ml for mice and rats, respectively) to wash away excess 4% PFA.
-
(iv)For whole-brain and whole-spinal cord clearing, ligate the arterial circulation, leaving the carotid arteries intact, and for rats or larger subjects remove tissue not directly perfused by these vessels to conserve reagents. Transfer the subject to the PARS chamber, positioning tissue atop the pipette tip wafer. For whole-brain clearing alone, if desired, decapitate the perfused subject and transfer only the head to the PARS chamber.
-
(v)Connect the PARS tubing to the cannula.
-
(i)
Formation of a tissue-hydrogel matrix ● TIMING 1 d
▲ CRITICAL The polymerization of tissue components with hydrogel monomers is crucial, as it ensures that SDS micelles preferentially solubilize and remove tissue lipids during clearing. We previously demonstrated that a minimal acrylamide-based network, which supports more rapid clearing, was nevertheless sufficient for stabilizing proteins and nucleic acid18. To increase the level of cross-linking without the addition of bis-acrylamide or PFA to the hydrogel monomer solution, the hydrogel-infused tissue should be carried through a rigorous degassing step.
-
6|
Infuse the sample with A4P0 via passive diffusion for PACT-based clearing (option A), or via continuous perfusion of A4P0 and then thermoinitiator for PARS and PARS-CSF (option B).
-
(A)Hydrogel-embedding of PACT samples
-
(i)Transfer the PFA-fixed samples into a Vacutainer or conical tube with a rubber stopper. Fill the container with ice-cold A4P0 hydrogel solution until the samples are fully submerged. Incubate the samples at 4 °C overnight.▲ CRITICAL STEP Once it is placed in monomer solution, the sample must remain at 2–8 °C. Warmer temperatures may cause premature polymerization of hydrogel monomers before they have uniformly diffused throughout the tissue sample.? TROUBLESHOOTING■ PAUSE POINT Samples may be incubated in A4P0 at 4 °C for 3 d.
-
(ii)Purge the samples and sample container of residual oxygen. Insert one 4-inch-long hypodermic needle into the stopper so that the needle reaches near the bottom of the container, fully submerged in the hydrogel solution. Insert a second 1-inch-long needle into the stopper—this needle should not touch the hydrogel solution; its sole purpose is to vent excess gas from the container to avoid pressure buildup.
-
(iii)Connect the hypodermic needle to the nitrogen gas source (Fig. 2b) and slowly turn on the flow of nitrogen. Allow the nitrogen gas to bubble through the hydrogel monomer solution for 1–10 min before turning off the flow of nitrogen, and then remove both needles.▲ CRITICAL STEP To form a more rigid tissue-hydrogel matrix, which imparts superior tissue cross-linking and only minor slowing of clearing and immunostaining steps, perform a more rigorous gas-exchange step. Place the sample container on ice and insert a 1-inch-long needle into the stopper. Connect the 1-inch-long needle to the house vacuum line and degas the sample for 5–10 min, depending on the sample size and volume of hydrogel. Gently tap or briefly vortex the sample container every minute to dislodge air bubbles from the tissue. Unhook the needle from the vacuum line, leaving the needle inserted in the stopper so that it may serve as a venting needle during nitrogen exchange. Remove the sample-container from ice, insert a 4-inch-long hypodermic needle that is connected to the nitrogen line into the stopper and bubble nitrogen gas into the hydrogel monomer solution for 5–10 min. Turn off the flow of nitrogen. For larger tissue samples, such as whole rat organs, repeat the degassing process (degassing the sample on ice, and then bubbling nitrogen through the hydrogel solution). When this step is completed, remove both needles and proceed to Step 6A(iv).
-
(iv)Place the sample container in a 37 °C water bath for 2–3 h.▲ CRITICAL STEP With rigorous degassing, the A4P0 solution will form a hydrogel having the consistency of honey or tacky silicone sealant, which is difficult to remove from the tissue. With a 1-min nitrogen gas exchange, the A4P0 solution will form a hydrogel with the consistency of syrup, which may be poured off easily.
-
(v)Remove the excess hydrogel from the tissue sample. Exercise caution when you are removing tacky hydrogel from the tissue: cut away excess hydrogel with a scalpel or small surgical scissors and then use a Kimwipe to carefully remove excess hydrogel from the tissue. Briefly rinse the samples in 1× PBS to wash away residual, syrupy-like, hydrogel from minimally degassed samples.! CAUTION Hydrogel waste disposal should be conducted according to federal, state and local regulations.
-
(vi)Hydrogel-embedded samples will have increased rigidity and structural integrity, and indeed this may be the primary goal for some users. In this case, it is possible to transfer hydrogel-embedded soft tissues (e.g., pancreas, spleen, thymus) and amorphous biological samples (e.g., sputum, mucus, organoid cell masses) that were prepared in Steps 1–6 to other lines of experimental evaluation without proceeding with the PACT clearing protocol. All other users should proceed directly to Step 7 for instructions on how to chemically clear PACT samples.? TROUBLESHOOTING
-
(i)
-
(B)Hydrogel-embedding of PARS samples
-
(i)Circulate 4% (wt/vol) acrylamide (A4P0) in 1× PBS through PARS tubing at RT overnight. Ensure that there is enough A4P0 pooled in the pipette tip box such that the tubing will not run dry during continuous recirculation.
-
(ii)Briefly perfuse the sample with 1× PBS to remove A4P0 and any residual PFA from the vasculature.▲ CRITICAL STEP Use only enough 1× PBS to flush the vasculature of A4P0 (e.g., <5 ml for a mouse; 10 ml for a rat); do not infuse 1× PBS for so long that it displaces the A4P0 from tissue.
-
(iii)Without disconnecting the perfusion lines, place the PARS chamber into a Ziploc bag and place the bag under nitrogen atmosphere (i.e., fill the bag with nitrogen gas, deflate the bag, refill it with nitrogen gas, seal the bag closed around the perfusion and set it aside for a few minutes while performing Step 6B(iv)).
-
(iv)Prepare 200 ml of 0.25% (wt/vol, final concentration) VA-044 initiator in 1× PBS; degas this solution via bubbling nitrogen gas through the solution for ~10 min. Add this solution to the pipette box within the Ziploc bag, and place the bag-encased pipette box into a 37–42 °C water bath. If necessary, place a lead weight on top of the perfusion chamber to prevent it from tipping over. Turn on the pump so that the initiator circulates through the sample and PARS tubing, and allow the sample to incubate for 2–3 h in the water bath, replacing the solution with freshly `nitrogen-degassed' solution every hour. Alternatively, if the setup permits, the nitrogen gas can be bubbled directly into the 200 ml of 0.25% VA-044 initiator in 1× PBS that is already loaded into the pipette box while slowly degassing the chamber, as shown in Figure 7i. This requires disassembly of the PARS chamber to make an airtight environment with the Ziploc bag.
-
(i)
Tissue delipidation with SDS ● TIMING 12 h–3 weeks
▲ CRITICAL The rate of tissue clearing depends on several parameters, including the inherent structural and biochemical properties of the tissue sample, the volume of the tissue sample, the hydrogel pore size and the density of tissue-hydrogel cross-linking, as well as the clearing setup (SDS concentration, incubation temperature and pH of clearing buffer). It is important for users to determine the clearing parameters for their specific tissue samples empirically, using these guidelines as a starting point for further optimization. Similarly, because the rate of clearing may vary greatly, tissues embedded in minimal hydrogel monomer compositions, such as the A4P0 hydrogel suggested here, are more susceptible to deteriorating when samples are left unattended in SDS.
-
7|
Clear the tissue samples at 37–42 °C using either gentle agitation (PACT, option A; or PACT-deCAL, option B) or perfusion (PARS, option C); the latter accelerates micelle diffusion for rapid whole-body clearing. Although 8% (wt/vol) SDS is sufficient to solubilize lipids in soft tissue, 10% (wt/vol) SDS and EDTA are required to clear and decalcify bone (PACT-deCAL, option B). The PACT clearing procedure (option A) may be paused for 1–2 d by transferring the sample to PBS or diluted SDS (37–42 °C water bath, or at RT), and then returning the sample to SDS to resume clearing. Although a primary benefit of PARS clearing (option C) is its efficiency, if it is necessary to delay the PARS procedure, whole organs of rodents may be cleared via PARS and then stored for up to 1 month in PBS or in diluted SDS. However, if it becomes necessary to abort PARS (option C) because of unforeseen technical difficulties (examples of such issues that we have encountered include antibodies being out of stock at the manufacturer, an out-of-order microscope, compromised vasculature or leaky pump lines and a damaged PARS pump head), partially cleared samples can be excised and transferred into diluted SDS for up to a month for storage or 8–10% (wt/vol) SDS to finish clearing via PACT (Step 7A(i)) or PACT-deCAL (Step 7B(i)). For delays within any of the methods of preparation (PACT, PACT-deCAL and PARS), it can be better for tissue quality to maintain excised cleared tissues in diluted SDS rather than storing samples after clearing/immunostaining in PBS or mountant. The latter case risks hydrogel disintegration, sample contamination and/or fluorescence signal loss.
? TROUBLESHOOTING
-
(A)PACT clearing
-
(i)Place each tissue-hydrogel sample into a 50-ml conical tube containing clearing buffer; gently rock the sample in a 37–42 °C shaking water bath until the tissue is optically transparent.
-
(ii)If you are using thin organ slices (<1 mm) embedded in A4P0, check the clearing progress every hour, as they should clear in <12 h. Similarly, porous tissue and samples with a high surface area–to-volume ratio may clear in less than 24 h. It is recommended that new users monitor the increasing transparency of such samples every 1–2 h during initial test runs. Once a sample's time to clear is determined empirically by the user, stringent monitoring is no longer necessary. Dense, highly myelinated or thick-sectioned (1–4 mm) tissue and whole organs should be checked daily during clearing, and it may require >96 h.▲ CRITICAL STEP Avoid overclearing. For certain organs, and for brain tissue in particular, variations in cell density and myelination cause specific regions to clear at different rates. Thus, some regions will become transparent, whereas the slower clearing regions will be only semitranslucent. Continuing to clear samples until all regions are uniformly transparent may lead to hydrogel softening, protein solubilization and/or structural deformity in the rapidly clearing areas. In addition, overclearing is deleterious to endogenous fluorescence. As tissue mounting in RIMS will lend an additional degree of optical transparency to tissues, it is crucial to remove tissues from SDS when the majority of tissue, or the portion of interest, is transparent, even if some regions appear undercleared. This will help ensure that the tissue macromolecular content is preserved. Alternatively, as opposed to terminating the incubation of tissue in 8% (wt/vol) SDS prematurely, one may lower the percentage of SDS (e.g., from 8% to 4% (wt/vol) SDS) in clearing buffer at the final stages of clearing.? TROUBLESHOOTING
-
(iii)Once the appropriate region of tissue appears optically transparent, wash the tissue extensively at RT with gentle shaking. For rapid tissue processing, conduct a minimal wash step of 4–5 buffer exchanges in 1× PBS over a 12- to 24-h period. Herein, residual SDS may precipitate, causing tissue cloudiness. To achieve more thorough removal of SDS, or to wash larger tissue blocks, wash the samples in either BBT or PBST for 1–2 d, with 4–5 buffer exchanges over the course of washing.▲ CRITICAL STEP It is almost always preferable to perform wash steps of cleared samples at RT. We have found that additional 37 °C incubations of cleared tissue or, in particular, alternating between RT and 37 °C incubations are hazardous to tissue structural integrity, as cleared tissue lacks the structural support previously offered by lipids and thus must be handled with care. However, a single 37 °C sample wash after clearing and/or a final 37 °C sample wash that precedes sample mounting (Step 10) may be beneficial to accelerating the diffusion of residual SDS from tissue and/or removing SDS precipitate, respectively.■ PAUSE POINT Cleared and washed samples can be stored in 1× PBS (or PBST) containing 0.01% (wt/vol) sodium azide at RT for 1–2 d. Tissues may become cloudy from salt precipitate, in which case wash them with a few changes of PB.
