Abstract
Single-particle tracking is increasingly used to extract quantitative parameters on single molecules and their environment, while advances in spatial and temporal resolution of tracking techniques inspire new questions and avenues of investigation. Correspondingly, sophisticated analytical methods are constantly developed to obtain more refined information from measured trajectories. Here we point out some fundamental limitations of these approaches due to the finite length of trajectories, the presence of localization error, and motion blur, focusing on the simplest motion regime of free diffusion in an isotropic medium (Brownian motion). We show that two recently proposed algorithms approach the theoretical limit of diffusion coefficient uncertainty. We discuss the practical performance of the algorithms as well as some important implications of these results for single-particle tracking.
I. INTRODUCTION
Single-particle tracking (SPT) methods are well established and widely used for studying the microscopic behavior and interactions of individual molecules or microscopic objects in soft matter and biological environments [1,2] and are part of the growing toolbox available to researchers studying nanoparticle and single-molecule dynamics. A single particle can be localized with nanoscale accuracy through optical microscopy—a fact that has been extensively analyzed and applied in recent years in general nanoscale measurement methods and specifically for super-resolution microscopy [3–5], high-resolution colocalization studies [6,7], or single-particle and single-molecule tracking [8–10]. However, the consequences of finite localization accuracy on measurements of properties of moving particles have not yet received much attention. As it turns out, this can lead to very large errors on dynamic parameter estimation, preventing in some cases any quantitative conclusion on the particle motion. The purpose of this article is to compare and summarize new theoretical and numerical developments, allowing a precise assessment of these effects, as well as to provide simple guidelines to adjust experimental control parameters (localization accuracy, exposure time, frame separation, and trajectory length) in order to ensure a desired precision on the measured parameters.
A basic dynamic characteristic of microscopic behavior is the diffusion coefficient D, which may be constant in an isotropic medium or vary spatially or temporally in complex environments [11,12]. For example, changes from slow to fast diffusion are sometimes observed in membrane proteins, and can in principle be characterized as changes in D [13–17]. Although not all motion regimes can be characterized by a single D value (a regime we will refer to as “free diffusion”) or even a few different D values—for example, diffusion may be superimposed to drift (directed motion), confined within hard boundaries (confined motion), or taking place within a fractal domain (anomalous diffusion) [2]—free diffusion is one of the most common models used to interpret experimental data, in particular when only short trajectories are available. In these complex situations, it is of fundamental importance to understand how accurately a free diffusion coefficient can be estimated from a finite-length trajectory. In particular, this knowledge is necessary to assess the statistical significance of apparent differences in diffusion coefficients between segments of a single trajectory, between multiple trajectories observed under identical conditions or between experiments obtained under different conditions.
The previous discussion raises the question of how well D can be obtained from the analysis of a finite length single-particle trajectory, but a no less important aspect of this question is the influence of localization error on the measured D. Surprisingly, although the precision of noise-limited single-particle localization has been the focus of significant attention due to its importance in super-resolution imaging [18,19], the consequence of a finite localization precision on D estimation (and its worsening due to diffusion) has until recently been scarcely addressed theoretically [20–22].
In this paper we answer two fundamental theoretical questions regarding the optimal estimation of free diffusion coefficients from SPT data.
What is the best theoretically achievable precision in estimating D from a single trajectory knowing its length (number of positions N), localization precision, and camera integration time? Since a theoretical bound without a practical means of attaining it is of limited use, we also address a second, more practical question.
What estimation procedure extracts D from the data in a way that achieves such precision?
Answering the first question provides relations allowing optimizing experimental data acquisition parameters in order to achieve a desired level of precision on the measured D.
The answer to the second question provides tools to extract an optimal estimation of the diffusion coefficient from the resulting experimental trajectories.
This work extends and ties together our two recent independent publications addressing partial aspects of these questions [20,21]. On one hand, asymptotic (large N) expressions for the optimal precision (or Cramér-Rao lower bound) as well as a maximum likelihood estimator (MLE) of D were obtained in Ref. [20]. On the other hand, an algorithm for optimized least-square fit (OLSF) of the familiar mean-square displacement curve, accounting for localization accuracy and trajectory length, was presented in Ref. [21]. Both results were incomplete in the following sense: the applicability of the asymptotic results of Ref. [20] to real data remained unknown (in particular, the question of how large must N be for the asymptotic results to apply), while the information theoretic optimality of the OLSF approach [21] remained to be addressed.
Here we give a complete theoretical analysis of diffusion coefficient estimation in SPT by first providing an exact Cramér-Rao lower bound formula for the diffusion coefficient uncertainty valid for all N, based on maximum likelihood estimation, thus answering question (1). We then briefly summarize the OLSF method and compare its theoretical performance with the Cramér-Rao lower bound, finding it to perform near optimally. We next compare numerical implementations of the MLE and OLSF approaches to the fundamental limit (Cramér-Rao bound) over a large simulated data set, verifying that both approach the theoretical performance. We conclude that (1) dynamic localization uncertainty σ sets a fundamental limit on the precision with which D can be determined, quantified by the Cramér-Rao lower bound; (2) both the MLE and OLSF provide a good approximation of the best possible diffusion coefficient estimate over a large portion of parameter space [thus answering question (2)]; and (3) in many experimental scenarios—including the estimation of short-time diffusion coefficients as might be encountered in complex or switching regimes—D cannot be measured precisely by any data analysis procedure. This, of course, should be of immediate practical interest, as it sets a practical limit to the claims that can be drawn from experimental data. We also hope that this work will motivate further studies of similar constraints existing in other diffusion or complex motion models.
This article is organized as follows. Section II briefly summarizes the maximum likelihood approach used in Ref. [20] and provides exact results for the Cramér-Rao lower bounds (CRLB) on fitted parameters (detailed in Appendix B), briefly discussing the general implications for parameter estimation in SPT. Section III provides a brief summary of the optimal least-square fit (OLSF) analysis of the mean-square displacement introduced in Ref. [21], while improvements introduced in this work are detailed in Appendix A. Section IV compares the results of OLSF with the CRLB, establishing that OLSF is nearly optimal. Section V then compares the performance of numerical implementations of the MLE and OLSF methods with the theoretical limits. Finally, Sec. VI summarizes our main results and discusses some applications.
II. MAXIMUM LIKELIHOOD ESTIMATION AND CRAMÉR-RAO LOWER BOUNDS ON THE VARIANCE OF D AND σ2
In a typical SPT experiment, a trajectory is obtained by determining a particle’s positions at equally spaced time intervals Δt, for instance using a series of N images (or frames) obtained by video microscopy [1,2]. The particle’s position at each time point iΔt is known with an uncertainty σ.
It is worth mentioning that the dynamic localization error σ involved in a tracking experiment is larger than the static localization error σ0 involved in super resolution or high-resolution colocalization studies [18,19,23]. Indeed, as discussed in Ref. [21], the effect of diffusion is to expand the single-particle image and thus “spread out” the total signal over a larger area, therefore decreasing the signal-to-noise ratio and correspondingly, the localization accuracy. A simple model to take this effect into account was proposed in Ref. [21], resulting in the following approximation:
| (1) |
where s0 is the standard deviation of a Gaussian approximation of the microscope point-spread function and tE is the camera exposure duration (tE ≤ Δt). From this expression, which involves the unknown parameter D, it is clear that σ should formally be considered as an unknown parameter of the model.
