Abstract
During aging, the mechanisms that normally maintain health and stress resistance strikingly decline, resulting in decrepitude, frailty, and ultimately death. Exactly when and how this decline occurs is unknown. Changes in transcriptional networks and chromatin state lie at the heart of age-dependent decline. These epigenomic changes are not only observed during aging but also profoundly affect cellular function and stress resistance, thereby contributing to the progression of aging. We propose that the dysregulation of transcriptional and chromatin networks is a crucial component of aging. Understanding age-dependent epigenomic changes will yield key insights into how aging begins and progresses and should lead to the development of new therapeutics that delay or even reverse aging and age-related diseases.
Introduction
Aging is characterized by progressive functional decline at the molecular, cellular, tissue, and organismal levels. As an organism ages, it becomes frail, its susceptibility to disease increases, and its probability of dying rises. In humans, age is the primary risk factor for a panoply of diseases including neurodegeneration, cardiovascular disease, diabetes, osteoporosis, and cancer. Over the past decades, a large body of research has shown that the molecular and cellular decline of aging can be organized into several evolutionarily conserved hallmarks or pillars of aging (Kennedy et al., 2014; López-Otín et al., 2013). For example, in yeast and animals, mitochondrial dysfunction increases with age and may contribute to the progression of aging (Sun et al., 2016). The hallmarks of aging are interconnected (Kennedy et al., 2014; López-Otín et al., 2013; Zhang et al., 2015a), and age-associated perturbations of one can affect others. While significant progress has been made in our understanding of aging, many outstanding questions remain: Which age-associated changes are causative? How are the hallmarks of aging related to each other, and are there “hubs” in this network? Which age-dependent changes occur first? When does aging begin? Can therapeutics slow aging or even rejuvenate some aging hallmarks in an animal at any stage during lifespan, or is there a “point of no return”?
The study of gene regulation is central to many of these questions. The regulation of gene expression is not only necessary for nearly every aspect of a cell’s function, but it can be sufficient to alter cellular fate. This is illustrated by the fact that in Saccharomyces cerevisiae, only three transcription factors control cell identity, and their mis-expression results in cell-cycle arrest and an inability to mate (Herskowitz, 1989). Furthermore, transient, ectopic expression of a small number of transcription factors in adult mammalian cells can convert terminally differentiated cells into pluripotent stem cells or into other differentiated states (Graf, 2011). In addition to transcription factors, chromatin state, which consists of a variety of histone marks, DNA methylation, and nucleosome positioning, helps to control the spatiotemporal expression of genes. Finally, non-coding RNAs have been found to play important roles in regulating chromatin states and gene expression (Rinn and Chang, 2012). Because of their crucial role in cellular function, we propose that age-associated changes in transcriptional factors and chromatin states represent a critical aspect of the aging process and an important hub that links many hallmarks of aging. In this review, we will use the term “epigenomic changes” to encompass alterations in transcription factor binding, histone marks, DNA methylation, nucleosome positioning, and non-coding RNAs, and we will focus on these aspects of gene regulation. However, in addition to these aspects of gene regulation, it is important to note that transcript stability, RNA splicing, and translation are important gene regulatory layers that can also become dysregulated with age.
While it is clear that many biological systems and hallmarks play a crucial role in the progression of aging, we propose that epigenomic changes are particularly important because of the following: (1) Changes in gene regulation (often through expression of a single transcription factor) have been shown to be key for cellular identity. Thus, age-associated changes in transcription regulatory networks are likely to impact the function of a cell or tissue and give rise to aging phenotypes and diseases. (2) Gene regulation is a natural “hub” in the cell. Transcription regulators and chromatin modifiers receive cytoplasmic and extracellular signals and, in turn, alter the responses of the cell in an orchestrated manner. For example, in response to proteostatic stress, protein chaperone expression increases. (3) Chromatin marks are long lasting and show a progressive change with age that persists through cellular divisions. Thus, they can act as a memory that helps to propagate age-associated cellular dysfunction. (4) Recent evidence suggests that epigenomic changes can occur extremely early in the aging process and be causative.
In this review, we will focus on the role of epigenomic mechanisms in aging, the way they interact with other hallmarks of aging, when these changes occur during aging, and how they are affected by interventions to delay or even reverse some aspects of aging and aging-related diseases.
The Aging Epigenomic Regulatory Landscape
Recent studies in diverse species and cell types have highlighted the pervasiveness of age-associated changes in transcription factor binding, histone marks, heterochromatin formation, and DNA methylation (recently reviewed by Benayoun et al., 2015; Zampieri et al., 2015) that contribute to changes in gene expression. Indeed, gene expression has been shown to change drastically during aging (for example, Baumgart et al., 2014; Budovskaya et al., 2008; Zahn et al., 2007). The gene regulatory landscape consists of many interconnected players that promote or repress gene expression (Figure 1). While it is clear that aging is associated with dysfunction at each layer of the gene regulatory landscape, it is relatively unknown which aspect of gene regulation is the first to become dysregulated during lifespan. The order and molecular mechanism of youthful gene regulation can give clues about the relationship between and order of age-associated epigenomic and gene expression changes.
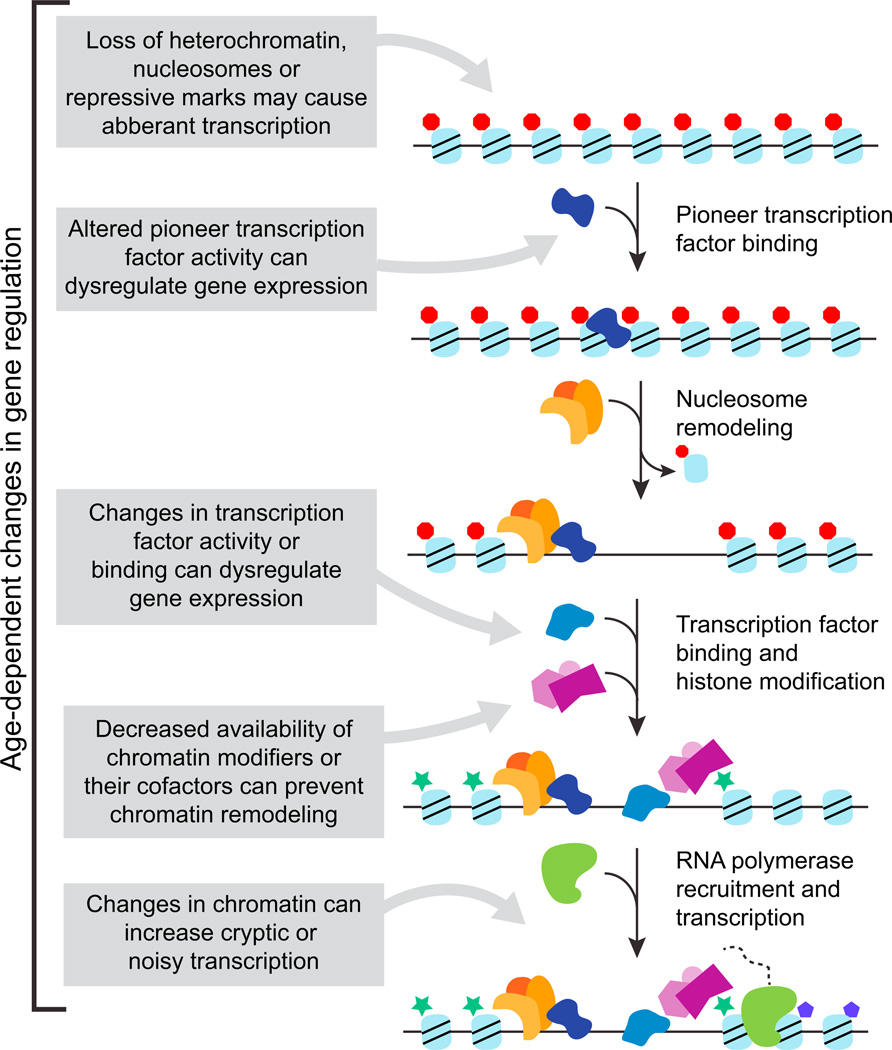
Figure 1. Transcription Factors and Chromatin Modifiers Act Together to Regulate Gene Expression.
The remodeling and transcription activation of a gene occurs in a stepwise fashion that begins with the binding of a sequence-specific DNA-binding protein called a pioneer transcription factor. Following recruitment by a pioneer transcription factor, nucleosome remodeling complexes (e.g., SWI/SNF) displace or move nucleosomes and open chromatin. This increases DNA accessibility and allows binding of additional transcription factors, recruitment of chromatin modifiers that remove repressive marks (red hexagons) and add activating marks (green stars), and finally, transcription of the gene. Age-associated changes have been observed at every step of this process, and changes in one step (for example, the activity of pioneer transcription factor) can have downstream consequences for gene regulation.
Transcription Factors
Transcription factors bind DNA in a sequence-specific manner and can activate or repress transcription. Some transcription factors (called pioneer factors) can also initiate remodeling of a genetic locus by binding to closed chromatin and mediating nucleosome remodeling and changes in histone marks (Zaret and Carroll, 2011) (Figure 1). The sequence specificity of transcription factors, their regulation by upstream signaling pathways and sensors, and their ability to recruit histone modifying enzymes and nucleosome remodelers (Figure 1) are all characteristics that contribute to the crucial role transcription factors play in the control of gene expression. Here, we focus on two well-conserved, pro-longevity transcription factors (FOXO/DAF-16 and NRF/SKN-1) as paradigms of the role of transcription factors in aging. It is important to mention that there are many other transcription factors, including HSF-1, XBP-1, REST/SPR-4, and p53/CEP-1, that are known to also play key roles during aging. The role of these transcription factors in aging will also be discussed in later sections of this review.
FOXO/DAF-16 promotes longevity and inhibits age-related diseases in species ranging from worms to humans (Eijkelenboom and Burgering, 2013; Salih and Brunet, 2008). Indeed, allele variants at the FOXO3 locus are associated with extreme longevity (centenarians) in humans (for example, Willcox et al., 2008). The activity of FOXO/DAF-16 is positively and negatively regulated by different molecular players (e.g., the insulin/IGF signaling pathway and the nutrient sensor AMPK) and stresses (e.g., oxidative and heat stress) (Eijkelenboom and Burgering, 2013; Salih and Brunet, 2008). Meta-analysis of the adult FOXO/DAF-16 transcription regulatory networks in human, mouse, Drosophila, and C. elegans has shown that FOXO/DAF-16 controls the expression of many genes, including those involved in stress response, metabolism, proteostasis, immunity, and neuronal function (Webb et al., 2016). Thus, FOXO/DAF-16 regulates a program of genes whose major function is to promote stress resistance, and this may be linked with their ability to extend lifespan. Interestingly, the role of FOXO/DAF-16 in longevity has been directly connected with chromatin state. Indeed, FOXO has been found to bind open chromatin, especially at enhancer regions (Eijkelenboom et al., 2013; Webb et al., 2013), and can act as a pioneer factor to open closed chromatin (Zaret and Carroll, 2011). Furthermore, in C. elegans, DAF-16 recruits the chromatin remodeler SWI/SNF to activate expression of age-related and stress-response genes including heat shock proteins and oxidative stress-response genes (Riedel et al., 2013). This interaction is required for the longevity-promoting action of DAF-16 (Riedel et al., 2013). Together, these studies identify FOXO/DAF-16 as a key regulatory hub that integrates diverse signals into the control of the chromatin landscape and the expression of genes important for longevity. However, much remains to be discovered about the ways that FOXO/DAF-16 activity (its expression, DNA binding ability, and interactions with chromatin modifiers) changes during normal, physiological aging.
