Abstract
Purpose
The present multimodal neuroimaging study examined whether amyloid pathology and glucose metabolism are related to cortical volume loss over time in Alzheimer’s disease (AD) patients and healthy elderly controls.
Methods
Structural MRI scans of eleven AD patients and ten controls were available at baseline and follow-up (mean interval 2.5 years). Change in brain structure over time was defined as percent change of cortical volume within seven a-priori defined regions that typically show the strongest structural loss in AD. In addition, two PET scans were performed at baseline: [11C]PIB to assess amyloid-β plaque load and [18F]FDG to assess glucose metabolism. [11C]PIB binding and [18F]FDG uptake were measured in the precuneus, a region in which both amyloid deposition and glucose hypometabolism occur early in the course of AD.
Results
While amyloid-β plaque load at baseline was not related to cortical volume loss over time in either group, glucose metabolism within the group of AD patients was significantly related to volume loss over time (rho=0.56, p<0.05).
Conclusion
The present study shows that in a group of AD patients amyloid-β plaque load as measured by [11C]PIB behaves as a trait marker (i.e., all AD patients showed elevated levels of amyloid, not related to subsequent disease course), whilst hypometabolism as measured by [18F]FDG changed over time indicating that it could serve as a state marker that is predictive of neurodegeneration.
Keywords: Alzheimer’s disease, Amyloid plaques, Hypometabolism, Atrophy, Longitudinal study
Introduction
Several biomarkers are in use for clinical research into Alzheimer’s disease (AD), both for diagnosis and for monitoring disease [1]. Recently, an updated version of the biomarker accumulation model has been proposed by Jack and colleagues [2], postulating a temporal cascade of several biomarkers, including accumulation of amyloid-beta and hyperphosphorylated tau, regional specific hypometabolism, structural loss, and finally cognitive decline. This model is based on the hypothesis that these biomarkers change in an ordered manner, eventually resulting in clinical AD. Hypometabolism and structural loss follow a similar temporal pattern, and continue to change substantially in the clinical phase of AD [2].
Understanding how AD biomarkers behave over time and how they are related to each other is crucial in understanding underlying mechanisms of the disease. In addition, it is relevant for the development of therapeutic interventions. Volumetric brain changes, measured using structural MRI, are well validated biomarkers in AD research. Indeed, volume loss has been found to correlate strongly with disease severity even at the later stages of the disease [3, 4]. However, little is known about the relationship between molecular markers of AD, such as amyloid-β plaque load and hypometabolism, on the one hand and brain volume loss over time on the other.
A leading theory proposes that amyloid-β plaque formation is the main initiator of AD [5, 6]. Amyloid-β can be measured in vivo using carbon-11 labelled Pittsburgh compound B ([11C]PIB) positron emission tomography (PET) [7]. Elevated levels of [11C]PIB binding in the posterior cingulate cortex/precuneus can be seen in prodromal AD, spreading throughout the entire association cortex at later stages [8, 9]. Amyloid-beta accumulation is commonly believed to stagnate in the clinical phase of the disease, i.e., in the stage where patients meet criteria for clinical AD [3, 10–12]. However, some studies do report slow, but continuous, build-up of amyloid-plaques in AD patients [13, 14].
Brain metabolism can be measured using fluorine-18 labelled fluorodeoxyglucose ([18F]FDG) and PET, and is an indicator of neuronal function [15, 16]. Hypometabolism in the precuneus is already observed in the preclinical stage of AD [17–19] and becomes even more pronounced in the clinical phase [20]. Both hypometabolism and brain volume loss progressively worsen throughout the different disease stages [2].
The purpose of the present multimodal neuroimaging study was to examine whether amyloid pathology and glucose metabolism are related to cortical volume loss over time in both AD patients and normal controls. As different biomarkers are believed to approach pathological levels at different times during the disease process, associations between biomarkers are expected to change with disease progression.
Methods
Participants
All participants received a standard dementia screening, which included obtaining a record of their medical history, neuropsychological testing, physical and neurological examination, structural MRI and screening laboratory tests. Patients were excluded when they had a history of major psychiatric or neurological illness other than AD, used non-steroidal anti-inflammatory drugs, or showed clinically significant abnormalities other than AD based on the MRI scan as determined by a neuroradiologist. Additional exclusion criteria for normal controls were subjective complaints and a positive [11C]PIB scan at baseline. The Mini Mental State Examination (MMSE) was part of neuropsychological testing [21]. Clinical diagnosis was established by a multidisciplinary team, both at baseline and at follow-up. Follow-up of patients was planned after approximately 2–3 years. There were three patients with mild cognitive impairment (MCI) [22] at baseline, who were diagnosed with probable AD [23] at follow-up. These patients were included in the final AD group (n=11) that was compared with an elderly control group (n=10). Longitudinal PET data were available for nine AD patients and ten controls, after an average of 2.5 years. Findings of this longitudinal PET study have been reported previously [11]. In the present study, only baseline PET data are reported, as the purpose was to assess whether baseline PET data are related to ongoing structural loss, i.e., can be used to predict future atrophy. Longitudinal MRI data were available for all subjects and were, on average, acquired 2.5 years after the baseline scan (Table 1). The study was approved by the Medical Ethics Review Committee of the VU University Medical Center. Written informed consent was obtained from subjects and/or subjects’ caregivers after complete written and verbal description of the study.
Table 1.