-
(i)
-
(B)
PACT-deCAL
▲ CRITICAL The following steps have been optimized for clearing the dissected tibia of an adult mouse. For other bone types, it is important to adjust the parameters of PACT-deCAL, such as the duration of bone incubations in clearing and decalcifying buffers, and the concentration of EDTA. Temperature fluctuations (e.g., from performing SDS or EDTA buffer changes with RT solutions rather than with prewarmed 37 °C solutions, or from a water bath that is unable to maintain a constant 37 °C environment) may adversely affect bone tissue morphology128.-
(i)Place each bone-hydrogel sample into a 50-ml conical tube containing 10% SDS-PBS (pH 8.0) clearing buffer; gently rock the sample in a 37 °C shaking water bath for 2 d.
-
(ii)Transfer the sample into 0.1 M EDTA in 1× PBS, pH 8.0, and incubate it for ≥2 d in the 37 °C shaking water bath.▲ CRITICAL STEP Bone becomes soft and flexible when decalcified. Larger samples may require up to 6 d and up to 0.5 M EDTA to decalcify.
-
(iii)Replace the EDTA-PBS with fresh 10% SDS-PBS (pH 8.0), and continue to clear the sample in a 37 °C shaking water bath for 2 d.
-
(iv)Wash the sample in an excess volume of 1× PBS (pH 7.4) for 24 h, by performing three or four buffer exchanges.? TROUBLESHOOTING■ PAUSE POINT Cleared and washed samples may be stored in 1× PBS containing 0.01% (wt/vol) sodium azide at RT for 1–2 d.
-
(i)
-
(C)PARS clearing
-
(i)After polymerization, wash the perfusion lines with 1× PBS, and then replace the wash buffer with 8% SDS-PBS (pH 7.5) clearing buffer. This procedure can be accomplished easily by removing the PARS chamber from the shaking water bath (optional), by turning off the pump, by removing excess initiator buffer from the PARS chamber and by replacing it with 100 ml of 1× PBS to perform the wash. Circulate the wash buffer through the sample for 10 min. Afterward, replace the buffer with 8% SDS-PBS (pH 7.5). Place the chamber back into the Ziploc bag and into the 37–42 °C water bath. Allow the SDS clearing buffer to recirculate through the system for 24 h. Perform a buffer exchange with fresh 8% SDS-PBS (pH 7.5), by clearing the buffer daily until the recirculated fluid is no longer yellowish, after which the SDS solution can be refreshed less frequently (every 48–72 h).■ PAUSE POINT Whole organs are rapidly cleared in situ using PARS. However, if the user requires a time delay between clearing and immunostaining steps or must discontinue the PARS procedure, hydrogel-perfused whole organs may be excised after hydrogel polymerization (Step 6B) or after the initiation of PARS clearing (Step 7C(i)), and then stored in 4–8% (wt/vol) SDS at 37 °C for up to 1 month. This allows whole organs to clear slowly during storage; their clearing progress must be monitored, albeit infrequently (e.g., weekly), as smaller, porous organs may become completely transparent in <1 month, wherein they should be transferred into 1× PBS (or PBST or BBT) containing 0.01% (wt/vol) sodium azide at RT. Ensure that all storage solutions contain 0.01% (wt/vol) sodium azide, and when you are ready to resume processing tissue follow the protocol steps for PACT-based clearing and labeling (Step 7A). Although this PACT-based clearing of PARS-prepared whole organs conserves reagents and minimizes the constant oversight required during PARS clearing, it negates the principal benefits of PARS: efficiency and uniform sample preparation.? TROUBLESHOOTING
-
(ii)Check on the clearing progress daily. Add additional SDS buffer to the PARS chamber if necessary, as depending on how well the Ziploc bag is sealed around the perfusion tubing, some buffer may evaporate over time.
-
(iii)The sample can be continuously perfused for up to 2 weeks until all desired organs have cleared, even if some organs appear clear within the first 24–48 h. Alternatively, if all but one or two organs appear sufficiently transparent after a few days, one may proceed directly to Step 7C(iv) to flush SDS from tissue, and then excise all organs. The one or two semiopaque excised organs are transferred into 8% (wt/vol) SDS to finish clearing via PACT (see Step 7A), whereas the organs that cleared more rapidly are immediately promoted to passive immunostaining (optional) and mounting (Step 10) without further delay.▲ CRITICAL STEP As with PACT clearing, it is possible to overclear PARS samples, wherein protein and other tissue components are solubilized and the stabilizing hydrogel matrix begins to disintegrate. Generally, most of the major organs are cleared within a similar timeline of 24–48 h, whereas the whole brain can take 1–2 weeks.
-
(iv)Once the tissue appears optically transparent, perfuse eight buffer changes of 200 ml of BBT or 1× PBS with 0.01% (wt/vol) sodium azide (pH 7.4) over a 2-d period in the 37–42 °C water bath.? TROUBLESHOOTING
-
(i)
-
8|
For samples that will be immunolabeled after clearing, the hydrogel matrix will be required to support the cleared tissue for several rounds of washing and multiday incubations with gentle shaking. If the already delicate tissue-hydrogel matrix seems precariously fragile after clearing (this usually only occurs with thin-sectioned tissue), it is advisable to repeat the hydrogel embedding and polymerization steps (Step 5A). This will stabilize tissue architecture during immunolabeling, prevent tissue loss or disintegration and counteract expansion in mounting medium. For cleared samples that will not undergo any immunohistochemical labeling steps before imaging, skip Steps 8 and 9, and proceed directly to Step 10.
Single-cell phenotyping of cleared tissues ● TIMING 12 h–2 weeks
▲ CRITICAL PACT and PARS prepared tissues are amenable to most standard immunohistochemical protocols; a list of validated small-molecule dyes, primary antibodies and secondary fluorescent labels is provided in Table 4.
▲ CRITICAL This PARS-histology protocol is sufficient to label molecular targets in the peripheral organs of mice and rats, with antibody amounts adjusted for body size. Individual users may need to adjust the incubation times and/or lengthen wash steps.
-
9|
Prepare the primary antibody cocktail in IHC buffer. An antibody dilution of 1:200–400 is recommended; however, a more or less concentrated antibody dilution may be required, depending on the tissue identity and bimolecular target. Perform passive labeling (option A) or perfusion-assisted labeling (option B). For passive labeling schemes (option A), thick-sectioned tissues should be incubated in enough of the antibody cocktail to fully bathe all surfaces, usually a few milliliters of antibody cocktail or less, if the tissue is placed in a minimally sized container. For example, 1.5- to 5-ml Eppendorf tubes are recommended. For perfusion-assisted labeling using the PARS setup (option B), ~20–100 ml of primary antibody cocktail or labeling solution is required, depending on the tissue volume to be perfused and the total volume of the perfusion system (PARS tubing volume plus an additional amount of solution to partially fill the perfusion chamber).
? TROUBLESHOOTING
-
(A)Passive histology
-
(i)Incubate the tissues in the primary antibody cocktail at RT with shaking for 3–7 d. For small-molecule stains or fluorescent dyes, a 1- to 3-d incubation is usually sufficient, and thin tissue sections (<0.5 mm) may be stained within a few hours.▲ CRITICAL STEP The duration of primary antibody incubation must be determined on a case-specific basis (see antibody penetration guidelines, Fig. 4d). It is highly recommended to use smaller antibody formats for thick-tissue staining, when available129. For A4P0-embedded rodent brain tissue, a full IgG will penetrate ~500 μm over a 3-d incubation at RT with shaking. This length of time is often sufficient for 1 mm tissue slices if the tissue can be imaged from either side. For A4P4-embedded rodent brain tissue, a full IgG will penetrate ~200 μm over a 3-d incubation at RT with shaking.? TROUBLESHOOTING
-
(ii)To remove unbound antibody or stain, wash the samples in an excess volume of 1× PBS buffer: transfer the samples to a larger container (e.g., a 15- to 50-ml conical tube) and perform four or five 1× PBS buffer exchanges over the course of 1 d. Larger tissue blocks, PACT-cleared whole organs, and samples in which high background or nonspecific antibody binding are common should be washed for 2 d in PBST, with four or five buffer exchanges.? TROUBLESHOOTING
-
(iii)Prepare the secondary antibody cocktail (1:200–400 recommended dilution) in IHC buffer. Fab fragment secondary antibodies are preferred.
-
(iv)Incubate the washed samples in the secondary antibody cocktail for 2–5 d at RT and with shaking. Again, samples may be transferred to 1.5- to 5-ml Eppendorf tubes in order to accomplish staining with a minimal volume of antibody.
-
(v)Wash labeled samples with four or five buffer exchanges of 1× PBS over 1 d.? TROUBLESHOOTING
-
(i)
-
(B)Perfusion-assisted labeling
-
(i)Replace the PBST in the perfusion tubing and PARS chamber with the primary antibody cocktail or stain and continuously perfuse the sample for 3 d.
-
(ii)Exchange the antibody cocktail for 1× PBS, and wash the sample by perfusing four buffer changes of 200 ml of 1× PBS over the course of 1 d.
-
(iii)Prepare the secondary antibody cocktail (1:200–400 recommended dilution) in IHC buffer. Again, Fab secondary antibodies are preferred.
-
(iv)Replace the 1× PBS in the perfusion tubing and PARS chamber with the secondary antibody cocktail (or stain), and continuously perfuse the sample for 3 d.
-
(v)Exchange the antibody cocktail for 1× PBS, and wash the sample by perfusing four buffer changes of 200 ml of 1× PBS over the course of 1 d.? TROUBLESHOOTING
-
(i)
RIMS for PACT and PARS samples ● TIMING 1 d
-
10|
Calculate the RI of the tissue to be mounted and imaged. Use a refractometer to measure the sample RI according to the manufacturer's instructions.
-
11|
Prepare a sample-optimized RIMS formulation by adjusting the amount of Histodenz dissolved in 0.02 M phosphate buffer (for common RI recipes see Fig. 6b, Supplementary Fig. 1b). For most tissues, a RIMS formulation with RI ~1.46–1.47 is optimal. For bone, prepare a graded series of RIMS formulations: RIMS with RI ~1.42, RIMS with RI ~1.46 and RIMS with RI ~1.48–1.49. For imaging thick tissue using immersion objectives corrected for immersion medium with an RI between 1.38 and 1.42, it is sometimes beneficial to match the RI of the RIMS to that of the immersion medium. For example, when using the LD-Plan Apochromat 20× 1.0 NA Scale objective (Zeiss), RIMS with RI ~1.42 will help reduce image distortion in the z-direction.
▲ CRITICAL STEP As discussed in the Experimental design section, different mounting solutions can be substituted for RIMS.
-
12|
Submerge the sample in excess RIMS (i.e., in a capped 15- or 50-ml conical tube, or in a 5-ml Eppendorf tube, filled three-fourths full with RIMS) and incubate it at RT until it reaches the desired transparency. Although thin tissue sections may become transparent in less than a single day, a whole rat brain requires a 1-week incubation in RIMS to achieve thorough RI homogenization throughout the sample. These incubation times may be shortened substantially by placing samples on a nutating mixer. Bone should be carried through a graded series of RIMS formulations, each with a progressively higher RI. For example, incubate cleared bone in RIMS-1.42, then in RIMS-1.46, and lastly in RIMS-1.48-1.49 for 1 day each before mounting. Similarly, perform a graded series of RIMS incubations for very fragile tissues, as this will prevent the unlikely event of tissue damage from rapid shrinking and swelling.