Like FOXO/DAF-16, NRF/SKN-1 is a “pro-longevity” transcription factor. Indeed, increasing the level of active NRF/SKN-1 extends lifespan in flies and worms (Sykiotis and Bohmann, 2008; Tullet et al., 2008). Consistent with a role in the progression of aging, the activity of NRF/SKN-1 is dysregulated late in life (Rahman et al., 2013). In response to elevated levels of reactive oxygen species (ROS) and environmental toxins or to nutrient and metabolic changes through mTOR signaling and insulin/IGF signaling, NRF/SKN-1 activates the expression of genes involved in cellular protection (Blackwell et al., 2015; Tebay et al., 2015). Classically considered to control the transcriptional response to oxidative stress, NRF/SKN-1 has also been implicated in general detoxification, proteostasis, and immunity (Blackwell et al., 2015). In mammals, NRF2 recruits BRG2, a component of the SWI/SNF nucleosome remodeling complex, to its regulatory targets (Zhang et al., 2006). This interaction results in increased transcriptional activation of stress-response genes (Zhang et al., 2006), presumably through changes in the chromatin landscape. Interestingly, the DNA-binding ability of the NRF/SKN-1 proteins in cultured embryonic stem cells appears to be sensitive to DNA methylation (Domcke et al., 2015). However, it is unknown if age-dependent changes in DNA methylation could impact NRF/SKN-1 binding during aging. Together, these studies show that NRF/SKN-1 is an important regulator of stress resistance and that its activity is intimately linked with chromatin state.
These two specific examples illustrate how changes in transcription factor activity could play a key role in aging, and likely apply to many other transcription factors (some examples of which are described in sections below). First, reduced levels of a pro-longevity transcription factor can lead to a loss of stress resistance and cellular protection due to decreased expression of stress-resistance genes. Second, age-associated changes in the activity of transcription factors could promote changes in other epigenomic regulatory layers. This can happen by either altering the expression of chromatin-modifying enzymes and complexes (e.g., histone methyltransferases or nucleosome remodelers) or by the differential recruitment of these enzymes and complexes. Both possibilities could result in widespread changes to the epigenomic landscape that could alter cellular function and contribute to the progression of aging. Importantly, both FOXO/DAF-16 and NRF/SKN-1 not only promote changes in chromatin state (Riedel et al., 2013; Zaret and Carroll, 2011; Zhang et al., 2006) but also are sensitive to them (Domcke et al., 2015; Eijkelenboom et al., 2013; Webb et al., 2013). Thus, the precise timing and the cause and effect relationship between changes in transcription factor binding and chromatin state are complex and in most cases have not been untangled yet.
Histone Marks
The core histone proteins that package DNA and comprise nucleosomes are subject to a variety of post-translational modifications. The differential recruitment of histone-modifying enzymes and complexes by transcription factors forms the patterns of histone marks in the genome (Figure 1). Histone marks, including H3K27me3, H3K4me3, H3K27ac, H3K14ac, and H3K36me3, help to control the expression of genes. The exact molecular mechanisms of histone mark-mediated gene regulation are not fully understood but likely involve promoting or inhibiting the recruitment of transcriptional machinery (Lauberth et al., 2013). Histone marks can also act to silence transcription through the formation of heterochromatin (i.e., H3K9me3, H4K20me2) and to regulate genome stability (i.e., H3K56ac, H3K14ac). Interestingly, the patterning of these marks in the genome changes during aging (Benayoun et al., 2015) (Figure 2).
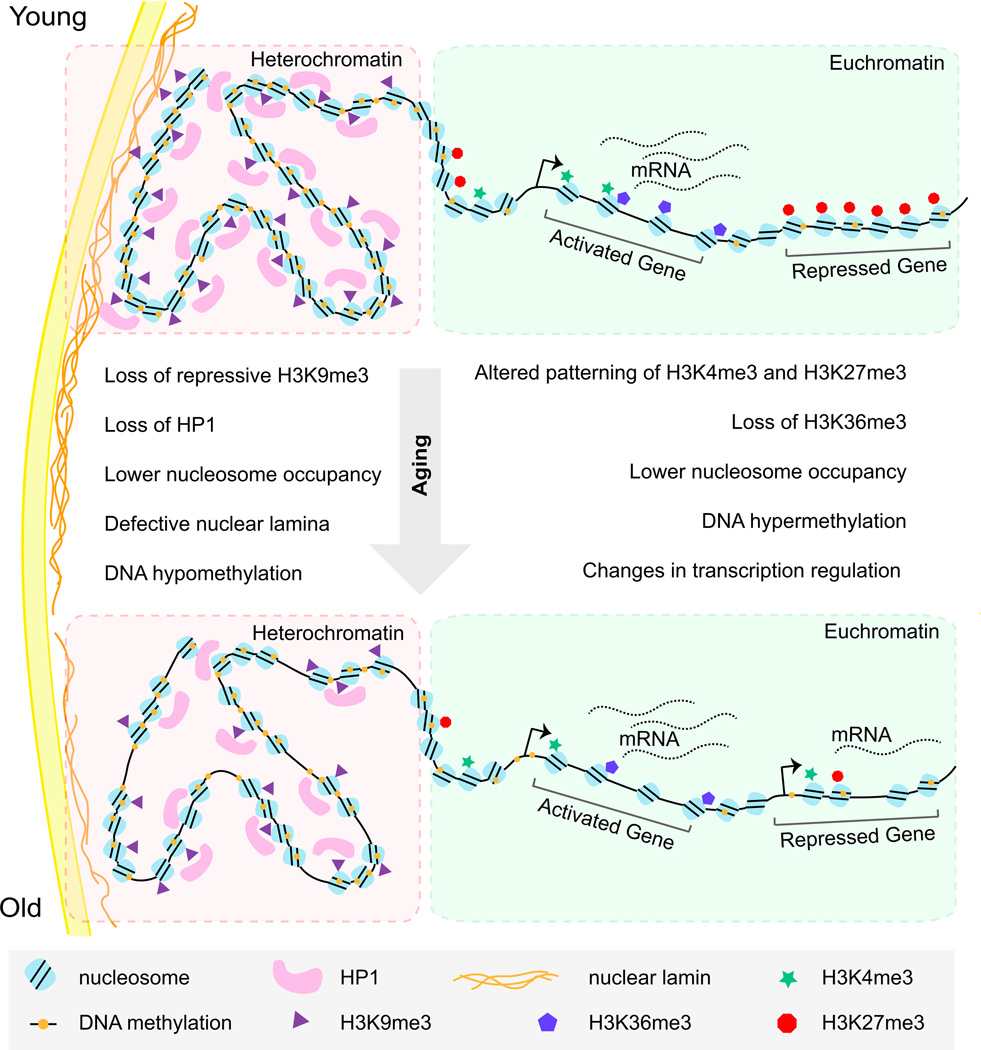
Figure 2. Chromatin States Change during Aging.
During aging and the emergence of cellular senescence, there is a general loss of heterochromatin that is characterized by a loss of repressive histone marks (H3K9me3), DNA methylation, nucleosome occupancy, and heterochromatin protein 1 (HP1) binding. These changes are associated with a loss of the nuclear lamina. In active regions of the genome (euchromatin), the patterning of histone marks changes (the active mark H3K4me3, the repressive mark H3K27me3, and the transcription elongation mark H3K36me3), DNA methylation increases at specific loci, and nucleosome remodeling occurs. Together, these changes can cause altered gene expression with age and contribute to the progression of aging.
Different enzymes that affect H3K27me3, a repressive histone mark, have opposing effects on lifespan. Knockdown of the H3K27me3 demethylase UTX-1 increases lifespan in C. elegans (Jin et al., 2011; Maures et al., 2011; Ni et al., 2012), suggesting that H3K27me3 levels are beneficial for longevity. In contrast, overexpression of the H3K27me3 demethylase KDM6B/JMJD-3.1 extends lifespan, suggesting that an increased level of H3K27me3 at specific loci (stress-response genes) is detrimental for lifespan (Labbadia and Morimoto, 2015). In addition, KDM6B/JMJD-3.1 and PHF8/JMJD-1.2 are necessary for lifespan extension induced by mitochondrial dysfunction, and overexpression of either of these genes extends lifespan in C. elegans (Merkwirth et al., 2016). Interestingly, mice with higher PHF8/JMJD-1.2 expression are longer lived, suggesting that the link between PHF8/JMJD-1.2 and longevity might be conserved from invertebrates to mammals (Merkwirth et al., 2016). Importantly, the Utx gene is located on the sex chromosomes in mammals and worms and appears to have some sex-specific functions. In mice, loss of Utx is associated with severe developmental defects in females, but not males, and a shortened male lifespan (Welstead et al., 2012). Furthermore, loss of components of Polycomb, the H3K27 methyltransferase, increases lifespan in male Drosophila, suggesting that increased levels of H3K27me3 are detrimental for male lifespan (Siebold et al., 2010). It is unknown if modulating H3K27me3 levels also affects female lifespan in mice and flies. Thus, an important area of future study will be to determine the specific roles of the different H3K27me3 enzymes, particularly their sex-specific functions in lifespan regulation.
Interestingly, age-associated changes in global H3K27me3 levels are also context dependent. Global H3K27me3 increases with age in the killifish brain and mouse quiescent satellite cells (Baumgart et al., 2014; Liu et al., 2013) but decreases with age during C. elegans aging and in human progeria models (Maures et al., 2011; Ni et al., 2012; Shumaker et al., 2006; Zhang et al., 2015b). A comparison of H3K27me3 deposition in young versus old murine hematopoietic stem cells revealed that while H3K27me3 levels increased at most loci, there were also loci with decreased H3K27me3 levels with age (Sun et al., 2014). In addition, cultured senescent human lung fibroblasts have large-scale gains and losses of H3K27me3 compared to early-passage, proliferating cells (Shah et al., 2013). Identifying the locus- and cell-type-specific roles of H3K27me3 in longevity will be key to untangling the effect of the H3K27me3-modifying enzymes on lifespan.
H3K4me3, a mark associated with active transcription, has also been implicated in longevity, and its deposition changes with age in murine hematopoietic stem cells and in Drosophila (Sun et al., 2014; Wood et al., 2010) and cellular senescence in human fibroblasts (Shah et al., 2013). Like the H3K27me3 modifiers, knockdown of the H3K4me3 enzymes has been shown to both increase and decrease lifespan, depending on the specific enzyme or context. In C. elegans, knockdown of the H3K4 methyltransferase SETD1A/SET-2 increases lifespan (Greer et al., 2010). Consistent with the idea that loss of H3K4me3 is beneficial to longevity, knockdown of the H3K4me3 demethylase KDM5B/RBR-2 in C. elegans hermaphrodites and Drosophila males shortens lifespan (Greer et al., 2010; Li et al., 2010). However, it has also been found that knockdown of the H3K4me3 demethylases (KDM5B/RBR-2 and KDM1A/LSD-1) can extend lifespan, often in the context of sterility (Lee et al., 2003; McColl et al., 2008; Ni et al., 2012), or has no effect on lifespan (Li et al., 2010). Together, these studies suggest that, like H3K27me3-modifying enzymes, H3K4me3-modifying enzymes can both promote and inhibit longevity, likely depending on environmental context and specific loci. Interestingly, H3K4me3 domains vary in breadth in a locus- and cell-type-specific manner (Benayoun et al., 2014; Chen et al., 2015), and these differences could provide clues about the context-dependent roles of this mark during aging. H3K4me3-modifying enzymes have also been shown to have long-lasting, transgenerational effects on C. elegans lifespan (Greer et al., 2011, 2016). However, the mechanisms underlying this phenomenon are not known.
