Abstract
The interactions between proteins and surfaces are critical to a number of important processes including biomineralization, the biocompatibility of biomaterials, and the function of biosensors. Although many proteins exist as monomers or small oligomers, amelogenin is a unique protein that self-assembles into supramolecular structures called “nanospheres,” aggregates of hundreds of monomers that are 20–60 nm in diameter. The nanosphere quaternary structure is observed in solution; however, the quaternary structure of amelogenin adsorbed onto hydroxyapatite (HAP) surfaces is not known even though it may be important to amelogenin’s function in forming highly elongated and intricately assembled HAP crystallites during enamel formation. We report studies of the interactions of the enamel protein, amelogenin (rpM179), with a well-defined (100) face prepared by the synthesis of large crystals of HAP. High-resolution in situ atomic force microscopy (AFM) was used to directly observe protein adsorption onto HAP at the molecular level within an aqueous solution environment. Our study shows that the amelogenin nanospheres disassemble onto the HAP surface, breaking down into oligomeric (25-mer) subunits of the larger nanosphere. In some cases, the disassembly event is directly observed by in situ imaging for the first time. Quantification of the adsorbate amounts by size analysis led to the determination of a protein binding energy (17.1kbT) to a specific face of HAP (100). The kinetics of disassembly are greatly slowed in aged solutions, indicating that there are time-dependent increases in oligomer–oligomer binding interactions within the nanosphere. A small change in the sequence of amelogenin by the attachment of a histidine tag to the N-terminus of rpM179 to form rp(H)M180 results in the adsorption of a complete second layer on top of the underlying first layer. Our research elucidates how supramolecular protein structures interact and break down at surfaces and how small changes in the primary sequence of amelogenin can affect the disassembly process.
Graphical Abstract
INTRODUCTION
The interactions between proteins and surfaces are important to a number of biological and technological processes. For example, interactions of proteins with inorganic surfaces are critical to the biomineralization of hard tissues such as bone, shell, and tooth.1 Protein layers adsorbed onto medical implants control cellular responses and dictate whether the implant will be successfully integrated into the body.2–4 The function of medical devices and biosensors can be interrupted by protein fouling.5 Protein adsorption to particulate and three-dimensional matrices is also of interest to drug delivery and tissue engineering.6,7 The adsorption of proteins onto surfaces is mediated by hydrophobic, electrostatic, and/or hydrogen bonding interactions between the protein and surface sites,8 and these interactions can control the adsorption coverage and induce changes in protein tertiary and secondary structure.3,9
Most of the research investigating the adsorption of proteins onto surfaces involves proteins that exist as monomers in solution. There has been little research on the adsorption of proteins that have quaternary structures and exist as large supramolecular assemblies in solution. An example of such a protein is amelogenin, a biomineralization protein involved in the formation of tooth enamel.10 Amelogenin forms aggregates called “nanospheres” consisting of hundreds of monomers and sizes ranging from 20 to 60 nm in diameter depending on the solution conditions.11–13 A number of in vitro studies have shown that the nanospheres are formed hierarchically, with monomers assembling into oligomers which in turn aggregate to form the larger nanospheres.14–16 Numerous researchers have observed the nanosphere structure within immature enamel tissue, indicating the presence of this quaternary structure in vivo.17–21 The nanosphere structures appear to be 15–30 nm in diameter, and they tend to be aligned in rows in between and parallel to the highly elongated developing calcium phosphate crystallites.
Amelogenin is excreted by ameloblasts into an extracellular mineralization front where it is believed to promote the formation of unusually elongated calcium phosphate crystallites and their parallel assembly into rods.18,22 Although the specific function of amelogenin is not well understood, roles in nucleation,23,24 growth,25 control of crystal size and habit,26,27 promotion of the orientation and spacing of crystallites,28,29 and acting as molds for crystallites22,30 have been proposed. Both in vivo18 and in vitro31 studies have suggested that amelogenin can interact directly with calcium phosphate, indicating that adsorption interactions are very important to its function. Since the function of amelogenin depends on its interactions with surfaces, the quaternary structure of amelogenin at interfaces is of great importance. Recent studies have shown that when amelogenin nanospheres in solution interact with model surfaces including self-assembled monolayers (SAMs) and mica surfaces, they disassemble or break down into monomers or oligomers, reversing the process of self-assembly.32–34 This unexpected result suggests that a detailed understanding of how amelogenin interacts with the biologically relevant calcium phosphate surface is necessary to further elucidate its biological function.
Here we describe studies of amelogenin adsorption onto the (100) face of single-crystal HAP using in situ high-resolution AFM. This method gave us the advantage of directly observing proteins interacting with surfaces at the molecular level, in real time and in aqueous solutions. Special methods were used to make HAP crystals with exposed (100) faces that were large enough and smooth enough to enable high-resolution spatial imaging. In contrast to other methods such as TEM,14 ellipsometry,32 ex situ AFM,32 and depletion methods,35 in situ AFM allowed us to simultaneously determine the adsorbed quaternary species, adsorbed microstructures, and adsorption amounts. For the first time, we show that amelogenin nanospheres disassemble onto HAP surfaces, and we observe the disassembly event in real time in some cases. Although previous research has determined binding energies onto high-surface-area HAP powders with undefined crystal faces,31 this work represents the successful determination of protein binding energies onto a specific HAP face. The advancement of using real-time in situ AFM to identify protein species and quantify adsorption kinetics and energetics has elucidated the interactions of a supramolecular protein on surfaces and can be applied to other protein–surface interactions important to interfacial processes.