After SPT data have been collected, estimating model parameters such as D and σ from the trajectory can be accomplished using various methods [24–26]. Mean-square displacement (MSD) analysis is among the most popular one [2,24], but has a number of fundamental limitations, which make it wholly inappropriate for reliable parameter estimation, unless special care is taken, as discussed in a later section. An alternative approach to diffusion coefficient estimation based on maximum likelihood estimation was introduced in Ref. [15] and updated in Ref. [20], where it was noted that, in contrast with the original assumption of Ref. [15], SPT displacements {Δi = r⃗i+1 − r⃗i} (i = 1, …, N − 1) calculated from realistic data are correlated due to localization error and motion blur, resulting in a multivariate Gaussian distribution of observed displacements with nondiagonal covariance matrix Σ [20,27,28]. For the one-dimensional case:
| (2) |
In this expression Δ is the (N − 1)-component column vector of observed displacements and Σ is the covariance matrix with (N − 1) × (N − 1) elements defined by [20]
| (3) |
where the reduced square localization error x is defined by
| (4) |
and R is the motion blur coefficient, a dimensionless parameter specifying the duration and type of camera exposure used in the experiment [20,21]:
| (5) |
In this expression, S(t) is the illumination percentage occurring before time t (between the beginning and end of a frame, separated by Δt). R ranges from 0 to 1/4 depending on the camera exposure mode and duration. For instance, in the common experimental case of a uniform exposure of duration tE ≤ Δt, R = 1/6(tE/Δt) [setting S(t) = t/tE in Eq. (5) for t < tE, S(t) = 1 otherwise] [15,20,21,29,30].
Using a standard assumption of statistical inference theory, the “best fit” parameters of the model are taken as those maximizing the probability distribution of the displacements, given the observed data. It is equivalent and in general simpler to maximize the log-likelihood, L(Δ) = log[p(Δ)], which can be done by diagonalizing the covariance matrix Σ. This computation can be done approximately (for large N) by a discrete Fourier transform, as discussed in Ref. [20]. From the diagonalized data, a numerical algorithm approximating the MLE for large N was given in Ref. [20], allowing an iterative search of the best fit parameters.
In Ref. [20] this approximation was used to compute the Fisher information matrix I in the two parameters D and σ2, whose matrix elements Ijk are equal to
| (6) |
where θj designates either parameter D or parameter σ and E indicates the mean value of the quantity in parentheses. From this matrix, the Cramér-Rao lower bounds (CRLB) on the model parameter variances for large N were obtained as the inverse of the diagonal elements of the Fisher information matrix. The CRLB is an information theoretic quantity that sets a lower limit on the variances of all unbiased estimation procedures (not just the MLE [31]). In other words, no practical algorithm used to extract parameters D and σ from the data can expect to achieve a smaller uncertainty on these parameters than the lower limit set by the CRLB. Expressed in yet another way, any algorithm achieving an error on the fitted parameters that approaches the CRLB result is performing optimally from an information theory point of view.
In Ref. [20] the number of points N required to make these approximations valid was not discussed. Here we present exact results for the MLE and CRLB for any of N, allowing an exhaustive comparison of the MLE and OLSF algorithms performance with the CRLB. As shown in Appendix A, the CRLB on D depends only on the number of trajectory points N and the reduced square localization error x.
The CRLB1 on σ2 also depends on the motion blur coefficient R. From the expressions for the CRLB on the variance of D and σ2 given in Appendix A, the relative uncertainties [ratio between the standard deviation (S) and the expected value of the parameter] satisfy
| (7) |
| (8) |
where d is the number of spatial dimensions of the trajectory and δ(x) [respectively, ς(x)] is an increasing (respectively, decreasing) function of x, whose exact expressions as a function of x, N, and R are given in Appendix A.
Equations (7) and (8) completely specify how accurately D and σ2 can be determined in SPT by an unbiased estimator. They are represented in Fig. 1, which shows contours of regions in the (x, N) plane within which each quantity can be estimated with a specified relative uncertainty ε.
FIG. 1.
(Color online) Contours of constant Cramér-Rao lower bounds ε [Eqs. (7) and (8)] for the relative uncertainty on D (plain curves) or σ2 (dashed curves), for R = 0 and d = 1. From top to bottom, ε = 0.1, 0.2, 0.5, and 1. Pairs of values (x, N) located above a given contour have a lower relative uncertainty than the value defined by this contour (reported in the legend). For instance, pairs in the top green hatched region have both relative uncertainties on D and σ2 smaller than 10%. By looking at the intercept of these contours with the N axis when x → 0 (respectively, x → ∞), the minimum number of trajectory points needed to achieve a desired level of precision for D (respectively, σ2) can be determined. For instance, a minimum of 600 (respectively, 150 or 24) trajectory points is needed to obtain a 10% (respectively, 20% or 50%) uncertainty on D in the absence of localization error (x = 0). Significantly more trajectory points are needed if x > 1. S is the standard deviation. Note that values of N reported to the left need to be replaced by 1 + (N − 1)/d in d > 1 dimensions.
It is worth noting that the dependence of the uncertainties on D and σ2 with respect to the reduced square localization error x are anticorrelated: better precision on D is achieved when the reduced square localization error x is small, whereas the opposite behavior is observed for the localization error σ2.
Equations (7) and (8) also express the fact that the standard deviation of both parameters vary as [d(N − 1)]−1/2, where d(N − 1) is the total number of one-dimensional displacements in the trajectory. In other words, an improvement of a factor of 2 of the precision on D requires 4 times more trajectory points (assuming the localization uncertainty remains constant). In particular, since both vary as d−1/2, where d is the number of spatial dimensions, this means that from a statistical point of view it is not recommended to perform a separate MSD analysis for each dimension, as sometimes seen in the literature [32], unless there is a reason to expect some diffusion anisotropy. Section VI will discuss different limiting cases of these results and their application in practical situations.
III. OPTIMIZED LEAST-SQUARE FIT OF THE MSD
As mentioned above, one of the most popular approaches to SPT data analysis involves computing the so-called mean-square displacement (MSD) curve of the trajectory and fitting it with the appropriate dynamic model. Among different possible definitions of the MSD, we consider the most commonly used:
| (9) |
where r⃗i, i ∈ {1, …, N} is the measured position of the particle in frame i. The MSD curve represents the average square distance traveled by the particle after different time lags iΔt. For an isotropic free diffusion regime in d dimensions, the theoretical MSD curve grows linearly with time, the slope of the curve being proportional to the diffusion coefficient D:
| (10) |
Equation (10) can be normalized by the average single-frame mean square displacement 2dDΔt to obtain
| (11) |
which identifies the reduced square localization error x defined in the previous section as the offset of the normalized MSD curve. When R = 0, a small value of x (x < 1) indicates a regime where the dynamic localization error σ is small compared to the mean displacement due to diffusion during a single frame duration, .
