Abstract
Cytosolic DNA-mediated activation of the transcription factor IRF3 is a key event in host antiviral responses. Here, we show that infection of DNA viruses induced the interaction of the mTOR downstream effector S6K1 (S6 kinase 1) and the signaling adaptor STING in a cGAS (cGAMP synthase)-dependent manner. We further demonstrate that the kinase domain, but not the kinase function of S6K1, was required for the S6K1-STING interaction and that the TBK1 critically promotes this process. The formation of a tripartite S6K1-STING-TBK1 complex was necessary for IRF3 activation and disruption of this signaling axis impaired the early-phase expression of IRF3 target genes and the induction of T cell responses and mucosal antiviral immunity. Thus, our results have uncovered a fundamental regulatory mechanism for IRF3 activation in the cytosolic DNA pathway.
In eukaryotic cells, double-stranded DNA is normally sequestered in the nucleus or the mitochondria, thus being prevented from direct contact with the cytosol. As such, cytosolic exposure of DNA resulting from microbial infections or damaged host cells turns on an evolutionary conserved ‘danger’ signal to alarm the host immune system1, 2. In particular, infection with a number of DNA virus families results in introduction of DNA into the cytosol. For example, adenovirus (Ad), a nonenveloped, double-stranded DNA virus extensively used as vectors for both gene therapy and DNA vaccines3, exposes its genomic DNA to the cytosol during the infection process and has been used as a prototypical DNA virus to study host innate immune DNA recognition pathways4, 5.
Cytosolic DNA is a key pathogen-associated molecular pattern (PAMP) and is sensed mainly through a cytosolic cyclic GMP-AMP (cGAMP) synthase termed cGAS6, 7, which triggers innate antiviral and autoimmune responses1, 2. Upon binding to cytosolic DNA, cGAS catalyzes the production of a secondary messenger, the noncanonical 2′,3′ cGAMP (c[G(2′,5′)pA(3′,5′)p]), which is distinct from the canonical bacterial 3′,3′ cGAMP (c[G(3′,5′)pA(3′,5′)p])6, 7, 8, 9. The cGAS product binds to and activates the downstream adaptor STING10, 11, also known as MPYS, MITA, ERIS and TMEM173 12, 13, 14, 15. Activated STING in turn recruits and activates IκB kinase and TBK1, which leads to the activation of NF-κB and IRF3, respectively6, 7, 9. Despite these advances in the understanding of DNA sensing mechanisms, the interaction between distinct regulatory molecules involved in cytosolic DNA signaling machinery remains to be further defined.
The two homologous isoforms of ribosomal protein S6 kinase, S6K1 and S6K2, are effector serine-threonine kinases downstream of mTOR, a central member in the PI(3)K-related kinase family that functions as a key signaling integrator of a variety of cues such as nutrients, stress, cell growth, apoptosis, transcriptional as well as translational co-ordination16. Of importance, recent studies show that the mTOR-S6K signaling axis modulates Toll-like receptor (TLR) pathways in a rapamycin-sensitive manner17, 18, 19. However, it is currently unknown whether mTOR-S6K may be integrated to cytosolic DNA-mediated STING signalosome.
Here, using Ad as a model DNA virus, we show that S6Ks, especially S6K1, are required for Ad-induced IRF3 activation independently of S6K’s kinase activity. We demonstrate that S6K1 interacts physically with activated STING in a cGAS-cGAMP-dependent manner, forming a tripartite S6K1-STING-TBK1 signaling complex. This S6K-dependent IRF3 signaling is important for inducing early innate immune responses as well as robust T cell immunity and mucosal antiviral defense. Thus, our report reveals a previously unrecognized regulatory function of S6Ks in the context of cytosolic DNA-elicited immune signaling.
Results
S6Ks are required for Ad-induced IRF3 activation
Ad induces IRF3 phosphorylation in a variety of immune cells4, 5. To characterize Ad-induced IRF3 phosphorylation profile in mouse bone marrow-derived myeloid dendritic cells (BMDCs), we transduced wild-type BMDCs using a recombinant human Ad serotype 5 vector with E1-E3 deletions to prevent undesirable infectious replication. Immunoblot analysis showed that Ad evoked sustained IRF3 serine 396 (S396) phosphorylation over a time course of 48 hours in BMDCs (Supplementary Fig. 1a) and that phosphorylated IRF3 was distributed in both nuclear and cytoplasmic fractions (Supplementary Fig. 1b), suggesting that Ad-induced IRF3 phosphorylation is a continual process in BMDCs
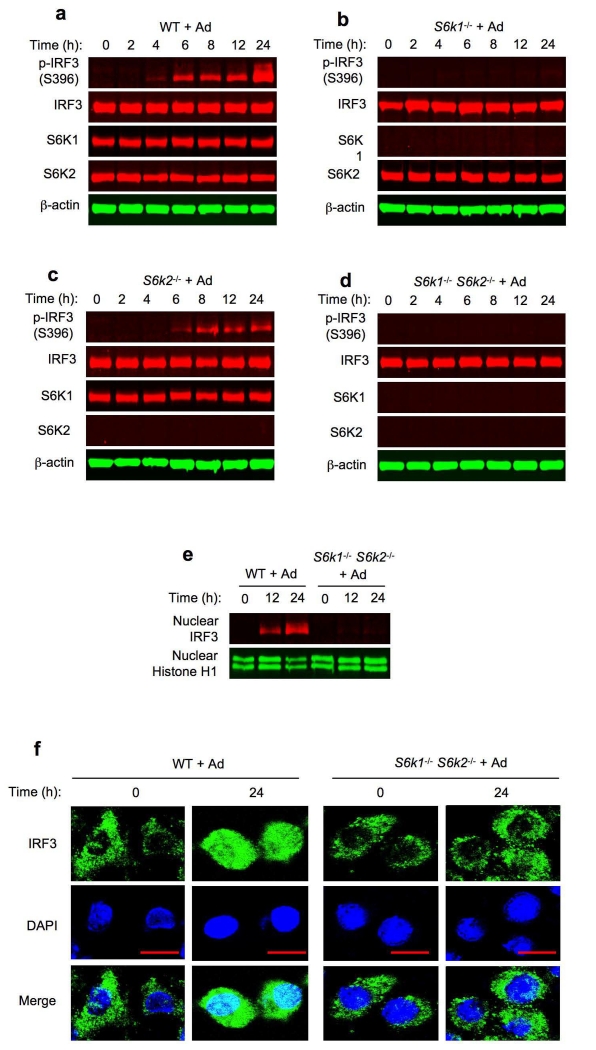
IRF3 signaling is pivotal in innate immune responses20. Because mTOR-S6Ks and mTOR-4E-BPs signaling modules, widely known as regulators for cell growth and metabolism, have been shown to shape host innate immunity17, 18, 19, 21, we asked whether S6Ks or 4E-BPs were involved in regulating cytosolic DNA-induced IRF3 phosphorylation by transducing Ad in S6k1−/−, S6k2−/−, S6k1−/− S6k2−/−, Eif4ebp1−/− Eif4ebp2−/− and Eif4ebp1−/− Eif4ebp2−/− Eif4ebp3−/− BMDCs. Compared to wild-type BMDCs (Fig. 1a), IRF3 phosphorylation was markedly reduced in S6k1−/− BMDCs (Fig. 1b), whereas the effect of S6K2 deletion was less severe (Fig. 1c). Significantly, IRF3 phosphorylation was profoundly impaired in S6k1−/− S6k2−/− BMDCs (Fig. 1d), while no such effect was observed in Eif4ebp1−/− Eif4ebp2−/− or Eif4ebp1−/− Eif4ebp2−/− Eif4ebp3−/− BMDCs (Supplementary Fig. 1c). Notably, IRF3 protein expression was not affected by S6K ablation (Fig. 1b-d), indicating that S6K regulation of Ad-induced IRF3 activation was not due to downregulation of IRF3 expression. Furthermore, immunoblot analysis showed that Ad promoted IRF3 nuclear translocation in wild-type but not S6k1−/− S6k2−/− BMDCs (Fig. 1e). Imunofluorescence microscopy confirmed that Ad-induced IRF3 nuclear translocation was significantly reduced in S6k1−/− S6k2−/− BMDCs (Fig. 1f). Together, these results indicate that S6Ks, particularly S6K1, are essential for Ad-triggered IRF3 activation. Additionally, we observed that herpes simplex virus-1 (HSV-1)-induced IRF3 phosphorylation was similarly reduced in S6k1−/− S6k2−/− BMDCs (Supplementary Fig. 2a).
Figure 1.
S6Ks are required for Ad-induced IRF3 activation. (a-d) Immunoblot analysis of p-IRF3(S396), IRF3, S6K1 and S6K2 in whole cell lysates of wild-type (a), S6k1−/− (b), S6k2−/− (c) and S6k1−/− S6k2−/− (d) BMDCs transduced with Ad. β-actin as loading control. (e) Immunoblot analysis of IRF3 in nuclear lysates of wild-type and S6k1−/− S6k2−/− BMDCs transduced with Ad. Histone H1 as a nuclear fraction marker. (f) Immunofluorescence microscopy of wild-type and S6k1−/− S6k2−/− BMDCs left untreated or transduced with Ad, immunostained for IRF3 (green) and the DNA-binding dye DAPI (blue). Bar represents 10 μM. Data are representative of four independent experiments (a-f).
In parallel experiments, we stimulated wild-type and S6k1−/− S6k2−/− BMDCs with TLR4 ligand lipopolysacchride (LPS) or vesicular stomatitis virus (VSV) known to activate RIG-I22. We observed no differences in IRF3 phosphorylation between wild-type and S6k1−/− S6k2−/− BMDCs after these treatments (Supplementary Fig. 2), suggesting that S6K is specifically required for viral DNA-induced IRF3 activation in the cytosolic DNA pathway. To extend this, we transfected purified Ad DNA or synthetic interferon stimulatory DNA (ISD) or non-canonical cGAMP, the mammalian cGAS cyclic dinucleotide8, 23. Transfection of Ad DNA (Supplementary Fig. 3a), ISD (Supplementary Fig. 3b) and cGAMP (Supplementary Fig. 3c) triggered robust IRF3 phosphorylation in wild-type but not S6k1−/− S6k2−/− BMDCs. Thus, S6K is essential for cytosolic DNA-induced IRF3 phosphorylation.
Next, we examined S6K-dependent induction of IFN-stimulated genes following ISD transfection in BMDCs. ISD stimulation triggered a robust induction of IFIT1, IFIT3 and ISG15 proteins in wild-type BMDCs (Supplementary Fig. 3d). In contrast, the 8 h induction of these IRF3 target genes was substantially impaired in both S6k1−/− S6k2−/− (Supplementary Fig. 3e) and Irf3−/− (Supplementary Fig. 3f) BMDCs. Together, these data suggest that S6K-IRF3 signaling is a crucial temporal regulator for the early phase induction of IRF3 target genes in the cytosolic DNA pathway.
IRF3 activation does not require S6K kinase function
S6Ks are known to regulate various cellular events through their kinase function16. To ascertain whether S6K kinase activity was critical in Ad-triggered IRF3 activation, we first characterized S6K1 phosphorylation on residues S371 and T38916, 24. Antibodies specific for phosphorylation at these sites indicated that S6K1 was phosphorylated in BMDCs even in the absence of Ad transduction (Fig. 2a). In addition, the prototypic S6K substrate rpS6 (ribosomal protein S6)16, 24 was also constitutively phosphorylated at residues S240 and S244 in unstimulated BMDCs (Fig. 2a), indicating that phosphorylated S6K1 signals to its downstream target rpS6 in BMDCs at steady-state.