EXPERIMENTAL SECTION
Amelogenin (rp(H)M180 and rpM179) Preparation and Purification
Full-length recombinant murine amelogenin was prepared either with a 12-residue,36 uncleavable, N-terminal histidine tag (RGSHHHHHHGS-) (rp(H)M180) or without the tag (rpM179) (Figure S1). Because the N-terminal methionine of the full-length untagged protein is removed in E. coli by methionine amino-peptidase,37 this protein is named rpM179. The tagged protein (rp(H)M180) was purified using metal-affinity chromatography under denaturing conditions,38 and the untagged protein was purified using a recently optimized protocol involving serial dialysis in 1 and 2% acetic acid.39 For both proteins, the last step of the purification involved reverse-phase chromatography on a XBridge Prep C18 (5 μm, 10 × 250 mm2) reverse-phase column (Waters, Milford, MA).38,39 The fractions containing murine amelogenin were pooled, frozen at −80 °C, lyophilized, weighed, and then stored at −4 °C until ready for use.
Chemicals
Tris-HCl, Ca(NO3)2, NH4H2PO4, and K2SO4 (99.99% purity) were purchased from Sigma-Aldrich. Poly-L-lysine (PLL) in H2O (0.1% w/v) was obtained from Ted Pella. The chemicals were used without further purification. Milli-Q water (resistivity = 18.2 MΩ· cm at 25 °C) was used in the experiments. All solutions were filtered (0.2 μm pore size;, Corning) prior to use.
Protein Solution Preparation
Solutions of rp(H)M180 and rpM179 were prepared by dissolving ~5–10 mg/mL protein in water at pH 3 to 4. After 1 to 3 days of dissolution, the concentrations of the protein solutions were determined using UV–vis spectroscopy and an experimentally determined extinction coefficient (23,100 M−1 cm−1). The stock protein solutions were diluted into 25 mM Tris-HCl buffer adjusted to pH 8.0. A pH 8.0 solution was used to keep the pH higher than amelogenin’s isoelectric point (~pH 6.8)40 and because the nanospheres have a narrow size distribution at this pH.13 The net charge and hydrophobicity of rp(H)M180 and rpM179 were calculated using the protein calculator at http://protcalc.sourceforge.net/ and the ProtParam tool at http://web.expasy.org/protparam/ as described in the Supporting Information (SI) section.
AFM Imaging
Micrometer-sized single crystals of HAP with six equivalent lateral (100) faces were prepared using a potassium sulfate molten salt synthesis technique as described previously41,42 and in the Supporting Information. The synthesized HAP crystals were hexagonal prisms with six equivalent lateral faces. As described in our recent publication,41 the lateral face index was determined to be (100) by the SEM morphology, enhanced (100), (200), and (300) peak intensities in the XRD pattern, and an AFM step height of 0.815 nm that matched the d spacing of (100). A muscovite mica disc (diameter 9.9 mm, Ted Pella, Inc.) was freshly cleaved and used as a supporting surface. A poly-L-lysine solution was placed on the mica surface for 3 min and was then thoroughly rinsed with water and dried by a stream of nitrogen gas. HAP crystals were transferred onto the poly-L-lysine-functionalized mica in Tris-HCl buffer solution (25 mM, pH 8.0) and were placed into an AFM equipped with a fluid cell. After thermal relaxation for 10 min, a small amount (200 μL) of Tris-HCl buffered solution (pH 8.0) containing amelogenin at different concentrations was injected into the buffer solution in the fluid cell while time-lapse AFM images were continuously collected on the HAP lateral (100) faces.
Experiments at each concentration of rpM179 and rp(H)M180 were repeated two to three times. All in situ AFM images were captured in tapping mode at room temperature (23 °C) with a NanoScope IIIa or 8 atomic force microscope (Digital Instruments J scanner, Bruker) and with hybrid probes consisting of silicon tips on silicon nitride cantilevers (HYDRA triangular lever, k = 0.088 N/m, tip radius <10 nm, resonance frequency 75 kHz in air, Applied Nanostructures, Inc., www.appnano.com). The drive amplitude was about 20 nm in fluid, and the signal-to-noise ratio was maintained above 10. The scanning speed was 1 to 2 Hz. The amplitude set point was carefully tuned to minimize the average loading force (~50 pN) during in situ imaging. The dependence of the measured amelogenin height on the drive amplitude was determined after amelogenin adsorption had stabilized (120 min). The images were analyzed using the SPIP 5.1.4 image processing software package (Image Metrology A/S, Hørsholm, Denmark). The particle size distribution and areal coverage were calculated by the particle and pore analysis module included in the SPIP 5.1.4 software.