Participant demographics, cortical volume in the AD-signature regions, [11C]PIB binding (BPND) and [18F]FDG uptake (SUVr) in the precuneus, and MMSE score at baseline in normal controls (NC) and patients with Alzheimer’s disease (AD)
| NC | AD | |
|---|---|---|
| N | 10 | 11 |
| Male/Female | 8/2 | 10/1 |
| Age at baseline (Years) | 65.80±7.39 | 62.82±6.23 |
| Time between baseline and follow- up (Years) | 2.44±0.28 | 2.64±0.59 |
| Cortical volume at baseline (mm3)* | 1006.89±151.97 | 931.02±91.81 |
| Cortical volume loss over time (%)* | 0.64±1.06 | 3.45±1.51 |
| Precuneus volume at baseline (mm3) | 907.1±124.80 | 902.20±177.30 |
| [11C]PIB BPND in precuneus at baseline* | 0.04±0.06 | 0.77±0.15 |
| [11C]PIB BPND (PVC) in precuneus at baseline* | 0.02±0.05 | 0.54±0.17 |
| [18F]FDG SUVr in precuneus at baseline | 1.07±0.08 | 1.01±0.12 |
| [18F]FDG SUVr (PVC) in precuneus at baseline | 1.03±0.08 | 0.97±0.13 |
| MMSE score at baseline* | 29.40±0.52 | 25.45±2.42 |
Data are presented as means ± standard deviations (SD) * represents p<0.05, [11C]PIB carbon-11 labeled Pittsburgh compound B, measuring amyloid-β. [18F]FDG fluorine-18 labeled fluorodeoxyglucose, measuring brain metabolism. PVC partial volume correction. PBND non-displaceable binding potential. SUVr standardized uptake value ratio
MRI protocol
All subjects underwent structural MRI using a 1.5 T Sonata scanner (Siemens Medical Solutions, Erlangen, Germany). The scanning protocol included a coronal T1-weighted 3-dimensional magnetization-prepared rapid-acquisition gradient echo (MPRAGE) image (echo time 3.97 ms; repetition time 2,700 ms; inversion time 950 ms; flip angle 8°; 160 coronal slices; voxel size 1×1.5×1 mm3).
PET protocol
Both [11C]PIB and [18F]FDG scans were acquired using an ECAT EXACT HR + scanner (Siemens/CTI, Knoxville, USA). The properties of this scanner have been reported elsewhere [24]. A head holder was used to restrict patient movement and head movement was checked on a regular basis. For the [11C]PIB scan, first a 10- min transmission scan was performed (to correct for photon attenuation), which was followed by a dynamic emission scan of 90 min. For the [18F]FDG scan, subjects rested for 10 min with eyes closed in a dimly lit room and with minimal background noise when [18F]FDG was injected. Thirty-five minutes later, patients underwent a 10-min transmission scan followed by a 15-min (static) emission scan. Further details of [11C]PIB and [18F]FDG scans can be found elsewhere [11]. The maximum interval between [18F]FDG and [11C]PIB scans was 1 month.
MRI analysis
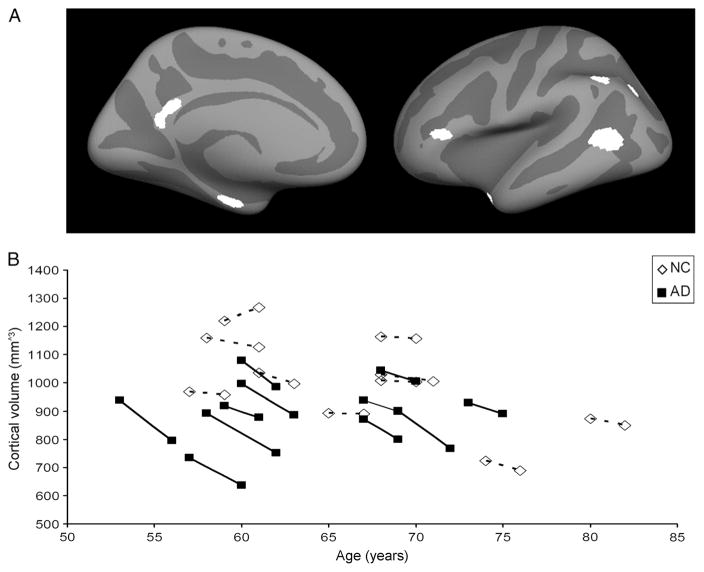
To extract reliable volume estimates, images were processed using the longitudinal stream in Freesurfer that is explained in detail elsewhere [25]. Briefly, an unbiased within-subject template space and image was created using robust inverse consistent registration. Several processing steps, such as skull stripping, and atlas registration as well as creation of spherical surface maps and grey matter segmentations were then initialized with common information from the within-subject template, significantly increasing reliability and statistical power. Volumetric brain changes were assessed in seven a-priori defined regions of interest (ROIs) that are known to be susceptible to AD related atrophy [26]. These regions were situated in the inferior frontal cortex, posterior cingulate cortex, temporal polar cortex, lateral temporal cortex, inferior parietal sulcus, inferior parietal cortex and medial temporal lobe (see Fig. 1a), which can be downloaded freely as Freesurfer surface labels (http://surfer.nmr.mgh.harvard.edu/fswiki/Chubs). The labels were mapped to the brain of each participant using surface based registration as implemented in Freesurfer [27]. The mean volume within these seven ROIs was calculated for both hemispheres of each subject at baseline and at follow-up. The percent change of volume within each ROI was calculated by dividing the rate of change in volume by the temporal average within these ROIs (symmetrized percent change). Initial results showed that the percent change within these seven regions for left and right hemispheres were highly correlated (rho=0.88, p<0.01) and, therefore, mean percent change across both hemispheres was used in all analyses. To test whether results for these regions were specific for AD, a second control analysis was performed measuring brain volume in an ROI consisting of primary motor and sensory regions (precentral and postcentral gyri), as these regions are thought to be relatively spared in early and mid-stages of AD [28].
Fig. 1.