▲ CRITICAL STEP Upon RIMS immersion, cleared tissue will shrink over the course of a few hours (e.g., A4P0-embedded coronal mouse brain sections shrink ~20%, size fluctuations are reduced in samples embedded in PFA-containing hydrogels). Continued incubation in RIMS will lead to gradual tissue expansion back to its starting size as RIMS penetrates the tissue. These size changes may confound the visualization of (sub)cellular morphology or introduce apparent tissue deformities. Thus, imaging should not be undertaken before the sample has equilibrated in RIMS. However, if the goal is coarse cellular phenotyping and/or rapid tissue visualization, a much shorter RIMS incubation may be performed (1–4 h, or until the sample is sufficiently transparent).
? TROUBLESHOOTING
■ PAUSE POINT Samples may be stored for the long term (~3 months) in RIMS. Herein, RIMS-submerged samples should be kept in an airtight container at RT and protected from light. Alternatively, when short-term sample storage at 4 °C is mandatory, samples may be mounted in cRIMS; store it in a dry, airtight container.
-
13|
Transfer the RI-homogenized tissue into an airtight container (e.g., Vacutainers or conical tubes with rubber stoppers) and fill the container with fresh RIMS (or with an alternative mounting medium such as sRIMS or 87% (vol/vol) glycerol) until it just covers the sample. Insert a 1-inch-long needle into the rubber stopper, connect the needle to the house vacuum line, and degas the sample for 5–10 min. When you are ready to image, proceed to the next step for mounting instructions.
▲ CRITICAL STEP Although RIMS outperforms sRIMS in our hands, the primary ingredient of sRIMS—sorbitol—not only offers a cost advantage over Histodenz but also is commonly available in research laboratories, owing to its broad use as a cell culture reagent. Importantly, sRIMS grants superior imaging resolution over glycerol.
▲ CRITICAL STEP For fine-scale analysis (e.g., of subcellular morphology) or lengthy image acquisition, do not image samples immediately after their placement in RIMS. Wait until their initial expansion after RIMS-mounting has plateaued (e.g., several days after mounting 1-mm slices).
▲ CRITICAL STEP It is crucial that RIMS or other mounting media be prepared with 0.01% (wt/vol) sodium azide to prevent microbial growth in mounted tissue. Limit the number of air bubbles in sealed slides.
? TROUBLESHOOTING
Acquisition ● TIMING 1 h to several days
-
14|
Mount and image tissues with a confocal microscope (option A) or with a light-sheet fluorescence microscope (option B).
-
(A)Confocal imaging
-
(i)Prepare glass slides with appropriately sized tissue wells, such as 0.5- to 1.0-mm thick iSpacers, which may be stacked to create deeper wells, or silicone sheets, which may be cut to size. If the silicone spacer is adhesive free, apply vacuum grease to the edge of the spacer. Place samples inside the spacer. Slightly overfill the spacer with RIMS and place a coverslip on top of the spacer. Gently press down to seal the coverslip. Remove overflow RIMS with a Kimwipe.
-
(ii)Place RI-homogenized, thick-sectioned tissues and small whole organs on a glass slide: place the tissue inside the sample well; overfill the well with fresh, degassed RIMS of an appropriate RI such that it forms a convex meniscus. Take care not to introduce bubbles into RIMS, or between the tissue and slide.
-
(iii)Place a cover glass over the sample well, using vacuum grease or nail polish to seal the cover glass onto the well edges. Again, avoid sealing bubbles into the sample well; if this occurs, remove the cover glass and repeat this step, adding more RIMS to the well as necessary.
-
(iv)To image PACT- and PARS-cleared samples with a standard microscopy setup (e.g., a single-photon confocal microscope), use a multi-immersion objective with a RI correction collar to match the RI of the mounted sample: RI ~1.46–1.47 for most tissues, or ~1.48–1.49 for bone. For immersion medium, use glycerol with the same RI as the mounting RIMS.
-
(v)Determine optimum acquisition parameters, and apply these. Acquisition parameters (e.g., photomultiplier tube (PMT) gain, laser power and scanning speed) need to be optimized for each sample on the basis of-the desired final image quality.▲ CRITICAL STEP To allow for accurate stitching, acquisition software should be set to acquire tiles with overlap (>10%). If the microscope has the option, it is useful to use the auto Z brightness correction (Fig. 4b). In Zen (Zeiss), Auto Z provides an automatic gradual adjustment of the detector gain, amplifier offset, amplifier gain and laser intensity setting between the first and last optical slice of a z-stack. This will help ensure that signal intensity is uniform throughout the sample, as even clear tissue will scatter at depth.
-
(i)
-
(B)Light-sheet microscopy
-
(i)Mix the glycerol in the immersion chamber using a pipette tip (Fig. 9) to prevent optical aberrations because of inhomogeneous medium, and let the glycerol settle for ~1 h.
-
(ii)Stabilize the sample in a quartz cuvette by submerging the sample in RIMS within the cuvette and by arranging gel pieces around the sample. We use 1% (wt/vol) low-melt agarose gel.
-
(iii)Position the cuvette on the custom sample holder, which may be 3D printed (Supplementary Data 2), and attach it to the sample translation stage, as in Figure 9b.▲ CRITICAL STEP Make sure that the cuvette is properly sealed (e.g., with Parafilm; see Fig. 9b), as evaporation of water from RIMS will cause severe aberrations.
-
(iv)Before lowering the sample in the immersion chamber, verify that the light sheet is centered in the field of view and that the objective lens is in focus.
-
(v)Lower the sample into the immersion chamber.
-
(vi)Re-adjust the light-sheet position to the center of the field of view and re-focus the objective lens.
-
(vii)Change the settings of the camera to a light-sheet mode, activate the driving voltage of the galvo scanner via the function generator and trigger the camera using the external trigger. The delay between the galvo scanner and the camera's external trigger signal should be fine-tuned in order to achieve optimal synchronization; here we used a custom MATLAB program to automatically find the optimum delay; the software is available upon demand.
-
(viii)Set the image acquisition parameters (e.g., laser power, scan depth and the scan resolution in z) and initiate the acquisition sequence.
-
(i)
3D image visualization ● TIMING 1 h
▲ CRITICAL We outline workflows for tracing and visualization using tools summarized in Table 3. Interactive processing and visualization software will perform best on a workstation with substantial RAM (>16 GB) and GPU memory (≥1 GB). The tools mentioned are multi-threaded, and they can often exploit multicore processors to further speed up computations.
-
15|
For the volumetric visualization of large images that do not fit in RAM, select one of the following workflows: (option A) using Imaris (Bitplane); (option B) using TeraStitcher and Vaa3D TeraFly; or (option C) using Vaa3D TeraConvert plug-in and Vaa3D TeraFly. Estimated timing is based on tests with a 30-GB image stack visualized on a 64-bit Windows 8 machine with Intel i7-3770 CPU and 16 GB of RAM.
-
(A)Imaris (v7.7.1)
-
(i)Stitch image tiles using the acquisition software.
-
(ii)Open the raw image stack (~12 min).
-
(iii)Save the loaded images in Imaris .ims format (~24 min). This is useful to streamline future loading and visualization. Reloading the resulting .ims file thereafter takes a few seconds.
-
(iv)Visualize the images using the Imaris volumetric view `Surpass', and then annotate and perform additional image processing using Imaris XTensions.
-
(i)
-
(B)
TeraStitcher and the Vaa3D TeraFly plug-in (v2.921/v0.999)
▲ CRITICAL STEP Stitch the tiles and save a multiresolution volume using TeraStitcher (Step 15B(i–iii), also see ref. 62); visualize the resulting image using the Vaa3D TeraFly plug-in in Step 15B(iv–v).-
(i)Store individual fields of view as separate .tif stacks in a hierarchical set of subdirectories as used by TeraStitcher using the layout specified in https://github.com/abria/TeraStitcher/wiki/User′s-guide.
-
(ii)Stitch images using either the TeraStitcher standalone program or the Vaa3D plug-in (`Plug-In→Image_stitching→TeraStitcher→Tera stitcher'). TeraStitcher requires that the user specify the voxel dimensions, tile size and tile overlap used during acquisition.
-
(iii)Save the stitched output as a tiled, multiresolution volume.
-
(iv)In Vaa3D, start the TeraFly plug-in (`Plug-In→TeramanagerTera→Fly') and select the directory containing the exported `multi-res tiled volume'.
-
(v)The 3D view window will now display the whole image data set and progressively load in higher-resolution subvolumes as the user zooms in to particular parts of the image. Use Vaa3D's color map, annotation and analysis tools on selected subvolumes.
-
(i)
-
(C)
Vaa3D TeraConvert and TeraFly plug-in (v2.921/v0.999)
▲ CRITICAL STEP Use the Vaa3D TeraConvert plug-in to convert an already stitched image to tiled, multi-res format (Step 15C(i–ii)), and then use the Vaa3D TeraFly plug-in to visualize the resulting image by following Step 15C(iii–iv).-
(i)Stitch images using the acquisition software. Save individual z-sections as a numbered sequence of files in a single directory using either acquisition software export options or by using the `BioFormats importer' plug-in in Fiji to load as a `Virtual Stack' and then using the `BioFormats exporter' plug-in to save as an OME-TIFF file130,131 (http://www.openmicroscopy.org/site/support/ome-model/ome-tiff/), by selecting the option to save each channel and z-section in a separate file.
-
(ii)Use TeraConvert plug-in (`Plug-In→Teramanager→TeraConverter') to convert the stored images into a multiresolution `tiled volume' by specifying the location of the image series and specifying `Vaa3D raw tiled format' and an output directory.▲ CRITICAL STEP Pay attention to `Estimated RAM usage' in the TeraConvert plug-in window. To reduce the memory needed by TeraConvert, deselect the checkboxes next to the lowest resolution output formats (starting from the one with the smallest x,y,z values) until the `estimated RAM' is less than the available RAM.
-
(iii)In Vaa3D, start the TeraFly plug-in (`Plug-In→Teramanager→TeraFly') and select the directory containing the exported multi-resolution `tiled volume'.
-
(iv)The 3D view window will now display the whole image data set and progressively load in higher-resolution subvolumes as the user zooms in to particular parts of the image. Use Vaa3D's color map and annotation and analysis tools on selected subvolumes.
-
(i)
3D image analysis ● TIMING variable
▲ CRITICAL A typical workflow is to first run automated tracing to generate initial estimates of morphology and then to perform more detailed semiautomated editing to refine the tracing. Automated tracing is computationally intensive, so it is essential to restrict processing to small ROIs or cropped-out subvolumes and to manually merge the traces afterward. In addition, it is worth noting that semiautomated and manual editing of traces can be greatly accelerated by taking time to learn keyboard shortcuts for a given software tool rather than clicking on graphical user interface elements such as menus or buttons.
-
16|
Automate the tracing of relevant image elements using either neuTube (option A) or Imaris (option B). Alternatively, one may perform tracing using the Vaa3D-Neuron2 plug-in, which has been reviewed previously62. The sample workflows in options A and B and the times reported here are based on our tracing of the test image (Supplementary Data 3) shown in Figure 10.
-
(A)neuTube (v1.0) ● TIMING 3 min
-
(i)Load the image into neuTube. For commercial image formats that are not recognized by neuTube, use the Fiji `BioFormats importer' and `BioFormats exporter' plug-ins to convert to .tif format.