RESULTS
Adsorption of rpM179 onto the HAP (100) Surface
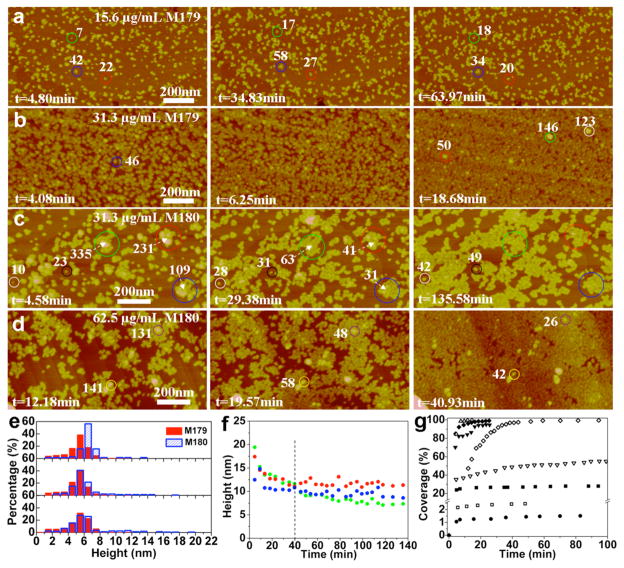
The recombinant form of native full-length mouse amelogenin, rpM179, was adsorbed onto HAP (100) from 7.8 to 250 μg/ mL solutions buffered with 25 mM Tris-HCl at pH 8. Images were captured at 4 min intervals for time periods of as long as 16 h. Figure 1a,b shows images of adsorbates at 15.6 and 31.3 μg/mL, and movies of time-lapse images are shown in the SI (Movies S1 (15.6 μg/mL) and S2 (31.3 μg/mL)). At these relatively low protein concentrations, individual adsorbate species could be resolved and the dominant species have heights of ~5.5 nm, with constant height distributions over time from 5 to 64 minutes (Figure 1e).
Figure 1.
In situ AFM images, height distributions, and adsorption kinetics of the first layer of rpM179 (labeled M179) and rp(H)M180 (labeled M180) adsorbed onto the HAP (100) face (pH 8.0) at different protein concentrations and time points: (a) [rpM179] = 15.6 μg/mL, (b) [rpM179] = 31.3 μg/mL, (c) [rp(H)M180] = 31.3 μg/mL, and (d) [rp(H)M180] = 62.5 μg/mL. (e) Height distributions of rpM179 in (a) (red bars) and rp(H)M180 in (c) (blue bars). Bottom distributions are at short times (~5 minutes), middle distributions are at 35 minutes for rpM179 and 29 minutes for rp(H)M180, and top distributions are at long times (64 minutes for rpM179 and 136 minutes for rp(H)M180). (f) Height versus time of circled nanospheres of rp(H)M180 in (c). (g) Kinetics of areal coverage for rpM179 (filled symbols) and rp(H)M180 (open symbols) adsorption at 7.8 (circles), 15.6 (squares), 31.3 (inverted triangles), 62.5 (diamonds), and 125 μg/mL (triangles) protein concentrations. The number of monomers inside various adsorbates is given in white. This figure shows that rpM179 and rp(H)M180 adsorb as oligomers onto HAP (100).
A method was developed to calculate the volume of the adsorbed species from the AFM height and AFM diameter corrected for tip size, assuming the protein had the shape of a spherical cap21 (Figure S2a–c, equation S1). The amelogenin species molecular weight and the number of monomers per species were determined using a calibration curve relating protein molecular weight to protein volume using known proteins (Figure S2d). According to the calibration, the adsorbates consist of 10–40 monomers with a mean of ~25 monomers, as indicated by the labels in Figure 1. DLS studies show that the rpM179 solutions consist of nanospheres with mean spherical diameters of ~25 nm and do not vary significantly in size over the range of solution concentrations studied (Figure S3), consistent with previous studies.13 The DLS sizes are also consistent with rpM179 nanosphere sizes determined by cryo-TEM analysis.14 If the nanospheres are modeled as spheres, then they correspond to species with ~200 monomers using the calibration curve in Figure S2d. A previous small-angle X-ray scattering study found that the rpM179 nanosphere is best modeled as an oblate ellipsoid with diameters of 24.6 and 11.4 nm in the long and short axes, respectively.43 Nanospheres of this shape contain ~100 monomers. The best estimates of the size and shape of rpM179 in the literature and our DLS data therefore suggest that the solution nanosphere consists of 100–200 monomers. This leads us to conclude that the dominant rpM179 species on HAP (100) consisting of ~25 monomers are oligomeric subunits of the larger nanospheres in solution.
The adsorbed structures vary from isolated oligomers at 7.8 μg/mL (Figure S4a) to small oligomer clusters at 15.6 μg/mL (Figure 1a) to oligomer monolayers at 31.3–62.5 μg/mL (Figures 1b and S4c). There are several isolated larger structures with AFM heights of 9–12 nm on top of the oligomer layer (Figure 1b). A plot of monomer number versus AFM height in Figure S5 shows that structures with 9–12 nm AFM heights contain 100–200 monomers, the monomer range expected for nanospheres. The height of the nanosphere adsorbate determined by AFM is smaller than the adsorbate diameter (~30 nm) and the nanosphere diameter in solution determined by DLS (~25 nm) suggesting that the nanosphere adsorbate has an asymmetric shape. At higher concentrations of up to 250 μg/mL, adsorbates form complete monolayers by 10 min of adsorption time, and it was not possible to resolve individual oligomers (Figure S4d). The studies performed at low concentrations were essential in identifying the adsorbed species of rpM179 on HAP (100). The coverage of the adsorbed oligomers was determined from the areal coverage of the oligomers (adjusted for tip diameter) relative to the entire area. The rate of oligomer adsorption displays a rapid increase at early times transitioning to a slow approach to full coverage (Figure 1g). The adsorption coverages increase with increasing protein concentration in solution.