The a priori selected regions of interest [27] are shown on an average Freesurfer surface, left hemisphere, and are situated in the inferior frontal cortex, posterior cingulate cortex, temporalpolar cortex, lateral temporal cortex, inferior parietal sulcus, inferior parietal cortex and the medial temporal lobe (a). Patients with Alzheimer’s disease (AD) showed reduced cortical volume when compared to normal controls (NC) at both baseline and follow-up. Cortical volume within the AD-signature regions is displayed for each subject on the y-axis. Age at baseline and follow-up is represented on the x-axis (b)
PET analysis
Standard preprocessing of PET data was performed [11]. Structural T1 images were aligned to corresponding PET images using a mutual information algorithm. Data were analyzed using PVE-lab, a software program that makes use of a probability map based on 35 defined ROIs [29]. Results are reported with and without partial volume correction (PVC), because at present there is no consensus on how to optimally perform PVC of PET data [30]. PVC based on iterative deconvolution [31] was used with a Gaussian kernel of 7 mm FWHM and ten iterations. The deconvolution method was applied with a Gibbs prior (neighbourhood of 1 voxel and weight of 0.15). Iterative deconvolution is PET based, does not make use of the MR images, and improves spatial resolution and contrast. It uses the point spread function (PSF) of the PET system to sharpen the PET scan iteratively. The advantage of the deconvolution method is that it is free of segmentation or co-registration errors and unaffected by tracer uptake heterogeneity [31]. Results with and without partial volume correction will further be referred to as “with PVC” and “without PCV”. Parametric images of non-displaceable binding potential (BPND), a quantitative measure of specific binding, of [11C]PIB were generated using a 2-step basis function implementation of the simplified reference tissue model with cerebellar grey matter as reference tissue (RPM2) [32, 33]. If global cortical [11C]PIB BPND was higher than 0.54, subjects were considered to be PIB-positive [9]. For [18F]FDG, parametric images of standardized uptake value ratio (SUVr), using cerebellar grey matter as reference, were extracted from the interval between 45 and 60 min after injection. Cerebellar grey matter lacks Congo red and thioflavin-S positive plaques and was, therefore, chosen as reference tissue for analysis of both [11C]PIB and [18F]FDG data [34].
[11C]PIB and [18F]FDG binding in ROIs
Both parametric PET images (i.e., [11C]PIB BPND and [18F]FDG SUVr) were registered to standard space (MNI 152), using 12° of freedom (DOF) linear translation (FLIRT, FSL version 4.1; www.fmrib.ox.ac.uk/fsl). Structural T1 scans were segmented into cerebrospinal fluid, grey matter and white matter using FSL [35]. Average binding of [11C]PIB and average uptake of [18F]FDG were then calculated for the precuneus (including only grey matter) based on the Harvard-Oxford cortical atlas (provided in FSL; http://www.cma.mgh.harvard.edu/fsl_atlas.html). The precuneus was chosen because both amyloid-plaque formation and glucose hypometabolism occur in this region relatively early in the course of the disease [8, 17]. At baseline, precuneus volume was not significantly associated with [18F]FDG uptake in the precuneus (AD no PVC; rho=0.36, p=0.14; AD with PVC; rho=0.32, p=0.17. NC no PVC; rho=0.46, p=0.09 NC with PVC; rho = 0.38, p = 0.14), suggesting that observed hypometabolism is not due to partial volume effects.
Statistical analysis
All other subsequent statistical analyses were performed using SPSS (version 15.0; SPSS, Chicago, IL, USA). Differences in gender distribution between AD patients and controls were assessed using a Chi-square test. Differences in age, time between baseline and follow-up, [11C]PIB BPND, [18F]FDG SUVr and MMSE score between AD patients and controls were examined using a one-way ANOVA. Differences in cross-sectional measures of cortical volume (baseline and follow-up) between controls and AD patients were assessed using an ANOVA with intracranial volume and age as covariates. Differences in percent change over time in cortical volume between controls and AD patients were assessed with an ANOVA with age as covariate. Spearman correlation coefficients between modalities were tested one-tailed because of the a-priori expected directionality of these associations. A p-value below 0.05 was considered statistically significant.
Results
Subject demographics are given in Table 1. Distribution of gender (X2(1)=0.51, p=0.48), age (F(1,20)=1.01, p=0.33), and time between baseline and follow-up (F(1,20)=1.04, p= 0.32) did not differ significantly between AD patients and controls. Cross sectional analysis indicated that cortical volume within a priori selected AD-signature regions was significantly lower in AD patients compared with normal controls at both baseline (F(1,17)=5.15, p<0.05) and follow-up (F(1,17)=6.78, p<0.05) (see Fig. 1b).
Longitudinal analysis revealed that AD patients showed significantly more cortical volume loss over time (mean volume loss 3.5 %) than normal controls (0.6 %) (F(1,18)=20.51, p<0.01), confirming expected progression of atrophy over time in the AD group. There was no significant difference between AD patients and controls in percent change of cortical volume in primary motor and sensory cortices (F(3,18)= 0.29, p=0.48), regions that are known to be relatively spared in early and mid-stages of Alzheimer’s disease [29], indicating that results were unlikely due to systematic measurement errors.