-
(ii)Select `View→3D View' to visualize a volumetric rendering. Click the `Transfer Function' under the `Control and Settings' panel to adjust contrast in the 3D view (i in Fig. 10a and Supplementary Fig. 7a). In a large volume containing many cells, center the cursor on an ROI and right-click and select `Open Zoom In View' to show only image data for a subregion.
-
(iii)Left-click on a cell body and select `Trace' to automatically trace a neurite from that point. Successively visit the remaining untraced neurites associated with the cell, and click `Trace' on each (ii in Fig. 10a).▲ CRITICAL STEP neuTube includes a fully automatic tracing option, but we found a semiautomatic tracing approach (which requires 1–2 clicks per neurite) to be faster and more reliable.? TROUBLESHOOTING
-
(i)
-
(B)Imaris (v7.7.1) ● TIMING 10 min for whole test image, less for smaller ROIs
-
(i)Load the image file. Make sure that the voxel dimensions are correct via the `Edit→Image Properties…' menu, and correct them if necessary. Use `Display Adjustment' to adjust the contrast.
-
(ii)Add a new `Filament' to the `Surpass Scene' session.
-
(iii)In the AutoPath tracing wizard, select the `AutoPath' algorithm and select the ROI checkbox.▲ CRITICAL STEP Specify the smallest possible ROI enclosing each neuron of interest to ensure fast processing (i in Fig. 10b and Supplementary Fig. 7b).
-
(iv)Adjust thresholds for automatic detection of starting points (i.e., soma) and seed points (ii in Fig. 10b). The goal is to have seed points distributed along the neurites, but avoid extra seed points in the background, which will slow tracing and produce errors that need to be corrected later. Depending on background noise and morphological complexity, it may be faster to identify starting and seed points by manual shift+clicking. The `Autopath wizard' will then find paths connecting seed points into a tree of neurite segments.? TROUBLESHOOTING
-
(i)
-
17|
Within the same program (neuTube or Imaris, respectively), manually refine the automated tracing results that were that were generated in Step 16. Stepwise instructions are presented for both neuTube (option A) and Imaris (option B).
▲ CRITICAL STEP We believe that the following guidelines for manipulating automated results will be broadly applicable to a variety of tracing projects. However, they do not represent an exhaustive list of the capabilities of the software, and the user should not feel limited to this set of keystrokes.
-
(A)neuTube (v1.0) ● TIMING 10 min
-
(i)After initial tracing yields good coverage of neurites, select and delete erroneous nodes, add connections and extend fibers by selecting nodes and using the right-click context menu (iii and iv in Fig. 10a). See the extensive tutorial on editing at http://www.neutracing.com/manual/.
-
(ii)To save tracing results, use `File→Save SWC'. Alternately, `File→Export Scaled SWC' allows the user to specify the relative scaling of the x,y and z axes to match acquisition parameters and to produce SWC data in physical units.? TROUBLESHOOTING
-
(i)
-
(B)Imaris (v7.7.1) ● TIMING 10 min
-
(i)Manually refine the automatically traced result from `AutoPath'. For example, users can extend individual traced paths by adding seed points and manually editing to correct errors.
-
(ii)If necessary, remove incorrect branches using `branch select mode' and the delete key. To split branches, use the `node select mode' and delete individual nodes. Select a pair of end points and the `Join' option to find a path between them (iii in Fig. 10b).
-
(iii)To extend neurites using the `AutoPath' method, select the `filament' corresponding to the traced component to be extended, and choose `Selection as Starting Point' to perform shortest path computation. Once the computation is done (~10 min for whole test image, less for smaller ROIs), the AutoPath mode will interactively trace from the cursor position back to the selected component. Shift+clicking will add the displayed candidate path (iv in Fig. 10b).
-
(iv)If necessary, use the filament editing tools to perform additional operations such as smoothing filaments, estimating neurite diameter, detection and annotation of spines, and fully manual tracing (refer to user manual for details).
-
(v)Visualize the resulting traces using Imaris Vantage or export tracing geometry in NEURON .hoc file format (see ref. 132, or for an up-to-date list of NEURON .hoc resources: http://www.neuron.yale.edu/neuron/publications) for use in other analysis tools.? TROUBLESHOOTING
-
(i)
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 5.
TABLE 5.
Troubleshooting PACT and PARS protocols.
| Step | Problem | Potential reason | Solution |
|---|---|---|---|
| 3, 5 (transcardial perfusion) | Incomplete exsanguination, or the absence of tissue stiffening with PFA perfusion | Catheter is not stably placed in the heart in order to deliver solutions into rodent vasculature; vasculature was compromised during initial hPBS flush because perfusion rate was too high; an insufficient amount of hPBS was pushed through vasculature such that blood remains in smaller vessels | Use a single suture (loop the thread around the aorta) or clip to secure the feeding needle in place at the level of the aortic arch; start the initial perfusion of hPBS at a slower rate, and flush twice the volume of hPBS through |
| 6 (hydrogel monomer (HM) embedding) | Tissue damage during clearing; tissue seems to be unnecessarily fragile | Inadequate infusion of HM solution throughout tissue | It may be necessary to leave large tissue samples such as whole rat organs in HM for >12 h so that the monomer may fully penetrate the tissue |
| Tissue is structurally fragile or delicate | Consider including PFA (1–4%) in HM formulation for subsequent sample preparations; extend the postfixation step | ||
| Poor HM polymerization after 37 °C incubation | Inadequate degassing | Repeat degassing step (10 min under vacuum, 10 min of nitrogen bubbling) and 37 °C incubation | |
| Bad reagents | Use fresh PFA for fixation; prepare HM solutions immediately before use and store the thermoinitiator, acrylamide and bis-acrylamide stock solutions at 4 °C | ||
| Embedded tissue or biological sample is too fragile for non-clearing applications (e.g., thin sectioning and imaging) | Insufficient density of tissue cross-linking | Increase the concentration of PFA (1–4%) and/or include bisacrylamide (0.05%) in the HM formulation | |
| 7 (tissue clearing) | Clearing rate appears to slow down before the tissue is clear | Clearing may slow down as the clearing buffer acidifies | Buffer-exchange the clearing solution |
| Dense cross-linking | If A4P1–4 was used, remove PFA from PACT hydrogel formulation in subsequent experiments; reduce the PFA postfixation incubation time by half | ||
| Tissue is dense, highly myelinated and/or otherwise difficult to clear | Continue incubating in clearing buffer while checking periodically. Consider PARS clearing rather than PACT clearing for peripheral organ samples, as perfusive force accelerates clearing rate | ||
| Tissue appears to degrade | Bacterial contamination | Buffer-exchange the clearing solution, adding 0.01–0.05% (wt/vol) sodium azide to PBS-based clearing solutions | |
| Poor hybridization of tissue to HMs | In subsequent clearing experiments, prepare the hydrogel monomer solution with fresh reagents, increase the PFA content by 1%, extend the tissue incubation in HM by 12–24 h and/or before polymerizing the tissue-hydrogel, perform two rounds of degassing (where one round equals 10 min under vacuum and 10 min nitrogen-bubbling) | ||
| Poor PFA cross-linking of tissues | Ensure that adequate fixation and postfixation steps are performed; use fresh 4% PFA | ||
| Hydrogel softening during clearing | Overclearing and/or initial poor hydrogel polymerization | Consider doubling the postfixation step or including PFA in the HM formulation in subsequent experiments; consider underclearing tissue, as RIMS incubation will cause translucent tissues to become transparent for imaging | |
| Difficulty obtaining complete bone decalcification | PACT-deCAL procedure requires further optimization by the user according to the bone size and density (guidelines provided are specific to the mouse femur and tibia) | Experiment with EGTA-based chelation and then 8% (wt/vol) SDS clearing. Alternate steps for `7B PACT-deCAL' are as follows: | |
| (i) Incubate bone-hydrogelin 0.1 M EGTA in 1× PBS (pH 9) for 72 h at 37 °C. | |||
| (ii) Rinse the sample in 1× PBS; clear it in 8% SDS-PBS (pH 7.4) for 7 d at 37 °C, performing one buffer exchange during clearing. | |||
| (iii) Wash the sample as usual: 24–48 h in 3–6 buffer changes of 1× PBS at RT | |||
| Dense, fibrous bone or larger samples may be resistant to decalcification by chelating reagents and SDS-based clearing alone | As bone consists of ~16% collagen196,197, consider incubating bone in collagenase before clearing in order to disrupt the collagen matrix | ||
| Tissue becomes turbid; white precipitate appears in the tissue | Incomplete washing after clearing, causing SDS and/or salts to precipitate in tissue when it is moved from 37 °C to RT | Double the time for of all wash steps, making sure to perform several exchanges of 1× PBS each day; wash with PBST or BBT instead of 1× PBS | |
| Tissue becomes white and nearly opaque upon transfer to 4 °C | Salts and, in particular, residual SDS will precipitate in tissue if it is moved to 4 °C. However, the precipitate should disappear upon gradual warming of tissue to RT or 37 °C. Consider performing more extensive wash steps in future experiments, particularly after SDS clearing | ||
| Slight tissue yellowing during clearing | Use of PFA-containing hydrogels or BB | We have not observed adverse effects from slight tissue yellowing on imaging results—tissue becomes clear upon RIMS mounting. However, very occasionally, some samples become very yellow during the first half of SDS clearing: these samples should be cleared for a longer length of time—until they are very transparent—or the yellowing will cause high background during imaging. Ensure that only fresh PFA is used in subsequent experiments | |
| Brain does not become transparent during PARS-based clearing | Insufficient perfusion with clearing buffer | Extend the clearing time: most rodent organs clear within 2 d via PARS; however, the brain requires an additional 1–2 weeks to clear. RIMS-mounting will also increase the transparency of `translucent' tissues | |
| A specific organ does not clear well via whole-body PARS | Vasculature becomes compromised during the clearing process | Identify and try to fix leakages in the vasculature; if unsuccessful, tie off the major vessels supplying that organ, excise the organ for PACT clearing and continue to perform PARS clearing with the remaining body. Starting over with a new PARS preparation should only be used as the last resort | |
| Poor flow to specific organ because of anatomic reasons (poorly vascularized) | If PACT is not a desirable option and the organ is sizable with accessible vasculature, consider PARS clearing the single organ, akin to published decellularization methods92 | ||
| 9 (histology) | Poor labeling, including faint signal | Shallow antibody penetration | Increase the antibody concentration in the primary antibody cocktail or replenish the antibody halfway through extended incubations, by either adding additional antibody directly to the original antibody cocktail or by preparing a fresh antibody dilution |
| Incomplete delipidation, which obstructs labeling | Increase the clearing time | ||
| High cross-linking density | High cross-link density in A4P1-4–hybridized tissues will slow antibody diffusion; thus, antibody incubations should be extended | ||
| Epitope loss or epitope masking (unlikely if adhering to protocol) | If tissue was damaged because of microbial contamination, consider adding 0.01–0.05% (wt/vol) sodium azide to all buffers and solutions that are used in long incubations; overfixation may lead to antigen masking, so postfixation steps should be decreased | ||
| In FISH experiments, degradation of nucleic acid transcripts, or diffusion of transcripts out of sample during clearing | Ensure that all hydrogel, clearing and labeling reagents are RNase-free; embed samples in a hydrogel formulation that contains PFA and/or bis-acrylamide (e.g., A4P1B0.05), and perform a rigorous degassing step to ensure thorough hydrogel-tissue hydridization | ||
| Poor quality of antibody or dye, which results in weak labeling | Only use high-quality antibodies that have been first verified in standard thin-section immunolabeling; experiment with a different antibody supplier—different antibodies against the same target may vary greatly in their labeling abilities, such as in their binding affinity and in their capacity to access intracellular compartments for cell-filling labeling versus only superficial or extracellular epitope binding. Finally, it can be helpful to simultaneously prepare a thin section (40–100 μm) alongside a thick, cleared section while troubleshooting to ensure that the visualization of a strong signal is possible | ||
| High background and/or autofluorescence | Tissue damage during processing | Review procedures carefully, and ensure that no reagents introduced bacterial contamination of sample; lengthen the wash steps to remove potential precipitate (SDS, donkey serum–antibody immunocomplexes) | |