Adsorption of rp(H)M180 onto the HAP (100) Surface
rp(H)M180 is the recombinant native mouse amelogenin with a short histidine tag added to the N-terminus (RGSHHH-HHHGS) to aid in the purification of amelogenin (Figure S1). This variant has been used extensively in the literature and is found to have similar nanosphere formation12 and calcium phosphate nucleation behavior23 compared to rpM179. AFM images of fresh rp(H)M180 adsorbed onto HAP (100) at low protein concentrations (31.3 to 62.5 μg/mL) (Figures 1c,d,e; Movies S3 (31.3 μg/mL) and S4 (62.5 μg/mL)) reveal particles with an average height of ~5.5 nm that increases over time to 6.5 nm. The volume analysis shows that these adsorbates contain ~25 monomers (Figure S5), consistent with oligomeric subunits of the larger nanospheres found in solution by DLS measurements. The microstructures vary from isolated islands of oligomers and coalesced islands for 31.3 μg/ mL solutions to a complete monolayer at 62.5 μg/mL (Figure 1d).
There are distinct differences in the adsorption behavior of rpM179 and rp(H)M180 as seen in the AFM images in Figure 1 and time-lapse Movies S1–S4. For rp(H)M180, oligomers tend to adsorb by attaching to the edges of islands of coalesced oligomers, resulting in islands that are much larger than the clusters observed for rpM179. The adsorption coverage is lower for rp(H)M180 than for rpM179 at the same solution concentration (Figure 1g). Also, nanospheres (10–15 nm height) occasionally adsorb from rp(H)M180 solutions (Figure 1c). The nanospheres decrease in height over time (Figure 1f) as the oligomer coverage in adjacent regions increases, implying a breakdown of the nanospheres into oligomeric subunits that spread into adjacent, unoccupied surface sites.
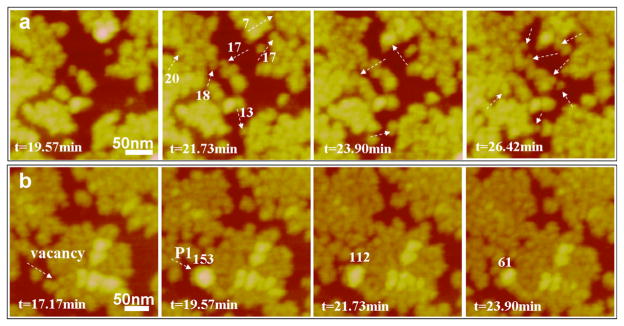
The mechanisms of rp(H)M180 adsorption were further studied at higher spatial and temporal resolution as shown in Figure 2, confirming the two different pathways seen in the lower-resolution images. Figure 2a shows time-lapse images of oligomers attaching to the edges of pre-existing islands. Figure 2b shows the adsorption of a nanosphere (P1 in Figure 2b) onto the “defective” region of an island followed by disassembly of the nanosphere into oligomers as evidenced by the decrease in the height of P1 and the increase in oligomer coverage adjacent to P1. Nanosphere disassembly can also be seen in the time-lapse images in Movie S4.
Figure 2.
Two types of adsorption behavior for rp(H)M180 at 62.5 μg/mL on the HAP (100) face: (a) most adsorption occurs by the attachment of oligomers and (b) occasionally nanospheres adsorb (P1) and break down into oligomers that spread into adjacent sites. Several adsorbates are labeled by their monomer number in white.
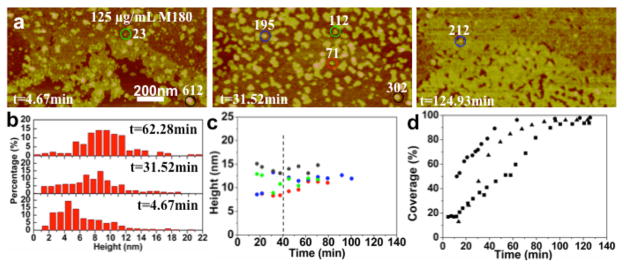
Increasing the rp(H)M180 protein concentration to 125 μg/ mL results in a striking change in adsorption behavior from the development of a single oligomer monolayer to a second protein layer as shown in Figure 3 and Movies S5 (125 μg/mL) and S6 (250 μg/mL). The first layer of oligomers is complete within ~10 min (Figure 1g) while the completion of the second layer takes ~90 min (Figure 3d). The particles in the second layer have a much broader distribution compared to those in the first layer and exhibit an increase in the average height over time as shown in Figure 3b. The large adsorbates in the second layer appear to be nanospheres based on the volume analysis. On the other hand, they might represent islands of oligomers that adsorb and coalesce over time. Overall, the second-layer absorbate structure and temporal evolution are more complex than the first layer behavior.
Figure 3.
In situ AFM images and height distributions of the second layer of rp(H)M180 adsorbed onto HAP (100) at different time points: (a) [rp(H)M180] = 125 μg/mL. (b) Height distribution of particles in the second layer in (a). (c) Height evolution of circled particles in corresponding colors in (a). (d) Kinetics of the second-layer areal coverage at 125 (squares), 200 (triangles), and 250 μg/mL (circles) protein concentrations. Several adsorbates are labeled by their monomer number in white.