H3K36me3, a mark associated with transcriptional elongation and splicing, has also recently been found to play an important role during aging. In yeast, loss of H3K36 trimethylation (a mark of transcription elongation) is correlated with a shorter lifespan (Sen et al., 2015). Consistent with this finding, deletion of the H3K36me3 demethylase extends lifespan in S. cerevisiae and C. elegans (Ni et al., 2012; Sen et al., 2015), whereas deletion of the H3K36 methyltransferase (met-1) in C. elegans shortens lifespan (Pu et al., 2015). Interestingly, low levels of H3K36me3 are correlated with increased cryptic transcription (Sen et al., 2015) and a greater degree of gene expression change with age (Pu et al., 2015). Thus, changes in the deposition of H3K36me3 appear to play an important role in the regulation of transcription during aging. These examples of age-associated changes in histone marks represent only the tip of the iceberg. Other histone marks are known to change during aging, and many more histone mark changes likely remain to be discovered.
How do changes in histone marks arise during aging? The enzymes that place and remove histone marks exist in large protein complexes that are differentially recruited to chromatin through interactions with transcription factors. Thus, the patterning of histone marks is reflective of the transcriptional activity of a locus and is as unique and specific to a given cell type or locus as the transcription networks themselves. During aging, the changes in histone mark patterning and levels could be the result of age-associated changes in the activity of transcription factors, either through changes in the expression of the components of histone-modifying complexes or through the differential recruitment of these complexes to chromatin. Delineating the specific mechanisms of action of these histone-modifying enzymes and their interactions with other gene regulatory layers (e.g., transcription factors) should provide important insights into the mechanisms of chromatin-mediated longevity.
DNA Methylation
Methylation of the cytosine of CpG dinucleotides (5-methylcytosine or 5mC) is a heritable and conserved regulatory mark that is generally associated with transcriptional repression. DNA methylation represses gene expression by inhibiting or promoting the binding of transcriptional activators and repressors (respectively) and by the differential recruitment of repressive histone-modifying enzymes. It is important to note that yeast and worms do not have classical (5mC) DNA methylation. However, another form of DNA methylation (6-methyladenosine or 6mA) was recently discovered to be present in C. elegans, Drosophila, and Chlamydomonas (reviewed by Luo et al., 2015). In C. elegans, 6mA DNA methylation is involved in H3K4me2-mediated transgenerational lifespan extension (Greer et al., 2016), suggesting a link between DNA methylation, histone marks, and aging.
In mammals, the genome-wide pattern of DNA methylation changes during aging (Figure 2), and these changes could play a role in the progression of aging (Benayoun et al., 2015; Zampieri et al., 2015). Intriguingly, DNA methylation patterns can serve as a biomarker for human chronological age (Horvath, 2013), and perhaps even biological age (i.e., how youthful or decrepit an individual actually is at a particular age). The gradual accumulation of differential DNA methylation with age is thought to act like an “epigenetic clock” (Horvath, 2013).
The specific genetic regions that have age-dependent differential DNA methylation may reveal the mechanism of the change during aging. While hypomethylation is common with age, some CpG islands and gene-rich regions become hypermethylated with age (reviewed by Benayoun et al., 2015; Zampieri et al., 2015). Interestingly, the loci that display age-dependent DNA hypermethylation are preferentially near tissue-specific genes, genes involved in differentiation and development, genes encoding transcription factors, and transcription factor binding sites (reviewed by Benayoun et al., 2015; Zampieri et al., 2015). These genes exist in active regions of the genome, and therefore it is possible that age-dependent changes in the regulation of these genes by transcription factors and histone-modifying enzymes result in aberrant recruitment of the DNA methylation enzymes. Interestingly, studies in stem cells have also demonstrated that loci with age-associated DNA hypermethylation are enriched for genes normally repressed by Polycomb repressive complex, the complex that methylates H3K27 (Beerman et al., 2013; Sun et al., 2014; Teschendorff et al., 2010). One model for normal DNA methylation patterning is that repression by H3K27 methylation occurs first and is “locked in” by the subsequent action of DNA methyltransferases. Thus, changes in the activity of chromatin modifiers (including Polycomb repressive complex) may occur first during aging and be followed by DNA methylation changes.
Another important consideration for the role of DNA methylation during aging is its effect on gene expression. For example, age-associated DNA hypermethylation of some transcription factor binding sites (Avrahami et al., 2015; Sun et al., 2014; Yuan et al., 2015) suggests that the ability of these proteins to bind DNA and regulate transcription could be disrupted. Surprisingly, several genome-wide studies have not found a correlation between age-dependent differential DNA methylation and altered gene expression (Beerman et al., 2013; Yuan et al., 2015). However, at least in the case of aged β cells, a significant correlation between DNA methylation and gene expression changes exists for distal regulatory regions (enhancers), but not proximal regulatory regions (promoters) (Avrahami et al., 2015). Thus, the potential gene expression effects of age-associated differential DNA methylation are likely to be context dependent and may require local changes in histones marks and transcription factor binding to occur before gene expression changes are observed.
Chromatin Structure
Chromosomes exist in higher-order structures that impact the regulation of gene expression through changes in the accessibility of transcriptional machinery and regulators to DNA. At the first structural level, the DNA is wrapped around nucleosomes, and their occupancy on DNA plays a role in the regulation of gene expression. Higher levels of chromatin structure create the three-dimensional architecture of the genome through associations between distal genomic regions as well as associations between the nuclear envelope and DNA (Pombo and Dillon, 2015). Genomic regions that are tightly packed around nucleosomes and are associated with the lamins of the nuclear envelope (lamin-associated domains) are less accessible and contain genes that are not actively transcribed. Interestingly, chromatin structure changes with age, and these changes have been linked with longevity.
Nucleosome occupancy can decrease with age (Figure 2), and ameliorating this loss in yeast increases lifespan (Feser et al., 2010; Hu et al., 2014; Ivanov et al., 2013). Two mechanisms could contribute to this loss: (1) a reduction in the amount of core histones made by the cell and/or (2) a change in the activity of histone chaperones (Feser et al., 2010; Ivanov et al., 2013; Liu et al., 2013; Ni et al., 2012). In some cases, post-transcriptional regulation (for example, the regulation of translation) appears to be playing a role in the age-associated loss of histones (Feser et al., 2010; O’Sullivan et al., 2010). However, in mammalian muscle stem cells, the loss of core histones with age appears to be due to changes in gene expression and is linked with the amount of the repressive H3K27me3 mark at the histone genes (Liu et al., 2013). This suggests that an altered chromatin state at specific loci (the histone genes) may precede the reduction of nucleosome occupancy during aging. Interestingly, nucleosome occupancy changes have also been demonstrated to be associated with the global upregulation of gene expression with age in yeast (Hu et al., 2014) and increased transcription factor (FOXA2) binding in aged mammalian livers (Bochkis et al., 2014). FOXA2 is a pioneer transcription factor (Zaret and Carroll, 2011), and therefore, the age-dependent increase in FOXA2 binding likely causes the eviction of nucleosomes from these regions. Together, these studies show that the positioning of nucleosomes on DNA changes with age and that changes in transcription factor activity and the chromatin landscape can trigger these changes during aging.
Heterochromatin is a transcriptionally inactive and inaccessible chromatin structure that is characterized by bound heterochromatin protein 1 (HP1) and H3K9me3 in animals and the Sir complex in budding yeasts (Oberdoerffer and Sinclair, 2007). The premature aging diseases Hutchinson-Gilford progeria syndrome (HGPS) and Werner syndrome are characterized by a reduction in heterochromatin and lower levels of H3K9me3, H3K27me3, and HP1 (Scaffidi and Misteli, 2006; Shumaker et al., 2006; Zhang et al., 2015b). Interestingly, mitochondrial stress can also trigger large-scale changes in heterochromatin and H3K9 methylation in C. elegans, suggesting a link between age-associated stress response and the formation of heterochromatin (Tian et al., 2016). Importantly, during normal aging in humans and invertebrates, HP1 and H3K9me3 levels also decline (Figure 2), suggesting that loss of heterochromatin is a common feature of aging (Larson et al., 2012; Ni et al., 2012; Scaffidi and Misteli, 2006; Wood et al., 2010).
What is the mechanism of age-associated heterochromatin loss? DNA damage appears to be a contributor to heterochromatin loss (Oberdoerffer and Sinclair, 2007) and is discussed in the next section of the review. Interestingly, the nuclear envelope may play a role in heterochromatin loss in both normal and progeroid aging: the levels of a cryptic splice isoform that results in a truncated nuclear envelope protein lamin A, and that mimics the mutation of HGPS, increase with age in humans and contribute to the loss of heterochromatin (Scaffidi and Misteli, 2006). Thus, there is a link between defects in the nuclear lamin, loss of heterochromatin, and aging. Intriguingly, it was recently discovered that the autophagosome can selectively degrade the endoplasmic reticulum and the nucleus (Mochida et al., 2015), and that this degradation can lead to a loss of the nuclear lamina (Dou et al., 2015). It will be interesting to determine if age-dependent dysfunction of autophagy could contribute to the loss of heterochromatin with age.
Non-coding RNAs
Non-coding RNAs are important regulators of transcriptional networks and chromatin states. For example, long non-coding RNAs (lncRNAs) such as XIST and HOTAIR promote gene silencing through interactions with chromatin-modifying enzymes (Rinn and Chang, 2012). These regulatory RNAs are emerging as important players in the progression of aging (Grammatikakis et al., 2014). One interesting example of an lncRNA that is associated with aging is the H19 lncRNA. This lncRNA interacts with MBD1 (a methyl-CpG-binding domain protein) to repress the expression of several genes, including a gene likely to have implications for aging—Igf2, which encodes an insulin-like growth factor (Monnier et al., 2013). Interestingly, with increased age, there is a loss of H19 lncRNA-mediated repression of the Igf2 locus in mice (Fu et al., 2008). Furthermore, H19-mediated repression of Igf2 has been demonstrated to be important for the maintenance of hematopoietic stem cell quiescence (Venkatraman et al., 2013). Quiescence is key to maintaining this stem cell population, and the loss of H19-mediated repression of Igf2 leads to hematopoietic stem cell exhaustion (Venkatraman et al., 2013). These data suggest the exciting possibility that changes driven by lncRNAs could play an important role in the progression of aging. Future studies of non-coding RNAs during aging are likely to reveal additional age-associated roles for these important gene regulatory players.
Epigenomic Regulation Is Connected to Other Aging Hallmarks
In addition to age-associated epigenomic changes, many other systems become dysfunctional with age (López-Otín et al., 2013). Excitingly, chromatin and transcription regulation have been found to play key roles in the age-dependent manifestation of these hallmarks of aging and in the response to their emergence (Figure 3). Epigenetic changes, such as DNA methylation, are relatively long-lived, and thus, one intriguing idea is that the epigenetic response to the emergence of a hallmark of aging could create a “memory” of the event, allowing it to be perpetuated over time and through cellular divisions (Rando and Chang, 2012). Here we discuss the relationship between epigenomic regulation and the hallmarks of aging.