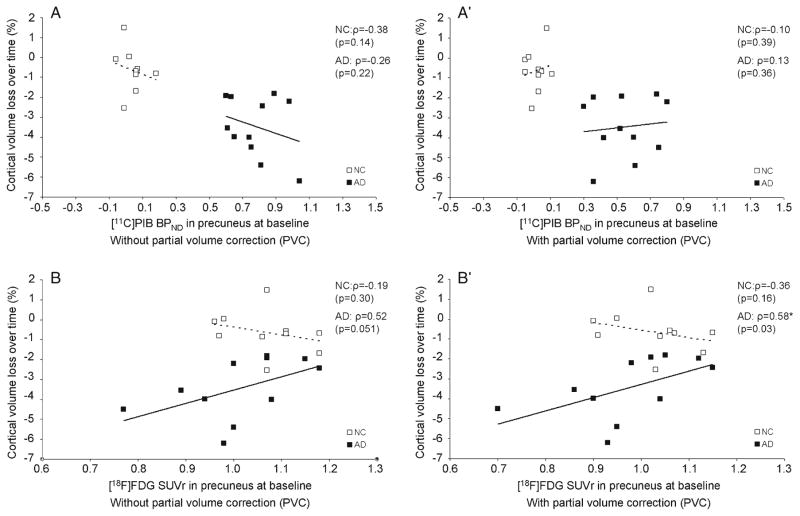
Table 1 provides baseline measures of [11C]PIB BPND, [18F]FDG SUVr in the precuneus (with and without PVC), together with MMSE scores, in AD patients and controls. As expected on the basis of the selection criteria for both groups, at baseline, [11C]PIB BPND was significantly higher (no PVC: F(1,20)=207.24, p<0.01) (with PVC: F(1,20)=82,66, p<0.01) and the MMSE score was significantly lower (F(1,20)=25.35, p<0.01) in AD patients than in controls. Although [18F]FDG SUVr at baseline was reduced in AD patients compared with controls, this was not significant (no PVC: F(1,20)=1.58, p=0.22) (with PVC: F(1,20)=1.24, p= 0.28). [11C]PIB binding at baseline was not related to cortical volume loss over time in either group (Fig. 2A and A′). In contrast, [18F]FDG SUVr with PVC at baseline was significantly related to volume loss over time in AD patients (rho= 0.58, p<0.05) (Fig. 2B′). Similar results were found when no PVC was applied, although this was not significant (rho= 0.52, p=0.05) (Fig. 2B). The relationship was such that AD patients with the lowest metabolism at baseline showed the most subsequent loss of cortical volume over time. No association of [18F]FDG SUVr with volume loss over time was found in controls (no PVC: rho=−0.19 p=0.30) (with PVC: rho=−0.36, p=0.16). An overview of various Spearman correlation coefficients for each group is given in Table 2.
Fig. 2.
Alzheimer’s disease (AD) patients with lowest metabolism at baseline showed the most subsequent loss of cortical volume over time. The associations of cortical volume loss over time with amyloid deposition and metabolism in the precuneus at baseline is displayed for normal controls (NC) and patients with AD. Results without and with partial volume correction (PVC) are displayed. Percent change in cortical volume over time is depicted on the y-axis of all panels. [11C]PIB binding in the precuneus at baseline without PVC (A) and with PVC (A′) and [18F]FDG retention in the precuneus at baseline without PVC (B) and with PVC (B′) is shown on the x-axis. [11C]PIB binding at baseline was not related to cortical volume loss over time in either group while [18]FDG uptake with PVC was significantly related to volume loss over time in AD patients only. Spearman correlation coefficients (ρ) are reported (p-value)
Table 2.
Associations of cortical volume change over time in AD-signature regions with cortical volume at baseline, [11C]PIB binding (BPND) and [18F]FDG uptake (SUVr) in the precuneus at baseline, and MMSE score at baseline in both normal controls (NC) and patients with Alzheimer’s disease (AD)
| Cortical volume change over time
|
||
|---|---|---|
| NC | AD | |
| Cortical volume at baseline | 0.69 (p=0.01)* | 0.26 (p=0.22) |
| Precuneus volume at baseline | 0.01 (p=0.49) | 0.09 (p=0.40) |
| [11C]PIB BPND in precuneus at baseline | −0.38 (p=0.14) | −0.26 (p=0.22) |
| [11C]PIB BPND (PVC) in precuneus at baseline | −0.10 (p=0.39) | 0.13 (p=0.36) |
| [18F]FDG SUVr in precuneus at baseline | −0.19 (p=0.30) | 0.52 (p=0.051) |
| [18F]FDG SUVr (PVC) in precuneus at baseline | −0.36 (p=0.16) | 0.58 (p=0.03)* |
| MMSE score at baseline | 0.14 (p=0.35) | −0.06 (p=0.43) |
Data are presented as 1-tailed Spearman correlation coefficients with corresponding p-values, * represents p<0.05, [11C]PIB carbon-11 labeled Pittsburgh compound B, measuring amyloid-β. [18]FDG fluorine-18 labeled fluorodeoxyglucose, measuring brain metabolism. PVC partial volume correction. PBND non-displaceable binding potential. SUVr standardized uptake value ratio
Discussion
The main finding of this study was the observed association between [18F]FDG uptake in the precuneus at baseline and volume loss over time in AD patients indicating that low metabolism at baseline is related to more subsequent cortical volume loss over time. This association was not observed in normal controls. In addition, [11C]PIB binding at baseline was not related to cortical volume loss over time in the AD group nor in the control group.
The lack of association between [11C]PIB binding and cortical volume loss over time in the AD group is in agreement with prior cross-sectional studies using [11C]PIB binding [36] and levels of amyloid in cerebral spinal fluid [37]. This indicates that amyloid-plaque formation already has reached a plateau in these patients [3, 10, 11, 38]. However, some research does indicate a continued build-up of amyloid-plaques in AD patients [13]. This might explain why some studies show a relationship between amyloid and atrophy rates [39]. Similar to this study, Tosun and colleagues [39] examined baseline amyloid formation with volumetric changes over time, but included MCI patients. Also in contrast to this study, they did not select a priori regions, but used parallel independent component analysis, allowing for an unbiased search for (spatially unrelated) associations between amyloid-β and atrophy rates. They concluded that the spatial distribution of increased amyloid-β and the related spatial atrophy rates were in line with regions known to be vulnerable for Alzheimer’s disease pathology. These results are conflicting with the present study. This could be explained by the fact that MCI patients might not have reached a plateau in amyloid-β formation unlike AD patients in this study, or by incorporating a more sensitive voxelwise approach. The absence of an association in our sample of AD patients could also be evidence that amyloid-plaque formation is not directly related to ongoing structural loss, but is mediated by other ‘downstream’ factors such as hypometabolism or neurofibrillary tangle formation [6, 36]. With respect to the control group, also no correlation between cortical volume loss over time and amyloid load was found. This was in agreement with our hypothesis; only healthy controls that were PIB negative at baseline were included and were, therefore, believed to show no early neurodegeneration. Regarding the relationship between amyloid and structural loss in healthy elderly there is no consensus in the literature; some studies find associations between atrophy and amyloid load in healthy elderly [36, 40, 41], while others do not [42, 43]. When PIB positive controls are included, controls with very early AD pathology might drive the association between amyloid load and structural loss. An association between amyloid load and regional atrophy has been reported in PIB positive healthy elderly, while this was not found in PIB negative elderly [44, 45].