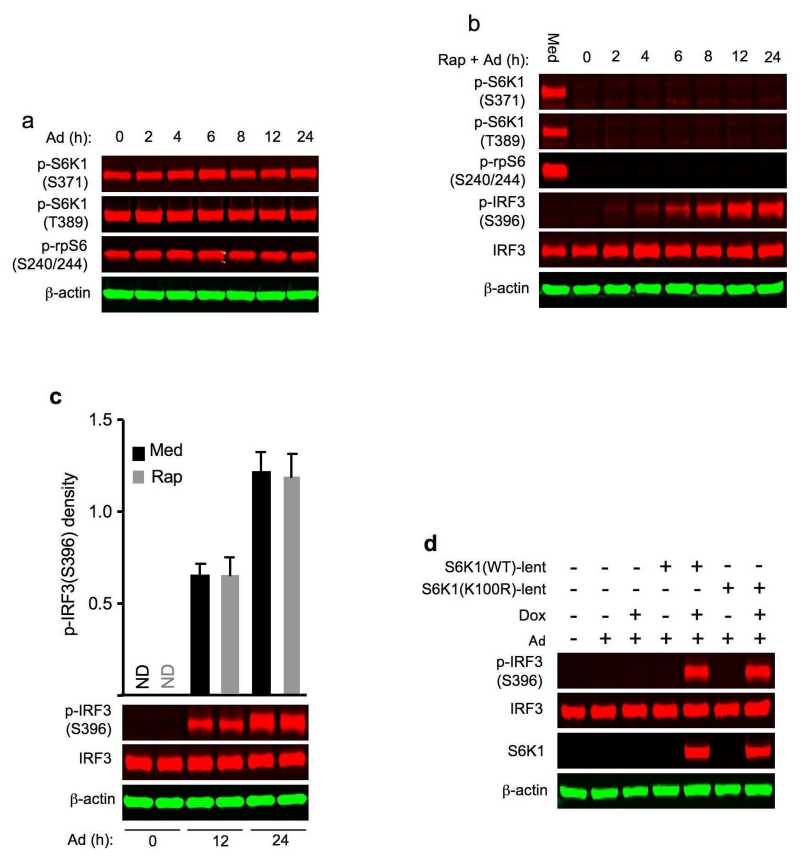
Figure 2.
S6K regulation of Ad-induced IRF3 activation is kinase function-independent. (a) Immunoblot analysis of p-S6K1(S371), p-S6K1(T389) and p-rpS6 (S240/244) in whole cell lysates of wild-type BMDCs transduced with Ad. β-actin as loading control. (b) Immunoblot analysis of p-S6K1(S371), p-S6K1(T389), p-rpS6 (S240/244), p-IRF3(S396) and IRF3 in whole cell lysates of wild-type BMDCs left untreated with medium (Med) or pre-treated with rapamycin (Rap) for 2 h and then transduced with Ad. β-actin as loading control. (c) Immunoblot analysis of p-IRF3(S396) and IRF3 in whole cell lysates of wild-type BMDCs left untreated with medium (Med) or rapamycin (Rap) for 2 h and then transduced with Ad. Upper, the relative densities of p-IRF3 to IRF3 in arbitrary units. Bottom, the representative immunoblot bands of p-IRF3(S396) and IRF3. ND: p-IRF3 not detected. β-actin as loading control. (d) Immunoblot analysis of p-IRF3(S396), IRF3 and S6K1 in whole cell lysates of S6k1−/− S6k2−/− BMDCs transduced with lentiviruss expressing either S6K1(WT) or S6K1(K100R) for 96 h, subsequently left un-induced or induced with doxycycline (Dox) for 24 h and then infected with Ad for 24 h. β-actin as loading control. Data are representative of three independent experiments (a-d).
Following Ad transduction, phosphorylation of S371 and T389 residues in S6K1 remained the same compared to non-transduced BMDCs (Fig. 2a). Pretreatment with the mTOR inhibitor rapamycin 16, 25 for two hours before Ad transduction abolished S6K1 and rpS6 phosphorylation in wild-type BMDCs (Fig. 2b). Despite the profound suppression of S6K1 phosphorylation, Ad-triggered IRF3 phosphorylation was not inhibited by rapamycin (Fig. 2b,c). Importantly, we observed that lentiviral reconstitution with a kinase-dead version of S6K1, hereafter S6K1(K100R), restored Ad-induced IRF3 phosphorylation in S6k1−/− S6k2−/− BMDCs (Fig. 2d). These observations indicate that S6K, but not its kinase activity, is required for Ad-triggered IRF3 activation.
STING-dependent TBK1 activation does not involve S6Ks
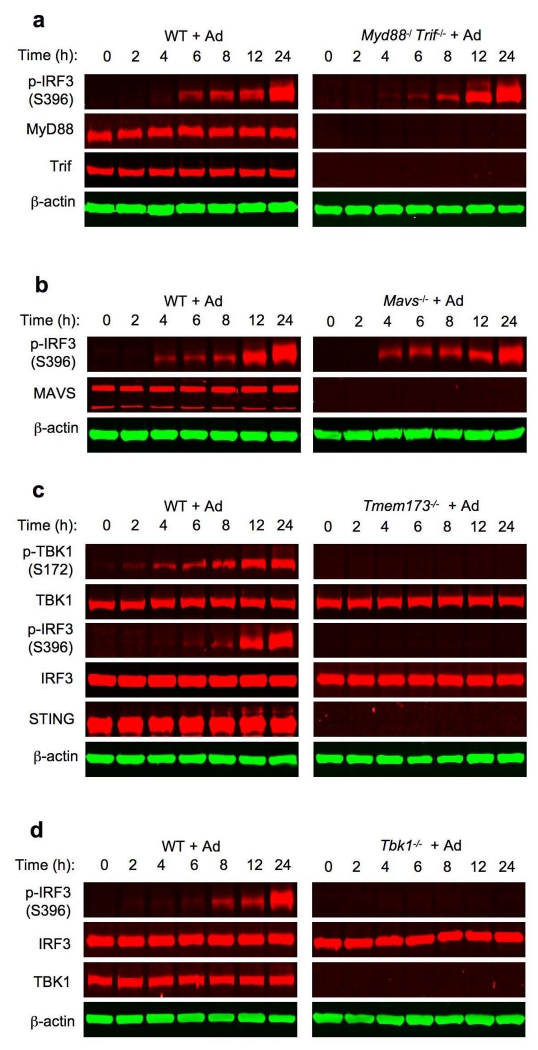
To define the mechanisms of S6K action in IRF3 signaling, we carried out Ad transduction of BMDCs in which key adaptors representing distinct signaling cascades were ablated genetically. We observed that Ad-triggered IRF3 phosphorylation was unimpaired in Myd88−/− Trif−/− BMDCs (Fig. 3a). Similarly, Ad-triggered IRF3 phosphorylation in Mavs−/− BMDCs was intact (Fig. 3b). In contrast, genetic ablation of STING, the adaptor representing the cytosolic DNA pathway12, 26, abolished Ad-induced IRF3 phosphorylation (Fig. 3c), confirming that the cytosolic DNA-mediated, STING-dependent pathway is responsible for Ad-induced IRF3 activation in BMDCs.
Figure 3.
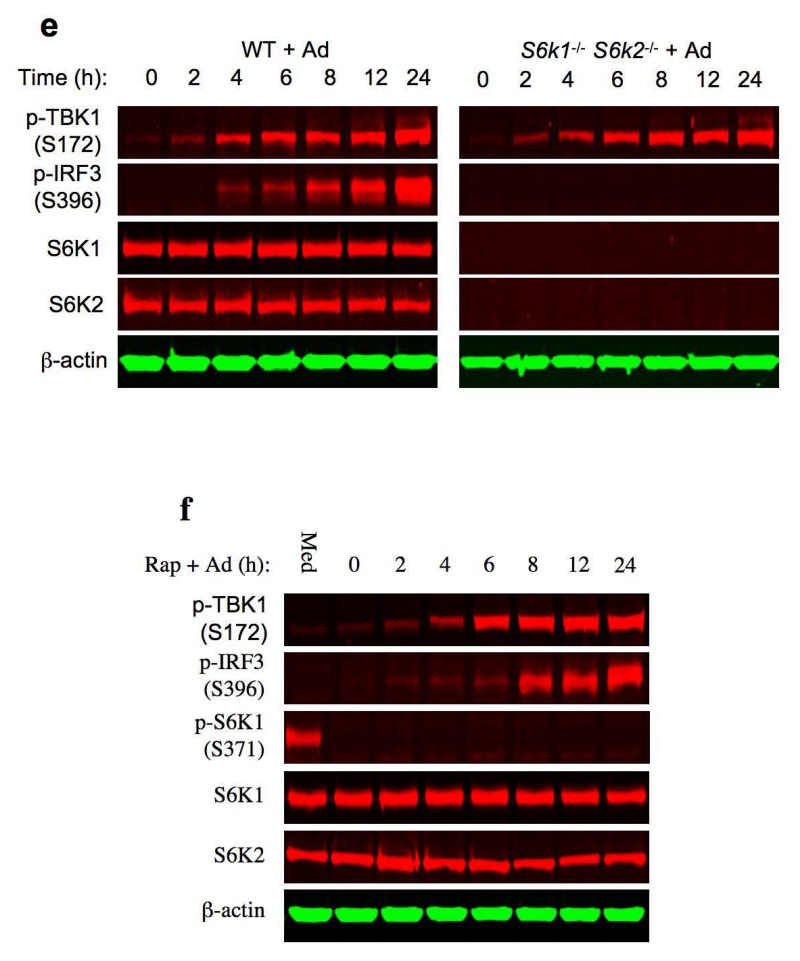
Ad-triggered, STING-dependent activation of TBK1 does not require S6Ks. (a) Immunoblot analysis of p-IRF3(S396), MyD88 and Trif in whole cell lysates of Myd88−/− Trif−/− BMDCs transduced with Ad. β-actin as loading control. (b) Immunoblot analysis of p-IRF3(S396) and MAVS in whole cell lysates of wild-type and Mavs−/− BMDCs transduced with Ad. β-actin as loading control. (c) Immunoblot analysis of p-TBK1(S172), TBK1, p-IRF3(S396), IRF3 and STING in whole cell lysates of wild-type and Tmem173−/− BMDCs transduced with Ad. β-actin as loading control. (d) Immunoblot analysis of p-IRF3(S396), IRF3 and TBK1 in whole cell lysates of wild-type and Tbk1−/− BMDCs transduced with Ad. β-actin as loading control. (e) Immunoblot analysis of p-TBK1(S172), p-IRF3(S396), S6K1 and S6K2 in whole cell lysates of wild-type and S6k1−/− S6k2−/− BMDCs transduced with Ad. β-actin as loading control. (f) Immunoblot analysis of p-TBK1(S172), p-IRF3(S396), p-S6K1(S371), S6K1 and S6K2 in whole cell lysates of wild-type and S6k1−/− S6k2−/− BMDCs transduced with Ad. β-actin as loading control. Data are representative of three independent experiments (a-f).