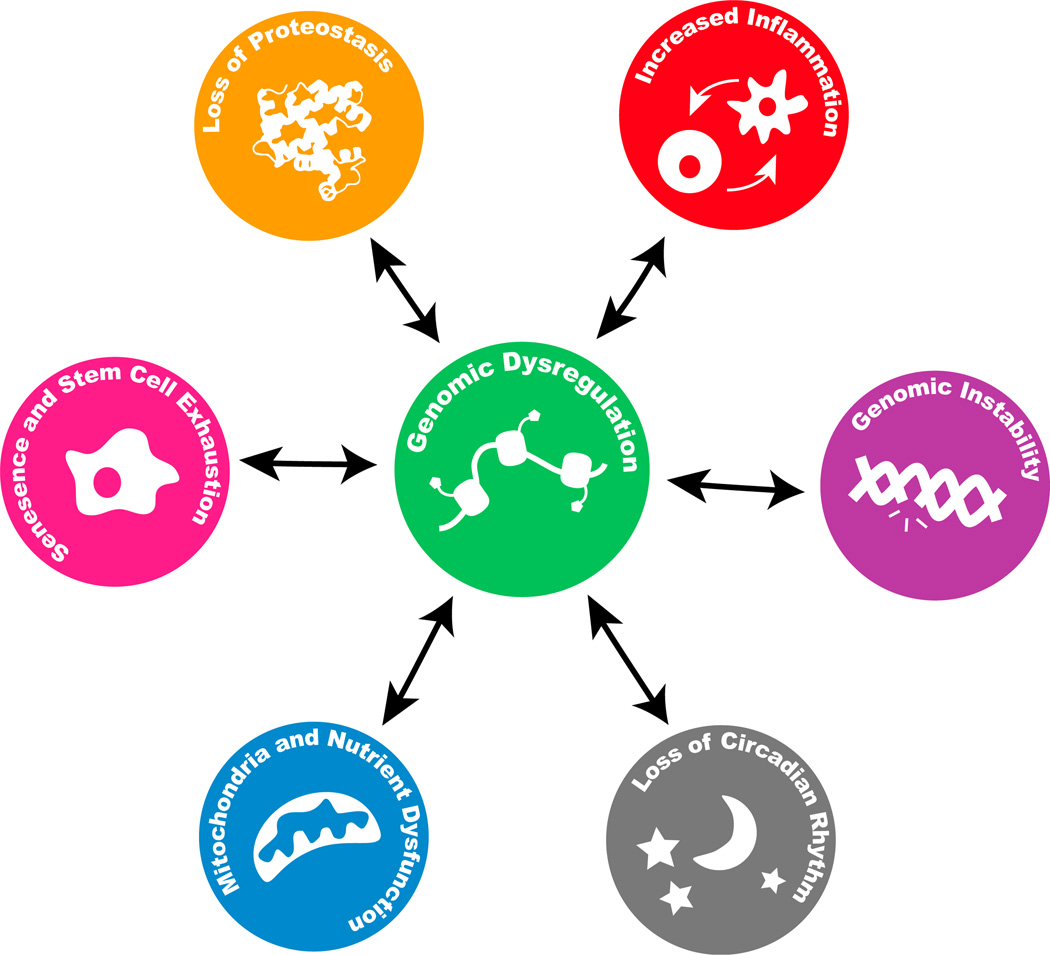
Figure 3. Epigenomic Changes Are a Hub in the Progression of Aging.
Epigenomic changes—changes in transcription factors, histone marks, nucleosome positioning, and DNA methylation—are connected with the other hallmarks of aging (Kennedy et al., 2014; López-Otín et al., 2013; Zhang et al., 2015a). Epigenomic changes can trigger the emergence of other hallmarks of aging and can also be affected by them.
Genome Stability and Telomeres
A loss of genomic integrity is a hallmark of aging (López-Otín et al., 2013; Vijg and Suh, 2013). With age, telomere attrition occurs, somatic DNA mutations accumulate, and transposons activate. There is also an increase in the rate of chromosomal abnormalities, some of which are oncogenic. The link between age-associated chromatin changes, aging, and genome stability was first discovered in yeast (reviewed by Sinclair et al., 1998). More recently, studies in many organisms (described below) have shown that the link between epigenomic changes and genomic integrity is well conserved.
Telomeres
Telomeres are normally maintained in a heterochromatic state by several chromatin modifiers. In yeast, histone deacetylases (sirtuins) are key for telomere silencing. Consistent with a link between longevity and telomere silencing, older yeast have less active Sir2 (a sirtuin) and, as a result, increased H4K16 acetylation at telomeres (Dang et al., 2009). Furthermore, increasing the level of Sir2 in yeast increases lifespan (Kaeberlein et al., 1999). In human cells, loss of SIRT6, an H3K9 deacetylase that silences telomeres, causes telomere dysfunction and premature cellular senescence (Michishita et al., 2008). These data suggest that altered chromatin states during aging may precede and cause age-associated telomere dysfunction. Interestingly, damaged telomeres can also lead to a reduction in the expression of core histones in a cell, suggesting that a loss of telomere integrity can cause changes in the aging chromatin landscape (O’Sullivan et al., 2010). Thus, epigenomic changes and telomere dysfunction may feedback and reinforce each other during aging.
Mutation Accumulation
In several species, there is an age-associated accumulation of mutations at varying frequencies depending on the species and rearing conditions (reviewed by Vijg and Suh, 2013). It is important to note, however, that in yeast the rate of age-associated mutation accumulation is low, and thereby is unlikely to play a major role in the progression of aging (Kaya et al., 2015). While the role of mutations in aging is debated and likely to be species specific, there is a link between increases in 5mC DNA methylation and mutation. For example, 5-methyl cytosine is inherently more mutagenic than its non-methylated counterpart because if it is deaminated, the base excision repair machinery replaces the nucleotide with a thymine, resulting in a C-to-T mutation (Schmutte et al., 1995). Interestingly, during human aging, mutations accumulate in the genome of blood cells and C-to-T mutations are the most prevalent (Welch et al., 2012). This raises the possibility that age-dependent DNA hypermethylation (increased DNA methylation) at specific loci contributes to the accumulation of point mutations during aging.
DNA Damage
DNA damage can result in transient gene repression (Kim et al., 2016) as well as more stable changes in gene expression that can be observed in humans as young as 40 years of age (Lu et al., 2004). Interestingly, sensing and responding to DNA damage involves the recruitment of chromatin modifiers, suggesting that an accumulation of DNA damage during life could be a source for age-associated epigenomic changes and that changes in chromatin state could affect DNA repair (Burgess et al., 2012; Oberdoerffer and Sinclair, 2007). For example, some sirtuins (e.g., SIRT6 and SIRT1, discussed below) are important players in the response to DNA damage. The recruitment of SIRT6 to sites of DNA damage helps to remodel the local chromatin environment through deacetylation of H3K56 and allow DNA repair machinery to access the DNA (Toiber et al., 2013). Deletion of SIRT6 causes more rapid aging in mice and increased genomic instability (Mostoslavsky et al., 2006), suggesting that changes in SIRT6 activity and H3K56 acetylation during normal aging could impact the cell’s ability to repair DNA damage. In murine embryonic stem cells, DNA damage relocates the transcriptional silencer SIRT1 from promoters to the site of DNA damage and changes gene expression in a way that resembles organismal aging (Oberdoerffer et al., 2008). This suggests that an accumulation of DNA damage and loss of SIRT1 repression may underlie age-associated epigenomic changes. Chromatin state can also impact the efficiency of DNA repair. For example, the presence of the H3K36me3 histone mark improves DNA repair efficiency (Fnu et al., 2011), but global levels of this mark and its distribution are altered with age (Ni et al., 2012; Pu et al., 2015; Sen et al., 2015). It will be interesting to determine the impact of chromatin state changes on the ability to repair DNA damage during aging.
Chromosomal Abnormalities
Aneuploidies and large-scale chromosomal rearrangements have an increased incidence with age (reviewed by Vijg and Suh, 2013). Reduced DNA methylation (hypomethylation) in mice increases the risk of aneuploidy and cancer (Gaudet et al., 2003), suggesting that the observed age-associated DNA hypomethylation could contribute to chromosome instability with age. It is important to note that these large-scale chromosomal abnormalities are distinct from other aspects of genome instability, such as mutation accumulation, in both their molecular origin and sensitivity to DNA methylation changes. Consistent with the link between chromosome abnormalities, epigenomic changes, and aging, patients with progeria diseases have an abnormally high rate of chromosome rearrangements and abnormalities (Vijg and Suh, 2013).
Transposon Activation
DNA methylation and heterochromatin help to maintain transposons in an inactive state (Slotkin and Martienssen, 2007). With age, the expression of transposons increases, and in Drosophila, this may be directly linked to age-dependent memory decline and mortality (Li et al., 2013). Because of the important role of repressive chromatin states in the maintenance of inactive transposons, age-associated transposon activation could be triggered by epigenomic changes during aging. For example, in yeast, the loss of nucleosomes with age is associated with increased expression (and perhaps increased retrotransposition) of the Ty retrotransposable element (Hu et al., 2014). In late-passage human cell culture, senescent cells undergo epigenomic changes (see “Cellular Changes: Senescence and Stem Cell Exhaustion” section) and the chromatin at transposable elements (Alu, L1, and SVA) becomes more open (De Cecco et al., 2013). Consistently, the expression of these repetitive DNA elements also increases in senescent cells (De Cecco et al., 2013). Together, these studies suggest that age-associated epigenomic changes may trigger transposon activation during aging.
Mitochondria
As cells age, mitochondrial turnover declines and mitochondrial dysfunction increases (Sun et al., 2016). Importantly, disruption of mitochondrial activity (for example, by knocking down components of the electron transport chain), mutations in mtDNA, and changes inmitochondrial biogenesis have been functionally implicated in the progression of aging (reviewed by Sun et al., 2016).
Communication between mitochondria and the nuclear regulatory genome plays an important role in the progression of aging. In yeast, signals from the mitochondria can trigger gene expression changes in the nucleus (retrograde regulation), and this regulation is required for the longer replicative lifespan of yeast lacking mtDNA (Kirchman et al., 1999). Importantly, nuclear-mitochondrial communication is also a key determinant of longevity in animals (Chang et al., 2015; Gomes et al., 2013). In worms, lifespan extension through knockdown of an electron transport chain component is dependent on a complex mitochondria-responsive transcription regulatory program that includes the ROS-sensitive transcription factors NRF2/SKN-1 and p53/CEP-1 (Chang et al., 2015). ROS signals from the mitochondria can also impact chromatin-mediated silencing in yeast (Schroeder et al., 2013). Furthermore, in C. elegans, mitochondrial dysfunction results in increased H3K9 methylation and global compaction of chromatin. This global repression of chromatin is important to allow the specific upregulation of mitochondrial stress-response genes and lifespan extension (Tian et al., 2016). Together, these studies show that changes in the efficiency or activity of mitochondria with age can result in epigenomic changes that alter lifespan.
The interaction between the nucleus and mitochondria is not one sided, and nuclear changes can also influence mitochondria. For example, changes in the activity of the transcription activator PGC-1α with age contribute to the loss of mitochondrial biogenesis and function (Rera et al., 2011; Sun et al., 2016). In aged mice, loss of SIRT1 activity (via a decline in NAD+ levels) leads to increased activity of the HIF-1α transcription factor that is linked with age-associated mitochondrial dysfunction and changes in oxidative phosphorylation (Gomes et al., 2013). Finally, changes in H3K4me2/3 deposition with age (Sun et al., 2014; Wood et al., 2010) may alter the ability of the H3K4me2/3 demethylase KDM5/RBR-2 to activate the transcription of key mitochondrial genes (Liu and Secombe, 2015). Together, these studies and others support the idea that communication between nuclear gene regulatory mechanisms and mitochondria contributes to the progression of aging. The investigation of the relative timing and order of age-dependent changes in mitochondrial function and epigenomic regulation as well as the general mechanisms of their communication will be important challenges for the future.
Proteostasis
Proteostasis (protein homeostasis) maintains a high-quality proteome by promoting the correct folding of proteins and by the degradation of proteins via the proteasome or lysosome. Many cellular and regulatory mechanisms work together to maintain proteome health in the cell, and these mechanisms can become dysregulated with age. Appropriate epigenomic regulation is key to proteostasis, and age-associated epigenomic changes can alter the ability of a cell to respond to proteostatic stress. For example, the FOXO/DAF-16 transcription factor controls the expression of many genes important for maintenance of a healthy proteome (Webb et al., 2016), and thus, dysregulation of FOXO/DAF-16 with age can disrupt proteostasis. Here we will discuss several of the transcription factors and chromatin modifiers that act to maintain the proteome.