The finding that [18F]FDG uptake was related to volume loss over time in AD patients is in line with prior cross-sectional studies [46–49]. Whilst these results do not establish a causal relationship between neuronal dysfunction and atrophy, the fact that metabolism at baseline is related the subsequent atrophy can be interpreted as support for the hypothesis that neuronal dysfunction, as indicated by low [18F]FDG, precedes structural loss [2]. Since this association was stronger after PVC, and baseline cortical volume was not associated with ongoing structural loss in AD patients, it is believed that hypometabolism provides more information than atrophy alone. This is supported by the finding that hypometabolism exceeds atrophy in AD patients when examining spatial overlap [50]. Since amyloid is shown to precede hypometabolism in early AD [51], this also explains why, in contrast to AD patients, we found no association between hypometabolism and volume loss over time in normal controls.
It has been reasoned that hypometabolism may be the mediating factor that links amyloid-plaque formation to ongoing structural loss. In cognitively normal elderly controls, it was found that abnormal levels of β-amyloid accompanied by a brain injury marker (hypometabolism or atrophy) resulted in higher rates of ongoing structural changes [52]. The associations of AD-biomarkers may be different within disease stages. It has been shown that metabolism (briefly) increases together with amyloid burden in MCI patients [53], perhaps serving as a compensatory mechanism. A different causal mechanism has been postulated for early AD patients: higher metabolism, especially in ‘hub’ regions, was linked to higher amyloid-β accumulation over time [54], which is in line with evidence from cellular investigations [55]. Future studies might focus on the longitudinal relationship between amyloid, metabolism and atrophy across different stages of AD, preferably incorporating multiple follow-up scans, in order to investigate the complex relationship between hypometabolism, amyloid accumulation and atrophy.
Future recommendations and limitations
Although the present results illustrate the feasibility of state of the art multi-modal longitudinal neuroimaging methods, the study also had limitations. The major limitation was the limited sample size. Still, even with the small sample, results are in line with the hierarchical biomarker model [2]. Another limitation was the fact that only AD patients with both a baseline and a follow-up MRI scan were included in the present study, which may have lead to exclusion of patients in more advanced stages of the disease and possibly limits the generalisability of the results. Also, the control group was specifically selected to be PIB negative in order ensure a clear difference between groups in terms of amyloid-β neuropathology, which limits generalisability to populations where amyloid-β status is unknown. To fully test current models of the development of different biomarkers, inclusion of PIB positive clinically normal older adults will be necessary and longer follow-up time with more measurement occasions are needed. Three patients in the AD group were initially defined as MCI patients. Since these MCI patients progressed to AD after clinical diagnosis at follow-up, these subjects were classified as probable AD. All MCI patients were PIB-positive at baseline, similar to all AD patients. Inclusion of these three MCI patients to our probable AD group may have influenced our results, since AD patients have more progressed neurode-generation than MCI patients. We chose for an inclusive approach for the present study to increase our sample size. In addition, in this longitudinal study structural images were acquired on a 1.5 T MR system. It is possible that accuracy of atrophy estimates could be improved by using an MR scanner with a higher field strength, as this is associated with higher spatial resolution. However, volumetry measurements on MR scanners with different field-strengths (1.5 T vs. 3 T) have been reported to be interchangeable [56], not exceeding within-scanner measurement error [57]. Finally, the precuneus was selected as ROI for both PET measures since this region is vulnerable for both amyloid-β formation and hypometabolism [8, 17]. It should be noted that this implies that results should not be generalised to other regions in the brain. A voxel-wise approach would be needed to assess whether other vulnerable regions exist. This will need to be carried out in future studies, as the sample size of the present study was too small for such an analysis.
Conclusion
In conclusion, the present study shows the feasibility of assessing significant relationships between multiple neuroim-aging modalities and can, therefore, be considered as a proof-of-concept study for multimodal neuroimaging initiatives in the spectrum of normal aging, mild cognitive impairment, and AD. The present results show that, in a group of AD patients, amyloid load as measured by [11C]PIB behaves as a trait marker (i.e., all AD patients show elevated levels of amyloid not related to rate of subsequent neurodegeneration). By contrast, rate of hypometabolism in (preclinical) AD as measured by [18F]FDG can serve as a state marker that is predictive of neurodegeneration.
Acknowledgments
This study was sponsored by Internationale Stichting Alzheimer Onderzoek (ISAO; project number #11539) and Hersenstichting Nederland (KS2011(1)-24).
We thank the support of the Athinoula A. Martinos Center for Biomedical Imaging at MGH including analysis methods developed by NIH grant P41RR14075.
Footnotes
Disclosure statement: Dr. Van Berckel receives research support from the American Health Assistance Foundation, Alzheimer Association, Internationale Stichting Alzheimer Onderzoek, the Center of Translational Molecular Medicine and the Dutch Organisation for Scientific Research.
Dr. Barkhof serves on the editorial boards of Brain, European Radiology, the Journal of Neurology, Neurosurgery & Psychiatry, the Journal of Neurology, Multiple Sclerosis and Neuroradiology and serves as a consultant for Bayer-Schering Pharma, Sanofi-Aventis, Biogen-Idec, UCB, Merck-Serono, Jansen Alzheimer Immunotherapy, Baxter, Novartis and Roche.