Although a linear fit to the experimental MSD curve should provide a good estimate of D, the problem is complicated by the increased variance and correlated values of the MSD at large time lags, as noted originally [24,33]. This suggests using only the first few points of the MSD curve in order to reduce the influence of the large variance of the later MSD points. How few points to use has remained largely an empirical decision and varies from author to author. It was recently recognized that the MSD variance is also severely affected at short time lags by dynamic localization error σ and motion blur R [20,21] (the explicit expression for the MSD variance and covariance for arbitrary dimension d including dynamic localization error and motion blur is provided in Appendix C for reference). The increased variance of the MSD curve at short time lags is particularly noticeable when the reduced square localization error x is larger than 1, and results in poor MSD fits if only small time lags are used for the fit, as is often practiced. This suggested that there exists an optimal number pmin of MSD points to use (depending on the reduced parameter x as well as on the number of trajectory points N) in order to obtain the smallest uncertainty on the fitted parameter D.
This heuristic argument was confirmed by computing the expected error on both parameters (D and σ2) as a function of number of fitting points p. As shown in Ref. [21], a minimal error on each fitted parameter can be achieved using an optimal number of MSD points pmin, whose exact value depends on x and N. Arguably the most important conclusion of this work was the fact that using any other number of MSD points (say 10% of the total number, or the first four points, or any other empirical choice) results in potentially much larger uncertainty on the fitted parameters. Although pmin can be computed without approximation as detailed in Ref. [21], it is more computationally efficient to use the approximate formulas for pmin(D) and pmin(σ2) provided in Appendix B [Eqs. (B3) and (B4)]. The resulting error on each parameter D and σ2 can then be computed using Eqs. (B11) and (B16).
The least-square fit of the unweighted MSD using the optimal number of points described above is what we will refer to in the following as the optimized least square fit (OLSF) of the MSD curve. Note that since this fit requires the a priori knowledge of x to compute the optimal number of points needed to fit D and σ2, it is not directly applicable to experimental data, for which, by definition, D and σ are not known. However, an iterative search of the optimal number of fitting points can be devised, as briefly discussed in Ref. [21]. An improved version of this algorithm and results of extensive study of its performance are presented in Appendix B, and discussed in Sec. V.
IV. COMPARISON OF THE THEORETICAL OLSF RESULTSWITH THE CRAMÉR-RAO LOWER BOUNDS
One of the unanswered questions regarding the theoretical OLSF approach is in what sense the fitted OLSF parameters are optimal. An indication that they might be more than optima for the least-square fit approach only, is given by the fact that uncertainties on fitted parameters [D and σ2, Eqs. (B11) and (B16)] appear numerically indistinguishable whether one used a weighted or an unweighted optimized least-square fit [21]. This is a nontrivial result, as both types of fit (weighted or unweighted) result in very different behavior of the parameter uncertainties as a function of the number of MSD points p used for the fit (see Ref. [21] for details). The fact that both fits yield the same minimum uncertainty on the parameters is thus a strong indication that this minimum is an absolute minimum. However, the only way to establish that the OLSF results are truly optimal in a information theoretic sense is by a direct comparison with the lowest statistical bounds on each parameter’s uncertainty.
We thus compared the OLSF theoretical results for the uncertainties on D and σ2 [Eqs. (B11) and (B16)] with the respective CRLB obtained in Sec. II.
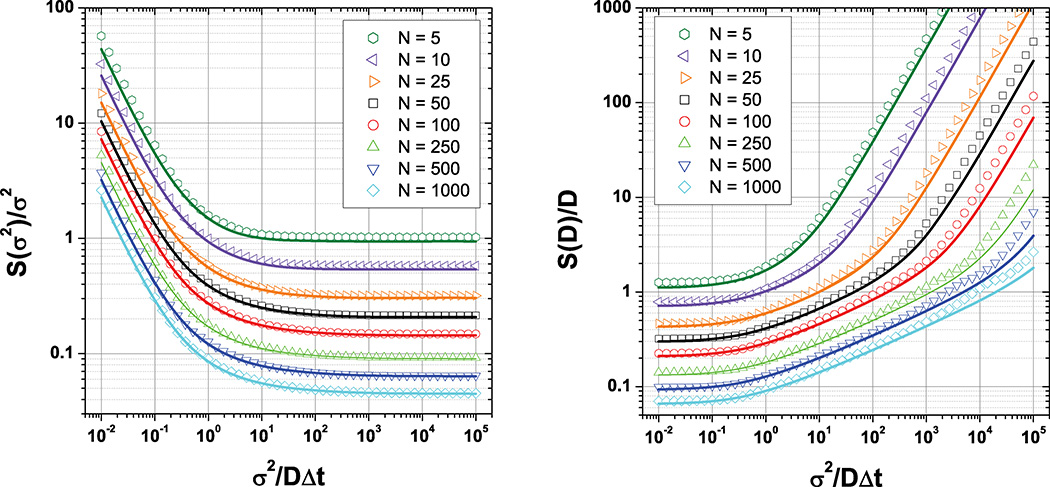
As shown in Fig. 2 for R = 1/6 (camera integration during the whole frame duration) and d = 1, there is an excellent agreement between the two approaches over several orders of magnitude for x and N. Similar results hold for R = 0 (no integration, see Fig. 8 in Appendix A) and values in between.
FIG. 2.
(Color online) Comparison between the relative error on OLSF fit parameters [open symbols, Eqs. (B11) and (B16)] and the corresponding Cramér-Rao lower bounds [plain curves, Eqs. (7) and (8)] for various number of trajectory points N, in the case R = 1/6 (camera integration during the whole frame duration) and d = 1. N = 5 to 1000 from top to bottom. There is a remarkable agreement between the two, demonstrating that OLSF provides close to optimal estimate of the parameters. S is the standard deviation.
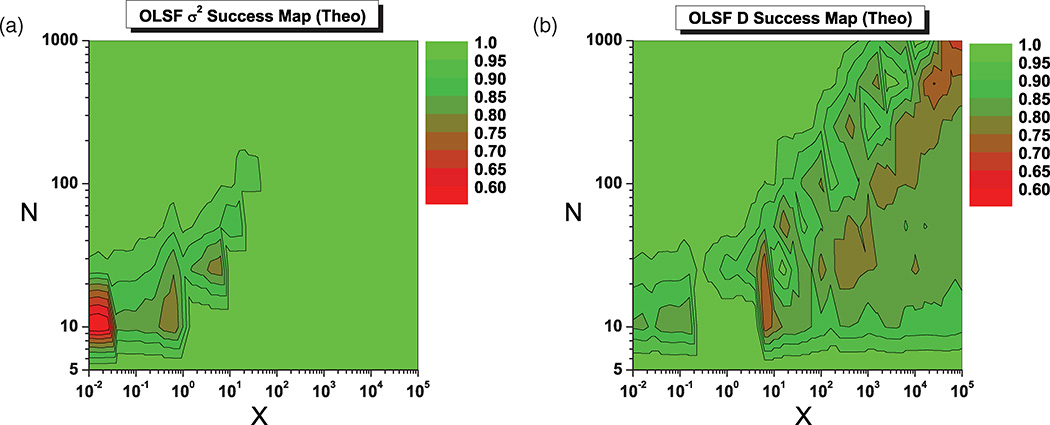
FIG. 8.