Effect of Solution Aging on Amelogenin Adsorption onto the HAP (100) Surface
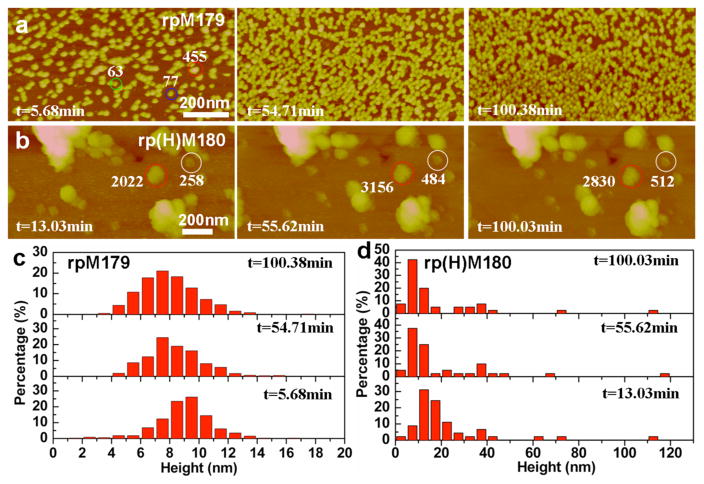
Interestingly, there are significant changes in the adsorption behavior of rp(H)M180 if the Tris-HCl buffered solutions (pH 8.0) were aged for 21 days. Figure 4b,d shows that the initial adsorbates at 62.5 μg/ mL were primarily nanospheres with heights of ~10–15 nm. There are increases in the percentages of larger structures (>20 nm heights), most likely aggregates of nanospheres, compared to the adsorbate distributions for fresh rp(H)M180 (Figure 1d). The DLS data show that there is no change in nanosphere size in aged solutions compared to fresh solutions (Figure S3). The adsorbates decrease in size over time, down to 7.5 nm height by 100 min as shown in Figure 3d, suggesting the disassembly of the nanospheres onto the surface; however, no coherent monolayer of oligomers was observed. In contrast, the “fresh” solutions at 62.5 μg/mL formed a complete oligomeric monolayer by 40 min of adsorption (Figure 1g). This result suggests that there are time-dependent changes in the rp(H)M180 nanosphere structure in solution at pH 8.0 that greatly slow the disassembly of the nanospheres on the HAP surface.
Figure 4.
In situ AFM images and height distributions of 21-day-aged rpM179 and rp(H)M180 on the HAP (100) face (pH 8.0) at different time points: (a) [rpM179] = 62.5 μg/mL; (b) [rp(H)M180] = 62.5 μg/mL; (c) the height distribution of rpM179 at different time points; and (d) the height distribution of rp(H)M180 at different time points. The data show that aging the rp(H)M180 and rpM179 solutions decreases the rate of nanosphere disassembly.
There is also an aging effect for the rpM179 solutions as shown in Figure 4a,c. Even though there is no change in the nanosphere size in solution (Figure S3), the adsorbates for aged 62.5 μg/mL solutions are more broadly distributed (Figure 4c) than for fresh solutions (red histogram in Figure 1e), and the dominant species is a nanosphere (N ≈ 150, 9.5 nm height). The average particle size decreases over time from ~9.5 to ~7.5 nm by 100 min. The kinetics of adsorption are slower with only 56% coverage at 55 min for the aged solution compared to 92% coverage at 6 min for the fresh solution. This result also suggests time-dependent increases in oligomer–oligomer interactions within the rpM179 nanosphere that lead to slower rates of nanosphere disassembly than for the fresh solutions.
Free Energy and Cooperativity of rp(H)M180 and rpM179 on the HAP (100) Face
The observed dependencies of adsorption coverage on protein concentration can be used to determine the binding constants and binding free energies for both the amelogenin oligomer–HAP and oligomer–oligomer interactions. The adsorption process of amelogenin onto HAP (100) can be represented by
| (1) |
where n is the number of amelogenin oligomers (or, in some cases, nanospheres), Amel is the free amelogenin oligomer, B is the available binding site on the surface, Amel·B is the adsorbed amelogenin oligomer, and Ka is the equilibrium binding constant.
The coverage in the equilibrium state based on eq 1 is
| (2) |
where θ represents the fraction of the surface covered by protein, [Amel] is the oligomer concentration in solution given in M (mol/L), and Ka has units of M−n.
A linear formulation of eq 2 can be derived as
| (3) |
Equation 3 is also known as the Hill equation, a relationship developed to model the phenomena of enhanced binding of ligands to macromolecules in the presence of other ligands,23 where n is the Hill coefficient describing cooperativity. The Hill coefficient provides a way to quantify a cooperative effect during binding processes. It describes the fraction of the macromolecule saturated by ligands as a function of the ligand concentration. A coefficient of 1 indicates completely independent binding, regardless of how many additional ligands are already bound. A Hill coefficient greater than 1 indicates positive cooperativity, while a number less than 1 indicates negative cooperativity. Once n and Ka have been determined from a fit of eq 3 to the oligomer coverage data, the oligomer-surface binding free energy can then be determined. For solution amelogenin at a standard concentration, normally 1 M, the standard binding free energy per mole of amelogenin is
| (4) |
where kB is Boltzmann’s constant and T is the absolute temperature.