Studies of the heat shock response have revealed much about proteostasis. In invertebrates and mammals, the transcription factor HSF1 activates expression of protein-folding chaperones in response to elevated levels of unfolded proteins (Labbadia and Morimoto, 2014; Mahat et al., 2016). In mammals, additional transcription factors, including SRF and HSF2, are important contributors to the transcriptional response to heat shock (Mahat et al., 2016). Proteostasis and the HSF-1 response decline with age (Labbadia and Morimoto, 2014). Interestingly, an increase in the amount of the repressive histone mark H3K27me3 at specific stress-response genes in C. elegans has been shown to lead to a loss of stress resistance and proteostasis (Labbadia and Morimoto, 2015). This change occurs at the very beginning of adulthood and, similar to DAF-16 regulation, may be linked to signals from the germline (Hsin and Kenyon, 1999; Labbadia and Morimoto, 2015). Consistent with a role for epigenomic changes in the decline of the heat shock response, overexpression of the H3K27me3 demethylase KDM6B/JMJD-3.1 extends C. elegans lifespan and improves stress tolerance (Labbadia and Morimoto, 2015). Thus, changes in the expression of histone-modifying enzymes and the downstream effect on chromatin state and gene expression can shape the ability of cells and animals to withstand stress early during aging (Labbadia and Morimoto, 2015). This suggests the exciting idea that epigenomic changes are one of the first to occur during aging and that these changes trigger the emergence of other hallmarks of aging (Labbadia and Morimoto, 2015).
Another means of responding to elevated levels of unfolded proteins is the unfolded protein response (UPR). There are two distinct UPRs—UPRmt and UPRER—that monitor and respond to the presence of unfolded proteins in the mitochondria and the endoplasmic reticulum, respectively. In addition to the presence of high levels of unfolded proteins in the mitochondria, the UPRmt responds to unbalanced levels of nuclear:mitochondrial-encoded oxidative phosphorylation enzymes, mtDNA depletion, and decline of the respiratory chain (Schulz and Haynes, 2015). In C. elegans, the UPRmt is activated primarily by a change in the subcellular localization (from the mitochondria to the nucleus) of the transcription factor ATFS-1. Once present in the nucleus, ATFS-1 regulates the expression of many genes, including genes important for proteostasis, detoxification, and metabolism (reviewed by Schulz and Haynes, 2015). In mammals, the transcription factor CHOP mediates the UPRmt gene expression changes in response to unfolded proteins in the mitochondria (Schulz and Haynes, 2015), although CHOP and ATFS-1 are not evolutionarily related. Interestingly, changes in the methylation of H3K9 and H3K27 have recently been found to be important for the induction of genes involved in the UPRmt in C. elegans, mice, and human cells (Merkwirth et al., 2016; Tian et al., 2016). Induction of the UPRER results in many transcriptional changes through the activation of the transcription regulators XBP1, NFκB, and ATF4 (Taylor, 2016). In C. elegans, the XBP-1 branch of the UPRER declines relatively early with age and overexpression of XBP-1 in neurons can extend lifespan, suggesting an important role for the UPRER in longevity (reviewed by Taylor, 2016). Importantly, XBP1 and the UPRER have also been found to play a role in human neurodegenerative diseases, including Alzheimer’s disease, where UPRER/XBP1 activation is thought to be neuroprotective early in the disease onset but then a contributor to neuronal death during later disease stages (reviewed by Taylor, 2016).
Neurodegenerative diseases, including Alzheimer’s disease, are characterized by a loss of youthful proteostasis and an increase in protein aggregation. Interestingly, a recent study using human brain tissue identified dysregulation of the transcription factor REST (also known as NRSF) as a key, early event in the development of Alzheimer’s disease (Lu et al., 2014). REST, and its invertebrate ortholog SPR-4, protects neurons from amyloid β-protein toxicity as well as oxidative stress (Lu et al., 2014). Interestingly, loss of REST activity during early Alzheimer’s disease was associated with a global gain of the activating mark H3K9ac (Lu et al., 2014), suggesting a potential link between changes in REST, chromatin state, and proteostasis. The identification of early molecular changes, such as REST activity, in neurodegenerative disease will be critical for the development of therapeutics that could act to halt or reverse the cognitive decline associated with neurodegenerative diseases.
Nutrient Sensing
Changes in nutrient sensing and signaling are important hallmarks of aging (López-Otín et al., 2013). Many of the nutrient-sensing mechanisms in the cell are dysregulated with age. Interestingly, nutrient sensing, mitochondria, and epigenomic regulation are interconnected (Berger and Sassone-Corsi, 2015), and these links are likely to be important for the progression of aging.
For example, sirtuins can regulate gene expression by deacetylating histone and non-histone substrates, and this activity depends on a cofactor linked to the cellular energy status, NAD+ (nicotinamide adenine dinucleotide) (Houtkooper et al., 2012). Changes in the availability of NAD+ during aging can affect the activity of the sirtuins, yielding altered gene expression and chromatin state with age. Because NAD+ levels are sensitive to diet and exercise (Cantó et al., 2015), the reliance of sirtuins on NAD+ can also be taken advantage of to slow or reverse aging using dietary changes or increased exercise (Guarente, 2013).
The cAMP-responsive element-binding protein (CREB) interacts with many co-activators (cAMP-regulated transcriptional co-activators, or CRTCs) to alter gene expression in response to fasting and feeding signals (Altarejos and Montminy, 2011). Consistent with an important role for this nutrient-sensing mechanism in longevity, knockdown of CRTC-1 in C. elegans extends lifespan (Burkewitz et al., 2015). Many upstream players modulate the activity of these regulatory elements via phosphorylation to allow this highly conserved regulatory system to respond to a diversity of nutrient signals. For example, in response to short-term fasting, CREB, CRTC2, TBP-association factor 4 (TAF-4), and p300/CREB-binding protein (CBP) bind together and activate gluconeogenesis gene expression, whereas following feeding, a different set of post-translational modifications disrupts binding, alters subcellular localization, and promotes degradation (Altarejos and Montminy, 2011). Interestingly, in mammals, p300/CBP also acts as a CREB co-activator through histone acetylation (e.g., H3K27ac and H3K14ac), suggesting a link between nutrient sensing, chromatin, and gene expression (Altarejos and Montminy, 2011).
AMPK, a pro-longevity protein kinase, is activated by low cellular energy (Burkewitz et al., 2014). Through the phosphorylation of transcription factors, AMPK links cellular energy status with transcription regulation. For example, AMPK-mediated phosphorylation can promote longevity and gene expression changes, in part by modulating the pro-longevity transcription factor FOXO/DAF-16 (Greer et al., 2007). In contrast, AMPK-mediated phosphorylation can also repress transcription factors (and their transcriptional target genes) that are detrimental to lifespan, such as the transcriptional co-activator CRTC (Mair et al., 2011). These examples suggest that changes in metabolism and cellular energy availability can trigger changes in transcription factor activity and gene expression during aging.
Cellular Changes: Senescence and Stem Cell Exhaustion
An increase in the number of senescent cells and a decline in tissue regeneration due to the loss of stem cell proliferation are both hallmarks of aging (López-Otín et al., 2013). Clearing senescent cells from adult mice by inducing apoptosis with the drug ABT263 or an inducible transgene rejuvenates hematopoietic and muscle stem cells, slows the onset of age-related disease, and increases lifespan (Baker et al., 2016; Chang et al., 2016). These studies strongly support a role for senescent cells in the progression of aging. Senescence can be triggered by several factors, including DNA damage and telomere dysfunction (Rai and Adams, 2012; Rodier and Campisi, 2011), two aspects of aging that are intimately linked with chromatin. Interestingly, knockdown of the histone acetyltransferase p300 results in hypoacetylation of histones H3 and H4 and an increase in senescence in human cells, providing evidence for a causative link between changes in chromatin state and senescence (Prieur et al., 2011). Importantly, this chromatin-based induction of senescence occurred independently of other mechanisms of inducing senescence, including DNA damage and activation of the CDK inhibition pathway (p53/p21/p16) (Prieur et al., 2011). The redistribution of heterochromatin is also emerging as a key player in increased senescence and stem cell loss with age. Patients with the progeroid diseases HGPS and Werner syndrome have abnormally high levels of senescent cells and low proliferative potential, and these changes are thought to be the result of a loss of lamin-associated heterochromatin (Shumaker et al., 2006; Zhang et al., 2015b). Senescence is also associated with heterochromatin gains in the form of H3K9me2/3-enriched senescence-associated heterochromatin foci (Rai and Adams, 2012). Importantly, senescence-associated heterochromatin foci are comprised of genomic regions distinct from lamin-associated genomic regions that experience a loss of heterochromatin in progeroid models (Rai and Adams, 2012; Zhang et al., 2015b). Together, these studies provide strong evidence that links epigenomic changes with senescence.
The transcription factor p53 is a key regulator of senescence and can respond to many of the known triggers of senescence. For example, nuclear lamin defects can activate p53 and induce expression of p53 target genes (Varela et al., 2005). p53 also promotes senescence in response to DNA damage and telomere attrition (Rufini et al., 2013). Interestingly, a mouse mutant that prevents the post-translational activating modification of p53 in response to DNA damage is short lived, suggesting that the activity of p53 is important for longevity. Importantly, too much p53 activity, for example, from constitutive activation, is also detrimental to longevity (reviewed by Rufini et al., 2013). Thus, appropriate regulation of the transcription factor p53 is key for longevity, and its dysregulation can lead to more rapid aging and increased senescence.
Stem cell exhaustion and a decline in stem cell function can result from the induction of senescence as well as from altered epigenomic regulation (Beerman and Rossi, 2015). Compared to youthful stem cells, aged stem cell populations have a unique chromatin state and gene expression profile (Beerman et al., 2013; Liu et al., 2013; Sun et al., 2014). Appropriate epigenomic regulation is key to stem cell function and differentiation, and thus, these epigenomic changes are likely to contribute to the progression of aging by altering the regenerative capacity of an animal (Beerman and Rossi, 2015).
Altered Intercellular Communication
Intercellular communication is important for the health of an animal and can become altered with age. This hallmark of aging includes hormonal signaling (e.g., insulin/IGF signaling) as well as changes to the immune system (López-Otín et al., 2013). Here, we will focus on the role of chromatin and transcription regulation in the age-dependent increase in the inflammatory response (“inflammaging”). Increased numbers of senescent cells with age are an important contributor to the age-associated increase in inflammation because senescent cells secrete a large number of pro-inflammatory proteins (senescence-associated secretory phenotype, or SASP) (Rodier and Campisi, 2011). Thus, age-dependent epigenomic changes associated with the induction of senescence may underlie inflammaging. Consistent with this idea, the H3K4 methyltransferase MLL1 is required for expression of SASP-related genes and promotes increased inflammation (Capell et al., 2016). Furthermore, the senescence-triggered appearance of H3K27me3-depleted “canyons” within genic and enhancer regions is associated with upregulation of SASP-related genes (Shah et al., 2013). Thus, epigenomic changes during senescence are important contributors to age-associated increased inflammation.