(Color online) Success rate of the OLSF algorithm in determining the optimal number of fitting point needed to optimally fit (a) σ2 and (b) D.
Since the CRLB are by definition the theoretical limits of the MLE approach, these results indicate than numerical implementations of both MLE and OLSF methods should be capable of approaching these theoretical limits in experimental situations. This, however, can only be determined by investigating the performance of actual algorithms implementing these approaches, which we do in the next section.
V. COMPARISON OF ACTUAL MLE AND OLSF ALGORITHMS WITH ONE ANOTHER AND WITH THE CRLB
To test the practical performance of MLE and OLSF algorithms we performed a series of simulations over a large range of reduced square localization error x and number of trajectory points N. For each (x, N) pair we simulated 1000 trajectories and ran both the MLE and OLSF estimators (provided as a Matlab routine in Supplemental Material [34]) on each of them, and analyzed the resulting distribution of fitted D and σ2 parameters.
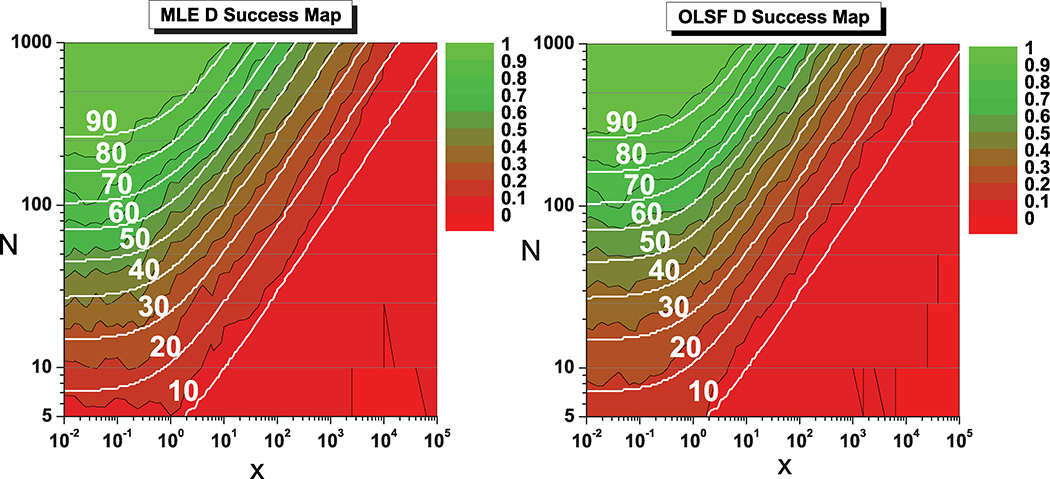
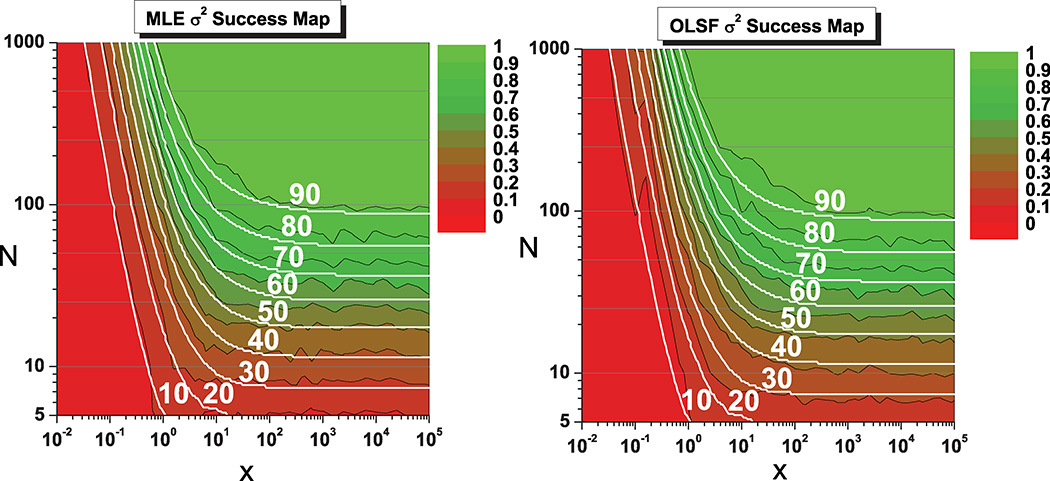
Figures 3 and 4 summarize the results of our numerical study in the form of “success maps” for both algorithms. For each (x, N) pair we computed the fraction of trajectories whose fitted D (Fig. 3) or σ2 (Fig. 4) was within 25% of the true value (the value used for the simulation), as a measure of the success rate of both approaches. We also overlaid smooth contours showing the expected success fraction (defined as above) for an optimal unbiased estimator that achieves the CRLB. Note that the choice of a 25% distance to the true value is arbitrary and does not change the fact that for any other choice the agreement between theory and algorithms is excellent.
FIG. 3.
(Color online) Success maps for the MLE and OLSF algorithms for d = 1, showing the fraction of trajectories resulting in an estimated D value within 25% of the true value, as a function of the reduced localization error x and the number of points N. Green indicates 100% success, while red indicates failure. White curves are contours where an optimal unbiased estimator reaches success 10% to 90% of the time (from bottom to top). Note that these curves are different from (though related) to those represented in Fig. 1. As in Fig. 1, values of N reported to the left need to be replaced by 1 + (N − 1)/d in d > 1 dimensions.
FIG. 4.
(Color online) Success map for the MLE and OLSF algorithms for d = 1, showing the fraction of trajectories resulting in an estimated σ2 value within 25% of the true value, as a function of the reduced localization error x and the number of points N. White curves are contours where an optimal unbiased estimator reaches success 10% to 90% of the time (from bottom to top). The same comment as for Fig. 3 applies.
From Figs. 3 and 4, which show a near perfect match between the success maps of both methods and the predicted success rates, it appears that both MLE and OLSF estimators achieve nearly optimal performance. That is, each estimator performs almost as well as an ideal unbiased estimator. In other words, no other unbiased estimator can significantly outperform the MLE or OLSF estimators presented here over the vast majority of the (x, N) space. A detailed comparison of the complete data set is available online as a hyperlinked PDF document [34] and is worth looking at to understand the difference between the situation in high success rate regions (green) and a low success rate regions (red). In the latter, the fitted parameters can be wildly off the exact result, casting doubt on the usefulness of even attempting to fit the data in these regions of the (x, N) plane.