Cell’s sensing of virally presented cytosolic DNA causes STING to recruit and activate TBK1 6, 7, 9. Activated TBK1 in turn phosphorylates its downstream substrate IRF36, 7, 9. As such, we investigated S6K1’s contribution in the STING signalosome that links to TBK1 and IRF3. Ad induced robust TBK1 serine 172 (S172) phosphorylation, a hallmark for TBK1 activation in wild-type BMDCs (Fig. 3c). Notably, genetic deletion of STING abrogated Ad-induced TBK1 phosphorylation (Fig. 3c), which correlated with an absence of IRF3 phosphorylation in Tbk1−/− BMDC (Fig. 3d). These data confirm that STING functions upstream of TBK1 and that TBK1 is the key kinase responsible for Ad-triggered IRF3 phosphorylation in BMDCs. However, despite the profound suppression of IRF3 phosphorylation in S6k1−/− S6k2−/− BMDCs (Fig. 3e), Ad-triggered TBK1 phosphorylation was normal in the same cells (Fig. 3e). Consistently, we observed that rapamycin did not inhibit Ad-triggered TBK1 phosphorylation in BMDCs (Fig. 3f). Rapamycin only inhibited S6K kinase activity, but did not deplete S6K proteins in BMDCs (Fig. 3f). As expected, rapamycin did not suppress IRF3 phosphorylation in BMDCs (Fig. 3f). Together, these results indicate that unlike IRF3, STING-dependent TBK1 activation occurs independently of S6Ks, suggesting that S6Ks modify the ability of activated TBK1 to phosphorylate IRF3 in the context of an Ad-triggered and STING-dependent recognition signal in BMDCs.
S6Ks are required for IRF3 binding to STING
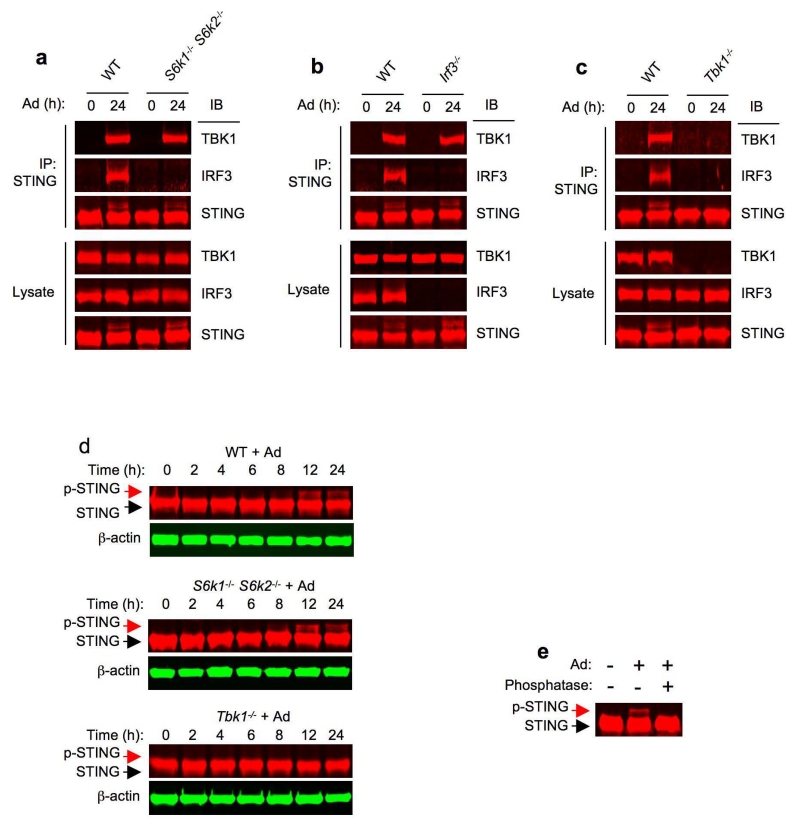
STING has a scaffold function and binds both TBK1 and IRF3 27. To test if S6K influences IRF3-TBK1 interaction through STING, we conducted co-immunoprecipitation studies and observed that endogenous STING bound to both TBK1 and IRF3 in Ad-transduced BMDCs (Fig. 4a). Although genetic deletion of S6Ks did not impair TBK1-STING interaction (Fig. 4a), IRF3 binding to STING was abrogated in S6k1−/− S6k2−/− BMDCs (Fig. 4a). These data suggest that S6Ks are required for IRF3 binding to the activated STING in Ad-triggered cytosolic DNA pathway.
Figure 4.
S6Ks are crucial for Ad-induced IRF3 binding to STING. (a-c) Immunoprecipitation and immunoblot analysis of the interaction between STING, TBK1 and IRF3 in wild-type vs S6k1−/− S6k2−/− (a), wild-type vs Irf3−/− (b), and wild-type vs Tbk1−/− BMDCs transduced with Ad. Lysates (below), immunoblot analysis of TBK1, IRF3 and STING. (d) Immunoblot analysis of STING in whole cell lysates of wild-type, S6k1−/− S6k2−/− and Tbk1−/− BMDCs transduced with Ad. β-actin as loading control. (e) Immunoblot analysis of STING in whole cell lysates of wild-type BMDCs transduced with Ad for 24 h, subsequently treated with (+) or without (−) calf intestine phosphatase. Data are representative of four independent experiments (a-e).
To extend these findings, we performed Ad transduction in Irf3−/− and Tbk1−/− BMDCs. STING immunoprecipitated TBK1 in Ad-transduced Irf3−/− BMDCs (Fig. 4b). In contrast, STING did not immunoprecipitate IRF3 in Ad-transduced Tbk1−/− BMDCs (Fig. 4c), indicating that Ad-triggered TBK1 binding to STING occurs independently of IRF3, which mechanistically explains the uncoupling of TBK1 and IRF3 activation (Fig. 3e and Fig. 4a). Phosphorylation of STING was previously shown to enhance IRF3 activation15, 27. Thus, we asked whether S6Ks would play a role in STING phosphorylation. In wild-type BMDCs, Ad transduction resulted in an apparent STING mobility shift in a time-dependent manner (Fig. 4d). Phosphatase treatment abrogated the higher molecular weight bands of STING after Ad transduction (Fig. 4e), indicating that the observed STING mobility shift was due to phosphorylation. Notably, Ad-triggered STING band shift was unimpaired in S6k1−/− S6k2−/− BMDCs (Fig. 4d). In contrast, Ad was unable to evoke this shift in Tbk1−/− BMDCs (Fig. 4d) demonstrating that Ad-triggered STING phosphorylation was TBK1-dependent. Remarkably, we observed that although VSV infection resulted in marked IRF3 phosphorylation, it did not cause any discernible STING mobility shift (Supplementary Fig. 4a). This is consistent with the finding that MAVS was required for VSV-induced IRF3 activation in BMDCs (Supplementary Fig. 4a), emphasizing that STING activation constitutes a hallmark for cytosolic DNA-mediated signaling12, 26. Collectively, these results suggest that S6K plays an important role in facilitating IRF3 binding to cytosolic DNA-activated STING.
S6K1 interacts with STING to promote IRF3 phosphorylation
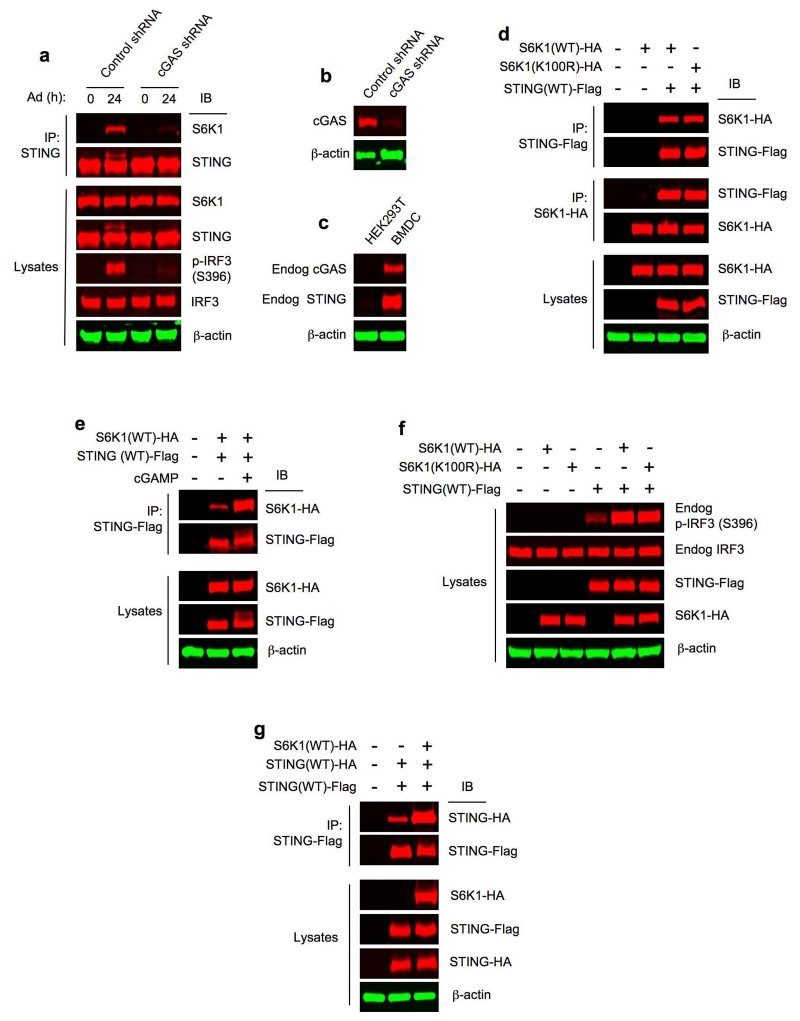
The requirement of S6Ks, particularly S6K1, for IRF3 binding to the activated STING suggests an interaction between S6K1 and STING in Ad-transduced BMDCs. To explore this possibility, we performed endogenous co-immunoprecipitation assays. While S6K1 and STING did not associate in mock-infected BMDCs, Ad induced endogenous S6K1 interaction with STING in BMDCs (Fig. 5a). Of significance, when the cytosolic DNA sensor cGAS was silenced with cGAS shRNA in BMDCs (Fig. 5a,b), endogenous S6K1-STING interaction was reduced (Fig. 5a) and IRF3 phosphorylation was inhibited (Fig. 5a). In contrast, LPS or VSV did not induce S6K1 and STING co-immunoprecipitation in BMDCs (Supplementary Fig. 4b) under the conditions when IRF3 was potently phosphorylated (Supplementary Fig. 4b). Collectively, these results indicate that S6K-STING interaction occurs only in the cytosolic DNA-cGAS-STING pathway to promote IRF3 phosphorylation.
Figure 5.
S6K1-STING interaction promotes IRF3 phosphorylation. (a) Immunoprecipitation and immunoblot analysis of S6K1-STING interaction in wild-type BMDCs treated with control shRNA or cGAS shRNA for 48 h, then infected with Ad. Lysates (below), immunoblot analysis of S6K1, STING, p-IRF3(S396), IRF3 and β-actin. (b) Immunoblot analysis of cGAS in whole cell lysates of wild-type BMDCs treated with control shRNA or cGAS shRNA for 48 h. β-actin as loading control. (c) Immunoblot analysis of endogenous (Endog) cGAS and STING in whole cell lysates of HEK293T cells and wild-type BMDCs. β-actin as loading control. (d) Immunoprecipitation and immunoblot analysis of the interaction between S6K1(WT)-HA or S6K1(K100R)-HA and STING(WT)-Flag in HEK293T cells transfected with respective plasmids for 24 h. Lysates (below), immunoblot analysis of S6K1-HA, STING-Flag and β-actin. (e) Immunoprecipitation and immunoblot analysis of the interaction between S6K1(WT)-HA and STING(WT)-Flag in HEK293T cells transfected with respective plasmids for 24 h, then transfected with 2′,3′ cGAMP for 6 h. Lysates (below), immunoblot analysis of S6K1-HA, STING-Flag and β-actin. (f) Immunoblot analysis of endogenous (Endog) p-IRF3(S396), endogenous IRF3, STING-Flag and S6K1-HA in whole cell lysates of HEK293T cells transfected with respective plasmids for 24 h. β-actin as loading control. (g) Immunoprecipitation and immunoblot analysis of the interaction between STING(WT)-HA and STING(WT)-Flag in HEK293T cells co-transfected with S6K1(WT)-HA plasmid for 24 h. Lysates (below), immunoblot analysis of S6K1-HA, STING-Flag and STING-HA. β-actin as loading control. Data are representative of three independent experiments (a-g).