| Sources of autofluorescence—part 1: fixative-induced autofluorescence, elastin, collagen | Many standard histological techniques for reducing autofluorescence, such as tissue bleaching23, performing wash steps in PBST containing 100 mM glycine to quench aldehydes and treating tissue with histology stains that quench or mask autofluorescence, may be adapted to thick-sectioned cleared tissues—typically by performing longer wash steps after the appropriate countermeasure; photobleaching the tissue before IHC at wavelengths that exhibit the highest autofluorescence may also help198 | ||
| Sources of autofluorescence—part 2: heme chromophores, lipofuscins | Thoroughly remove all blood during initial cardiac perfusion; to elute heme, incubate hydrogel-embedded PACT sections and in particular PACT-deCAL sections in aminoalcohol (CUBIC reagent-1 (refs. 11,21) for 12–24 h at 37 °C with shaking, and then transfer the sections directly into 8% (wt/vol) SDS for clearing; lipofuscin autofluorescence is partially combatted by tissue clearing; however, thick tissue sections may be incubated in 0.2%199 to 1.0% ((wt/vol) final concentration) Sudan Black B for 1–3 hours immediately before Step 5 (PACT hydrogel-embedding) in order to reduce high autofluorescent background—tissue clearing will allow Sudan Black B–treated sections to become sufficiently transparent for imaging (Supplementary Fig. 3) | ||
| High background, but with high signal of correctly labeled epitopes | Nonspecific antibody binding | Extend the wash steps after both primary and secondary antibody incubations an additional day, by performing four or five buffer exchanges each day, and wash the samples in PBST instead of 1× PBS; in rodent tissue samples, avoid using antibodies that require anti-mouse secondary antibody labeling23; also some chicken antibodies show strong staining with high background and/or aggregation—these antibodies should be diluted to 1:400 to 1:1,000 | |
| 12, 13 (tissue mounting and imaging) | Poor image quality and/or poor imaging depth | Tissue is of insufficient transparency for light to penetrate | Extend the tissue incubation time in RIMS to several days before imaging; for bone, incubate for an additional 1 d in RIMS-1.48 or RIMS-1.49 before imaging |
| Morphological distortion | Tissue size fluctuations | Immediately before RIMS incubation, postfix cleared, immunolabeled tissue in 4% PFA for a few hours at RT, and then wash and incubate in RIMS for at least several days to one week before imaging; consider preparing future samples in hydrogel that contains PFA (e.g., A4P1–4, depending on the degree of swelling) and/or consider a longer postfixation step after transcardial perfusion | |
| Bubbles in mounted tissue | Air trapped in tissue or dissolved air in RIMS; sample mounted with insufficient RIMS, causing the introduction of air bubbles between the RIMS meniscus and cover glass | Purge RIMS of excess air via degassing the tissue in fresh RIMS before mounting (e.g., using the vacuum line, akin to the hydrogel polymerization of Step 5; do not bubble nitrogen through the sample following its placement under vacuum)—use this degassed RIMS to mount the degassed sample | |
| Sample appears turbid or white | RIMS-mounted sample was placed at 4 °C, causing salts, etc., to precipitate | The precipitate should disappear upon gradual warming of tissue to RT or 37 °C. Store RIMS-mounted tissue at RT, protected from light, or mount tissue in cRIMS for cold storage | |
| 16, 17 (3D image analysis) | Imaging software and/or computer crashes; unable to load acquired images | Insufficient RAM for large images | Troubleshoot with a different option in the step 15 workflow: option A using Imaris, option B using TerraStitcher, or option C Vaa3D TerraFly; consider upgrading computer workstation and/or adding RAM and/or new graphics card; downsample the data set (note that compression cannot be used with Imaris); process the images in tiles (i.e., analyze each tile individually) |
● TIMING
Steps 1–5, tissue preparation for PACT and PARS: 2 h–1 d
Step 6, formation of a tissue-hydrogel matrix: 1 d
Steps 7 and 8, tissue delipidation with SDS: passive, 10–240 h (PACT on thin- and thick-sectioned tissue), 1 week (PACT-deCAL), or up to 1 month (whole-brain clearing via PACT); active: 1–4 d (PARS, all organs except brain) or 1–2 weeks (whole-brain clearing via PARS)
Step 9, single-cell phenotyping of cleared tissues: 12 h (labeling with small-molecule dyes) to 2 weeks (primary and secondary antibody labeling and nuclear staining with 24- to 48-h wash steps)
Steps 10–13, RIMS for PACT and PARS samples: 1 d
Step 14, acquisition: 1 h to several days (variable depending on tissue volume, the number of distinct color labels or channels, the desired resolution and the microscope setup)
Step 15, 3D image visualization: ~1 h
Steps 16 and 17, 3D image analysis: variable based on analysis goals
ANTICIPATED RESULTS
PACT, PARS and RIMS collectively form a tissue clearing toolkit that is versatile, user-friendly and sample-friendly across tissue types. Building on past research18, we detail here how both PACT and PARS methodologies are amenable to rapid, high-throughput histopathology of rodent (Fig. 1a–e) and human (Fig. 1f,g) tissue samples alike, whether through the visualization of natively expressed fluorescence markers (Figs. 1a–e, 5c and 9c; Supplementary Fig. 3d,e) or through immunolabeling whole-organ and thick tissue samples (Figs. 1f,g, 4b,c, 5d and 6a; Supplementary Fig. 6b) after clearing. Furthermore, the formation of a cross-linked, tissue-hydrogel matrix allows for rigorous detergent-based clearing with only minimal leaching of proteins into the clearing buffer (Fig. 3a and Supplementary Fig. 2a,b) and no detectable loss in YFP fluorescence between uncleared and cleared samples (see Yang et al.18). In addition to its RI-matching capability, RIMS also serves to preserve the molecular content of mounted samples: no protein was measured to leach out of mounted samples after a 1-week incubation, and YFP fluorescence was readily detected in cleared samples that were stored in RIMS for 3 months18. The enhanced optical transparency of delipidized and RI-matched tissues permits high-resolution detection of endogenously expressed fluorescent proteins, antibody-labeled proteins and nucleic acid transcripts at the single-molecule level (FISH18), usually with similar intensity to and lower background signals than are seen in uncleared tissues (Fig. 5c,d, confocal images for control versus cleared samples; Supplementary Fig. 4).
Among the recently developed PACT variations summarized here are dedicated protocols for specialty cases within tissue clearing. These include PACT-processing of fragile tissue samples (Figs. 3–5), pre-PACT tissue staining with Sudan Black B to mask autofluorescence in thick tissues (Supplementary Fig. 3 and Supplementary Methods), PACT-deCAL for clearing and imaging fluorescently labeled bone (Fig. 6a) and ePACT for tissue clearing through expansion (Supplementary Fig. 4 and Supplementary Methods). With respect to the latter, PACT tissues have previously been carried through to FISH studies, in which clearing and slight tissue swelling benefitted the visualization of single, labeled transcripts18. In ePACT, cellular components that were once poorly resolved in uncleared sections become visible (Supplementary Fig. 4b). Combining these techniques offers the potential to optically distinguish multiple transcripts or otherwise packed multicolor labels (e.g., neuronal positioning system, NPS133) within the expanded sample space of a cell.
To accompany these methods for chemically clearing a variety of tissue types, we extended our RIMS formulation guidelines to include recipes for different tissue types (i.e., to better match the RIs of different samples; see Fig. 6b and Supplementary Fig. 1b). As PACT and PARS have now been optimized to clear a wide variety of tissues, the method can further benefit from exploring alternative labeling schemes for visualizing protein and nucleic acid targets in thick tissues (Supplementary Fig. 6). Although traditional antibody-based labeling methods have been used very effectively to illuminate cell phenotype and tissue morphology (Figs. 1g and 5d and previous work18), they can be both cumbersome and costly. The slow penetration of full-format immunoglobulins in thick tissue necessitates long incubations (Fig. 4b–d), to the detriment of sample integrity. Herein, camelid nanobodies (Supplementary Fig. 6) and protein affinity tags (i.e., SNAP-tag134, Halo-tag135, CLIP-tag136, TMP-tag137, and Spy Tag Catcher138) present chemically stable and potentially cost-effective alternatives. With the ability to easily penetrate thick tissue, these reagents can recognize and bind their respective targets, either a cognate antigen or tagged protein, with high specificity and rapid kinetics. In addition, by using dyes that are several-fold brighter, highly photostable and easier to separate spectrally than fluorescent proteins, protein affinity reagents can provide an excellent signal-to-noise ratio in labeled tissues139.
Two major bottlenecks in the translation of fixed, unprocessed tissue banks into analyzable image databases are (i) acquisition time of thick tissues at high resolution and (ii) the computational demands to convert raw image stacks into manageable data sets for phenotypical and morphological study. Light-sheet microscopy has recently been applied to imaging large cleared volumes as it substantially reduces the acquisition time15. Here we provide a basic scheme for relatively inexpensive design of a light-sheet microscope that enables fast and high-resolution imaging of cleared samples (Fig. 9a,b and Supplementary Table 1), and we show its compatibility with PACT. A representative volume (1 mm depth) PACT-cleared mouse brain slice imaged at 45 frames per second can be seen in Figure 9c. In comparison with traditional confocal microscopes, a frame rate of 45 frames per second shows ~10–100 times improvement in image acquisition speeds and thus allows for rapid imaging of large cleared samples, in addition to its recognized utility for live-cell imaging140. In addition to increased speed, light-sheet microscopy substantially reduces photobleaching141, which is crucial for imaging dim samples and especially for conducting smFISH experiments, in which the 20–100 single fluorophore–labeled probes are used to visualize individual transcripts.
Regardless of the microscopy setup, image acquisitions for large tissue samples generate raw data files that are on the order of gigabytes or even terabytes in size. These data must be converted into a file format that allows data handling and visualization on computer workstations that are available to standard research laboratories. Many software packages for image analysis were not designed to accommodate such file sizes and inevitably crash partway through the computational process. Thus, we have endeavored to present a broadly applicable workflow for image data handling (Fig. 10 and Table 3), which will guide the user through the process of transferring raw image files into the image analysis software packages that we feel are most capable of performing basic functions such as tract-tracing and cell mapping.
In summary, we have validated the ability of PACT, PARS and RIMS to prepare a variety of tissue samples for imaging via confocal and light-sheet microscopy. With these methods in hand, biologists may tackle the broad spectrum of scientific demands, from the meticulous analysis of isolated cell niches to the global interrogation of intact biological systems.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. McBride, D.K. Newman and J. Allman for useful discussions on uses of tissue clearing across disciplines. We thank M. Brissova and A.C. Powers from Vanderbilt University for providing fixed human pancreas tissue used in Figure 1 and guidance with pancreatic markers and anatomy. This work was funded by grants to V.G.: the US National Institutes of Health (NIH) Director's New Innovator IDP20D017782-01; the NIH/National Institute on Aging (NIA) 1R01AG047664-01; the Beckman Institute for Optogenetics and CLARITY; the Pew Charitable Trust; and the Kimmel Foundation. Work in the Gradinaru Laboratory at Caltech is also funded by awards from the following (to V.G.): the NIH Brain Research through Advancing Innovative Neurotechnologies (BRAIN) 1U01NS090577; the NIH/National Institutes of Mental Health (NIMH) 1R21MH103824-01; the Human Frontiers in Science Program; the Mallinckrodt Foundation; the Gordon and Betty Moore Foundation through grant GBMF2809 to the Caltech Programmable Molecular Technology Initiative; the Michael J. Fox Foundation; Caltech-GIST; and the Caltech-City of Hope Biomedical Initiative. This work was also supported by grants to P.J.B. from the NIH (2 P50 GM082545-06; W.I. Sundquist, principal investigator) and gifts from the Gordon and Betty Moore Foundation and the Agouron Institute to support electron microscopy at Caltech; and by National Science Foundation (NSF) IIS-1253538 and DBI-1262547 grants to C.C.F. K.Y.C. and N.C.F. were supported by the NIH Predoctoral Training in Biology and Chemistry (2T32GM007616-36).