In particular, it is important to realize that, although MLE and OLSF are nearly optimal, they cannot give reliable estimates of D for small numbers N of trajectory points (N < 10). Indeed, it is impossible for any estimator to obtain reliable estimates in this case, even when the reduced square localization error x is fairly modest (x < 1). This can be seen from Eq. (7) and Figs. 1–4, and suggests that a number of approaches using short trajectory segments to infer subtle changes of diffusion regimes may need to be revisited (e.g., Refs. [35,36]). We discuss this important point further in Sec. VI.
Although they approach optimality, MLE and OLSF do not give identical results. Indeed, in contrast to OLSF, MLE cannot return an unphysical negative value for D or σ2. Although this seems initially to be a desirable attribute, it means that estimates of a parameter with a large variance (compared to the true value) tend to have an asymmetric distribution with positive values results “piling up” at D = 0 and σ2 = 0 (see Supplemental Material [34] for examples). This results in a positive bias (overestimation of the mean), which is particularly marked for x > 1. OLSF on the contrary is not constrained to return positive values and thus results in symmetrically distributed, unbiased results. However, in the same regime of large parameter variance (compared to the true value), this can result in unphysical negative fitted values. Each of these phenomena (biased results and unphysical estimates) are undesirable, but it is important to note that both issues appear in “red zones” of the parameter space (Figs. 3 and 4), where the CRLB is large compared to the mean, and even an optimal unbiased estimator performs rather poorly. In these regimes there is simply not enough information in the data to obtain high-quality estimates by any estimation procedure.
Another noteworthy difference between the two algorithms (implemented in the Matlab routine provided as Supplemental Information [34]) is their execution speed. A simple estimate of computation time as a function of the number of points in the trajectory shows an approximately linear behavior for MLE versus a quadratic dependence for the OLSF (Fig. S1). However, the OLSF algorithm appears to outperform the MLE algorithm for N < 3000 points. As the computer speed steadily improves, these differences might become irrelevant.
So far we only discussed results for cases where σ is unknown and must be estimated along with D. If σ is already known, the CRLB on D is improved and a smaller number of trajectory points are needed to obtain a given uncertainty (Fig. 5). It is common to attempt to measure this quantity using immobilized particles. However, σ depends on all aspects of the experiment including motion blur contributions not present for static particles [Eq. (1)]. We therefore consider that the most important case of practical interest is that in which σ is unknown, as discussed in the text.
FIG. 5.
(Color online) Curves of constant Cramér-Rao lower bounds (CRLB) for the relative uncertainty on D for d =1 dimension in different experimental situations: unknown σ and arbitrary R (plain curves), known and uniform exposure (R = 1/6, dashed curves), and known σ and instantaneous exposure (R = 0, dotted curves). The first experimental situation requires more points to obtain a given uncertainty on D, but this disadvantage is negligible for large relative localization uncertainty. More precisely, noting Nk the number of trajectory points to achieve a given relative uncertainty on D when σ is known and Nuk that number when it is unknown, Nuk/Nk(R = 0,x = 0) = 3 and Nuk/Nk(R = 1/6,x = 0) = 3/2. Note that for R > 0, negative values of x can be obtained but are not represented here. S is the standard deviation.
VI. DISCUSSION
In the previous sections we have presented exact results for the MLE and OLSF approaches and established that the two corresponding numerical algorithms perform optimally in most practical situations. In this section we discuss some possible applications of these results for SPT analysis.
A. Estimating the uncertainty on D
A first application of these results consists, after using either one of these algorithms to obtain the best possible estimates of D and σ2 from a given trajectory, of estimating the uncertainty on these parameters using the CRLB results [Eqs. (7) and (8)] computed using the fitted parameters. However, since Eqs. (7) and (8) depend on the reduced localization error x, which is very sensitive to the values of D and σ2, a large relative uncertainty on either one or both fitted parameters may yield artificially small or large x value, resulting in erroneous estimates of their relative uncertainties. Although analytically computable using the results of Appendix A, the exact distribution of x values is complex and can moreover be bimodal (with positive or negative values) [37], making a general discussion very tedious.
It is possible, however, to discuss a few common experimental cases. For instance, in the presence of negligible dynamic localization error (σ2/DΔt < 1) and assuming the exposure time tE is equal to the frame duration Δt (R = 1/6), the results of Fig. 2 show that a better estimate of D than of σ2 is expected. Supposing for instance that the true value of the right-hand side of Eq. (8) defining the minimum relative error on σ2 is of the order of 1, we could potentially obtain a fitted value of σfit2: (i) a few times larger than the true value [e.g., σfit2 ~ σ2 + V (σ2)1/2 ~λσ2, with λ = O(1)] or (ii) a few orders of magnitude smaller than the true value [e.g., σfit2 ~ σ2 − V (σ2)1/2 ~ σ2/λ, with λ = O(1)].
In the first case (i) the value xfit of x computed using the fitted values of D and σ2 [Eq. (4)], Dfit ~ D, and σfit2 > σ2 would be such that xfit > x, and would result in an overestimation of the error on D and an underestimation of the error on σ2. Note however, that if xfit + 2R = σfit2/DfitΔt ≪ 1, the overestimation of the error on D will be negligible (Fig. 2). Indeed, in this regime, the relative error on D is approximately independent of x.
In the second case (ii) [e.g., σfit2 ~ σ2 − V (σ2)1/2], the severe underestimation of σ2 could result in xfit + 2R ≪ 1, which, as we have just seen, would have little effect on the estimated error on D.
Inversely, in presence of significant dynamic localization error (σ2/DΔt > 1), the opposite would be expected: larger error on D and small error on σ2. Suppose the fitted Dfit is underestimated and a few orders of magnitude smaller than its true value [e.g., Dfit ~ D − V (D)1/2 ~ D/λ, with λ = O(1)]. Then xfit would be much larger than its true value, and the inferred relative error on D much larger than it is really. If, on the contrary, Dfit is a few times larger than its true value [e.g., Dfit ~ D + V (D)1/2 ~ λD, with λ = O(1)], xfit + 2R = σfit2/DfitΔt would be a few times smaller than its true value. If xfit + 2R ≪ 1, the effect on the estimated relative error on D would be negligible, whereas in the other cases, it would be difficult to determine how bad this estimate is.
In summary, it appears that the CRLB result for the relative error on D [Eq. (7), calculated using the fitted values] can be used reliably provided the fitted x value (xfit = σfit2/Dfit Δt − 2R) is smaller than 1, whereas no reliable error estimation can be made if xfit > 1. The opposite conclusion holds for the relative error on σ2 [Eq. (8)].
Naturally, if a large series of trajectories of identical particles can be studied, a reliable estimate of the mean value of the diffusion constant D may be obtained (law of large numbers), from which an estimate of σ could be made using Eq. (1) and an estimate of x using Eq. (4)). The resulting estimate of the relative uncertainty on D [Eq. (7)] could then be compared to the relative statistical standard deviation of D for self-consistency. Significant discrepancy between the two might indicate an underlying distribution of diffusion coefficients for the particles studied.