Dr. Scheltens serves/has served on the advisory boards of: Genentech, Novartis, Roche, Danone, Nutricia, Baxter and Lundbeck. He has been a speaker at symposia organised by Lundbeck, Merz, Danone, Novartis, Roche and Genentech. For all his activities he receives no personal compensation. He serves on the editorial board of Alzheimer’s Research & Therapy and Alzheimer Disease and Associated Disorders, is a member of the scientific advisory board of the EU Joint Programming Initiative and the French National Plan Alzheimer.
Dr. Reuter receives funding from several grants: NINDS 5R01NS052585-05, NIBIB 5R01EB006758-04, NINDS 2-R01-NS042861-06A1, NINDS 5-P01-NS058793-03, and NICHD R01-HD071664.
There are no other actual or potential conflicts of interest to disclose. All authors have read and agreed with the contents of the manuscript. The results of the study have not been published before and they are not under consideration to be published by another journal.
Contributor Information
Sofie M. Adriaanse, Email: s.adriaanse@vumc.nl, Department of Radiology and Nuclear Medicine, VU University Medical Center, Amsterdam, The Netherlands. Department of Radiology and Nuclear Medicine, Alzheimer Center, Neuroscience Campus Amsterdam, VU University Medical Center, P.O. Box 7057, 1007 MB Amsterdam, The Netherlands
Koene R. A. van Dijk, Department of Psychology, Center for Brain Science, Harvard University, Cambridge, MA, USA. Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, USA
Rik Ossenkoppele, Department of Radiology and Nuclear Medicine, Alzheimer Center, Neuroscience Campus Amsterdam, VU University Medical Center, Amsterdam, The Netherlands.
Martin Reuter, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, USA. Computer Science and Artificial Intelligence Laboratory, Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, USA.
Nelleke Tolboom, Department of Radiology and Nuclear Medicine, Alzheimer Center, Neuroscience Campus Amsterdam, VU University Medical Center, Amsterdam, The Netherlands.
Marissa D. Zwan, Department of Radiology and Nuclear Medicine, Alzheimer Center, Neuroscience Campus Amsterdam, VU University Medical Center, Amsterdam, The Netherlands
Maqsood Yaqub, Department of Radiology and Nuclear Medicine, Neuroscience Campus Amsterdam, VU University Medical Center, Amsterdam, The Netherlands.
Ronald Boellaard, Department of Radiology and Nuclear Medicine, Neuroscience Campus Amsterdam, VU University Medical Center, Amsterdam, The Netherlands.
Albert D. Windhorst, Department of Radiology and Nuclear Medicine, Neuroscience Campus Amsterdam, VU University Medical Center, Amsterdam, The Netherlands
Wiesje M. van der Flier, Department of Neurology, Alzheimer Center, Neuroscience Campus Amsterdam, VU University Medical Center, Amsterdam, The Netherlands
Philip Scheltens, Department of Neurology, Alzheimer Center, Neuroscience Campus Amsterdam, VU University Medical Center, Amsterdam, The Netherlands.
Adriaan A. Lammertsma, Department of Radiology and Nuclear Medicine, Neuroscience Campus Amsterdam, VU University Medical Center, Amsterdam, The Netherlands
Frederik Barkhof, Department of Radiology and Nuclear Medicine, Alzheimer Center, Neuroscience Campus Amsterdam, VU University Medical Center, Amsterdam, The Netherlands.
Bart N. M. van Berckel, Department of Radiology and Nuclear Medicine, Alzheimer Center, Neuroscience Campus Amsterdam, VU University Medical Center, Amsterdam, The Netherlands
References
- 1.Jack CR., Jr Alliance for aging research AD biomarkers work group: structural MRI. Neurobiol Aging. 2011;32(Suppl 1):S48–57. doi: 10.1016/j.neurobiolaging.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jack CR, Jr, Knopman DS, Jagust WJ, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–16. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caroli A, Frisoni GB. The dynamics of Alzheimer’s disease bio-markers in the Alzheimer’s Disease Neuroimaging Initiative cohort. Neurobiol Aging. 2010;31:1263–74. doi: 10.1016/j.neurobiolaging.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frisoni GB, Fox NC, Jack CR, Jr, Scheltens P, Thompson PM. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. 2010;6:67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fjell AM, Walhovd KB. New tools for the study of Alzheimer’s disease: what are biomarkers and morphometric markers teaching us? Neuroscientist. 2011;17:592–605. doi: 10.1177/1073858410392586. [DOI] [PubMed] [Google Scholar]
- 6.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–6. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 7.Klunk WE, Engler H, Nordberg A, et al. Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound-B. Ann Neurol. 2004;55:306–19. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 8.Rabinovici GD, Jagust WJ. Amyloid imaging in aging and dementia: testing the amyloid hypothesis in vivo. Behav Neurol. 2009;21:117–28. doi: 10.3233/BEN-2009-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tolboom N, Yaqub M, van der Flier WM, et al. Detection of Alzheimer pathology in vivo using both 11C-PIB and 18F-FDDNP PET. J Nucl Med. 2009;50:191–7. doi: 10.2967/jnumed.108.056499. [DOI] [PubMed] [Google Scholar]
- 10.Jack CR, Jr, Lowe VJ, Weigand SD, et al. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer’s disease: implications for sequence of pathological events in Alzheimer’s disease. Brain. 2009;132:1355–65. doi: 10.1093/brain/awp062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ossenkoppele R, Tolboom N, Foster-Dinsley J, et al. Longitudinal imaging of Alzheimer pathology using [11C]PIB, [18F]FDDNP and [18F]FDG PET. Eur J Nucl Med Mol Imaging. 2012;39:990–1000. doi: 10.1007/s00259-012-2102-3. [DOI] [PubMed] [Google Scholar]