Next, we transfected S6K1 and STING expression plasmids into HEK293T cells, human embryonic kidney cells widely used as transfection hosts, which lack endogenous cGAS and STING (Fig. 5c)6, 28. Co-transfection resulted in association between HA-tagged mouse S6K1 and Flag-tagged mouse STING (Fig. 5d). Notably, S6K1(K100R)-HA29 bound to STING-Flag in a similar manner compared to wild-type S6K1-HA (Fig. 5d), indicating that S6K1-STING interaction in mammalian cells does not require S6K1 kinase function. Next, we examined the influence of STING activation on S6K1-STING interaction using the STING activator cGAMP 8, 23 in HEK293T cells, which do not express endogenous cGAS (Fig. 5c) and do not produce endogenous cGAMP 6. Because ectopically expressed STING was reported to be auto-activated in a ligand-independent manner28, 30, we transfected HEK293T cells with a pre-titrated amount of STING-Flag plasmid together with S6K1-HA such that STING-Flag remained responsive and treated the co-transfected HEK293T cells with cGAMP. We observed that cGAMP increased S6K1-HA and STING-Flag interaction in HEK293T cells (Fig. 5e). To evaluate the physiological relevance of this interaction, we transfected HEK293T cells with a pre-tested amount of STING-Flag plasmid such that STING-Flag induced only low IFR3 phosphorylation (Fig. 5f). Co-transfection of wild-type S6K1-HA or S6K1(K100R)-HA similarly augmented STING-Flag-dependent phosphorylation of endogenous IRF3 in HEK293T cells (Fig. 5f).
Next, we explored whether S6K1 affected STING dimerization or oligomerization, as this is a key process for signal propagation in innate immune signaling14, 26, 27. In HEK293T cells co-transfected with STING-HA and STING-Flag, STING existed partially as a dimer (Fig. 5g). However, STING dimer fraction was increased substantially when S6K1-HA was co-transfected (Fig. 5g), suggesting that S6K1-STING interaction augments the formation or stabilization of STING dimers or oligomers.
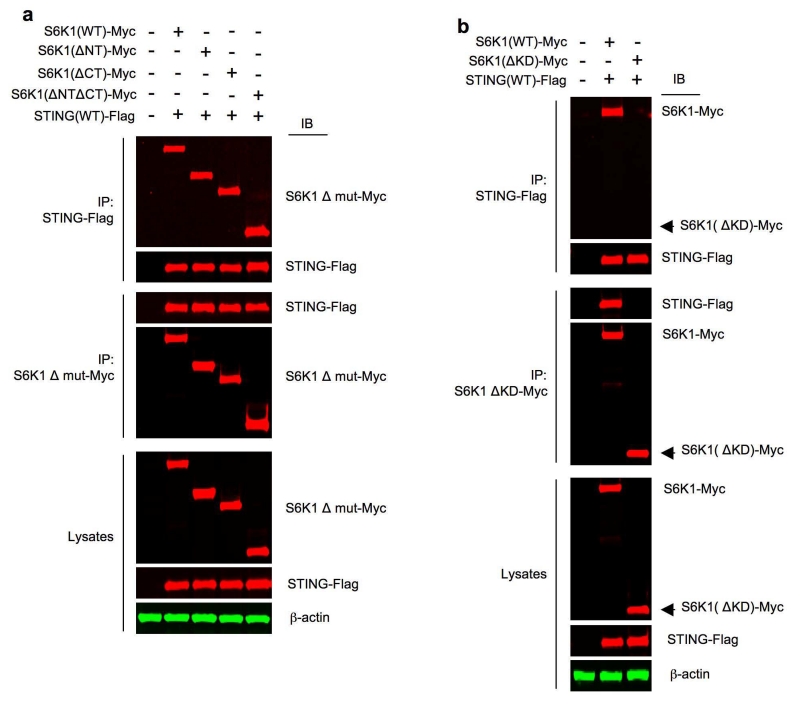
S6K1 N- and C-terminal regions are known to regulate S6K1 activity16, 31. To determine whether these regions are involved in S6K1-STING interaction, wild-type STING-Flag was transfected into HEK293T cells together with Myc-tagged wild-type S6K1 or various S6K1 truncation mutants. Truncation of N-terminus had no impact on S6K1-Myc and STING-Flag interaction (Fig. 6a). Notably, the deletion of C-terminus did not exhibit an inhibitory effect either (Fig. 6a). C-termini of signaling proteins are often involved in protein-protein interactions15, 27, 32. Thus, to further confirm the noninvolvement of S6K1 C-terminus in S6K1-STING interaction, we used a construct expressing only S6K1 C-terminus, S6K1(CT). S6K1(CT) was unable to associate with STING in HEK293T cells (Supplementary Fig. 5a), demonstrating that the regulatory domains of S6K1 required for canonical mTOR-S6K1 signaling are not required for S6K1 binding to STING. In contrast, deletion of the S6K1 kinase domain S6K1(ΔKD) abrogated S6K1-STING interaction in HEK293T cells (Fig. 6b).
Figure 6.
The kinase domain but neither the N-nor the C-terminus of S6K1 is involved in mediating S6K1 interaction with STING. (a) Immunoprecipitation and immunoblot analysis of the interaction between STING(WT)-Flag and S6K1 truncation mutants (Δ mut)-Myc in HEK293T cells transfected with respective plasmids for 24 h. Lysates (below), immunoblot analysis of S6K1 Δ mut-Myc, STING-Flag and β-actin as loading control. (b) Immunoprecipitation and immunoblot analysis of the interaction between STING(WT)-Flag, S6K1(WT)-Myc or S6K1(ΔKD)-Myc in HEK293T cells transfected with respective plasmids for 24 h. Lysates (below), immunoblot analysis of S6K1-Myc, S6K1(ΔKD)-Myc, STING-Flag and β-actin as loading control. Data are representative of three independent experiments (a-b).
Next, we determined which S6K1 domain(s) is involved in augmenting STING-induced IRF3 phosphorylation, as STING is the key structure for anchoring IRF3 to be phosphorylated by TBK127, 32. Following co-transfection of various Myc-tagged S6K1 mutants with STING-Flag in HEK293T cells, wild-type S6K1-Myc substantially enhanced STING-Flag-dependent IRF3 phosphorylation (Supplementary Fig. 5b). Notably, this S6K1 enhancement of IRF3 phosphorylation remained unchanged when either S6K1(ΔNT) or S6K1(ΔCT) or S6K1(ΔNTΔCT) was co-transfected with STING(WT)-Flag (Supplementary Fig. 5b). In contrast, S6K1(ΔKD) was unable to promote the STING-dependent IRF3 phosphorylation (Supplementary Fig.5b). Collectively, these data show that S6K is a new STING binding partner to promote IRF3 phosphorylation.
S6K1 forms a tripartite complex with STING and TBK1
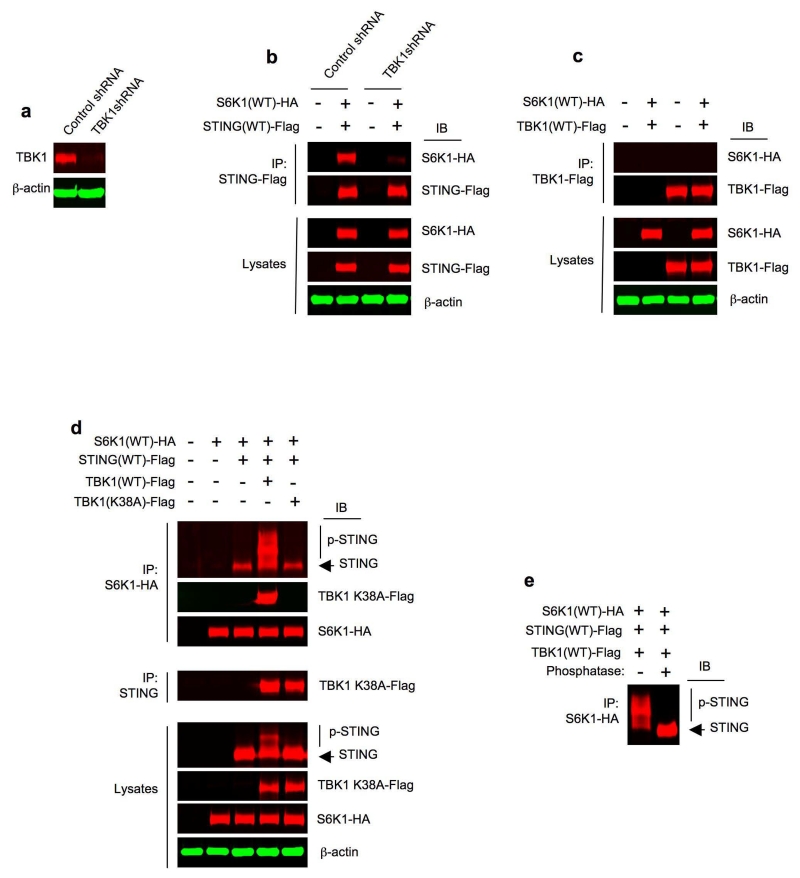
To further investigate how TBK1 modulated S6K1-STING interaction, we silenced TBK1 in HEK293T cells with TBK1 shRNA (Fig. 7a). TBK1 silencing markedly inhibited S6K1-HA and STING-Flag interaction (Fig. 7b), indicating that TBK1 plays a pivotal role in S6K1-STING association. To address whether S6K1 and TBK1 interact directly, we took advantage of the lack of endogenous STING in HEK293T cells6 and co-transfected S6K1-HA and TBK1-Flag into these cells. We did not observe an appreciable direct interaction between S6K1-HA and TBK1-Flag in the absence of endogenous STING (Fig. 7c), suggesting that TBK1 might modify STING activity by unveiling a docking site on STING for S6K1.
Figure 7.
S6K1 interacts with STING and TBK1 to form a tripartite complex. (a) Immunoblot analysis of TBK1 in control shRNA or TBK1 shRNA HEK293T cells. β-actin as loading control. (b) Immunoprecipitation and immunoblot analysis of the interaction between S6K1(WT)-HA and STING(WT)-Flag in control shRNA or TBK1 shRNA HEK293T cells. Lysates (below), immunoblot analysis of S6K1-HA, STING-Flag and β-actin as loading control. (c) Immunoprecipitation and immunoblot analysis of the interaction between S6K1(WT)-HA and TBK1(WT)-Flag in HEK293T cells transfected with respective plasmids. Lysates (below), immunoblot analysis of S6K1-HA, TBK1-Flag and β-actin as loading control. (d) Immunoprecipitation and immunoblot analysis of the interaction between S6K1(WT)-HA, STING(WT)-Flag, TBK1(WT)-Flag or TBK1(K38A)-Flag in HEK293T cells transfected with respective plasmids for 24 h. Lysates (below), immunoblot analysis of STING, S6K1-HA, TBK1-Flag and β-actin as loading control. (e) Immunoblot analysis of STING in the immunoprecipitates pulled down by anti-S6K1-HA from the whole cell lysates of HEK293T cells triply transfected with S6K1(WT)-HA, STING(WT)-Flag and TBK1(WT)-Flag for 24 h, treated with (+) or without (−) calf intestine phosphatase. Data are representative of four independent experiments (a-e).