Further evidence for the role of inflammation in aging comes from gene expression studies. Inflammatory pathways and genes are upregulated in older animals, including humans (Tilstra et al., 2011). The pro-inflammatory transcription factors NF-κB, STAT1, and STAT3 are mediating many of these changes (O’Brown et al., 2015; Tilstra et al., 2011). Consistent with a causative role in aging, the activity of these transcription factors increases with age in humans, and reducing NF-κB activity ameliorates mouse models of progeria and reverses age-associated pathology (O’Brown et al., 2015; Tilstra et al., 2011). NF-κB is regulated by many upstream factors and regulators including cytokines, growth factors, the deacetylases SIRT1 and SIRT6, and the transcription factor FOXO (Kawahara et al., 2009; Salminen et al., 2008). Together, these upstream regulators and increased pro-inflammatory cytokines from senescent cells lead to increased NF-κB activity and gene expression changes with age. Moreover, NF-κB activates the expression of the H3K27me3 demethylase Jmjd3 in immune cells, leading to a loss of H3K27me3 (De Santa et al., 2007). Thus, age-associated increased NF-κB activity could alter the chromatin state of an aging cell by directly increasing the expression of a histone-modifying enzyme. It will be interesting to determine the extent to which chromatin state is altered as a result of changes in the inflammatory milieu with age.
Environmental Signals
Recently, several other important effectors of the aging process have emerged. Inter-individual (social) interactions and social caste may be linked with longevity through chromatin marks. In C. elegans, the presence of males shortens hermaphrodite lifespan (Maures et al., 2014; Shi and Murphy, 2014), and this phenomenon may involve male-induced changes in chromatin via the H3K27me3 demethylase UTX-1 (Maures et al., 2014). Social insects may also provide unique insights into the regulation of longevity; the social castes of ants and bees have distinct behaviors, reproductive capacity, and life-spans that could serve as a model for the role of chromatin in aging. Comparisons between individuals from different castes have revealed that they have caste-specific DNA methylation and H3K27ac patterns that are functionally important for their behavior (Simola et al., 2016; Yan et al., 2015). It is unknown if caste-specific chromatin states affect lifespan, but this will be an exciting question to address in the future.
The dysregulation of circadian rhythm has recently been identified as a hallmark of aging (Kondratov, 2007; Orozco-Solis and Sassone-Corsi, 2014). Genetic and environmental manipulations that perturb circadian rhythm shorten lifespan and result in metabolic defects and diseases including obesity and diabetes (Kondratov, 2007). Consistent with a link between metabolism and circadian rhythm, the ability of caloric restriction to extend Drosophila lifespan and improve fat metabolism depends on circadian regulation (Katewa et al., 2016). Interestingly, the NAD+-dependent deacetylase SIRT1 may be a molecular link between metabolism, circadian rhythm, and aging (Orozco-Solis and Sassone-Corsi, 2014). SIRT1 activity is circadian, and through an interaction with components of the circadian transcription regulatory network and an H3K4 methyltransferase, SIRT1 helps to control circadian gene expression patterns (Aguilar-Arnal et al., 2015; Orozco-Solis and Sassone-Corsi, 2014). Independently of metabolism, chromatin and nucleosome remodeling are important contributors to the control of circadian rhythm (Kim et al., 2014; Tamayo et al., 2015). Because of the importance of precise gene regulation for circadian rhythm, age-dependent epigenomic alterations could result in a disruption of circadian rhythm. In addition, the dysregulation of circadian gene expression could itself disrupt the epigenome.
Anti-aging Interventions
Once aging has begun and age-associated changes accumulate, is it possible to stop the progression of aging or even to return to a more youthful state? Given the interconnectedness of the hallmarks of aging, a therapeutic that directly impacts only one contributor to aging could ameliorate other hallmarks of aging. As described above, epigenomic regulation is highly connected to the other hallmarks of aging, and dysregulation of gene expression and the epigenome promote the progression of aging. Thus, a therapeutic that restores the regulatory landscape could have a far-reaching impact in slowing or reversing the progression of aging (Rando and Chang, 2012). Consistent with this idea, genetic manipulations that affect chromatin state and chromatin modifiers can increase lifespan (reviewed by Benayoun et al., 2015). If an intervention or therapeutic is able to rejuvenate an organism, it would be able to reverse the appearance of some or all of the hallmarks of aging, even when initiated in an older organism. For example, the intervention might restore the patterning of histone marks to a more youthful state. Several interventions have shown promise for rejuvenation and amelioration of the hallmarks of aging: parabiosis, increased exercise, changes in diet, and reprogramming. Recently developed drugs targeting epigenetic modifiers may also provide a targeted way to slow aging. Some of these interventions can rejuvenate some hallmarks in an old animal and restore it to a more youthful state, while others may primarily act to slow the progression of aging without reversing it (Figure 4). Here we will discuss the potential effect of the major interventions on the aging epigenome.
Figure 4. The Aging Snowball Effect.
Aging is the result of the accumulation of dysregulation and damage in a snowball effect that eventually ends in death. A small change or dysfunction of a cellular protective mechanism (for example, transcriptional networks or chromatin state) can begin the aging process by triggering the emergence of other age-associated changes. However, the aging snowball effect and the appearance of age-associated disease and dysfunction can be delayed or even reversed using aging therapeutics and interventions (reprogramming, parabiosis, epigenetic drugs, exercise, and diet). Eventually, the accumulation of age-associated changes may become so great that a “point of no return” is reached and interventions cannot extend lifespan or healthspan.
Parabiosis
Heterochronic parabiosis (a technique that links the circulatory systems of an old and young animal) can “rejuvenate” several hallmarks of aging in an old mouse (Conboy et al., 2005; Sinha et al., 2014). Many of the molecular signatures of aging, including stem cell exhaustion, altered intercellular signaling, and a loss of genome integrity, are reversed in an old mouse as a result of heterochronic parabiosis, suggesting that the epigenome and gene expression could also be restored to a more youthful state by this treatment (Rando and Chang, 2012; Sinha et al., 2014) (Figure 4). Epigenetic state (including chromatin marks and gene expression) are highly responsive to the environment, and, therefore, one might expect that exposure to more youthful circulating factors could reverse or slow the appearance of age-related epigenetic changes. In the future, it will be interesting to determine if heterochronic parabiosis can restore a more youthful epigenome and if the previously observed, rejuvenating effects of parabiosis are dependent on epigenetic changes.
Exercise and Diet
Exercise and dietary changes, including caloric restriction and intermittent fasting, can slow and even reverse the progression of aging (Brandhorst et al., 2015; Fontana and Partridge, 2015; Garatachea et al., 2015; Mercken et al., 2012) (Figure 4). There are also drugs, such as rapamycin and metformin, that are able to mimic the effects of exercise and diet changes (Mercken et al., 2012). Changes in diet and exercise impact the activity of various longevity regulatory mechanisms, including insulin/IGF signaling and the nutrient-sensing kinase AMPK, and can alter the expression of many genes (Fontana and Partridge, 2015; Garatachea et al., 2015). The effects of caloric restriction can be long lasting and even be passed on to the next generation (Fontana and Partridge, 2015). Recently, it was reported that caloric restriction in yeast involves nucleosome remodeling (Dang et al., 2014). This result may point toward a possible chromatin-based mechanism for the persistence of the pro-longevity effects of dietary changes. Importantly, the initiation timing and duration of these metabolic interventions (exercise and diet changes) can alter their effect on longevity, suggesting that context is an important factor and that these are not “one size fits all” interventions (Fontana and Partridge, 2015).
Reprogramming
The ability of an old nucleus to become pluripotent and youthful again is a phenomenon that occurs as a result of fertilization and that can be mimicked in the lab through nuclear transfer or by inducing pluripotency through the expression of particular transcription factors (Rando and Chang, 2012). While it is clear that fertilization completely resets the aging clock, the extent to which laboratory induction of pluripotency can reset the aging clock is an ongoing question. Reprogramming cells from elderly humans (as old as 101 years of age) into pluripotent cells erases many of the hallmarks of aging and restores a youthful gene expression profile that is maintained during differentiation (Lapasset et al., 2011; Mertens et al., 2015). This suggests that reprogramming can reset the aging clock even for very old cells and could indicate that there is no point during aging when this therapeutic would not be effective (Figure 4). However, induced neurons from old cells retain some age-associated phenotypes (Mertens et al., 2015), suggesting that passage through a fully pluripotent state may be necessary to completely reset the aging clock. The induction of pluripotency and the trans-differentiation of cells require different sets of transcription factors (Graf, 2011) and are thus likely to result in distinct changes in transcriptional networks and chromatin state. Comparing the transcriptional networks and chromatin states of cells that have passed through a fully pluripotent state versus trans-differentiation may provide exciting insights into which epigenomic changes are necessary to reset the aging clock by reprogramming.
Epigenetic Drugs: Potential Therapeutics?
Several chemical inhibitors of chromatin-modifying enzymes have been recently developed (Helin and Dhanak, 2013). Given the important role of chromatin modifications in the progression of aging, it is possible that these drugs could be utilized to slow the progression of aging or to treat age-related diseases. One example of an epigenetic targeting drug is Remodelin, a small molecule that inhibits the acetyl-transferase protein NAT10 (Larrieu et al., 2014). Excitingly, when cells derived from patients with the progeria disease HGPS were treated with the drug Remodelin, the cells had improved nuclear architecture and increased chromatin compaction, consistent with a gain of heterochromatin (Larrieu et al., 2014). While the ability of this drug to slow the progression of normal aging has not been tested, epigenetic targeting drugs hold promise as new therapeutics for slowing or reversing the hallmarks of aging. Identifying compounds that specifically target the chromatin features that are most affected with age, and identifying ways to deliver these compounds in a specific manner to target tissues and cells, will be critical.
Outlook
Aging is a gradual process that ultimately ends in the death of an organism. Together, numerous studies in different model organisms have revealed that aging is the result of dysfunction of a complex network that maintains organismal health, tissue homeostasis, and stress resistance. The elucidation of exactly how this network is connected and how it begins to go awry will be key to understanding aging. Gene regulation is a crucial hub in this network; epigenetic marks and transcription factors are pivotal for nearly every cellular process, and age-associated changes in gene regulation could in turn trigger the emergence of other hallmarks of aging in a snowball effect (Figure 4).
While it is clear that epigenomic changes are an important aspect of the progression of aging, it is unclear exactly how these changes arise. Because most genomic profiling experiments have been performed using whole tissues or whole organisms, two mechanisms could underlie the observed, age-associated epigenomic changes: (1) the exact cellular composition of a tissue or organism may change with age (even within highly purified populations of cells), and/or (2) the gene expression and chromatin state within the cells that normally reside in a particular tissue may change. Newly developed technologies that profile single cells (e.g., assay for transposase-accessible chromatin sequencing [ATAC-seq] and RNA sequencing [RNA-seq]) will help to address this crucial question (Wang and Navin, 2015). Indeed, age-associated gene expression changes have recently been identified at the single-cell level in hematopoietic stem and progenitor cells (Kowalczyk et al., 2015), suggesting that at least for some tissues, changes in the cellular composition of a tissue with age cannot fully explain age-associated epigenomic dysregulation. Untangling cell composition changes from cell-intrinsic changes will be particularly important for understanding which epigenomic features could serve as reliable biomarkers of age.
Significant progress has been made in increasing our understanding of the epigenomic changes that occur with age. However, mysteries still abound. First, the molecular players in gene regulation are numerous and many more chromatin marks and even transcription factor families are still being discovered (Lohse et al., 2010; Luo et al., 2015). A more systematic understanding of the role and precise timing of age-associated changes in well-known and newly discovered or poorly understood transcription factors, histone marks, and DNA methylation states could provide unique insight into aging. Simple model organisms, such as S. cerevisiae, C. elegans, and Drosophila, provide an essential experimental platform for the detailed and systematic probing of the role of epigenomic changes with age. The ability to perform precise genome editing and to subtly manipulate these simple model systems will allow us to gain a better understanding of the extent to which the small epigenomic changes that accumulate during normal aging could overwhelm the cellular “buffering” mechanisms and throw cellular pathways into disequilibrium. Additionally, their short lifespan and ease of cultivation make it possible to profile the progression of aging at a very fine timescale and precisely identify which hallmarks of aging are the first to go awry.