Coverages in the first adsorbate layer were used to obtain Hill parameters for protein-HAP binding (rp(H)M180-HAP (100) and rpM179-HAP (100)), and coverages in the second protein layer were used to obtain Hill parameters for oligomer–oligomer binding (rp(H)M180-rp(H)M180 and rpM179-rpM179). A Hill plot based on the linear formulation of the Hill equation is shown in Figure 5, and the resulting parameters are shown in Table 1. An analysis using the Langmuir equation, which is a reduced form of the Hill equation where n = 1, was also performed but did not result in good linear fits. The larger Hill coefficient for rp(H)M180-HAP (100) indicates that there is a larger degree of cooperativity during HAP binding, consistent with the higher extent of island formation compared to rpM179. rp(H)M180–rp(H)M180 interactions have a relatively high Hill coefficient and a high binding constant suggesting a stronger oligomer binding interaction compared to that for rpM179–rpM179. For both rp(H)M180 and rpM179, the oligomer–HAP binding interactions are stronger than the oligomer–oligomer interactions and would promote a disassembly mechanism resulting in the peeling off of oligomers at the surface.
Figure 5.
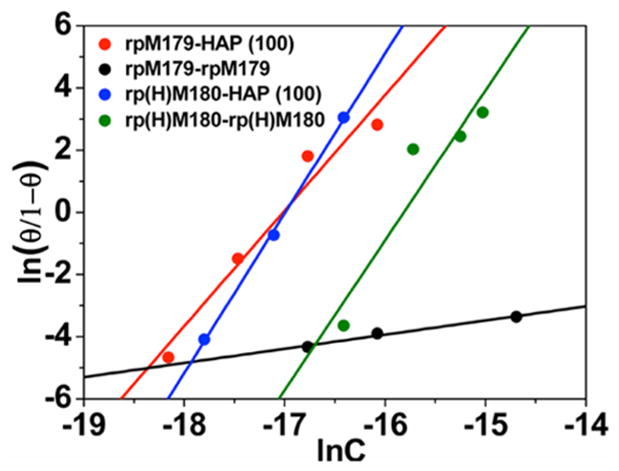
Hill cooperativity plots for oligomer–HAP(100) (rp(H)-M180–HAP(100) and rpM179–HAP (100)) and oligomer–oligomer (rp(H)M180–rp(H)M180 and rpM179–rpM179) interactions. The straight lines are the linear fits to eq 3.
Table 1.
Hill Coefficients (n), Binding Constants (Ka), and Binding Energies (ΔG) for Oligomer–HAP and Oligomer– Oligomer Interactions
| interaction | n | ln Ka | ΔG (kBT) |
|---|---|---|---|
| rp(H)M180-HAP (100) | 5.14 ± 0.18 | 87.34 ± 3.06 | −17.0 ± 1.2 |
| rpM179-HAP (100) | 3.71 ± 0.55 | 63.21 ± 4.47 | −17.1 ± 4.4 |
| rp(H)M180-rp(H)M180 | 4.82 ± 1.24 | 76.17 ± 4.39 | −15.8 ± 6.7 |
| rpM179-rpM179 | 0.46 ± 0.06 | 3.35 ± 0.97 | −7.3 ± 3.5 |
DISCUSSION
Adsorption Mechanism
Our study reveals that the rpM179 and rp(H)M180 amelogenin nanospheres disassemble onto the HAP surface, breaking down into oligomeric subunits of the larger nanosphere as shown in the schematic in Figure 6. The HAP surface induces the breakdown of nanospheres into oligomers, reversing the hierarchical self-assembly process. In most cases, the disassembly event occurs very quickly, so quickly that only the resulting oligomers could be detected by AFM (Figure 6, pathway 1). There are several cases, however, where the nanosphere breakdown is slow enough to be directly observed as shown in Figure 6, pathway 2. For example, nanospheres occasionally adsorb from fresh rp(H)M180 solutions and disassemble into oligomers that spread onto adjacent HAP surface sites as imaged in real time (Figure 2, Movie S4). Also, the disassembly process is greatly slowed for aged rp(H)M180 and rpM179 solutions, which allowed us to capture the adsorption of nanospheres and their breakdown into smaller structures on the surfaces (Figure 3).
Figure 6.
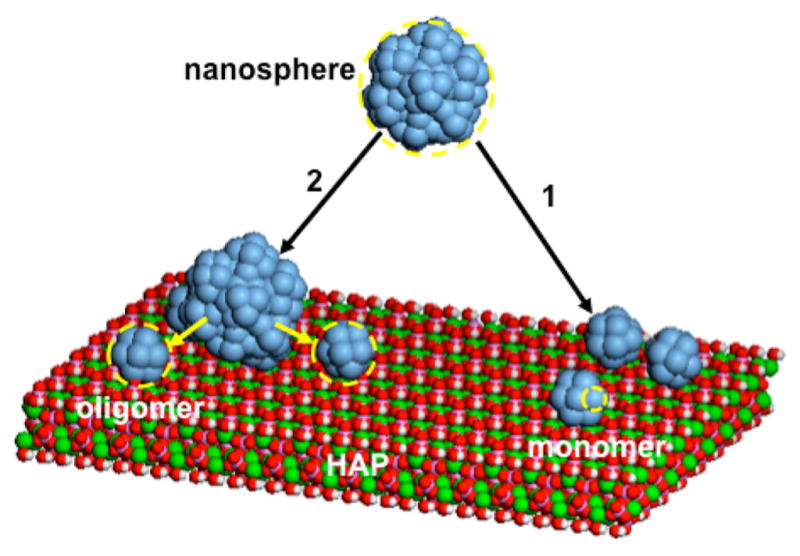
Schematic depicting nanospheres in solution consisting of oligomeric subunits and nanosphere disassembly onto the HAP (100) face. In most cases, nanospheres interact with the HAP surface and leave behind oligomers without the detection of the nanosphere (pathway 1). In some cases (aged rpM179 and rp(H)M180 solutions and occasionally for fresh rp(H)M180), nanospheres adsorb onto the surface and then slowly disassemble into oligomers which spread into unoccupied surface sites (pathway 2).