B. Adjusting experimental parameters to improve the precision on D
A second application of these results consists in optimizing SPT experimental parameters (N, Δt, R, and σ) to achieve a desired precision on the measured diffusion constant D (assuming one has an approximate idea of its order of magnitude). As shown in Appendix A and Fig. 6, when the desired relative uncertainty is smaller than 1, it is sufficient to use the asymptotic forms of Eqs. (A7) and (A8) for the CRLB. In this approximation, the relative uncertainty on D can be expressed as
| (12) |
showing that when N and x are large (compared to 1), , and the relative uncertainty on D increases as x1/4 (i.e., σ1/2) but decreases as N−1/2. In other words, when the reduced square localization error x is large, halving the dynamic localization error σ (for instance by increasing the brightness of the particle’s PSF) allows halving the number of data points N needed to obtain a given precision on D.
FIG. 6.
(Color online) Contours of constant Cramér-Rao lower bounds for the relative uncertainty on D, for d = 1. Open symbols correspond to the exact formula (7) [right-hand side of Eq. (7) = ε], whereas the plain curves correspond to the approximate formula [right-hand side of Eq. (12) = ε]. As in Fig. 1, values of N reported to the left need to be replaced by 1 + (N − 1)/d in d > 1 dimensions. S is the standard deviation.
When N is large and the reduced square localization error x is small, δ(x)2 ~ 2, indicating that the relative uncertainty on D varies as . For instance, in order to obtain a 10% precision on D in two dimensions, N > 300 trajectory points are necessary. A 30 points trajectory with negligible dynamic localization error would only allow at best a ~30% precision on D, while a 10 points trajectory only a ~54% precision. These figures would be increased by a factor ~2 for a reduced square localization error x = 1 and significantly more for larger x values (see also Fig. 5 for a visual guide). Note that none of these figures are recommendable goals since a 30% relative uncertainty translates into a three-sigma confidence range going approximately from zero to twice the actual value of the diffusion constant. In other words, the fitted value has potentially very little resemblance to the true value.
Clearly there is quite some flexibility in adjusting x or N in order to reduce the uncertainty on D. For instance, x can always be decreased by increasing the frame interval Δt or reducing the static localization error σ0 [Eq. (1)]. The latter can for instance be obtained by increasing the particle brightness (number of collected photons). This, however, may reduce the lifetime of the particle due to premature photobleaching, thus limiting the number of observable trajectory points N, which is always detrimental to the precision of the measurement.
On the other hand, if one is interested in measuring D over short time scales (in order to detect changes in diffusion coefficient), the total duration of the measurement is constrained by the time scale of interest: N Δ t = T, therefore increasing Δt would reduce N and vice versa.
We can quantify these different statements using the approximate expression for the dynamic localization error σ [Eq. (1)].
The reduced square localization error x can be expressed as
| (13) |
In the absence of pixilation, background, detector noise, and amplifier excess noise, the static localization error σ0 itself can be approximated by [21]
| (14) |
where P is the total number of photons in the PSF image and Q is the average photon rate emitted by the particle. From these two equations, it is straightforward to verify that x is minimized when tE = Δt (R = 1/6) and when Δt is maximized. In fact, according to Fig. 2, given a number of trajectory points N, a quasi-optimal precision on D can be obtained as soon as x + 1/3 < 1, which reads
| (15) |
Using a PSF size s0 ~ 100 nm and a typical membrane protein diffusion coefficient D ~ 0.1 µm2/s, we see that the number of photons required to satisfy Eq. (15) is easily achieved in practice (Δt = 100 ms → P > 2, Δt = 1 ms → P > 100). For particles diffusing more slowly, accurate measurements of D will require longer integration times or brighter images (D ~ 10−3 µm2/s, Δt = 100 ms → P > 100, Δt = 10 ms → P > 1000).
So far we have not specified the precision desired for D. According to Fig. 2 it can be improved either by decreasing x + 2R or by increasing the number of trajectory points N. If we are interested in measuring the diffusion coefficient rapidly with a relative precision ε, we can use the asymptotic behavior provided the number of frames N is sufficiently large (N > 10) and x < 1. The latter condition can be ensured by adjusting Δt as discussed previously and therefore the minimum number of frames needed to achieve a precision ε will be given by N > 6/dε2. For d = 2 and ε = 0.5 (a 50% precision on D, which again is not a recommendable goal), we obtain N > 12. For a better precision (for instance, to distinguish between D and D/2), ε = 0.1 would require N > 300. Since the total duration of the measurement T = N Δ t might be constrained by other considerations, Δt might become relatively short, requiring stronger signal to keep x small.
VII. CONCLUSION
In conclusion, we show that both MLE and OLSF estimators achieve nearly optimal performance in estimating D and σ2. Exact expressions for the CRLB on parameter uncertainties provided here can be used for determining when an experiment can reasonably be expected to give any useful information at all. It could also be used to determine a priori, based on experimental parameters such as integration time, frame rate, signal-to-noise ratio, maximum experiment duration, etc., what maximum precision can be expected for an estimate of the diffusion coefficient D. These results will also allow benchmarking other analysis methods with respect to the exact Cramér-Rao lower bound for various (x, N) values. In regimes where experimental data contains sufficient information to give a useful estimate, MLE and OLSF can be implemented to achieve near-optimal results with high confidence. Extension of these results to more sophisticated dynamic models will obviously be of great practical interest but are beyond the scope of this study.
Supplementary Material
Acknowledgments
This work was supported in part by Grants No. NIH EB006353 and No. GM084327 (XM).
APPENDIX A
CRAMÉR-RAO LOWER BOUND ON THE UNCERTAINTY OF D AND σ2
We briefly extend results from Ref. [20] to find exact, rather than approximate, expressions for the maximum likelihood estimator (MLE) and Cramér-Rao (CR) bounds for D and σ2. For a one-dimensional (d = 1), N-element trajectory (X1, …, XN), we define the (N − 1)-element displacement (column) vector to be Δ = [X2 − X1, X3 − X2, …, XN − XN − 1]T. For free diffusion with diffusion coefficient D and zero drift velocity, recorded at a frame interval Δt with noise σ in each frame, the (N − 1) × (N − 1) covariance matrix of Δ was found to be tridiagonal with matrix elements [20]:
| (A1) |
where x is the reduced square localization error [21] including a motion blur term [Eq. (5)].
The displacement covariance matrix is diagonal if and only if x = 0. R is the motion blur coefficient defined in Eq. (5). In Ref. [20] approximate expressions for the eigenvalues and eigenvectors of Σ were used to derive the MLE, Fisher information matrix, and Cramér-Rao lower bound on estimates of D and σ. Those results hold asymptotically for large N. Here we update those results with exact expressions. Let v(k) and Ψk be the kth eigenvector and corresponding eigenvalue of Σ. These are found to be [38]
| (A2) |
| (A3) |
and satisfy Σv(k) = Ψk v(k) and v(k) ·v(l) = δkl. Equation (A2) corresponds to the eigenvectors of the discrete sine transform (DST), for which a fast numerical implementation is known, similar to the fast Fourier transform [39]. In the terminology of stochastic processes, we have observed that the DST is the Karhunen-Loève transform [38] of the single-particle-tracking displacement process Δ. Consequently, the DST of Δ has a diagonal covariance matrix (its components are uncorrelated), and an exact numerical implementation of the MLE can be efficiently computed in this basis using Eq. (18) of Ref. [20] but setting Δ̃ = DST (Δ). The exact Fisher information matrix I in the two parameters D and σ2 can also be found from the eigenvalues given above using the formula in Eq. (19) of Ref. [20]. Its inverse, which sets a lower bound on the covariance matrix of estimates of D and σ2 is found after algebraic manipulation to be (in d dimensions)
| (A4) |
where
| (A5) |
| (A6) |
| (A7) |
| (A8) |
These formulas can be used to compute the exact CRLB of Eqs. (7) and (8). The approximate results indicated by “~” arise from approximating the sum over N as a continuous integral. Plugging the asymptotic forms of Eqs. (A7) and (A8) into Eqs. (A5) and (A6), and in turn plugging these into Eq. (A4) gives the asymptotic form of the CRLB.