Another key point in the role of epigenomic changes with age is how malleable versus stable epigenomic networks are. Understanding how environmental stimuli can modulate these networks will not only increase our understanding of aging, but could also lead to the discovery of new (or repurposed) compounds that can slow or even reverse the progression of aging. Indeed, while compounds targeting transcription factor activity are notoriously difficult to generate and use, compounds that affect chromatin-modifying enzymes have recently been used to successfully treat cancer (Helin and Dhanak, 2013). It would be interesting to test if some of these compounds that take advantage of the malleability of epigenomic networks could be repurposed for aging or other aging-related diseases. On the other side of the spectrum, the stable aspect of some chromatin changes raises the important possibility that some changes that occur very early (for example, during development) could affect the course of aging in adults. In fact, changes in one generation could even influence the gametes and perhaps be passed on to next generations (Lim and Brunet, 2013). It will be important to understand further the role of chromatin modifiers in the transgenerational effects of lifespan.
Finally, we do not fully understand the molecular basis of interand intra-species lifespan differences. The diversity of lifespans between species, even closely related species, illustrates that aging and longevity evolve and suggests that unique molecular mechanisms may underlie the exceptionally long or short lifespans found in nature. Interestingly, the evolution of transcription regulatory networks is a rich source of phenotypic diversity (Carroll, 2008). Whether similar evolutionary changes could underlie the diversity of lifespans is an open question. However, it seems likely that this will be the case because not only are changes in gene expression and regulation common throughout evolution, but these changes can also profoundly impact the ability of a cell or organism to withstand stress in a changing environment and throughout an animal’s lifespan.
Lifespan differences also exist between individuals of the same species. For example, sexual dimorphisms in genetics (due to the presence of a different chromosome) and epigenetics (due to the induction of different gene expression programs) could inform the molecular basis of sex-specific lifespans, agerelated disease susceptibility, and responses to interventions. The study of “extreme” versions of individual-to-individual lifespan variation (for example, in social insect castes) is likely to reveal that this variation is primarily the result of epigenetic differences, and could serve as a great model to provide key insights into the role of epigenomic changes in longevity variation. A better understanding of individual-to-individual variations in lifespan trajectories, even among genetically identical animals, and how epigenomic changes could contribute to these different trajectories, will be critical to our understanding of mysteries of aging.
Acknowledgments
Our apologies to the authors of work we could not cite due to space constraints. We are grateful to Ashley Webb and the members of the Brunet lab, especially Bérénice Benayoun and Itamar Harel, for helpful comments and discussion. This work was supported by the NIH grant DP1AG044848 (A.B.) and the Helen Hay Whitney Foundation (L.N.B.).
REFERENCES
- Aguilar-Arnal L, Katada S, Orozco-Solis R, Sassone-Corsi P. NAD(+)-SIRT1 control of H3K4 trimethylation through circadian deacetylation of MLL1. Nat. Struct. Mol. Biol. 2015;22:312–318. doi: 10.1038/nsmb.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrahami D, Li C, Zhang J, Schug J, Avrahami R, Rao S, Stadler MB, Burger L, Schübeler D, Glaser B, Kaestner KH. Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved β cell function. Cell Metab. 2015;22:619–632. doi: 10.1016/j.cmet.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, Saltness RA, Jeganathan KB, Verzosa GC, Pezeshki A, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart M, Groth M, Priebe S, Savino A, Testa G, Dix A, Ripa R, Spallotta F, Gaetano C, Ori M, et al. RNA-seq of the aging brain in the short-lived fish N. furzeri—conserved pathways and novel genes associated with neurogenesis. Aging Cell. 2014;13:965–974. doi: 10.1111/acel.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I, Rossi DJ. Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell. 2015;16:613–625. doi: 10.1016/j.stem.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I, Bock C, Garrison BS, Smith ZD, Gu H, Meissner A, Rossi DJ. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell. 2013;12:413–425. doi: 10.1016/j.stem.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158:673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015;16:593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Sassone-Corsi P. Metabolic signaling to chromatin. Cold Spring Harb. Perspect. Biol. 2015 doi: 10.1101/cshperspect.a019463. Published online August 20, 2015. http://dx.doi.org/10.1101/cshperspect.a019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, Isik M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 2015;88(Pt B):290–301. doi: 10.1016/j.freeradbiomed.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkis IM, Przybylski D, Chen J, Regev A. Changes in nucleosome occupancy associated with metabolic alterations in aged mammalian liver. Cell Rep. 2014;9:996–1006. doi: 10.1016/j.celrep.2014.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandhorst S, Choi IY, Wei M, Cheng CW, Sedrakyan S, Navarrete G, Dubeau L, Yap LP, Park R, Vinciguerra M, et al. A periodic diet that mimics fasting promotes multi-system regeneration, enhanced cognitive performance, and healthspan. Cell Metab. 2015;22:86–99. doi: 10.1016/j.cmet.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RC, Misteli T, Oberdoerffer P. DNA damage, chromatin, and transcription: the trinity of aging. Curr. Opin. Cell Biol. 2012;24:724–730. doi: 10.1016/j.ceb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkewitz K, Zhang Y, Mair WB. AMPK at the nexus of energetics and aging. Cell Metab. 2014;20:10–25. doi: 10.1016/j.cmet.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkewitz K, Morantte I, Weir HJ, Yeo R, Zhang Y, Huynh FK, Ilkayeva OR, Hirschey MD, Grant AR, Mair WB. Neuronal CRTC-1 governs systemic mitochondrial metabolism and lifespan via a catecholamine signal. Cell. 2015;160:842–855. doi: 10.1016/j.cell.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Menzies KJ, Auwerx J. NAD(+) metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 2015;22:31–53. doi: 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell BC, Drake AM, Zhu J, Shah PP, Dou Z, Dorsey J, Simola DF, Donahue G, Sammons M, Rai TS, et al. MLL1 is essential for the senescence-associated secretory phenotype. Genes Dev. 2016;30:321–336. doi: 10.1101/gad.271882.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Chang HW, Shtessel L, Lee SS. Collaboration between mitochondria and the nucleus is key to long life in Caenorhabditis elegans. Free Radic. Biol. Med. 2015;78:168–178. doi: 10.1016/j.freeradbiomed.2014.10.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Wang Y, Shao L, Laberge RM, Demaria M, Campisi J, Janakiraman K, Sharpless NE, Ding S, Feng W, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Chen Z, Wu D, Zhang L, Lin X, Su J, Rodriguez B, Xi Y, Xia Z, Chen X, et al. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat. Genet. 2015;47:1149–1157. doi: 10.1038/ng.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Sutphin GL, Dorsey JA, Otte GL, Cao K, Perry RM, Wanat JJ, Saviolaki D, Murakami CJ, Tsuchiyama S, et al. Inactivation of yeast Isw2 chromatin remodeling enzyme mimics longevity effect of calorie restriction via induction of genotoxic stress response. Cell Metab. 2014;19:952–966. doi: 10.1016/j.cmet.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell. 2013;12:247–256. doi: 10.1111/acel.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schübeler D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527:105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelenboom A, Burgering BM. FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 2013;14:83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- Eijkelenboom A, Mokry M, Smits LM, Nieuwenhuis EE, Burgering BM. FOXO3 selectively amplifies enhancer activity to establish target gene regulation. Cell Rep. 2013;5:1664–1678. doi: 10.1016/j.celrep.2013.11.031. [DOI] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol. Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. USA. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L. Promoting health and longevity through diet: from model organisms to humans. Cell. 2015;161:106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu VX, Dobosy JR, Desotelle JA, Almassi N, Ewald JA, Srinivasan R, Berres M, Svaren J, Weindruch R, Jarrard DF. Aging and cancer-related loss of insulin-like growth factor 2 imprinting in the mouse and human prostate. Cancer Res. 2008;68:6797–6802. doi: 10.1158/0008-5472.CAN-08-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garatachea N, Pareja-Galeano H, Sanchis-Gomar F, Santos-Lozano A, Fiuza-Luces C, Morán M, Emanuele E, Joyner MJ, Lucia A. Exercise attenuates the major hallmarks of aging. Rejuvenation Res. 2015;18:57–89. doi: 10.1089/rej.2014.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T. Historical origins of transdifferentiation and reprogramming. Cell Stem Cell. 2011;9:504–516. doi: 10.1016/j.stem.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Grammatikakis I, Panda AC, Abdelmohsen K, Gorospe M. Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging (Albany, N.Y.) 2014;6:992–1009. doi: 10.18632/aging.100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Becker B, Latza C, Antebi A, Shi Y. Mutation of C. elegans demethylase spr-5 extends transgenerational longevity. Cell Res. 2016;26:229–238. doi: 10.1038/cr.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Calorie restriction and sirtuins revisited. Genes Dev. 2013;27:2072–2085. doi: 10.1101/gad.227439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K, Dhanak D. Chromatin proteins and modifications as drug targets. Nature. 2013;502:480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989;342:749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Hu Z, Chen K, Xia Z, Chavez M, Pal S, Seol JH, Chen CC, Li W, Tyler JK. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014;28:396–408. doi: 10.1101/gad.233221.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Pawlikowski J, Manoharan I, van Tuyn J, Nelson DM, Rai TS, Shah PP, Hewitt G, Korolchuk VI, Passos JF, et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 2013;202:129–143. doi: 10.1083/jcb.201212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Li J, Green CD, Yu X, Tang X, Han D, Xian B, Wang D, Huang X, Cao X, et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14:161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa SD, Akagi K, Bose N, Rakshit K, Camarella T, Zheng X, Hall D, Davis S, Nelson CS, Brem RB, et al. Peripheral circadian clocks mediate dietary restriction-dependent changes in lifespan and fat metabolism in Drosophila. Cell Metab. 2016;23:143–154. doi: 10.1016/j.cmet.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, Chua KF. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya A, Lobanov AV, Gladyshev VN. Evidence that mutation accumulation does not cause aging in Saccharomyces cerevisiae. Aging Cell. 2015;14:366–371. doi: 10.1111/acel.12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, Franceschi C, Lithgow GJ, Morimoto RI, Pessin JE, et al. Geroscience: linking aging to chronic disease. Cell. 2014;159:709–713. doi: 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Kwak PB, Weitz CJ. Specificity in circadian clock feedback from targeted reconstitution of the NuRD corepressor. Mol. Cell. 2014;56:738–748. doi: 10.1016/j.molcel.2014.10.017. [DOI] [PubMed] [Google Scholar]