The smallest oligomers observed on the surface contain 10–15 monomers, similar in size to the dodecameric oligomers found to be subunits of nanospheres by cryo-TEM.14 Computer reconstructions of the oligomer images showed that they are composed of two rings of six amelogenin monomers joined by the C-terminus, which is located on the outside of the ring structure. As the nanosphere contacts the surface, binding interactions between the oligomer and the HAP surface may cause the oligomer to break off from the larger nanosphere. These interactions may be promoted by the acidic and basic residues in the oligomer surface interacting with acidic and basic sites on the HAP crystal.
The experimental determination of the oligomer–oligomer and oligomer–HAP binding energies using AFM is unprecedented and provides new insights into how the adsorbed protein microstructures evolve. A small change in the sequence of amelogenin by the attachment of a histidine tag to the N-terminus in rpM179 results in large changes in the oligomer–oligomer binding energy, from 7.3kBT for rpM179 to 15.8kBT for rp(H)M180, which in turn leads to large effects on the adsorption pathways and microstructural evolution. This increase in oligomer–oligomer binding energy leads to the attachment of oligomers onto the edges of oligomeric islands due to the increased protein cooperativity effect, fewer oligomers bound at the same concentration, and the adsorption of a complete second protein layer at higher protein concentrations.
The histidine tag must promote the stronger oligomer–oligomer binding interactions found in rp(H)M180 (Figure S1). The histidine imidazole ring is easily protonated with a pKa around physiological pH (pH 6 to 7),44 indicating that histidine residues can greatly affect the net charge of the protein as shown in Figure S5. In addition, the aromatic rings can interact by π– π stacking.45 Calculations show that the charge on the rpM179 monomer at pH 8.0 is −3.2e while the charge on the rp(H)M180 monomer is −2e (Figure S5). Increased oligomer–oligomer binding for rp(H)M180 at pH 8.0 may be promoted by the lower net charge as well as histidine π– π stacking. The aging studies show time-dependent changes in oligomer–oligomer binding which may involve increasing degrees of stacking between histidine imidazole rings in adjacent oligomers, which would be promoted in rings with lower charge at pH 8.0.45 Since aging effects also occur for rpM179 even though it lacks the histidine tag, time-dependent changes in oligomer–oligomer binding may also be occurring, promoted by π– π stacking between the 12 histidine residues in the central domain of amelogenin. It may also be possible that differences in protein folding increase oligomer–oligomer binding interactions for rp(H)M180; however, there is no experimental comparison of the secondary structure of amelogenins with and without the histidine tag within the nanosphere state. Previous 1H–15N HSQC NMR studies showed no differences in folding between rpM179 and rp(H)M180 in the monomer quaternary structure.39
The effect of the histidine tag on the net charge of amelogenin is completely reversed at lower pH values. Here, amelogenin monomers were adsorbed from solutions at pH 3.8, and the aggregation of monomers was induced by increasing the pH to 6.5 and 8.0 as shown in Figure S7. Figure S5 shows that rp(H)M180 has a higher positive charge at pH 6.5 (+8e) and pH 3.8 (+24.5e) compared to rpM179 (+4e and +17.5e, respectively). Under these pH conditions, the larger Coulombic repulsion between rp(H)M180 monomers promotes less aggregation and smaller oligomer sizes than rpM179.
Our current studies have primarily involved amelogenin solutions in Tris buffer and at pH 8.0, solution conditions that have been used in numerous previous in vitro studies of amelogenin structure and amelogenin’s role in calcium phosphate mineralization.11,13,14,32,34 At pH 8.0, nanospheres have a narrow size distribution11 and do not agglomerate, unlike solutions near the isoelectric point of pH ~6.8.40 Also, the pH of enamel fluid can vary from as low as pH 5.846 to as high as pH 8.5,47,48 depending on the tissue location and degree of enamel maturity, indicating that a pH value of 8.0 has physiological relevance. Although our solution system is useful for studies on the interfacial behavior of supramolecular proteins described here, future studies will involve solutions containing sodium chloride, calcium, and phosphate at pH 7.4 and 37 °C, conditions that more closely mimic the enamel environment.