Footnotes
Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS J.B.T. and V.G. wrote the manuscript with input from all coauthors. J.B.T., K.Y.C., N.C.F., B.Y., B.E.D. and V.G. designed and performed experiments, analyzed the data and prepared figures. C.C.F. wrote the data analysis section, including associated figures and data analysis; C.X. assisted with tissue clearing and imaging for data sets in this section. A.G., A.L., L.C. and V.G. planned for and built the light sheet and collected and analyzed the associated data. M.S.L., P.J.B. and V.G. planned and performed TEM tissue processing and imaging and prepared the EM figure. V.G. supervised all aspects of the project. All authors edited and approved the manuscript.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akerboom J, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peters AJ, Chen SX, Komiyama T. Emergence of reproducible spatiotemporal activity during motor learning. Nature. 2014;510:263–267. doi: 10.1038/nature13235. [DOI] [PubMed] [Google Scholar]
- 4.White RM, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaletta T, Hengartner MO. Finding function in novel targets: C. Elegans as a model organism. Nat. Rev. Drug Discov. 2006;5:387–399. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- 6.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 7.Ke M-T, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 2013;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 8.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hama H, et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 10.Kuwajima T, et al. ClearT: a detergent-and solvent-free clearing method for neuronal and non-neuronal tissue. Development. 2013;140:1364–1368. doi: 10.1242/dev.091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Susaki EA, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 12.Becker K, Jährling N, Saghafi S, Weiler R, Dodt H-U. Chemical clearing and dehydration of GFP-expressing mouse brains. PLoS ONE. 2012;7:e33916. doi: 10.1371/journal.pone.0033916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodt H-U, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 14.Ertürk A, et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- 15.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced clarity for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang MD, et al. Neuronal calcium-binding proteins 1/2 localize to dorsal root ganglia and excitatory spinal neurons and are regulated by nerve injury. Proc. Natl. Acad. Sci. USA. 2014;111:E1149–1158. doi: 10.1073/pnas.1402318111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou B, et al. Scalable and DiI-compatible optical clearance of the mammalian brain. Front. Neuroanat. 2015;9:19. doi: 10.3389/fnana.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang B, et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakhalkar HS, Dewhirst M, Oliver T, Cao Y, Oldham M. Functional imaging in bulk tissue specimens using optical emission tomography: fluorescence preservation during optical clearing. Phys. Med. Biol. 2007;52:2035–2054. doi: 10.1088/0031-9155/52/8/001. [DOI] [PubMed] [Google Scholar]
- 20.Aoyagi Y, Kawakami R, Osanai H, Hibi T, Nemoto T. A rapid optical clearing protocol using 2,2′-thiodiethanol for microscopic observation of fixed mouse brain. PLoS ONE. 2015;10:e0116280. doi: 10.1371/journal.pone.0116280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tainaka K, et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell. 2014;159:911–924. doi: 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 22.Ke MT, Imai T. Optical clearing of fixed brain samples using SeeDB. Curr. Protoc. Neurosci. 2014;66:2.22.21–22.22.19. doi: 10.1002/0471142301.ns0222s66. [DOI] [PubMed] [Google Scholar]
- 23.Renier N, et al. Idisco: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat. Methods. 2013;10:508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- 25.Kim SY, Chung K, Deisseroth K. Light microscopy mapping of connections in the intact brain. Trends Cogn. Sci. 2013;17:596–599. doi: 10.1016/j.tics.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Ertürk A, Bradke F. High-resolution imaging of entire organs by 3-dimensional imaging of solvent cleared organs (3DISCO) Exp. Neurol. 2013;242:57–64. doi: 10.1016/j.expneurol.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Deisseroth KA, Gradinaru V. Functional targeted brain endoskeletonization. US patent US2014030192. 2012
- 28.Richardson DS, Lichtman JW. Clarifying tissue clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ragan T, et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mccormick BH. Brain tissue scanner enables brain microstructure surveys. Neurocomputing. 2002;44:1113–1118. [Google Scholar]
- 31.Kuan L, et al. Neuroinformatics of the Allen mouse brain connectivity atlas. Methods. 2015;73:4–17. doi: 10.1016/j.ymeth.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 32.Rah J-C, et al. Thalamocortical input onto layer 5 pyramidal neurons measured using quantitative large-scale array tomography. Front. Neural Circuits. 2013;7:177. doi: 10.3389/fncir.2013.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zingg B, et al. Neural networks of the mouse neocortex. Cell. 2014;156:1096–1111. doi: 10.1016/j.cell.2014.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bohland JW, et al. A proposal for a coordinated effort for the determination of brainwide neuroanatomical connectivity in model organisms at a mesoscopic scale. PLoS Comput. Biol. 2009;5:e1000334. doi: 10.1371/journal.pcbi.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Craddock RC, et al. Imaging human connectomes at the macroscale. Nat. Methods. 2013;10:524–539. doi: 10.1038/nmeth.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh SW, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hintiryan H, et al. Comprehensive connectivity of the mouse main olfactory bulb: analysis and online digital atlas. Front. Neuroanat. 2012;6:30. doi: 10.3389/fnana.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.George MS, et al. Vagus nerve stimulation: a new tool for brain research and therapy. Biol. Psychiatry. 2000;47:287–295. doi: 10.1016/s0006-3223(99)00308-x. [DOI] [PubMed] [Google Scholar]
- 40.Berthoud H-R, Neuhuber WL. Functional and chemical anatomy of the afferent vagal system. Auton. Neurosci. 2000;85:1–17. doi: 10.1016/S1566-0702(00)00215-0. [DOI] [PubMed] [Google Scholar]
- 41.Birmingham K, et al. Bioelectronic medicines: a research roadmap. Nat. Rev. Drug Discov. 2014;13:399–400. doi: 10.1038/nrd4351. [DOI] [PubMed] [Google Scholar]
- 42.Ertürk A, et al. Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nat. Med. 2012;18:166–171. doi: 10.1038/nm.2600. [DOI] [PubMed] [Google Scholar]
- 43.Bucher D, Scholz M, Stetter M, Obermayer K, Pflüger HJ. Correction methods for three-dimensional reconstructions from confocal images: I. Tissue shrinking and axial scaling. J. Neurosci. Methods. 2000;100:135–143. doi: 10.1016/s0165-0270(00)00245-4. [DOI] [PubMed] [Google Scholar]
- 44.Staudt T, Lang MC, Medda R, Engelhardt J, Hell SW. 2,2′-thiodiethanol: a new water-soluble mounting medium for high-resolution optical microscopy. Microsc. Res. Tech. 2007;70:1–9. doi: 10.1002/jemt.20396. [DOI] [PubMed] [Google Scholar]
- 45.Ott HC, et al. Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart. Nat. Med. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 46.Rahman A, Brown CW. Effect of pH on the critical micelle concentration of sodium dodecyl sulphate. J. Appl. Polym. Sci. 1983;28:1331–1334. [Google Scholar]
- 47.Otzen DE. Protein unfolding in detergents: effect of micelle structure, ionic strength, pH, and temperature. Biophys. J. 2002;83:2219–2230. doi: 10.1016/S0006-3495(02)73982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piatkevich KD, et al. Extended Stokes shift in fluorescent proteins: chromophore-protein interactions in a near-infrared TagRFP675 Variant. Sci. Rep. 2013;3:1847. doi: 10.1038/srep01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kragh-Hansen U, Le Maire M, Møller JV. The mechanism of detergent solubilization of liposomes and protein-containing membranes. Biophys. J. 1998;75:2932–2946. doi: 10.1016/S0006-3495(98)77735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammouda B. Temperature effect on the nanostructure of SDS micelles in water. J. Res. Natl. Inst. Stand. Technol. 2013;118:151–167. doi: 10.6028/jres.118.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Desmyter A, Spinelli S, Roussel A, Cambillau C. Camelid Nanobodies: killing two birds with one stone. Curr. Opin. Struct. Biol. 2015;32:1–8. doi: 10.1016/j.sbi.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Pardon E, et al. A general protocol for the generation of Nanobodies for structural biology. Nat. Protoc. 2014;9:674–693. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lichtman JW, Sanes JR. Ome sweet Ome: what can the genome tell us about the connectome? Curr. Opin. Neurobiol. 2008;18:346–353. doi: 10.1016/j.conb.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 55.Lehrer J. Neuroscience: making connections. Nature. 2009;457:524–527. doi: 10.1038/457524a. [DOI] [PubMed] [Google Scholar]
- 56.Peng H, et al. Bigneuron: large-scale 3D neuron reconstruction from optical microscopy images. Neuron. 2015;87:252–256. doi: 10.1016/j.neuron.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kasthuri N, et al. Saturated reconstruction of a volume of neocortex. Cell. 2015;162:648–661. doi: 10.1016/j.cell.2015.06.054. [DOI] [PubMed] [Google Scholar]
- 58.Burns R, et al. Sci. Stat. Database Manag. 2013. The open connectome project data cluster: scalable analysis and vision for high-throughput neuroscience. doi:10.1145/2484838.2484870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costa M, Ostrovsky AD, Manton JD, Prohaska S, Jefferis GSXE. Biorxiv. 2014. Nblast: rapid, sensitive comparison of neuronal structure and construction of neuron family databases. doi:10.1101/006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng H, Bria A, Zhou Z, Iannello G, Long F. Extensible visualization and analysis for multidimensional images using Vaa3D. Nat. Protoc. 2014;9:193–208. doi: 10.1038/nprot.2014.011. [DOI] [PubMed] [Google Scholar]
- 63.Peng H, Ruan Z, Long F, Simpson JH, Myers EW. V3d Enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat. Biotechnol. 2010;28:348–353. doi: 10.1038/nbt.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Longair MH, Baker DA, Armstrong JD. Simple neurite tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- 65.Dercksen VJ, Hege HC, Oberlaender M. The Filament Editor: an interactive software environment for visualization, proof-editing and analysis of 3D neuron morphology. Neuroinformatics. 2014;12:325–339. doi: 10.1007/s12021-013-9213-2. [DOI] [PubMed] [Google Scholar]
- 66.Peng H, et al. Virtual finger boosts three-dimensional imaging and microsurgery as well as terabyte volume image visualization and analysis. Nat. Commun. 2014;5:4342. doi: 10.1038/ncomms5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng L, Zhao T, Kim J. Eneuro. 2015. Neutube 1.0: a new design for efficient neuron reconstruction software based on the Swc Format. doi:10.1523/Eneuro.0049-1514.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Glaser JR, Glaser EM. Neuron imaging with Neurolucida—a PC-based system for image combining microscopy. Comput. Med. Imaging Graph. 1990;14:307–317. doi: 10.1016/0895-6111(90)90105-k. [DOI] [PubMed] [Google Scholar]
- 69.Bria A, Iannello G. TeraStitcher—a tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinformatics. 2012;13:316. doi: 10.1186/1471-2105-13-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu Y, Peng H. Automated high speed stitching of large 3D microscopic images. Proc. of IEEE 2011 International Symposium on Biomedical Imaging: From Nano to Macro.2011. pp. 238–241. [Google Scholar]