- 12.Engler H, Forsberg A, Almkvist O, et al. Two-year follow-up of amyloid deposition in patients with Alzheimer’s disease. Brain. 2006;129:2856–66. doi: 10.1093/brain/awl178. [DOI] [PubMed] [Google Scholar]
- 13.Villain N, Chetelat G, Grassiot B, et al. Regional dynamics of amyloid-beta deposition in healthy elderly, mild cognitive impairment and Alzheimer’s disease: a voxelwise PiB-PET longitudinal study. Brain. 2012;135:2126–39. doi: 10.1093/brain/aws125. [DOI] [PubMed] [Google Scholar]
- 14.Villemagne VL, Pike KE, Chetelat G, et al. Longitudinal assessment of Abeta and cognition in aging and Alzheimer disease. Ann Neurol. 2011;69:181–92. doi: 10.1002/ana.22248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rocher AB, Chapon F, Blaizot X, Baron JC, Chavoix C. Resting-state brain glucose utilization as measured by PET is directly related to regional synaptophysin levels: a study in baboons. Neuroimage. 2003;20:1894–8. doi: 10.1016/j.neuroimage.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz WJ, Smith CB, Davidsen L, Savaki H, Sokoloff L, Mata M, et al. Metabolic mapping of functional activity in the hypothalamo-neurohypophysial system of the rat. Science. 1979;205:723–5. doi: 10.1126/science.462184. [DOI] [PubMed] [Google Scholar]
- 17.Drzezga A, Becker JA, Van Dijk KR, et al. Neuronal dysfunction and disconnection of cortical hubs in non-demented subjects with elevated amyloid burden. Brain. 2011;134:1635–46. doi: 10.1093/brain/awr066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Rinne JO, Mosconi L, et al. Regional analysis of FDG and PIB-PET images in normal aging, mild cognitive impairment, and Alzheimer’s disease. Eur J Nucl Med Mol Imaging. 2008;35:2169–81. doi: 10.1007/s00259-008-0833-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosconi L, Sorbi S, de Leon MJ, et al. Hypometabolism exceeds atrophy in presymptomatic early-onset familial Alzheimer’s disease. J Nucl Med. 2006;47:1778–86. [PubMed] [Google Scholar]
- 20.Drzezga A, Lautenschlager N, Siebner H, et al. Cerebral metabolic changes accompanying conversion of mild cognitive impairment into Alzheimer’s disease: a PET follow-up study. Eur J Nucl Med Mol Imaging. 2003;30:1104–13. doi: 10.1007/s00259-003-1194-1. [DOI] [PubMed] [Google Scholar]
- 21.Folstein MF, Robins LN, Helzer JE. The mini-mental state examination. Arch Gen Psychiatry. 1983;40:812. doi: 10.1001/archpsyc.1983.01790060110016. [DOI] [PubMed] [Google Scholar]
- 22.Albert MS, DeKosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:270–9. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–9. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brix G, Zaers J, Adam LE, et al. Performance evaluation of a whole-body PET scanner using the NEMA protocol. National Electrical Manufacturers Association. J Nucl Med. 1997;38:1614–23. [PubMed] [Google Scholar]
- 25.Reuter M, Schmansky NJ, Rosas HD, Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage. 2012;61:1402–18. doi: 10.1016/j.neuroimage.2012.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabuncu MR, Desikan RS, Sepulcre J, et al. The dynamics of cortical and hippocampal atrophy in Alzheimer disease. Arch Neurol. 2011;68:1040–8. doi: 10.1001/archneurol.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999;8:272–84. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitwell JL, Jack CR., Jr Comparisons between Alzheimer disease, frontotemporal lobar degeneration, and normal aging with brain mapping. Top Magn Reson Imaging. 2005;16:409–25. doi: 10.1097/01.rmr.0000245457.98029.e1. [DOI] [PubMed] [Google Scholar]
- 29.Svarer C, Madsen K, Hasselbalch SG, et al. MR-based automatic delineation of volumes of interest in human brain PET images using probability maps. Neuroimage. 2005;24:969–79. doi: 10.1016/j.neuroimage.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 30.Nissen IA, Boellaard R, Ossenkoppele R, et al. Impact of partial volume corrections on quantitative brain PET studies [abstract]. SNM Conference; 2012. [Google Scholar]
- 31.Teo BK, Seo Y, Bacharach SL, Carrasquillo JA, Libutti SK, Shukla H, et al. Partial-volume correction in PET: validation of an iterative postreconstruction method with phantom and patient data. J Nucl Med. 2007;48:802–10. doi: 10.2967/jnumed.106.035576. [DOI] [PubMed] [Google Scholar]
- 32.Wu Y, Carson RE. Noise reduction in the simplified reference tissue model for neuroreceptor functional imaging. J Cereb Blood Flow Metab. 2002;22:1440–52. doi: 10.1097/01.WCB.0000033967.83623.34. [DOI] [PubMed] [Google Scholar]
- 33.Yaqub M, Tolboom N, Boellaard R, van Berckel BN, van Tilburg EW, Luurtsema G, et al. Simplified parametric methods for [11C]PIB studies. Neuroimage. 2008;42:76–86. doi: 10.1016/j.neuroimage.2008.04.251. [DOI] [PubMed] [Google Scholar]
- 34.Yamaguchi H, Hirai S, Morimatsu M, Shoji M, Nakazato Y. Diffuse type of senile plaques in the cerebellum of Alzheimer-type dementia demonstrated by beta protein immunostain. Acta Neuropathol. 1989;77:314–9. doi: 10.1007/BF00687584. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 36.Chetelat G, Villemagne VL, Bourgeat P, et al. Relationship between atrophy and beta-amyloid deposition in Alzheimer disease. Ann Neurol. 2010;67(3):317–24. doi: 10.1002/ana.21955. [DOI] [PubMed] [Google Scholar]