To test this hypothesis, we co-transfected wild-type S6K1-HA and wild-type STING-Flag into HEK293T cells with or without wild-type TBK1-Flag or a kinase inactive mutant TBK1(K38A)-Flag33. Wild-type TBK1-Flag and TBK1(K38A)-Flag were expressed at similar levels (Fig. 7d). Notably, the co-transfected wild-type TBK1-Flag, but not TBK1(K38A)-Flag, resulted in higher molecular weight STING band shifts (Fig. 7d), which disappeared after phosphatase treatment (data not shown), indicating a TBK1-dependent STING phosphorylation. Importantly, when cells were co-transfected with wild-type TBK1-Flag, the STING-Flag precipitated by S6K1-HA appeared largely phosphorylated, as differentiated by its slower mobility (Fig. 7d) and phosphatase treatment (Fig. 7e). In contrast, no mobility shifted STING was precipitated by S6K1-HA in the presence of TBK1(K38A)-Flag (Fig. 7d). The faster migrating band of STING pulled down by S6K1-HA in the presence of TBK1(K38A)-Flag (Fig. 7d) can be attributed to the endogenous TBK1, as that band further diminished when the triple transfection of S6K1-HA, STING-Flag and TBK1(K38A)-Flag was conducted in TBK1 knockdown cells (data not shown).
Of importance, S6K1-HA also precipitated wild-type TBK1-Flag but not TBK1(K38A)-Flag (Fig. 7d) even though both bound to STING (Fig. 7d), indicating that TBK1 binding to STING alone may not be sufficient for promoting an interaction event between S6K1 and STING and suggesting that TBK1-mediated STING phosphorylation plays an important role in facilitating S6K1-STING interaction. To further confirm this, we found that isolated recombinant TBK1-Flag but not S6K1-HA caused phosphorylation of isolated STING-Flag in the in vitro kinase assay as demonstrated by slower mobility (Supplementary Fig. 6). Isolated S6K1-HA pulled down both isolated STING-Flag and TBK1-Flag following the kinase reaction (Supplementary Fig. 6), Collectively, these results further demonstrate that S6K1 can interact directly with phosphorylated STING and form a S6K1-STING-TBK1 tripartite complex in mammalian cells.
S6K-IRF3 signaling is required for in vivo antiviral immunity
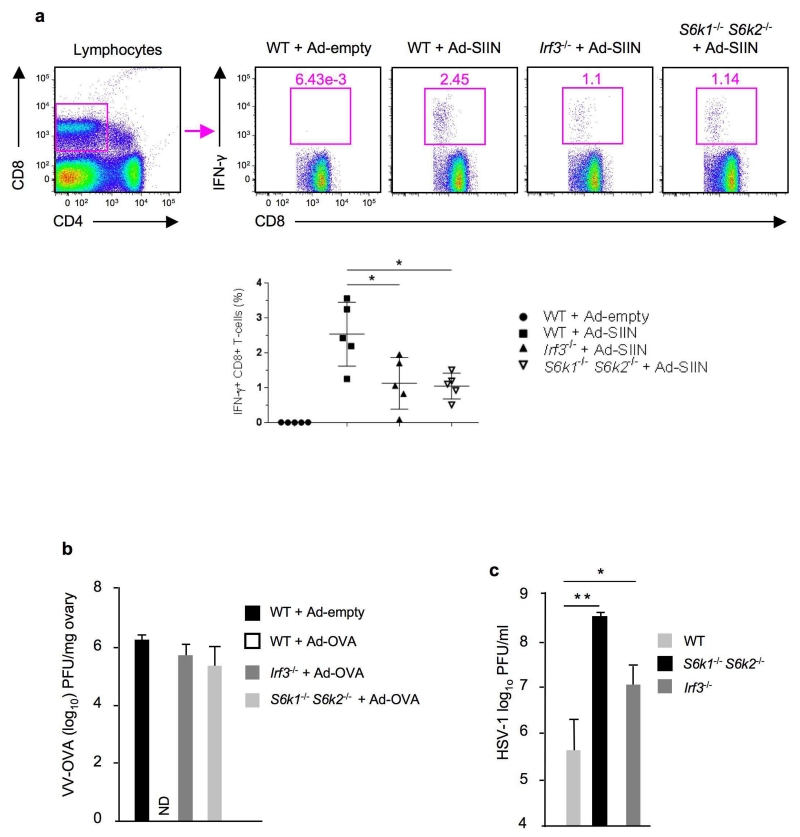
To test S6K’s physiological significance in Ad-induced IRF3 signaling in MBDCs, we used an Ad-transduced DC-based vaccination as an in vivo system. We transduced wild-type, S6k1−/− S6k2−/− and Irf3−/− BMDCs with either an empty Ad vector or Ad vector encoding chicken ovalbumin peptide SIINFEKL (Ad-OVA) as a model foreign antigen and injected these transduced BMDCs into the footpads of wild-type recipient mice. We observed that transfer of Ad-OVA transduced S6k1−/− S6k2−/− or Irf3−/− BMDCs resulted in significantly impaired OVA-specific CD8+ T cell responses compared to those inoculated with similarly transduced wild-type BMDCs (Fig. 8a). Subsequent challenge of the DC-vaccinated mice with a single i.p. injection of recombinant vaccinia virus expressing OVA (VV-OVA) showed that the mice vaccinated with Ad-OVA-transduced S6k1−/− S6k2−/− or Irf3−/− BMDCs failed to control vaccinia virus infection (Fig. 8b).
Figure 8.
Disruption of DNA virus-induced S6K-IRF3 signaling in donor’s BMDCs impairs the induction of the recipient host’s T cell responses. (a) Flow cytometry analysis of OVA-specific IFNγ+ CD8+ T-cells in wild-type recipient mice vaccinated with Ad-empty-transduced wild-type BMDCs or Ad-OVA-transduced wild-type, S6k1−/− S6k2−/− or Irf3−/− BMDCs. Wild-type vs knockout BMDCs significant, * P <0.001, one way ANOVA (mean ± s.d.). (b) Plaque assay of VV-OVA in ovaries of the mice vaccinated with Ad-empty-transduced wild-type BMDCs or Ad-OVA-transduced wild-type, S6k1−/− S6k2−/− or Irf3−/− BMDCs (n=5 per group). ND: VV-OVA not detected. (c) Plaque assay of HSV-1 in the vaginal washes of wild-type mice (n=8), S6k1−/− S6k2−/− mice (n=3) and Irf3−/− mice (n=5) infected intravaginally with HSV-1 for 48 h. * P <0.05 and ** P <0.01 for wild-type vs Irf3−/− mice and S6k1−/− S6k2−/− mice, respectively, by one-way ANOVA (mean ± s.d.). Data are representative of three independent experiments (a-b) and two independent experiments (c).
S6Ks are known for their role in mRNA translation 34. Importantly, S6k1−/− S6k2−/− BMDCs showed similar expression of Ad transgene-encoded antigens as wild-type BMDCs (Supplementary Fig. 7a,b), indicating that S6k1−/− S6k2−/− BMDCs were not impaired in their ability to induce T cell responses due to global defects in protein expression. Similarly, S6k1−/− S6k2−/− BMDCs exhibited comparable levels of SIINFEKL peptide presentation (SIIN-H-2Kb) as Irf3−/− and wild-type BMDCs (Supplementary Fig. 7c). In addition, S6k1−/− S6k2−/− BMDCs had a comparable ability to migrate to draining lymph nodes (Supplementary Fig. 7d,e), where they displayed a similar surface expression of CD11c (data not shown) compared to Irf3−/− and wild-type BMDCs. Collectively, these data indicate that S6K-IRF3 signaling in the donor BMDCs is important for DNA vector-transduced DCs to invoke optimal antigen-specific T cell responses in the recipient hosts.
Furthermore, the innate antiviral response to HSV-1 is STING-dependent12. This virus can cause productive infection in mice via intravaginal route12. We observed that HSV-1 replication was markedly increased in the vaginal tracts of both S6k1−/− S6k2−/− and Irf3−/− mice compared to wild-type mice (Fig. 8c), indicating that HSV-1 infection in the vaginal mucosa also is restricted by S6K-dependent IRF3 signaling. Together, these findings suggest that S6K-IRF3 signaling is an important defense component in the host’s immune network.
Discussion
S6Ks are known to play extensive roles in important cellular processes such as mitochondrial metabolism, cell-cycle progress and ribosome biogenesis16. In this study, we show that S6Ks are required for DNA virus-triggered IRF3 activation in BMDCs. We provide evidence that S6K1 associates physically with the activated STING and forms a tripartite S6K1-STING-TBK1 signaling complex. We further demonstrate that S6K-mediated IRF3 signaling contributes to the early phase expression of IRF3 target genes and the induction of robust T cell responses and mucosal antiviral defense. Thus, our report reveals a crucial function of S6Ks in regulating the cytosolic DNA-cGAS-STING pathway to activate IRF3.
S6Ks are serine-threonine kinases, whose function is subject to rapamycin inhibition16. Previous reports have shown that rapamycin treatment substantially suppressed the induction of IFN and proinflammatory cytokines in a variety of cell types including plasmacytoid DCs, glioma cells and keratinocytes17, 18, 19, thus suggesting that the kinase activity of S6Ks is required for regulating these signaling processes. Here, we found that S6K-dependent activation of IRF3 was resistant to rapamycin inhibition in BMDCs following Ad transduction. In addition, the kinase-dead version of S6K1 restored Ad-induced IRF3 phosphorylation in S6k1−/− S6k2−/− BMDCs at levels similar to wild-type S6K1. Thus, the kinase-independent role of S6Ks in IRF3 activation provides an example of a context-specific switch for the regulatory roles of S6Ks. Of significance, we have not observed any difference in the expression of the related endogenous signaling proteins STING, TBK1 or IRF3 between wild-type and S6k1−/− S6k2−/− BMDCs. Moreover, we have confirmed that DNA virus-mediated transgene products or their antigen-presentation was not inhibited in S6k1−/− S6k2−/− BMDCs. These observations support the notion that S6K protein, but not its kinase function, may directly modulate innate antiviral immunity as a result of being an IRF3 signaling regulator.
IRF3 activation is a key effector event, upon which diverse regulatory signals converge in a context-specific way. On the basis of our molecular interaction data, we suggest the following model for S6K1-promoted IRF3 activation in the cytosolic DNA-cGAS-STING pathway: upon activation by cGAS-cGAMP, STING recruits TBK1 to form a STING-TBK1 signaling complex. This event occurs independently of S6Ks and IRF3. Subsequently, S6K1 interacts with phosphorylated STING to form a tripartite S6K1-STING-TBK1 signaling complex. Because the kinase-inactive mutant S6K1(K100R) interacted with STING similarly compared to wild-type S6K1, our working model further suggests that, independently of its kinase function, S6K1 acts as an “auxiliary adaptor” or “scaffold” in promoting IRF3 to STING-TBK1 complex to facilitate TBK1-mediated IRF3 phosphorylation.