The asymptotic formulas can be used for a large range of (x, N) values as shown in Fig. 6, which represent curves of constant relative CRLB for D using either the exact formula (open symbols) or the approximate ones (plain curves). The asymptotic formulas result in an underestimation of the minimum number of trajectory points N needed to obtain a given relative error ε on D only if ε > 1 (which corresponds to situations where D cannot be estimated reliably and therefore should be avoided).
APPENDIX B
OPTIMIZED LEAST-SQUARE FIT (OLSF) OF THE MSD
As discussed in detail in Ref. [21], for each value of x [Eq. (4)], there is an optimal number of MSD points pmin to use in order to obtain the “best” least-square fit parameters a (intercept) and b (slope) of the MSD, where
| (B1) |
in which d is the space dimension. The fitted parameters are “optimal” fits in the sense that their standard deviation (or uncertainty) is minimum among all fitted parameters obtained with a different number of MSD points p. Reference [21] only considered the blur-free case (R = 0) in the derivation of the fit parameters uncertainty and provided only a very short discussion of a practical algorithm to obtain pmin. This Appendix provides additional details on the algorithm and extends the analysis to the general case 0 ≤ R ≤ 1/4.
Optimal number of fitting points
Ref. [21] provided an approximate formula to estimate the optimal number of MSD points to use for an unweighted fit of a and b:
| (B2) |
where x is given by Eq. (4). Note that if x < 0, Eq. (B2) should read pmin = 2. This approximation is poor for large x values (x ≫ 1) and N < 1000 values and does not reflect the fact that slightly different optimal number of points are needed to fit the slope b (i.e., D) or the intercept a (i.e., σ2).2
Figure 7 (open symbols) represents the number of points to use to obtain the optimal intercept a [Fig. 7(a)] or slope b [Fig. 7(b)] using an unweighted LSF of the MSD curve, as a function of both x and N. These values were obtained by direct calculation of the relative fitted parameter uncertainties as a function of number of fitting points p and determination of the minimum location [21]. They are therefore exact results of the OLSF algorithm with prior information on x. The corresponding relative uncertainties on each parameter can be computed using Eqs. (B16) and (B11) [21].
FIG. 7.
(Color online) Optimal number of points for least-square fitting of (left) the intercept a (i.e., σ2) and (right) the slope b (i.e., D) of the MSD. Each curve corresponds to a different number of trajectory points N (N = 5 to 10000 from bottom to top). The plain curves correspond to Eq. (B3) (left) and Eq. (B4) (right). These results are valid for any value of the blur factor R. For x < 0, pmin = 2.
To avoid computing the relative uncertainties for all p (2 ≤ p < N) in order to find the minimum uncertainty, an iterative search of the minimum can be used since the relative uncertainties are, in general, convex curves of p. Although this approach is very fast for modest values of trajectory points N, computation of the uncertainties can become slow for large N (N > 1000). Since this procedure needs to be repeated at each step of the iterative algorithm mentioned in Sec. III and described next, using a heuristic formula can significantly speed up the approach.
Plain curves in Fig. 7 represent heuristic formulas providing a good approximation of pmin for x ∈ [10−2, 107] and N ≤ 104. The heuristic formulas represented on these graphs are
| (B3) |
and
| (B4) |
La,b(N) are the limits for x → ∞ of pmin(a,b)(x, N). Note that these formulas are ad hoc formulas and could be replaced by any other algebraic form fitting the exact results sufficiently well. In practice, the fitted a and b obtained using these approximate formulas are indistinguishable from those obtained using the exact results.
Determining the optimum number of fitting points without a priori knowledge
In the absence of any a priori knowledge on x, an iterative search of pmin(a,b)(x,N) [yielding a series of pi(a) and pi(b) values for each parameter a and b] is needed. The algorithm proposed in Ref. [21] (modified here to account for the improved formulas (B3) and (B4)) starts with pi(a) = pi(b) = N/10 and proceed as follows:
Using the new pi, fit the MSD curve and obtain new fit parameters ai and bi.
Using the new fit parameters ai and bi compute a new xi.
Using the new xi, compute new pi+1 using Eqs. (B3) and (B4).
Repeat steps 1–4 until a previously found set of pi is repeated.
In the latter case, use the previous set of pi.
In practice, this algorithm does not function well for very small or very large x, because in these cases, either the relative error on a or the relative error on b is very large (and the computed x is therefore extremely unreliable) if the optimum p are not used. These cases are however easily detected. When the relative error on a is very large, a will occasionally have a negative value. In this case, the algorithm is stopped and a value of x = 0 is assumed (in other words, pmin = 2 is used for both parameters).
When the relative error on b is large, b will occasionally have a negative value. In this case, the algorithm is stopped and a value of x = ∞ is assumed [in other words, pmin(a,b) = La,b(N) are used].
This simple algorithm is surprisingly successful and very rapid (only a few iterations are necessary).
Figure 8 shows the percentage of success as a function of x and N in determining the optimal pmin using simulated data described in the text. Success if defined here as computing a pmin with the previous algorithm, which, if used to perform the MSD least-square fit, would result in an error no more than 25% larger than the optimal error. The algorithm is successful in more than 50% of cases even in the most unfavorable regions of the (x, N) plane (x ≫ 1 for D and x ≪ 1 for σ2).
Relative error on D and σ2 in the presence of motion blur (R > 0)
This section expands the results of Ref. [21] in the presence of blur, limiting it to the case of unweighted linear fit. We explicitly introduce the number of spatial dimensions d in the final results.
The first p ≤ N − 1 points of the MSD curve are fitted by the equation
| (B5) |
where a and b are related to the variable parameters σ2 and D by Eq. (B1). The best fit values of an unweighted LSF limited to the first p points of the MSD are obtained by minimization of the χ square:
| (B6) |
with respect to σ2 and D. Here we have used the notations of Ref. [21]:
| (B7) |
After some straightforward algebra we obtain
| (B8) |
where we have introduced the intermediate quantities
| (B9) |
and Δ is defined by
| (B10) |
The results of Ref. [21] for the relative variance (Var) of the intercept a and slope b need to be multiplied by a factor 2/d for an arbitrary number of spatial dimensions d:
| (B11) |
where functions f (i, N, x) and g(i, j, N, x) have been introduced in Ref. [21] and are defined by
| (B12) |
where the quantities in the above equation are the covariance (variance when i = j) of the MSD derived in Refs. [20,21] and reported in Appendix C.