- 37.Fagan AM, Head D, Shah AR, et al. Decreased cerebrospinal fluid Abeta[42] correlates with brain atrophy in cognitively normal elderly. Ann Neurol. 2009;65(2):176–83. doi: 10.1002/ana.21559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villemagne VL, Burnham S, Bourgeat P, et al. Amyloid β deposition, neurodegeneration and cognitive decline in sporadic Alzheimer’s disease: a prospective cohort study. Lancet Neurol. 2013;12(4):357–67. doi: 10.1016/S1474-4422(13)70044-9. [DOI] [PubMed] [Google Scholar]
- 39.Tosun D, Schuff N, Mathis CA, Jagust W, Weiner MW. Spatial patterns of brain amyloid-beta burden and atrophy rate associations in mild cognitive impairment. Brain. 2011;134(Pt 4):1077–88. doi: 10.1093/brain/awr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mormino EC, Kluth JT, Madison CM, et al. Episodic memory loss is related to hippocampal-mediated beta-amyloid deposition in elderly subjects. Brain. 2009;132(Pt 5):1310–23. doi: 10.1093/brain/awn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrews KA, Modat M, Macdonald KE, et al. Atrophy rates in asymptomatic amyloidosis: implications for Alzheimer prevention trials. PLoS One. 2013;8(3):e58816. doi: 10.1371/journal.pone.0058816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Driscoll I, Zhou Y, An Y, et al. Lack of association between 11C-PiB and longitudinal brain atrophy in non-demented older individuals. Neurobiol Aging. 2011;32(12):2123–30. doi: 10.1016/j.neurobiolaging.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Josephs KA, Whitwell JL, Ahmed Z, et al. Beta-amyloid burden is not associated with rates of brain atrophy. Ann Neurol. 2008;63(2):204–12. doi: 10.1002/ana.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chetelat G, Villemagne VL, Villain N, et al. Accelerated cortical atrophy in cognitively normal elderly with high beta-amyloid deposition. Neurology. 2012;78(7):477–84. doi: 10.1212/WNL.0b013e318246d67a. [DOI] [PubMed] [Google Scholar]
- 45.Doré V, Villemange VL, Bourgeat P, et al. Cross-sectional and longitudinal analysis of the relationship between Aβ deposition, cortical thickness, and memory in cognitively unimpaired individuals and in Alzheimer disease. JAMA Neurol. 2013;70(7):903–11. doi: 10.1001/jamaneurol.2013.1062. [DOI] [PubMed] [Google Scholar]
- 46.DeSanti S, de Leon MJ, Rusinek H, et al. Hippocampal formation glucose metabolism and volume losses in MCI and AD. Neurobiol Aging. 2001;22(4):529–39. doi: 10.1016/s0197-4580(01)00230-5. [DOI] [PubMed] [Google Scholar]
- 47.Jagust W, Gitcho A, Sun F, Kuczynski B, Mungas D, Haan M. Brain imaging evidence of preclinical Alzheimer’s disease in normal aging. Ann Neurol. 2006;59(4):673–81. doi: 10.1002/ana.20799. [DOI] [PubMed] [Google Scholar]
- 48.Meguro K, LeMestric C, Landeau B, Eustache F, Baron J-C. Relations between hypometabolism in the posterior association neocortex and hippocampal atrophy in alzheimer’s disease: a PET/MRI correlative study. J Neurol Neurosurg Psychiatry. 2001;71:315–21. doi: 10.1136/jnnp.71.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaguchi S, Meguro K, Itoh M, et al. Decreased cortical glucose metabolism correlates with hippocampal atrophy in Alzheimer’s disease as shown by MRI and PET. J Neurol Neurosurg Psychiatry. 1997;62:596–600. doi: 10.1136/jnnp.62.6.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chetelat G, Desgranges B, Landeau B, Mezenge F, Poline JB, de la Sayette V, et al. Direct voxel-based comparison between grey matter hypometabolism and atrophy in Alzheimer’s disease. Brain. 2008;131:60–71. doi: 10.1093/brain/awm288. [DOI] [PubMed] [Google Scholar]
- 51.Förster S, Grimmer T, Miederer I, et al. Regional expansion of hypometabolism in Alzheimer’s disease follows amyloid deposition with temporal delay. Biol Psychiatry. 2012;71:792–7. doi: 10.1016/j.biopsych.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 52.Knopman DS, Jack CR, Jr, Wiste HJ, et al. Selective worsening of brain injury biomarker abnormalities in cognitively normal elderly persons with β-amyloidosis. JAMA Neurol. 2013;70(8):1030–8. doi: 10.1001/jamaneurol.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen AD, Price JC, Weissfeld LA, et al. Basal cerebral metabolism may modulate the cognitive effects of aβ in mild cognitive impairment: an example of brain reserve. J Neurosci. 2009;29(47):14770–8. doi: 10.1523/JNEUROSCI.3669-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Förster S, Yousefi BH, Wester HJ, et al. Quantitative longitudinal interrelationships between brain metabolism and amyloid deposition during a 2-year follow-up in patients with early Alzheimer’s disease. Eur J Nucl Med Mol Imaging. 2012;39(12):1927–37. doi: 10.1007/s00259-012-2230-9. [DOI] [PubMed] [Google Scholar]
- 55.Cirrito JR, Kang JE, Lee J, et al. Endocytosis is required for synaptic activity-dependent release of amyloid-beta in vivo. Neuron. 2008;58:42–51. doi: 10.1016/j.neuron.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Briellmann RS, Syngeniotis A, Jackson GD. Comparison of hippocampal volumetry at 1.5 Tesla and at 3 Tesla. Epilepsia. 2001;42(8):1021–4. doi: 10.1046/j.1528-1157.2001.0420081021.x. [DOI] [PubMed] [Google Scholar]
- 57.Scorzin JE, Kaaden S, Quesada CM, Müller CA, Fimmers R, Urbach H, et al. Volume determination of amygdale and hippocampus at 1.5 and 3.0 T MRI in temporal lobe epilepsy. Epilepsy Res. 2008;82(1):29–37. doi: 10.1016/j.eplepsyres.2008.06.012. [DOI] [PubMed] [Google Scholar]