- 71.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emmenlauer M, et al. XuvTools: free, fast and reliable stitching of large 3D datasets. J. Microsc. 2009;233:42–60. doi: 10.1111/j.1365-2818.2008.03094.x. [DOI] [PubMed] [Google Scholar]
- 73.Model MA, Blank JL. Concentrated dyes as a source of two-dimensional fluorescent field for characterization of a confocal microscope. J. Microsc. 2008;229:12–16. doi: 10.1111/j.1365-2818.2007.01880.x. [DOI] [PubMed] [Google Scholar]
- 74.Smith K, et al. CIDRE: an illumination-correction method for optical microscopy. Nat. Methods. 2015;12:404–406. doi: 10.1038/nmeth.3323. [DOI] [PubMed] [Google Scholar]
- 75.Edelstein AD, et al. Advanced methods of microscope control using μManager software. J. Biol. Methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmid B, Schindelin J, Cardona A, Longair M, Heisenberg M. A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics. 2010;11:274. doi: 10.1186/1471-2105-11-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.De Chaumont F, et al. Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods. 2012;9:690–696. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- 78.Kankaanpaa P, et al. Bioimagexd: an open, general-purpose and high-throughput image-processing platform. Nat. Methods. 2012;9:683–689. doi: 10.1038/nmeth.2047. [DOI] [PubMed] [Google Scholar]
- 79.Kvilekval K, Fedorov D, Obara B, Singh A, Manjunath BS. Bisque: a platform for bioimage analysis and management. Bioinformatics. 2010;26:544–552. doi: 10.1093/bioinformatics/btp699. [DOI] [PubMed] [Google Scholar]
- 80.Pietzsch T, Saalfeld S, Preibisch S, Tomancak P. Bigdataviewer: visualization and processing for large image data sets. Nat. Methods. 2015;12:481–483. doi: 10.1038/nmeth.3392. [DOI] [PubMed] [Google Scholar]
- 81.Benmansour F, Cohen LD. Tubular structure segmentation based on minimal path method and anisotropic enhancement. Int. J. Comput. Vis. 2011;92:192–210. [Google Scholar]
- 82.Chothani P, Mehta V, Stepanyants A. Automated tracing of neurites from light microscopy stacks of images. Neuroinformatics. 2011;9:263–278. doi: 10.1007/s12021-011-9121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ming X, et al. Rapid reconstruction of 3D neuronal morphology from light microscopy images with augmented rayburst sampling. PLoS ONE. 2013;8:e84557. doi: 10.1371/journal.pone.0084557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luisi J, Narayanaswamy A, Galbreath Z, Roysam B. The FARSIGHT trace editor: an open source tool for 3-D inspection and efficient pattern analysis aided editing of automated neuronal reconstructions. Neuroinformatics. 2011;9:305–315. doi: 10.1007/s12021-011-9115-0. [DOI] [PubMed] [Google Scholar]
- 85.Myatt DR, Hadlington T, Ascoli GA, Nasuto SJ. Neuromantic—from semi-manual to semi-automatic reconstruction of neuron morphology. Front. Neuroinform. 2012;6:4. doi: 10.3389/fninf.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gleeson P, et al. NeuroML: a language for describing data driven models of neurons and networks with a high degree of biological detail. PLoS Comput. Biol. 2010;6:e1000815. doi: 10.1371/journal.pcbi.1000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Parekh R, Ascoli GA. Neuronal morphology goes digital: a research hub for cellular and system neuroscience. Neuron. 2013;77:1017–1038. doi: 10.1016/j.neuron.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meijering E. Neuron tracing in perspective. Cytometry A. 2010;77:693–704. doi: 10.1002/cyto.a.20895. [DOI] [PubMed] [Google Scholar]
- 89.Scorcioni R, Polavaram S, Ascoli GA. L-Measure: a web-accessible tool for the analysis, comparison and search of digital reconstructions of neuronal morphologies. Nat. Protoc. 2008;3:866–876. doi: 10.1038/nprot.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gleeson P, Steuber V, Silver RA. Neuroconstruct: a tool for modeling networks of neurons in 3D space. Neuron. 2007;54:219–235. doi: 10.1016/j.neuron.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ascoli GA, Donohue DE, Halavi M. Neuromorpho.Org: a central resource for neuronal morphologies. J. Neurosci. 2007;27:9247–9251. doi: 10.1523/JNEUROSCI.2055-07.2007. Neuromorpho.Org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guyette JP, et al. Perfusion decellularization of whole organs. Nat. Protoc. 2014;9:1451–1468. doi: 10.1038/nprot.2014.097. [DOI] [PubMed] [Google Scholar]
- 93.Uygun BE, et al. Decellularization and recellularization of whole livers. J. Vis. Exp. 2011 doi: 10.3791/2394. doi:10.3791/2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen F, Tillberg PW, Boyden ES. Optical imaging. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bach SP, Renehan AG, Potten CS. Stem cells: the intestinal stem cell as a paradigm. Carcinogenesis. 2000;21:469–476. doi: 10.1093/carcin/21.3.469. [DOI] [PubMed] [Google Scholar]
- 96.Barry ER, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nat. Rev. Immunol. 2006;6:93–106. doi: 10.1038/nri1779. [DOI] [PubMed] [Google Scholar]
- 98.Vakoc BJ, et al. Three-dimensional microscopy of the tumor microenvironment in vivo using optical frequency domain imaging. Nat. Med. 2009;15:1219–1223. doi: 10.1038/nm.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reardon S. Electroceuticals spark interest. Nature. 2014;511:18. doi: 10.1038/511018a. [DOI] [PubMed] [Google Scholar]
- 100.Grace AA, Llinas R. Morphological artifacts induced in intracellularly stained neurons by dehydration: circumvention using rapid dimethyl sulfoxide clearing. Neuroscience. 1985;16:461–475. doi: 10.1016/0306-4522(85)90018-1. [DOI] [PubMed] [Google Scholar]
- 101.Kasthuri N, Lichtman JW. Neurocartography. Neuropsychopharmacology. 2010;35:342–343. doi: 10.1038/npp.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Henry AM, Hohmann JG. High-resolution gene expression atlases for adult and developing mouse brain and spinal cord. Mamm. Genome. 2012;23:539–549. doi: 10.1007/s00335-012-9406-2. [DOI] [PubMed] [Google Scholar]
- 103.Dodt HU. Microscopy. The superresolved brain. Science. 2015;347:474–475. doi: 10.1126/science.aaa5084. [DOI] [PubMed] [Google Scholar]
- 104.Betzig E, Trautman JK, Harris TD, Weiner JS, Kostelak RL. Breaking the diffraction barrier: optical microscopy on a nanometric scale. Science. 1991;251:1468–1470. doi: 10.1126/science.251.5000.1468. [DOI] [PubMed] [Google Scholar]
- 105.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 106.Gustafsson MG. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc. Natl. Acad. Sci. USA. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 108.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sumbul U, Zlateski A, Vishwanathan A, Masland RH, Seung HS. Automated computation of arbor densities: a step toward identifying neuronal cell types. Front. Neuroanat. 2014;8:139. doi: 10.3389/fnana.2014.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Videen TO, et al. Validation of a fiducial-based atlas localization method for deep brain stimulation contacts in the area of the subthalamic nucleus. J. Neurosci. Methods. 2008;168:275–281. doi: 10.1016/j.jneumeth.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gutman B, et al. Registering cortical surfaces based on whole-brain structural connectivity and continuous connectivity analysis. Med. Image Comput. Comput. Assist. Interv. 2014;17:161–168. doi: 10.1007/978-3-319-10443-0_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pantazis D, et al. Comparison of landmark-based and automatic methods for cortical surface registration. Neuroimage. 2010;49:2479–2493. doi: 10.1016/j.neuroimage.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuwajima M, Mendenhall JM, Harris KM. Large-volume reconstruction of brain tissue from high-resolution serial section images acquired by SEM-based scanning transmission electron microscopy. Methods Mol. Biol. 2013;950:253–273. doi: 10.1007/978-1-62703-137-0_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Masich S, Östberg T, Norlén L, Shupliakov O, Daneholt B. A procedure to deposit fiducial markers on vitreous cryo-sections for cellular tomography. J. Struct. Biol. 2006;156:461–468. doi: 10.1016/j.jsb.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 115.Kuwajima M, Mendenhall JM, Lindsey LF, Harris KM. Automated transmission-mode scanning electron microscopy (tSEM) for large volume analysis at nanoscale resolution. PLoS ONE. 2013;8:e59573. doi: 10.1371/journal.pone.0059573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Choi HMT, Beck VA, Pierce NA. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano. 2014;8:4284–4294. doi: 10.1021/nn405717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Choi HM, et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huisken J, Stainier DYR. Selective plane illumination microscopy techniques in developmental biology. Development. 2009;136:1963–1975. doi: 10.1242/dev.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Baumgart E, Kubitscheck U. Scanned light-sheet microscopy with confocal slit detection. Opt. Express. 2012;20:21805–21814. doi: 10.1364/OE.20.021805. [DOI] [PubMed] [Google Scholar]
- 122.Gage GJ, Kipke DR, Shain W. Whole-animal perfusion fixation for rodents. J. Vis. Exp. 2012 doi: 10.3791/3564. doi:10.3791/3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tremblay M-È, Riad M, Majewska AK. Preparation of mouse brain tissue for immunoelectron microscopy. J. Vis. Exp. 2010 doi: 10.3791/2021. doi:10.3791/2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dominguez E, et al. Non-invasive in vivo measurement of cardiac output in C57BL/6 mice using high-frequency transthoracic ultrasound: evaluation of gender and body weight effects. Int. J. Cardiovasc. Imaging. 2014;30:1237–1244. doi: 10.1007/s10554-014-0454-4. [DOI] [PubMed] [Google Scholar]
- 125.Tournoux F, et al. Validation of non invasive measurements of cardiac output in mice using echocardiography. J. Am. Soc. Echocardiogr. 2011;24:465–470. doi: 10.1016/j.echo.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Janssen B, Debets J, Leenders P, Smits J. Chronic measurement of cardiac output in conscious mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002;282:R928–935. doi: 10.1152/ajpregu.00406.2001. [DOI] [PubMed] [Google Scholar]
- 127.Delp MD, Evans MV, Duan CP. Effects of aging on cardiac output, regional blood flow, and body composition in Fischer-344 rats. J. Appl. Physiol. 1998;85:1813–1822. doi: 10.1152/jappl.1998.85.5.1813. [DOI] [PubMed] [Google Scholar]
- 128.Reineke T, et al. Ultrasonic decalcification offers new perspectives for rapid FISH, DNA, and RT-PCR analysis in bone marrow trephines. Am. J. Surg. Pathol. 2006;30:892–896. doi: 10.1097/01.pas.0000213282.20166.13. [DOI] [PubMed] [Google Scholar]
- 129.Fridy PC, et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods. 2014;11:1253–1260. doi: 10.1038/nmeth.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Goldberg IG, et al. The Open Microscopy Environment (Ome) data model and XML file: open tools for informatics and quantitative analysis in biological imaging. Genome Biol. 2005;6:R47. doi: 10.1186/gb-2005-6-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Linkert M, et al. Metadata matters: access to image data in the real world. J. Cell Biol. 2010;189:777–782. doi: 10.1083/jcb.201004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Carnevale NT, Hines ML. The Neuron Book. Cambridge University Press; 2006. [Google Scholar]
- 133.Tsuriel S, Gudes S, Draft RW, Binshtok AM, Lichtman JW. Multispectral labeling technique to map many neighboring axonal projections in the same tissue. Nat. Methods. 2015;12:547–552. doi: 10.1038/nmeth.3367. [DOI] [PubMed] [Google Scholar]