STING phosphorylation by TBK1 was shown to have an essential role in recruiting IRF3 32. Here, we show that TBK1-mediated phosphorylation of STING is crucial for promoting a tri-molecular S6K1-STING-TBK1 interaction, particularly in the intact cells, which is required for propagating IRF3 signal transduction. Thus, one may hypothesize that the phosphorylation site(s) on STING targeted by TBK1 that is used for IRF3 recruitment may also be involved in S6K1-STING interaction. This hypothesis awaits substantiation in future studies.
Upon ligand activation in the cytosol, STING triggers both NF-κB and IRF3 signaling cascades6, 7, 9, 23. Ad transduction promoted both IRF3 and NF-κB phosphorylation in BMDCs. However, in contrast to IRF3 phosphorylation, Ad-elicited NF-κB phosphorylation was not S6K-dependent in BMDCs (data not shown). This observation demonstrates the specificity of S6Ks as cellular signaling molecules.
DNA virus-transduced DC-based vaccines hold promises for immunotherapy of cancer as well as certain viral diseases35. We observed that the Ad-transduced donor BMDCs required S6K-dependent IRF3 signaling to achieve optimal T cell responses in the recipient hosts. This finding not only demonstrates the utility of understanding the innate immune signaling pathways of virally transduced DCs, but also provides a rationale for exploring manipulation of virally transduced DCs ex vivo for the desired outcomes of DC-based vaccines in vivo. Importantly, the response to HSV-1, another DNA virus that induced a STING-dependent antiviral response 12 also depends on S6K-dependent IRF3 signaling. In the absence of S6Ks, this virus exhibited increased replication in the mucosa.
IRF3 is a key transcription factor that mediates an array of pivotal immune processes20. Congruently, a sophisticated regulatory mechanism for IRF3 activation might be evolutionarily advantageous as it offers the host possible context-specific regulation so that a given response can be timely initiated, finely tuned and properly terminated. Our discovery of S6Ks as crucial IRF3 regulators provides new insight into the IRF3 signaling network, particularly in its linkage to the STING-TBK1 signaling complex, a key signalosome in cellular innate immune responses.
ONLINE METHODS
Mice
Wild-type C57BL/6 and Goldenticket STING (Tmem173)-deficient mice were obtained from Charles River Laboratories and The Jackson Laboratory, respectively. Irf3−/− mice and Myd88−/− Trif−/− mice were bred in a specific pathogen-free environment within the animal facility at McMaster University. All other genotypic mice used for generating bone marrows were as follows: S6k1−/−, S6k2−/−, S6k1−/− S6k2−/−, Eif4ebp1−/− Eif4ebp2−/− and Eif4ebp1−/− Eif4ebp2−/− Eif4ebp3−/− mice and control wild-type mice17, 36, 37, 38: McGill University; Irf3−/−, Myd88−/−, Mavs−/− and control wild-type mice39: Washington University School of Medicine in St. Louis; Mavs−/− and control wild-type mice: University of Washington in Seattle; Tnfr1−/− Tbk1−/− and Tnfr1−/− (control for Tbk1−/−) mice40: University of Massachusetts Medical School. All animal experiments were approved by the Institutional Animal Research Ethics Board of McMaster University and concurred with the guidelines established by the Canadian Council on Animal Care. No specific exclusion criteria were employed in mouse experiments nor randomization of the allocation of mice to experimental groups was conducted.
BMDCs preparation
BMDCs were prepared as previously described41. Briefly, bone marrows were flushed from mouse femurs and tibiae and the dissociated bone marrow cells were resuspended in complete RPMI 1640 in the presence of 55 μM of β-mercaptoethanol (Gibco) and recombinant murine GM-CSF (40 ng/ml; PeproTech). On days 3 and 6, the cultures were provided with new media and fresh GM-CSF. From day 6 to day 8, BMDCs were harvested for various experimental purposes as specified.
Viruses, viral transduction, LPS stimulation and reagents
Recombinant human serotype 5 Ad with E1/E3-deletion (Ad-BHGdE1E3) harboring no transgene, Ad-OVA expressing chicken ovalbumin peptide SIINFEKL fused to luciferase, and VSV with an M protein mutant (VSV-ΔM51) were described42. Ad and Ad-OVA were used at an MOI of 100 whereas VSV was used at an MOI of 25 for the various times as indicated. These MOIs were chosen because they were shown to be effective in transducing DCs for use in DC-based vaccination42. HSV-1 (strain KOS) was used at an MOI of 5 for BMDC infection for various time points as specified. Intravaginal infection and titration of infectious HSV-1 progenies were conducted as previously described43. Briefly, mice were anaesthetized by injectable anaesthetic given intraperitoneally and inoculated intravaginally with 10 μl containing 104 PFU of wild-type HSV-1. Vaginal washes were collected 48 h after infection and subjected to standard plaque assays using Vero cells. LPS (E. coli O111:B4, Sigma) was used at 100 ng/ml for various time points as specified. Rapamycin was from Calbiochem. Calf intestinal phosphatase (CIP) was from New England Biolabs. Noncanonical 2′,3′ cGAMP (c[G(2′,5′)pA(3′,5′)p]) was purchased from BioLog.
Expression constructs and transfection
HA or Flag-tagged STING was amplified by PCR and cloned into pcDNA3.1 plasmid for expression in mammalian cells. Mammalian expression plasmids encoding HA-tagged wild-type S6K1, kinase-inactive mutant S6K1(K100R) and Myc-tagged wild-type S6K1 and S6K1 truncated versions, as well as expression plasmids for Flag-tagged IRF3, wild-type TBK1 and kinase-inactive mutant TBK1(K38A) were described previously29, 31, 44, 45. The expression plasmid for S6K1 with kinase domain deletion, S6K1(ΔKD)-Myc, was made in GenScript. To construct S6K1 C-terminus fragment (411-512), the region containing the N-terminus, the kinase domain and the linker domain was removed using deletion site directed mutagenesis on pRK5-S6K1-Myc 45. HEK293T cells were cultured in DMEM supplemented with 10% (v/v) FBS and were transfected using XtremeGENE HP DNA transfection reagent (Roche) according to the manufacturer’s instructions for various times as specified.
Lentiviruses
Lentiviral vectors for shRNA silencing of TBK1, cGAS and a scrambled shRNA sequence were obtained from Sigma. The Sigma MISSION shRNA vectors accession numbers were: human TBK1 (TRCN0000314840), mouse cGAS (TRCN0000178625) and the non-targeting scrambled shRNA control (SHC002). Each shRNA vector was co-transfected into HEK293T cells with the lentivirus packaging plasmids PLP1, PLP2, and PLP-VSVG (Invitrogen) using Lipofectamine 2000 (Invitrogen). Viral supernatants were collected 48 and 72 hours post-transfection and were filtered through a 0.45 μm nitrocellulose filter before use. For silencing of TBK1 gene expression in HEK293T cells, TBK1 shRNA lentiviral particles were incubated with the cells for 24 h before selection with puromycin and the drug-resistant cells were used for various experiments as indicated. For cGAS silencing, BMDCs collected on day 6 were transduced with cGAS lentiviral particles and were used 48 h later for various experiments as specified. Silencing efficiency was verified by Western blotting.
For lentiviral-mediated transgene expression, wild-type S6K1 and the kinase-inactive version S6K1(K100R) were amplified by PCR and cloned into the lentiviral vector pCW57.1 (Addgene) using NheI and BamHI. Lentiviruses were produced using the PLP1, PLP2 and PLP-VSVG packaging vectors on HEK293T cells and concentrated on sucrose cushion. For the reconstitution experiments, S6k1−/− S6k2−/− BMDCs were transduced on day 3 with lentivirus expressing either wild-type S6K1 or S6K1(K100R). The expressions of the transgenes were induced on day 7 with doxycycline (1 μg/ml). On day 8, the lentivirus-transduced S6k1−/− S6k2−/− BMDCs were used for Ad infection for 24 h.
In vivo immunization, T cell preparation and intracellular staining for flow cytometry and VV-OVA challenge
Following transduction with either Ad-empty or Ad-OVA, the transduced wild-type, Irf3−/− or S6k1−/− S6k2−/− BMDCs were injected subcutaneously into the footpads of wild-type recipient mice at 0.5×106 cells per footpad. T cell response analysis was conducted as previously described46. Briefly, 14 days after immunization, T cells were prepared and incubated with SIINFEKL peptide (OVA257-264, Biomer Technologies) at 1μg/ml for specific T cell restimulation. Incubation was performed for 5 h, in the presence of brefeldin A (GolgiPlug, BD Pharmingen) at 1 μg/ml for the last 4 h. Then, PBMCs were incubated with antibodies against CD16/CD32 (clone 2.4G2, dilution 1:200, BD Biosciences 553142, 1DB-001-0000807496) to block Fc receptors, before T cell surface staining with anti-CD3-APC-Cy7 (clone 145-2C11, dilution 1:100, BD Biosciences 557596, 1DB-001-0000869695), anti-CD8-PE (clone 53-6.7, dilution 1:400, BD Biosciences 553033, 1DB-001-0000868920) and anti-CD4-PerCP-Cy5.5 (clone RM4-5, dilution 1:800, eBioscience 45-0042-82, 1DB-001-0000884856). Cells were permeabilized and fixed with Cytofix/Cytoperm (BD Pharmagen) before intracellular cytokine staining with anti-IFN-γ-APC (clone XMG1.2, dilution 1:100, BD Biosciences 554413, 1DB-001-0000874263) and anti-TNF-α-FITC (clone MP6-XT22, dilution 1:300, BD Biosciences 554418, 1DB-001-0000871919). All these antibodies were previously cited. Data were acquired using a FACSCanto flow cytometer with FACSDiva software (BD Pharmingen) and analyzed with FlowJo Mac software (Treestar, Ashland, OR).
On day 26 after immunization with Ad-empty or Ad-OVA-transduced BMDCs, the recipient mice were challenged by i.p. injection of 107 pfu of recombinant vaccinia virus encoding an endoplasmic reticulum-targeted SIINFEKL epitope (VV-OVA). Seven days later, VV-OVA titers were determined by plaque assay in the ovaries of the challenged mice47.
Flow cytometry analysis of BMDCs and lymph node cells
CFSE (5-(and -6)-carboxyfluorescein diacetate succinimidyl ester) was obtained from Sigma. BMDC labeling with CFSE was conducted as previously described48. The popliteal lymph nodes were collected 48 h after mock or the CFSE-labeled BMDCs injection at the footpad. Stained BMDCs and lymph node cell suspensions were prepared for cytometry analysis using the flow cytometer and software as stated above. The monoclonal antibody 25-D1.16 was obtained from eBioscience (#17-5743-80) and CD11c antibody was purchased from BD Biosciences (PE-Cy7 CD11c, #558079).
Immunofluorescence microscopy
For immunofluorescence staining, BMDCs were grown on coverslips. After Ad transduction for 24 h, the transduced cells were fixed in cold methanol for 10 min, washed in PBS and blocked in 5% normal goat serum for 45 min. A rabbit anti-mouse IRF3 antibody (#4302, Cell Signaling Technology) was then applied at room temperature for 1 h. After washing in PBS, the cells were incubated with fluorescence-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories) for 45 min. Thereafter, the nuclear DNA of the immunolabeled cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride, Molecular Probes) according to the product specification. The fluorescence staining was examined with an LSM 510 META inverted confocal microscope (63 oil objective; Carl Zeiss).
