Abstract
We herein describe the main characteristics of ‘Neglecta timonensis’ strain SN17 (CSUR P2265) that was isolated from the stool of an 88-year-old woman with type 2 diabetes.
Keywords: Culturomics, emerging bacteria, gut microbiota, Neglecta timonensis, taxonomy
In 2015, the bacterial strain SN17 (CSUR P2265) was cultivated from the stool of an 88-year-old woman with type 2 diabetes. This study was part of the culturomics project that we are conducting on the human microbiota [1], [2]. The patient provided signed informed consent and the study was validated by the ethics committee of the Institut Federatif de Recherche 48 under number 09-022. Growth was obtained in anaerobic atmosphere at 37°C after 3 days of culture on 5% sheep blood-enriched Columbia agar (bioMérieux, Marcy l'Etoile, France). Agar-grown colonies were translucent white and had a diameter of 0.5–1 mm. Cells were Gram-positive rods ranging in length from 1.5 to 6 μm. Strain SN17 was strictly anaerobic, catalase-positive and oxidase-negative. The bacterium could not be identified using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Microflex, Bruker Daltonics, Bremen, Germany) [3].
As a consequence, we sequenced the complete 16S rRNA gene using a 3130-XL sequencer (Applied Biotechnologies, Villebon sur Yvette, France) and compared it to GenBank. The complete 16S rRNA gene of strain SN17 exhibited a sequence identity of 92.67% with Clostridium sporosphaeroides strain DSM 1294T (GenBank Accession number M59116), the phylogenetically closest species with standing in nomenclature (Fig. 1). Clostridium sporosphaeroides is an anaerobic bacterium isolated for the first time in 1948, and recently identified in biogas plants in Germany [4]. Clostridium sporosphaeroides produces butyric acid, acetic acid and propionic acid from the fermentation of the biomass [4]. Another closely related species, Clostridium leptum, has been proposed to play a role in the pathogenesis of inflammatory bowel diseases [5] as well as an immunomodulatory role in diseases such as asthma [6].
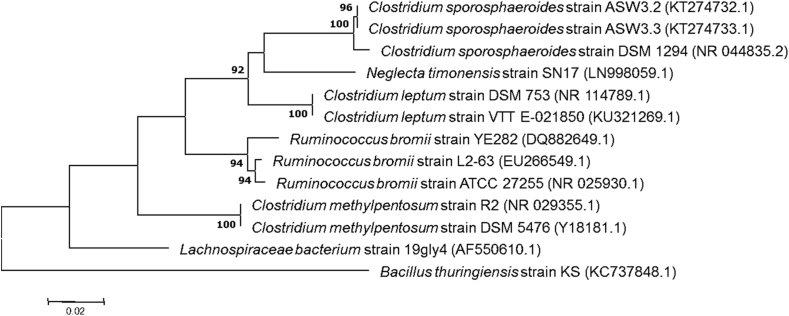
Fig. 1.
Phylogenetic tree showing the position of ‘Neglecta timonensis’ strain SN17Trelative to other phylogenetically close members of the order Clotridiales. GenBank Accession numbers are indicated in parentheses. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using the maximum-likelihood method within MEGA software. Numbers at the nodes are percentages of bootstrap values obtained by repeating the analysis 500 times to generate a majority consensus tree. Only bootstrap scores of at least 90% were retained. The scale bar indicates a 2% nucleotide sequence divergence.
As the 16S rRNA identity rate of strain SN17 with the C. sporosphaeroides type strain was lower than the 95% cut-off gene sequence suggested to delineate bacterial genera [7], we considered it as a representative of a putative new genus within the order Clostridiales. We propose to name this new genus ‘Neglecta gen. nov.’ and the new species ‘Neglecta timonensis’ gen. nov., sp. nov. Strain SN17T is the type species of ‘Neglecta timonensis’ gen. nov., sp. nov.
MALDI-TOF MS Spectrum
The MALDI-TOF MS spectrum of N. timonensis is available at http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database
Nucleotide Sequence Accession Number
The 16S rRNA gene sequence was deposited in GenBank under accession number LN998059.
Deposit in a Culture Collection
Strain SN17T was deposited in the collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under number P2265.
Conflicts of interest
The authors certify that they have no conflict of interest in relation to this research.
Acknowledgements
This research is funded by Mediterrannee-Infection Foundation.
References
- 1.Lagier J.-C., Hugon P., Khelaifia S., Fournier P.-E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagier J.-C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 3.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.-C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cibis K.G., Gneipel A., König H. Isolation of acetic, propionic and butyric acid-forming bacteria from biogas plants. J Biotechnol. 2016;220:51–63. doi: 10.1016/j.jbiotec.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Kedia S., Rampal R., Paul J., Ahuja V. Gut microbiome diversity in acute infective and chronic inflammatory gastrointestinal diseases in North India. J Gastroenterol. 2016 doi: 10.1007/s00535-016-1193-1. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 6.Huang F., Qiao H., Yin J., Gao Y., Ju Y., Li Y. Early-Life Exposure to Clostridium leptum causes pulmonary immunosuppression. PloS One. 2015;10:e0141717. doi: 10.1371/journal.pone.0141717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim M., Oh H.-S., Park S.-C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]