- 134.Keppler A, et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 135.Los GV, et al. Hatotag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 136.Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 137.Miller LW, Cai YF, Sheetz MP, Cornish VW. In vivo protein labeling with trimethoprim conjugates: a flexible chemical tag. Nat. Methods. 2005;2:255–257. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 138.Bedbrook CN, et al. Genetically encoded spy peptide fusion system to detect plasma membrane-localized proteins in vivo. Chem. Biol. 2015;22:1108–1121. doi: 10.1016/j.chembiol.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kohl J, et al. Ultrafast tissue staining with chemical tags. Proc. Natl. Acad. Sci. USA. 2014;111:E3805–E3814. doi: 10.1073/pnas.1411087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Keller PJ, Ahrens MB. Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron. 2015;85:462–483. doi: 10.1016/j.neuron.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 141.Stelzer EHK. Light-sheet fluorescence microscopy for quantitative biology. Nat. Methods. 2015;12:23–26. doi: 10.1038/nmeth.3219. [DOI] [PubMed] [Google Scholar]
- 142.Paxinos G, Franklin KBJ. The Mouse Brain In Stereotaxic Coordinates Compact. 2nd edn Elsevier Academic Press; 2004. [Google Scholar]
- 143.Connell BJ, Lortat-Jacob H. Human immunodeficiency virus and heparan sulfate: from attachment to entry inhibition. Front. Immunol. 2013;4:385. doi: 10.3389/fimmu.2013.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jones CT, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat. Biotechnol. 2010;28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sattentau Q. Avoiding the void: cell-to-cell spread of human viruses. Nat. Rev. Microbiol. 2008;6:815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- 146.Wu Z, Asokan A, Samulski RJ. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol. Ther. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 147.Fiege JK, Langlois RA. Investigating influenza A virus infection: tools to track infection and limit tropism. J. Virol. 2015;89:6167–6170. doi: 10.1128/JVI.00462-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Foust KD, et al. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kollarik M, et al. Transgene expression and effective gene silencing in vagal afferent neurons in vivo using recombinant adeno-associated virus vectors. J. Physiol. 2010;588:4303–4315. doi: 10.1113/jphysiol.2010.192971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang H, et al. Several rAAV vectors efficiently cross the blood–brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system. Mol. Ther. 2011;19:1440–1448. doi: 10.1038/mt.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tapia JC, et al. High-contrast en bloc staining of neuronal tissue for field emission scanning electron microscopy. Nat. Protoc. 2012;7:193–206. doi: 10.1038/nprot.2011.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Magnon C, et al. Autonomic nerve development contributes to prostate cancer progression. Science. 2013;341:1236361. doi: 10.1126/science.1236361. [DOI] [PubMed] [Google Scholar]
- 154.Colomba A, Ridley AJ. Analyzing the roles of Rho GTPases in cancer cell migration with a live cell imaging 3D-morphology-based assay. Methods Mol. Biol. 2014;1120:327–337. doi: 10.1007/978-1-62703-791-4_21. [DOI] [PubMed] [Google Scholar]
- 155.Fukamachi K, et al. Total-circumference intraoperative frozen section analysis reduces margin-positive rate in breast-conservation surgery. Jpn. J. Clin. Oncol. 2010;40:513–520. doi: 10.1093/jjco/hyq006. [DOI] [PubMed] [Google Scholar]
- 156.Louveau A, et al. Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523:337–341. doi: 10.1038/nature14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Goodell MA, Nguyen H, Shroyer N. Somatic stem cell heterogeneity: diversity in the blood, skin and intestinal stem cell compartments. Nat. Rev. Mol. Cell. Biol. 2015;16:299–309. doi: 10.1038/nrm3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sharp FR, Liu JL, Bernabeu R. Neurogenesis following brain ischemia. Brain Res. Dev. Brain Res. 2002;134:23–30. doi: 10.1016/s0165-3806(01)00286-3. [DOI] [PubMed] [Google Scholar]
- 159.Nakatomi H, et al. Regeneration of hippocampal pyramidal neurons after ischemic brain injury by recruitment of endogenous neural progenitors. Cell. 2002;110:429–441. doi: 10.1016/s0092-8674(02)00862-0. [DOI] [PubMed] [Google Scholar]
- 160.Ross JD, Cullen DK, Harris JP, Laplaca MC, Deweerth SP. A three-dimensional image processing program for accurate, rapid, and semi-automated segmentation of neuronal somata with dense neurite outgrowth. Front. Neuroanat. 2015;9:87. doi: 10.3389/fnana.2015.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 162.Alitalo K. The lymphatic vasculature in disease. Nat. Med. 2011;17:1371–1380. doi: 10.1038/nm.2545. [DOI] [PubMed] [Google Scholar]
- 163.Bajénoff M, et al. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 2006;25:989–1001. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hanoun M, Maryanovich M, Arnal-Estape A, Frenette PS. Neural regulation of hematopoiesis, inflammation, and cancer. Neuron. 2015;86:360–373. doi: 10.1016/j.neuron.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gibson EM, et al. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science. 2014;344:1252304. doi: 10.1126/science.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bartzokis G, et al. Multimodal magnetic resonance imaging assessment of white matter aging trajectories over the lifespan of healthy individuals. Biol. Psychiatry. 2012;72:1026–1034. doi: 10.1016/j.biopsych.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 167.Callaway E, et al. The discovery of Homo floresiensis: tales of the hobbit. Nature. 2014;514:422–426. doi: 10.1038/514422a. [DOI] [PubMed] [Google Scholar]
- 168.Taupin P, Gage FH. Adult neurogenesis and neural stem cells of the central nervous system in mammals. J. Neurosci. Res. 2002;69:745–749. doi: 10.1002/jnr.10378. [DOI] [PubMed] [Google Scholar]
- 169.Hsiao EY, Patterson PH. Placental regulation of maternal-fetal interactions and brain development. Dev. Neurobiol. 2012;72:1317–1326. doi: 10.1002/dneu.22045. [DOI] [PubMed] [Google Scholar]
- 170.Zoukos Y, Leonard JP, Thomaides T, Thompson AJ, Cuzner ML. Beta-adrenergic receptor density and function of peripheral blood mononuclear cells are increased in multiple sclerosis: a regulatory role for cortisol and interleukin-1. Ann. Neurol. 1992;31:657–662. doi: 10.1002/ana.410310614. [DOI] [PubMed] [Google Scholar]
- 171.Hsiao EY, Mcbride SW, Chow J, Mazmanian SK, Patterson PH. Modeling an autism risk factor in mice leads to permanent immune dysregulation. Proc. Natl. Acad. Sci. USA. 2012;109:12776–12781. doi: 10.1073/pnas.1202556109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hsiao EY, Patterson PH. Activation of the maternal immune system induces endocrine changes in the placenta via Il-6. Brain Behav. Immun. 2011;25:604–615. doi: 10.1016/j.bbi.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kaya F, et al. Live imaging of targeted cell ablation in Xenopus: a new model to study demyelination and repair. J. Neurosci. 2012;32:12885–12895. doi: 10.1523/JNEUROSCI.2252-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kim BJ, et al. Bacterial induction of Snail1 contributes to blood-brain barrier disruption. J. Clin. Invest. 2015;125:2473–2483. doi: 10.1172/JCI74159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lucas SM, Rothwell NJ, Gibson RM. The role of inflammation in CNS injury and disease. Br. J. Pharmacol. 2006;147:S232–S240. doi: 10.1038/sj.bjp.0706400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bianco P, Riminucci M, Gronthos S, Robey PG. Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells. 2001;19:180–192. doi: 10.1634/stemcells.19-3-180. [DOI] [PubMed] [Google Scholar]
- 178.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 180.Omatsu Y, Seike M, Sugiyama T, Kume T, Nagasawa T. Foxc1 is a critical regulator of haematopoietic stem/progenitor cell niche formation. Nature. 2014;508:536–540. doi: 10.1038/nature13071. [DOI] [PubMed] [Google Scholar]
- 181.Mendez-Ferrer S, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Greenbaum A, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Singh PK, et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 184.Ernst RK, et al. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- 185.Ramsey DM, Wozniak DJ. Understanding the control of pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 186.Tan SY, Chew SC, Tan SY, Givskov M, Yang L. Emerging frontiers in detection and control of bacterial biofilms. Curr. Opin. Biotechnol. 2014;26:1–6. doi: 10.1016/j.copbio.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 187.Basser PJ, Mattiello J, Lebihan D. MR diffusion tensor spectroscopy and imaging. Biophys. J. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Schain AJ, Hill RA, Grutzendler J. Label-free in vivo imaging of myelinated axons in health and disease with spectral confocal reflectance microscopy. Nat. Med. 2014;20:443–449. doi: 10.1038/nm.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Huppi PS, et al. Microstructural development of human newborn cerebral white matter assessed in vivo by diffusion tensor magnetic resonance imaging. Pediatr. Res. 1998;44:584–590. doi: 10.1203/00006450-199810000-00019. [DOI] [PubMed] [Google Scholar]
- 190.Alexander AL, Lee JE, Lazar M, Field AS. Diffusion tensor imaging of the brain. Neurotherapeutics. 2007;4:316–329. doi: 10.1016/j.nurt.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Seehaus A, et al. Histological validation of high-resolution DTI in human postmortem tissue. Front. Neuroanat. 2015;9:98. doi: 10.3389/fnana.2015.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ertürk A, Lafkas D, Chalouni C. Imaging cleared intact biological systems at a cellular level by 3DISCO. J. Vis. Exp. 2014 doi: 10.3791/51382. doi:10.3791/51382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Epp JR, et al. Optimization of CLARITY for clearing whole-brain and other intact organs. Eneuro. 2015;2 doi: 10.1523/ENEURO.0022-15.2015. doi:10.1523/Eneuro.0022-1515.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Li T, et al. Cell-penetrating Anti-GFAP VHH and corresponding fluorescent fusion protein VHH-GFP spontaneously cross the blood-brain barrier and specifically recognize astrocytes: application to brain imaging. FASEB J. 2012;26:3969–3979. doi: 10.1096/fj.11-201384. [DOI] [PubMed] [Google Scholar]
- 195.Perruchini C, et al. Llama VHH antibody fragments against GFAP: better diffusion in fixed tissues than classical monoclonal antibodies. Acta Neuropathol. 2009;118:685–695. doi: 10.1007/s00401-009-0572-6. [DOI] [PubMed] [Google Scholar]
- 196.Pifferi A, et al. Optical biopsy of bone tissue: a step toward the diagnosis of bone pathologies. J. Biomed. Opt. 2004;9:474–480. doi: 10.1117/1.1691029. [DOI] [PubMed] [Google Scholar]
- 197.Genina EA, Bashkatov AN, Tuchin VV. Optical clearing of cranial bone. Adv. Opt. Technol. 2008;2008 doi:10.1155/2008/267867. [Google Scholar]
- 198.Duong H, Han M. A multispectral led array for the reduction of background autofluorescence in brain tissue. J. Neurosci. Methods. 2013;220:46–54. doi: 10.1016/j.jneumeth.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kupferschmidt DA, Cody PA, Lovinger DM, Davis MI. Brain BLAQ: post hoc thick-section histochemistry for localizing optogenetic constructs in neurons and their distal terminals. Front. Neuroanat. 2015;9 doi: 10.3389/fnana.2015.00006. doi:10.3389/fnana.2015.0000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.