Immunoblot and immunoprecipitation
For immunoblot (IB) analysis, the treated cells were lysed in radioimmunoprecipitation buffer (RIPA) (10 mM phosphate pH 7.4, 137 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS) supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail II and III (Sigma). The preparation of cytoplasmic and nuclear extracts was conducted as previously described49. Total or fractionated cellular extracts were resolved on 7.5-12% SDS-PAGE and transferred to nitrocellulose membrane (Santa Cruz). Blots were blocked in Odyssey blocking buffer (Li-cor) unless otherwise specified and detection was performed with respective primary antibodies, and bands were visualized with infrared dye-conjugated secondary antibodies using Odyssey scanner (Li-cor). For immunoprecipitations (IP), the treated cells were lysed in IP buffer (20 mM Tris-HCl pH 7.4, 137 mM NaCl, 1 mM EDTA, 1% NP-40, 10% glycerol and protease/phosphatase inhibitor cocktail from ThermoFisher Scientific, #78441). The cellular extracts were incubated with respective IP antibodies or IP antibodies conjugated to Sepharose beads at 4°C for 1-3 h or overnight as specified. The immuoprecipitates were washed thoroughly and boiled in SDS sample buffer for Western blot analysis.
In vitro kinase assay for S6K1-STING-TBK1 complex formation
To obtain the required recombinant proteins, single transfections of S6K1-HA, STING-Flag and TBK1-Flag were conducted with Lipofectamine 2000 in HEK293T cells for 56 h. The respective recombinant proteins were isolated using HA tagged protein purification kit (MBL, #3320A) and Flag tagged protein purification kit (MBL, #3325A) according to the product specifications. S6K1-HA or STING-Flag or TBK1-Flag was released from respective tag antibody-Sepharose beads with free HA or Flag peptides to retain maximal biological activities. S6K1-HA alone or the reaction mixtures of S6K1-HA and STING-Flag with or without TBK1-Flag were then incubated in 1 × kinase buffer (Cell Signaling Technology, #9802) at 30°C for 1 h in the presence of ATP (Cell Signaling Technology, #9804) at a final concentration of 200 μM. In some experiments, the kinase reactione with STING-Flag and TBK1-Flag were conducted at 30°C first and S6K1-HA was added at 4°C later for 1 h. Thereafter, IP buffer (300 μl containing protease/phosphatase inhibitor cocktail from ThermoFisher Scientific, #78441) was added at 4°C. To avoid the interference with the subsequent IP reactions by free HA or Flag peptides in the reaction mixtures from the competitive elution process, the IP reactions were conducted with rabbit anti-S6K1 antibody (Millipore, #05-781R) for 1 h and the S6K1-antibody complexes were pulled down by anti-rabbit IgG F(ab’)2 fragment Sepharose (Cell Signaling Technology #3400) for 1 h at 4°C. The immunoprecipitates were analyzed by immunoblot analysis using anti-Flag or anti-S6K1 antibody as indicated.
Antibodies
The primary antibodies used for immunoblot analysis and IP were from the following sources: abcam: IFIT1 (ab11821); Cell Signaling Technology: phospho-TBK1 (#5483), TBK1 (3504), phospho-IRF3 (S396, #4947), IRF3 (#4302), STING (#3337), STING (#13647), p70S6K1 (#9202), phospho-p70S6K1 (S371, #9208), phospho-p70S6K1 (T389, #9234), phospho-S6 ribosomal protein (S240/244, #5364), S6K2 (#14130), MyD88 (#4283), IPS-1(#4983), ISG15 (#2743), cGAS antibody (#31659), HA-Tag antibody-Sepharose (#3956), HA-Tag antibody (#3724), HA-Tag antibody (#2367), Flag-Tag antibody-Sepharose (#5750), Flag-Tag antibody (#2368), Flag-Tag antibody (#8146), anti-rabbit IgG F(ab’)2 fragment Sepharose (#3400),, Myc-Tag antibody (#2278), GFP-Tag antibody (#2956); Abiocode: cGAS (R-3252-1); Santa Cruz Biotechnology: IRF3 (sc-9082 and sc-15991), TBK1 (sc-9910), histone H1 (sc-10806), p70S6Kβ (S6K2, sc-9381), rabbit anti-goat IgG-HRP (sc-2768), HA-probe (sc-7392), Myc-Tag antibody-Sepharose (sc-40 AC), luciferase antibody (sc-74548); Sigma: β-actin antibody (A5441); Millipore: p70S6K1 (05-781R); Novus Biologicals: Trif (NB-120-13810); and Zymed Laboratories: IRF3 (ZM3, 51-3200); Rockland Immunochemicals: anti-Flag antibody (#600-401-383).
Supplementary Material
Supplementary Figure 1 Ad-induced IRF3 phosphorylation does not require 4E-BPs. (a) Immunoblot analysis of p-IRF3(S396) and IRF3 in whole cell lysates of wild-type BMDCs transduced with Ad. β-actin as loading control. (b) Immunoblot analysis of p-IRF3(S396) and IRF3 in the cytoplasmic and nuclear lysates of wild-type BMDCs transduced with Ad. α-tubulin and histone H1 as cytoplasmic fraction and nuclear fraction markers, respectively. (c) Immunoblot analysis of p-IRF3(S396) and IRF3 in whole cell lysates of wild-type, Eif4ebp1−/− Eif4ebp2−/− and Eif4ebp1−/− Eif4ebp2−/− Eif4ebp3−/− BMDCs transduced with Ad. β-actin as loading control. Data are representative of four independent experiments (a-b) and three independent experiments (c).
Supplementary Figure 2 HSV-1 but not LPS or VSV requires S6K for IRF3 phosphorylation. (a-c) Immunoblot analysis of p-IRF3(S396) and IRF3 in whole cell lysates of wild-type and S6k1−/− S6k2−/− BMDCs infected with HSV-1 (a), treated with LPS (b) or infected with VSV (c). β-actin as loading control. Data are representative of three independent experiments (a, b) and four independent experiments (c).
Supplementary Figure 3 Transfection of DNA or cGAMP triggers S6K-dependent IRF3 phosphorylation and early phase IRF3 target gene induction. (a-c) Immunoblot analysis of p-IRF3(S396), IRF3, S6K1 and S6K2 in whole cell lysates of wild-type and S6k1−/− S6k2−/− BMDCs transfected with purified Ad DNA (a), ISD (b) or cGAMP (c). β-actin as loading control. (d-f) Immunoblot analysis of IFIT1, IFIT3, ISG15, p-IRF3(S396), IRF3, S6K1 and S6K2 in whole cell lysates of wild-type (d), S6k1−/− S6k2−/− (e) and Irf3−/− (f) BMDCs transfected with ISD. β-actin as loading control. Data are representative of four independent experiments (a-c) and four independent experiments (d-f).
Supplementary Figure 4 STING does not regulate VSV or LPS-triggered IRF3 phosphorylation. (a) Immunoblot analysis of p-IRF3(S396) and STING or p-IRF3(S396) alone in whole cell lysates of wild-type, Tmem173−/− and Mavs−/− BMDCs infected with VSV. β-actin as loading control. (b) Immunoprecipitation and immunoblot analysis of STING-S6K1 association in wild-type BMDCs treated with LPS or infected with VSV. Lysates (below), immunoblot analysis of S6K1, STING, p-IRF3(S396) and IRF3. β-actin as loading control. Data are representative of four independent experiments (a) and three independent experiments (b).
Supplementary Figure 5 The kinase domain but not the C-terminus of S6K1 is involved in promoting STING-dependent phosphorylation of endogenous IRF3. (a) Immunoprecipitation and immunoblot analysis of the interaction between S6K1(WT)-Myc or S6K1(CT)-Myc and STING(WT)-Flag in transfected HEK293T cells. Lysate (below), immunoblot analysis of S6K1-Myc, S6K1(CT)-Myc, STING-Flag. β-actin as loading control. (b) Immunoblot analysis of endogenous p-IRF3(S396), IRF3, STING(WT)-Flag, S6K1(WT)-Myc, S6K1(ΔNT)-Myc, S6K1(ΔCT)-Myc, S6K1(ΔNTΔCT)-Myc and S6K1(ΔKD)-Myc in transfected HEK293T cells. β-actin as loading control. Data are representative of three independent experiments (a-b).
Supplementary Figure 6 S6K1 interacts with STING and TBK1 to form the tri-molecular S6K1-STING-TBK1 complex in vitro. Upper, immunoprecipitation and immunoblot analysis of the interaction between purified recombinant STING-Flag, S6K1-HA, and TBK1-Flag after the kinase reaction. Middle, immunoblot analysis of purified recombinant STING-Flag, TBK1-Flag, and S6K1-HA after the kinase reaction but before Immunoprecipitation. Bottom, immunoblot analysis of purified recombinant STING-Flag, TBK1-Flag, and S6K1-HA before the kinase reaction. Data are representative of three independent experiments.
Supplementary Figure 7 S6K knockout does not suppress transgene expression or antigen presentation. (a) Immunoblot analysis of OVA-Luc (a) and GFP (b) in whole cell lysates of wild-type, Irf3−/− and S6k1−/− S6k2−/− BMDCs transduced with Ad-OVA-Luc (a) or Ad-GFP (b). β-actin as loading control. (c) Flow cytometry analysis of the cell surface complexes of SIINFEKL and H-2Kb (SIIN-H-2Kb) in wild-type, S6k1−/− S6k2−/− and Irf3−/− BMDCs transduced with Ad-OVA-Luc for 8 h. (d) Flow cytometric confirmation of CFSE labeling in wild-type, S6k1−/− S6k2−/− and Irf3−/− BMDCs transduced with Ad-OVA followed by CFSE staining before the footpad injection. (e) Flow cytometric identification of the CFSE-labeled wild-type, S6k1−/− S6k2−/− and Irf3−/− BMDCs migrated to popliteal lymph nodes (n=6 per group). Data are representative of two independent experiments (a-e).
ACKNOWLEDGMENTS
We would like to thank the other members of the McMaster Immunology Research Centre (MIRC). We would also like to thank N. Kazhdan, S. Collins, D. Cummings (McMaster University) and C. Shao (Western University) for their technical help. We are grateful to M. Noll, J. Govero (Washington University School of Medicine, St Louis), K. McCoy (University of Bern), V. Fensterl and G. Sen (Cleveland Clinic) and X. Feng (McMaster University) for reagents, which helped during the development of this project. We thank M. Orr-Asman, C.Mercer (University of Cincinnati) and R. Lin (McGill University) for plasmids. TA was supported by a grant from the Cancer Research Society/Steven E. Drabin Research Fund. Supported by grants to BDL from the Terry Fox Foundation and to YW and MIRC from the Canadian Institutes of Health Research.
Footnotes
AUTHOR CONTRIBUTIONS
F.W., Y.W. and B.D.L conceived and designed the study and wrote the paper. F.W. did most work. T.A. involved in initiating the collaborative work and experimental designing, prepared S6K KO mouse bones, made shRNAs and various expression plasmids. K.J.S. secured and prepared various KO mouse bones. K.S. made STING and S6K1 pcDNA3 plasmids. M.J.A. prepared plasmids. J.G.P. performed in vivo immunization, T cell flow cytometry and VV-OVA challenge. H.D.H., B.D.F. and T.A. made S6K1 mutant plasmids and prepared lentiviruses expressing S6K1(K100R). L.C. conducted flow cytometry of CFSE-labeled BMDCs and lymph node cells. A.H., E.S.Y.C. and C.T. prepared BMDCs and performed gel electrophoresis during the initial stage of the project. Z.R. conducted HSV-1 mouse experiments. C.Z. conducted genotyping of S6K KO mice. M.S.S. prepared Mavs−/− mouse bones. Z.J. prepared Tbk1−/− mouse bones and cells. M.S.D. provided experimental materials and edited manuscript. K.M., N.S. and A.A.A. provided experimental materials and work support. K.A.F., G.T., S.C.K. and M.G.J. provided experimental materials.