The variance of 2dσ2 can be either computed directly or using the properties of multivariate normal distributions:
| (B13) |
where Cov(a, b) is the covariance of a and b defined by
| (B14) |
We obtain
| (B15) |
The relative variance of σ2 can thus be rewritten:
| (B16) |
where the expressions for Var(a)/a2 and Var(b)/b2 are given by Eq. (B11) and U(p,N,x) by Eq. (B15).
The relative variance of D is simply given by the relative variance of b [Eq. (B11)]. These results were used to plot the graphs of Figs. 2 and 9.
FIG. 9.
(Color online) Comparison between the relative error on OLSF fit parameters and the corresponding Cramér-Rao lower bounds [plain curves, Eqs. (8) and (7) of the main text] for various number of trajectory points N, in the case R = 0 (no camera integration) and d = 1. N = 5 to 1000 from top to bottom.
APPENDIX C
VARIANCE AND COVARIANCE OF THE MSD FOR ARBITRARY SPATIAL DIMENSION d
The variance and covariance of the MSD were computed in Refs. [20,21]. However, Ref. [20] only provided a compact form for d = 1, whereas Ref. [21] provided an expanded form for d = 2. Both expressions are equivalent and can be easily generalized for arbitrary dimension d (assuming an identical dynamic localization uncertainty σ for all spatial dimensions) as follows:
| (C1) |
Footnotes
To facilitate comparison of the CRLB results with those of the optimized least square fit approach presented in the next section, it is easier to consider the CRLB on the variance of σ2 rather than of σ.
The values of pmin given here correspond to the fit of the MSD curve as defined by Eq. (9). Any other definition would require a new computation of the variance and covariance of the MSD, and consequently, of the theoretical uncertainty of the fitted parameters.
References
- 1.Crocker JC, Grier DG. J. Colloid Interface Sci. 1996;179:298. [Google Scholar]
- 2.Saxton MJ, Jacobson K. Annu. Rev. Biophys. Biomol. Struct. 1997;26:373. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 3.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 4.Hess ST, Girirajan TPK, Mason MD. Biophys. J. 2006;91:4258. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang B, Wang W, Bates M, Zhuang X. Science. 2008;319:810. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antelman J, Wilking-Chang C, Weiss S, Michalet X. Nano Lett. 2009;9:2199. doi: 10.1021/nl901163k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pertsinidis A, Zhang YX, Chu S. Nature (London) 2010;466:647. doi: 10.1038/nature09163. [DOI] [PubMed] [Google Scholar]
- 8.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Science. 2003;300:2061. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 9.Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A. Science. 2003;302:442. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 10.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, Spector DL, Singer RH. Science. 2004;304:1797. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao J. Phys. Rev. E. 2001;63:041101. [Google Scholar]
- 12.Witkoskie JB, Yang S, Cao J. Phys. Rev. E. 2002;66:051111. doi: 10.1103/PhysRevE.66.051111. [DOI] [PubMed] [Google Scholar]
- 13.Dietrich C, Yang B, Fujiwara T, Kusumi A, Jacobson K. Biophys. J. 2002;82:274. doi: 10.1016/S0006-3495(02)75393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T. Annu. Rev. Biophys. Biomol. Struct. 2005;34:351. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 15.Montiel D, Cang H, Yang H, Phys J. Chem. B. 2006;110:19763. doi: 10.1021/jp062024j. [DOI] [PubMed] [Google Scholar]
- 16.Huet S, Karatekin E, Tran VS, Fanget I, Cribier S, Henry J-P. Biophys. J. 2006;91:3542. doi: 10.1529/biophysj.105.080622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinaud F, Michalet X, Iyer G, Margeat E, Moore H-P, Weiss S. Traffic. 2009;10:691. doi: 10.1111/j.1600-0854.2009.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson RE, Larson DR, Webb WW. Biophys. J. 2002;82:2775. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ram S, Ward ES, Ober RJ. Proc. Natl. Acad. Sci. USA. 2006;103:4457. doi: 10.1073/pnas.0508047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berglund AJ. Phys. Rev. E. 2010;82:011917. doi: 10.1103/PhysRevE.82.011917. [DOI] [PubMed] [Google Scholar]
- 21.Michalet X. Phys. Rev. E. 2010;82:041914. doi: 10.1103/PhysRevE.82.041914. 83, 059904(E) (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong Y, Lin ZP, Ober RJ. IEEE Trans. Signal Process. 2011;59:895. doi: 10.1109/TSP.2010.2098403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michalet X, Lacoste TD, Weiss S. Methods. 2001;25:87. doi: 10.1006/meth.2001.1218. [DOI] [PubMed] [Google Scholar]
- 24.Qian H, Sheetz MP, Elson EL. Biophys. J. 1991;60:910. doi: 10.1016/S0006-3495(91)82125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schütz GJ, Schindler H, Schmidt T. Biophys. J. 1997;73:1073. doi: 10.1016/S0006-3495(97)78139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuoka S, Shibata T, Ueda M. Biophys. J. 2009;97:1115. doi: 10.1016/j.bpj.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong WP, Halvorsen K. Opt. Express. 2006;14:12517. doi: 10.1364/oe.14.012517. [DOI] [PubMed] [Google Scholar]
- 28.Savin T, Doyle PS. Phys. Rev. E. 2007;76:021501. doi: 10.1103/PhysRevE.76.021501. [DOI] [PubMed] [Google Scholar]
- 29.Goulian M, Simon SM. Biophys. J. 2000;79:2188. doi: 10.1016/S0006-3495(00)76467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savin T, Doyle PS. Biophys. J. 2005;88:623. doi: 10.1529/biophysj.104.042457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zacks S. The Theory of Statistical Inference. New York: Wiley; 1971. [Google Scholar]
- 32.Kusumi A, Sako Y, Yamamoto M. Biophys. J. 1993;65:2021. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saxton MJ. Biophys J. 1997;72:1744. doi: 10.1016/S0006-3495(97)78820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.See Supplemental Material at http://link.aps.org/supplemental/10.1103/PhysRevE.85.061916 for a PDF file showing a comparison of the MLE and OLSF algorithms performance and a Matlab routine implementing both algorithms.
- 35.Saxton MJ. Biophys. J. 1993;64:1766. doi: 10.1016/S0006-3495(93)81548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umemura YM, Vrljic M, Nishimura SY, Fujiwara TK, Suzuki KGN, Kusumi A. Biophys. J. 2008;95:435. doi: 10.1529/biophysj.107.123018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marsaglia G, Stat J. Software. 2006;16:1. http://www.jstatsoft.org/v16/i04/paper. [Google Scholar]
- 38.Jain AK. IEEE Trans. Commun. 1976;24:1023. [Google Scholar]
- 39.Wang Z. IEEE Trans. Acoust. 1984;32:803. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.