- Kim J, Sturgill D, Tran AD, Sinclair DA, Oberdoerffer P. Controlled DNA double-strand break induction in mice reveals post-damage transcriptome stability. Nucleic Acids Res. 2016;44:e64. doi: 10.1093/nar/gkv1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV. A role of the circadian system and circadian proteins in aging. Ageing Res. Rev. 2007;6:12–27. doi: 10.1016/j.arr.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, Regev A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI. Proteostasis and longevity: when does aging really begin? F1000Prime Rep. 2014;6:7. doi: 10.12703/P6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI. Repression of the heat shock response is a programmed event at the onset of reproduction. Mol. Cell. 2015;59:639–650. doi: 10.1016/j.molcel.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Aït-Hamou N, Leschik J, Pellestor F, Ramirez JM, De Vos J, et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011;25:2248–2253. doi: 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014;344:527–532. doi: 10.1126/science.1252651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson K, Yan SJ, Tsurumi A, Liu J, Zhou J, Gaur K, Guo D, Eickbush TH, Li WX. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012;8:e1002473. doi: 10.1371/journal.pgen.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- Li L, Greer C, Eisenman RN, Secombe J. Essential functions of the histone demethylase lid. PLoS Genet. 2010;6:e1001221. doi: 10.1371/journal.pgen.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Prazak L, Chatterjee N, Grüninger S, Krug L, Theodorou D, Dubnau J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 2013;16:529–531. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JP, Brunet A. Bridging the transgenerational gap with epigenetic memory. Trends Genet. 2013;29:176–186. doi: 10.1016/j.tig.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Secombe J. The histone demethylase KDM5 activates gene expression by recognizing chromatin context through its PHD reader motif. Cell Rep. 2015;13:2219–2231. doi: 10.1016/j.celrep.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Zordan RE, Cain CW, Johnson AD. Distinct class of DNA-binding domains is exemplified by a master regulator of phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA. 2010;107:14105–14110. doi: 10.1073/pnas.1005911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature. 2014;507:448–454. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo GZ, Blanco MA, Greer EL, He C, Shi Y. DNA N(6)-methyladenine: a new epigenetic mark in eukaryotes? Nat. Rev. Mol. Cell Biol. 2015;16:705–710. doi: 10.1038/nrm4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol. Cell. 2016;62:63–78. doi: 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Morantte I, Rodrigues AP, Manning G, Montminy M, Shaw RJ, Dillin A. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470:404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10:980–990. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maures TJ, Booth LN, Benayoun BA, Izrayelit Y, Schroeder FC, Brunet A. Males shorten the life span of C. elegans hermaphrodites via secreted compounds. Science. 2014;343:541–544. doi: 10.1126/science.1244160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl G, Killilea DW, Hubbard AE, Vantipalli MC, Melov S, Lithgow GJ. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J. Biol. Chem. 2008;283:350–357. doi: 10.1074/jbc.M705028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercken EM, Carboneau BA, Krzysik-Walker SM, de Cabo R. Of mice and men: the benefits of caloric restriction, exercise, and mimetics. Ageing Res. Rev. 2012;11:390–398. doi: 10.1016/j.arr.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, et al. Two conserved histone demethylases regulate mitochondrial stress-induced longevity. Cell. 2016 doi: 10.1016/j.cell.2016.04.012. Published online April 27, 2016. http://dx.doi.org/10.1016/j.cell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Paquola AC, Ku M, Hatch E, Böhnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Herdy JR, et al. Directly reprogrammed human neurons retain aging-associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell Stem Cell. 2015;17:705–718. doi: 10.1016/j.stem.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, Nakatogawa H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L. H19 lncRNA controls gene expression of the imprinted gene network by recruiting MBD1. Proc. Natl. Acad. Sci. USA. 2013;110:20693–20698. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Ni Z, Ebata A, Alipanahiramandi E, Lee SS. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell. 2012;11:315–325. doi: 10.1111/j.1474-9726.2011.00785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brown ZK, Van Nostrand EL, Higgins JP, Kim SK. The inflammatory transcription factors NFκB, STAT1 and STAT3 drive age-associated transcriptional changes in the human kidney. PLoS Genet. 2015;11:e1005734. doi: 10.1371/journal.pgen.1005734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Sinclair DA. The role of nuclear architecture in genomic instability and ageing. Nat. Rev. Mol. Cell Biol. 2007;8:692–702. doi: 10.1038/nrm2238. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Solis R, Sassone-Corsi P. Circadian clock: linking epigenetics to aging. Curr. Opin. Genet. Dev. 2014;26:66–72. doi: 10.1016/j.gde.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 2015;16:245–257. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- Prieur A, Besnard E, Babled A, Lemaitre JM. p53 and p16(INK4A) independent induction of senescence by chromatin-dependent alteration of S-phase progression. Nat. Commun. 2011;2:473. doi: 10.1038/ncomms1473. [DOI] [PubMed] [Google Scholar]
- Pu M, Ni Z, Wang M, Wang X, Wood JG, Helfand SL, Yu H, Lee SS. Trimethylation of Lys36 on H3 restricts gene expression change during aging and impacts life span. Genes Dev. 2015;29:718–731. doi: 10.1101/gad.254144.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MM, Sykiotis GP, Nishimura M, Bodmer R, Bohmann D. Declining signal dependence of Nrf2-MafS-regulated gene expression correlates with aging phenotypes. Aging Cell. 2013;12:554–562. doi: 10.1111/acel.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai TS, Adams PD. Lessons from senescence: chromatin maintenance in non-proliferating cells. Biochim. Biophys. Acta. 2012;1819:322–331. doi: 10.1016/j.bbagrm.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando TA, Chang HY. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 2012;148:46–57. doi: 10.1016/j.cell.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jr, Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel CG, Dowen RH, Lourenco GF, Kirienko NV, Heimbucher T, West JA, Bowman SK, Kingston RE, Dillin A, Asara JM, Ruvkun G. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 2013;15:491–501. doi: 10.1038/ncb2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Campisi J. Four faces of cellular senescence. J. Cell Biol. 2011;192:547–556. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: the critical roles of p53. Oncogene. 2013;32:5129–5143. doi: 10.1038/onc.2012.640. [DOI] [PubMed] [Google Scholar]
- Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr. Opin. Cell Biol. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Huuskonen J, Ojala J, Kauppinen A, Kaarniranta K, Suuronen T. Activation of innate immunity system during aging: NF-kB signaling is the molecular culprit of inflamm-aging. Ageing Res. Rev. 2008;7:83–105. doi: 10.1016/j.arr.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutte C, Yang AS, Beart RW, Jones PA. Base excision repair of U;G mismatches at a mutational hotspot in the p53 gene is more efficient than base excision repair of T;G mismatches in extracts of human colon tumors. Cancer Res. 1995;55:3742–3746. [PubMed] [Google Scholar]
- Schroeder EA, Raimundo N, Shadel GS. Epigenetic silencing mediates mitochondria stress-induced longevity. Cell Metab. 2013;17:954–964. doi: 10.1016/j.cmet.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz AM, Haynes CM. UPR(mt)-mediated cytoprotection and organismal aging. Biochim. Biophys. Acta. 2015;1847:1448–1456. doi: 10.1016/j.bbabio.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen P, Dang W, Donahue G, Dai J, Dorsey J, Cao X, Liu W, Cao K, Perry R, Lee JY, et al. H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev. 2015;29:1362–1376. doi: 10.1101/gad.263707.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013;27:1787–1799. doi: 10.1101/gad.223834.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Murphy CT. Mating induces shrinking and death in Caenorhabditis mothers. Science. 2014;343:536–540. doi: 10.1126/science.1242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebold AP, Banerjee R, Tie F, Kiss DL, Moskowitz J, Harte PJ. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc. Natl. Acad. Sci. USA. 2010;107:169–174. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simola DF, Graham RJ, Brady CM, Enzmann BL, Desplan C, Ray A, Zwiebel LJ, Bonasio R, Reinberg D, Liebig J, Berger SL. Epigenetic (re)programming of caste-specific behavior in the ant Camponotus floridanus. Science. 2016;351:aac6633. doi: 10.1126/science.aac6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Mills K, Guarente L. Molecular mechanisms of yeast aging. Trends Biochem. Sci. 1998;23:131–134. doi: 10.1016/s0968-0004(98)01188-8. [DOI] [PubMed] [Google Scholar]
- Sinha M, Jang YC, Oh J, Khong D, Wu EY, Manohar R, Miller C, Regalado SG, Loffredo FS, Pancoast JR, et al. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science. 2014;344:649–652. doi: 10.1126/science.1251152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Sun D, Luo M, Jeong M, Rodriguez B, Xia Z, Hannah R, Wang H, Le T, Faull KF, Chen R, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Youle RJ, Finkel T. The mitochondrial basis of aging. Mol. Cell. 2016;61:654–666. doi: 10.1016/j.molcel.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D. Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo AG, Duong HA, Robles MS, Mann M, Weitz CJ. Histone monoubiquitination by Clock-Bmal1 complex marks Per1 and Per2 genes for circadian feedback. Nat. Struct. Mol. Biol. 2015;22:759–766. doi: 10.1038/nsmb.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC. Aging and the UPR(ER) Brain Res. 2016 doi: 10.1016/j.brainres.2016.04.017. Published online April 8, 2016. http://dx.doi.org/10.1016/j.brainres.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Tebay LE, Robertson H, Durant ST, Vitale SR, Penning TM, Dinkova-Kostova AT, Hayes JD. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic. Biol. Med. 2015;88(Pt B):108–146. doi: 10.1016/j.freeradbiomed.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, Dillin A. Mitochondrial stress induces chromatin reorganization to promote longevity and UPR(mt) Cell. 2016 doi: 10.1016/j.cell.2016.04.011. Published online April 28, 2016. http://dx.doi.org/10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilstra JS, Clauson CL, Niedernhofer LJ, Robbins PD. NF-κB in aging and disease. Aging Dis. 2011;2:449–465. [PMC free article] [PubMed] [Google Scholar]
- Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Cadiñanos J, Pendás AM, Gutiérrez-Fernández A, Folgueras AR, Sánchez LM, Zhou Z, Rodríguez FJ, Stewart CL, Vega JA, et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, Zhao M, Christenson MK, Sanchez R, Yu JY, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500:345–349. doi: 10.1038/nature12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J, Suh Y. Genome instability and aging. Annu. Rev. Physiol. 2013;75:645–668. doi: 10.1146/annurev-physiol-030212-183715. [DOI] [PubMed] [Google Scholar]
- Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol. Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AE, Pollina EA, Vierbuchen T, Urbán N, Ucar D, Leeman DS, Martynoga B, Sewak M, Rando TA, Guillemot F, et al. FOXO3 shares common targets with ASCL1 genome-wide and inhibits ASCL1-dependent neurogenesis. Cell Rep. 2013;4:477–491. doi: 10.1016/j.celrep.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AE, Kundaje A, Brunet A. Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging Cell. 2016 doi: 10.1111/acel.12479. Published online April 8, 2016. http://dx.doi.org/10.1111/acel.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, Jaenisch R. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc. Natl. Acad. Sci. USA. 2012;109:13004–13009. doi: 10.1073/pnas.1210787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. USA. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Hillenmeyer S, Lawrence C, Chang C, Hosier S, Lightfoot W, Mukherjee E, Jiang N, Schorl C, Brodsky AS, et al. Chromatin remodeling in the aging genome of Drosophila. Aging Cell. 2010;9:971–978. doi: 10.1111/j.1474-9726.2010.00624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Bonasio R, Simola DF, Liebig J, Berger SL, Reinberg D. DNA methylation in social insects: how epigenetics can control behavior and longevity. Annu. Rev. Entomol. 2015;60:435–452. doi: 10.1146/annurev-ento-010814-020803. [DOI] [PubMed] [Google Scholar]
- Yuan T, Jiao Y, de Jong S, Ophoff RA, Beck S, Teschendorff AE. An integrative multi-scale analysis of the dynamic DNA methylation landscape in aging. PLoS Genet. 2015;11:e1004996. doi: 10.1371/journal.pgen.1004996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, Weeraratna AT, Taub DD, Gorospe M, Mazan-Mamczarz K, et al. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampieri M, Ciccarone F, Calabrese R, Franceschi C, Bürkle A, Caiafa P. Reconfiguration of DNA methylation in aging. Mech. Ageing Dev. 2015;151:60–70. doi: 10.1016/j.mad.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ohta T, Maruyama A, Hosoya T, Nishikawa K, Maher JM, Shibahara S, Itoh K, Yamamoto M. BRG1 interacts with Nrf2 to selectively mediate HO-1 induction in response to oxidative stress. Mol. Cell. Biol. 2006;26:7942–7952. doi: 10.1128/MCB.00700-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Chen H-Z, Liu D-P. The four layers of aging. Cell Syst. 2015a;1:180–186. doi: 10.1016/j.cels.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, et al. Aging stem cells. a Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015b;348:1160–1163. doi: 10.1126/science.aaa1356. [DOI] [PMC free article] [PubMed] [Google Scholar]