Competing interest statement: The authors declare that they have no competing financial interests.
References
- 1.Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 2.Rathinam VA, Fitzgerald KA. Cytosolic surveillance and antiviral immunity. Curr Opin Virol. 2011;1:455–462. doi: 10.1016/j.coviro.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes E, et al. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci Transl Med. 2012;4:115ra111. doi: 10.1126/scitranslmed.3003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nociari M, Ocheretina O, Murphy M, Falck-Pedersen E. Adenovirus induction of IRF3 occurs through a binary trigger targeting Jun N-terminal kinase and TBK1 kinase cascades and type I interferon autocrine signaling. J Virol. 2009;83:4081–4091. doi: 10.1128/JVI.02591-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stein SC, Falck-Pedersen E. Sensing adenovirus infection: activation of interferon regulatory factor 3 in RAW 264.7 cells. J Virol. 2012;86:4527–4537. doi: 10.1128/JVI.07071-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ablasser A, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa H, Barber GN. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell Mol Life Sci. 2011;68:1157–1165. doi: 10.1007/s00018-010-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin L, et al. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun W, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Fenton TR, Gout IT. Functions and regulation of the 70kDa ribosomal S6 kinases. Int J Biochem Cell Biol. 2011;43:47–59. doi: 10.1016/j.biocel.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 17.Alain T, et al. Vesicular stomatitis virus oncolysis is potentiated by impairing mTORC1-dependent type I IFN production. Proc Natl Acad Sci U S A. 2010;107:1576–1581. doi: 10.1073/pnas.0912344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao W, et al. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol. 2008;9:1157–1164. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J, et al. Mammalian target of rapamycin (mTOR) regulates TLR3 induced cytokines in human oral keratinocytes. Mol Immunol. 2010;48:294–304. doi: 10.1016/j.molimm.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 21.Mohr I, Sonenberg N. Host translation at the nexus of infection and immunity. Cell Host Microbe. 2012;12:470–483. doi: 10.1016/j.chom.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loo YM, Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao P, et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dann SG, Selvaraj A, Thomas G. mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med. 2007;13:252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Soliman GA. The mammalian target of rapamycin signaling network and gene regulation. Curr Opin Lipidol. 2005;16:317–323. doi: 10.1097/01.mol.0000169352.35642.06. [DOI] [PubMed] [Google Scholar]
- 26.Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheatham L, Monfar M, Chou MM, Blenis J. Structural and functional analysis of pp70S6k. Proc Natl Acad Sci U S A. 1995;92:11696–11700. doi: 10.1073/pnas.92.25.11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diner EJ, et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schalm SS, Blenis J. Identification of a conserved motif required for mTOR signaling. Curr Biol. 2002;12:632–639. doi: 10.1016/s0960-9822(02)00762-5. [DOI] [PubMed] [Google Scholar]
- 32.Liu S, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 33.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 34.Fonseca BD, et al. The ever-evolving role of mTOR in translation. Semin Cell Dev Biol. 2014;36C:102–112. doi: 10.1016/j.semcdb.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 35.Senesac J, Gabrilovich D, Pirruccello S, Talmadge JE. Dendritic cells transfected with adenoviral vectors as vaccines. Methods Mol Biol. 2014;1139:97–118. doi: 10.1007/978-1-4939-0345-0_10. [DOI] [PubMed] [Google Scholar]
- 36.Colina R, et al. Translational control of the innate immune response through IRF-7. Nature. 2008;452:323–328. doi: 10.1038/nature06730. [DOI] [PubMed] [Google Scholar]
- 37.Le Bacquer O, et al. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest. 2007;117:387–396. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McWhirter SM, et al. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci U S A. 2004;101:233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daffis S, Suthar MS, Szretter KJ, Gale M, Jr., Diamond MS. Induction of IFN-beta and the innate antiviral response in myeloid cells occurs through an IPS-1-dependent signal that does not require IRF-3 and IRF-7. PLoS Pathog. 2009;5:e1000607. doi: 10.1371/journal.ppat.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonnard M, et al. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 2000;19:4976–4985. doi: 10.1093/emboj/19.18.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boudreau JE, et al. IL-15 and type I interferon are required for activation of tumoricidal NK cells by virus-infected dendritic cells. Cancer Res. 2011;71:2497–2506. doi: 10.1158/0008-5472.CAN-10-3025. [DOI] [PubMed] [Google Scholar]
- 42.Boudreau JE, et al. Recombinant vesicular stomatitis virus transduction of dendritic cells enhances their ability to prime innate and adaptive antitumor immunity. Mol Ther. 2009;17:1465–1472. doi: 10.1038/mt.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao T, et al. The NEMO adaptor bridges the nuclear factor-kappaB and interferon regulatory factor signaling pathways. Nat Immunol. 2007;8:592–600. doi: 10.1038/ni1465. [DOI] [PubMed] [Google Scholar]
- 45.Dennis PB, Pullen N, Pearson RB, Kozma SC, Thomas G. Phosphorylation sites in the autoinhibitory domain participate in p70(s6k) activation loop phosphorylation. J Biol Chem. 1998;273:14845–14852. doi: 10.1074/jbc.273.24.14845. [DOI] [PubMed] [Google Scholar]
- 46.Pol JG, et al. Maraba virus as a potent oncolytic vaccine vector. Mol Ther. 2014;22:420–429. doi: 10.1038/mt.2013.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bassett JD, et al. CD8+ T-cell expansion and maintenance after recombinant adenovirus immunization rely upon cooperation between hematopoietic and nonhematopoietic antigen-presenting cells. Blood. 2011;117:1146–1155. doi: 10.1182/blood-2010-03-272336. [DOI] [PubMed] [Google Scholar]
- 48.Christopher R, Parish MHG, Quah Ben J. C., Warren Hilary S. Use of the Intracellular Fluorescent Dye CFSE to Monitor Lymphocyte Migration and Proliferation. 2009. pp. 4.9.1–4.9.13. [DOI] [PubMed] [Google Scholar]
- 49.Manel N, et al. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 2010;467:214–217. doi: 10.1038/nature09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Ad-induced IRF3 phosphorylation does not require 4E-BPs. (a) Immunoblot analysis of p-IRF3(S396) and IRF3 in whole cell lysates of wild-type BMDCs transduced with Ad. β-actin as loading control. (b) Immunoblot analysis of p-IRF3(S396) and IRF3 in the cytoplasmic and nuclear lysates of wild-type BMDCs transduced with Ad. α-tubulin and histone H1 as cytoplasmic fraction and nuclear fraction markers, respectively. (c) Immunoblot analysis of p-IRF3(S396) and IRF3 in whole cell lysates of wild-type, Eif4ebp1−/− Eif4ebp2−/− and Eif4ebp1−/− Eif4ebp2−/− Eif4ebp3−/− BMDCs transduced with Ad. β-actin as loading control. Data are representative of four independent experiments (a-b) and three independent experiments (c).
Supplementary Figure 2 HSV-1 but not LPS or VSV requires S6K for IRF3 phosphorylation. (a-c) Immunoblot analysis of p-IRF3(S396) and IRF3 in whole cell lysates of wild-type and S6k1−/− S6k2−/− BMDCs infected with HSV-1 (a), treated with LPS (b) or infected with VSV (c). β-actin as loading control. Data are representative of three independent experiments (a, b) and four independent experiments (c).
Supplementary Figure 3 Transfection of DNA or cGAMP triggers S6K-dependent IRF3 phosphorylation and early phase IRF3 target gene induction. (a-c) Immunoblot analysis of p-IRF3(S396), IRF3, S6K1 and S6K2 in whole cell lysates of wild-type and S6k1−/− S6k2−/− BMDCs transfected with purified Ad DNA (a), ISD (b) or cGAMP (c). β-actin as loading control. (d-f) Immunoblot analysis of IFIT1, IFIT3, ISG15, p-IRF3(S396), IRF3, S6K1 and S6K2 in whole cell lysates of wild-type (d), S6k1−/− S6k2−/− (e) and Irf3−/− (f) BMDCs transfected with ISD. β-actin as loading control. Data are representative of four independent experiments (a-c) and four independent experiments (d-f).
Supplementary Figure 4 STING does not regulate VSV or LPS-triggered IRF3 phosphorylation. (a) Immunoblot analysis of p-IRF3(S396) and STING or p-IRF3(S396) alone in whole cell lysates of wild-type, Tmem173−/− and Mavs−/− BMDCs infected with VSV. β-actin as loading control. (b) Immunoprecipitation and immunoblot analysis of STING-S6K1 association in wild-type BMDCs treated with LPS or infected with VSV. Lysates (below), immunoblot analysis of S6K1, STING, p-IRF3(S396) and IRF3. β-actin as loading control. Data are representative of four independent experiments (a) and three independent experiments (b).
Supplementary Figure 5 The kinase domain but not the C-terminus of S6K1 is involved in promoting STING-dependent phosphorylation of endogenous IRF3. (a) Immunoprecipitation and immunoblot analysis of the interaction between S6K1(WT)-Myc or S6K1(CT)-Myc and STING(WT)-Flag in transfected HEK293T cells. Lysate (below), immunoblot analysis of S6K1-Myc, S6K1(CT)-Myc, STING-Flag. β-actin as loading control. (b) Immunoblot analysis of endogenous p-IRF3(S396), IRF3, STING(WT)-Flag, S6K1(WT)-Myc, S6K1(ΔNT)-Myc, S6K1(ΔCT)-Myc, S6K1(ΔNTΔCT)-Myc and S6K1(ΔKD)-Myc in transfected HEK293T cells. β-actin as loading control. Data are representative of three independent experiments (a-b).
Supplementary Figure 6 S6K1 interacts with STING and TBK1 to form the tri-molecular S6K1-STING-TBK1 complex in vitro. Upper, immunoprecipitation and immunoblot analysis of the interaction between purified recombinant STING-Flag, S6K1-HA, and TBK1-Flag after the kinase reaction. Middle, immunoblot analysis of purified recombinant STING-Flag, TBK1-Flag, and S6K1-HA after the kinase reaction but before Immunoprecipitation. Bottom, immunoblot analysis of purified recombinant STING-Flag, TBK1-Flag, and S6K1-HA before the kinase reaction. Data are representative of three independent experiments.
Supplementary Figure 7 S6K knockout does not suppress transgene expression or antigen presentation. (a) Immunoblot analysis of OVA-Luc (a) and GFP (b) in whole cell lysates of wild-type, Irf3−/− and S6k1−/− S6k2−/− BMDCs transduced with Ad-OVA-Luc (a) or Ad-GFP (b). β-actin as loading control. (c) Flow cytometry analysis of the cell surface complexes of SIINFEKL and H-2Kb (SIIN-H-2Kb) in wild-type, S6k1−/− S6k2−/− and Irf3−/− BMDCs transduced with Ad-OVA-Luc for 8 h. (d) Flow cytometric confirmation of CFSE labeling in wild-type, S6k1−/− S6k2−/− and Irf3−/− BMDCs transduced with Ad-OVA followed by CFSE staining before the footpad injection. (e) Flow cytometric identification of the CFSE-labeled wild-type, S6k1−/− S6k2−/− and Irf3−/− BMDCs migrated to popliteal lymph nodes (n=6 per group). Data are representative of two independent experiments (a-e).