Relevance to in Vivo Enamel Formation
Recent studies have shown that both the absence of amelogenin in amelogenin-null mice49 and the presence of amelogenin mutations19 result in enamel that is very thin and has a disorganized rod pattern. These results suggest that amelogenin may play an important role in promoting the elongation and organization of the enamel rods. Early enamel tissue consists of very long, thin, parallel crystallites of amorphous calcium phosphate50 separated by organic matter.17,51–53 The calcium phosphate crystallites are oriented perpendicular to the mineralization front.22 Once amelogenin is enzymatically degraded by Mmp-20 at the end of the secretory stage, crystallites grow in the lateral directions until they coalesce into a single rod.10,54 Amelogenin, therefore, may adsorb to the early crystallite surfaces and limit growth in the axial directions in order to control growth and prevent early crystallite fusion.
Our studies reveal that an ~25-mer oligomer is the dominant quaternary structure of amelogenin adsorbed onto HAP surfaces, and this suggests that the oligomer may be the most important amelogenin structure involved in controlling crystallite growth in vivo. Previous studies have shown that the hydrophobic amelogenin polymer appears to be white and electron-deficient in TEM images of early enamel tissue because it does not stain with typical TEM stains. Small electron-deficient structures, with sizes similar to the oligomer sizes we observed in our in vitro studies, were found directly adjacent to calcium phosphate crystallites in immature enamel.18 This suggests that our in vitro results may be consistent with previous in vivo studies.
In addition to oligomers, the nanosphere quaternary structure is also seen in TEM images of early enamel with ~20 nm nanospheres forming rows in between and parallel to the elongated calcium phosphate crystallites but not directly interacting with the crystallites.10,18 Both oligomers and nanospheres, therefore, may be part of the amelogenin matrix or mold. It has been proposed that the nanospheres may function as spacers between crystallites, promoting the parallel alignment and organization of the crystallites and essentially defining the final width of the crystallites.19,28,55 As we have suggested previously,32 amelogenin may have several different quaternary structures in vivo, and each quaternary structure may have a different biological function. For example, the oligomer may function to interact directly with the crystallites in order to control the lateral growth of calcium phosphate crystallites, and the nanosphere may function to promote the parallel alignment and organization of crystallites. Amelogenin has also been found to form higher-order protein ribbon structures in vitro,15,56,57 suggesting that the ribbon quaternary structure may also be important, perhaps forming the organic sheaths surrounding each enamel rod in vivo.
Given that small changes in the amelogenin primary sequence by adding a histidine tag led to significant changes in protein adsorption pathways and microstructures, small changes in the amelogenin sequence caused by point mutations might also be expected to greatly affect amelogenin adsorption processes and adsorbed protein microstructures. Single amino acid mutations within amelogenin cause diseases known as amelogenesis imperfecta that result in malformed enamel phenotypes.58 These point mutations might significantly alter protein–protein and protein–HAP interactions and lead to changes in enamel microstructures. Future studies on the adsorption of amelogenin nanospheres containing point mutations will be of great interest in understanding how supramolecular structures interact at surfaces.
CONCLUSIONS
High-resolution in situ AFM was used to study the adsorption of amelogenins onto the (100) face of single-crystal HAP. The crystals were large enough and smooth enough to allow the simultaneous determination of adsorbate quaternary species, adsorption amounts, and adsorption microstructures in real time. Although DLS shows that amelogenin exists as monodisperse nanospheres in solution at pH 8.0, the nanospheres disassembled onto the HAP surface to form oligomeric adsorbates. The surface-triggered disassembly mechanism reverses the process of oligomer self-assembly to form nanospheres. The adsorption behavior of rp(H)M180 is strikingly different than that of rpM179 even though the two proteins differ by a small histidine tag at the N-terminus. The adsorption of rp(H)M180 involves higher oligomer–oligomer binding energies, the formation of island morphologies, and the adsorption of a complete second protein layer. We propose that the histidine tag promotes oligomer–oligomer binding interactions by changing the protein charge and promoting imidazole ring-stacking interactions. These interactions lead to time-dependent increases in oligomer–oligomer binding that greatly reduce the disassembly kinetics in aged solutions. Our use of AFM to observe nanosphere disassembly, identify the adsorbate species, and quantify adsorption kinetics on HAP (100) has led to important insights on how self-assembled protein structures interact at surfaces and how small changes in the protein sequence can affect those interactions.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding from the National Institutes of Health, NIH-NIDCH grant number DE-015347. The research was primarily performed at Pacific Northwest National Laboratory (PNNL), a facility operated by Battelle for the U.S. Department of Energy. A portion of the research was performed using EMSL, a national scientific user facility sponsored by the Department of Energy’s Office of Biological and Environmental Research, located at Pacific Northwest National Laboratory. Part of the research was performed as a user project at the Molecular Foundry, Lawrence Berkeley National Laboratory, with support from the Office of Science, Office of Basic Energy Sciences of the U.S. Department of Energy, under contract number DE-AC02-05CH11231.
Footnotes
Supporting Information: The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.lang-muir.5b02549.
- Protein sequences (Figure S1), size calibration curve (Figure S2), DLS data (Figure S3), additional AFM images of adsorbates (Figure S4), monomer number versus AFM height (Figure S5), summary of adsorbate structures (Table S1), charge calculations (Figure S6), and monomer aggregation studies (Figure S7) (PDF)
- Movie 1, time-lapse AFM images of rpM179 adsorption at 15.6 μg/mL (AVI)
- Movie 2, time-lapse AFM images of rpM179 adsorption at 31.3 μg/mL (AVI)
- Movie 3, time-lapse AFM images of rp(H)M180 adsorption at 31.3 μg/mL (AVI)
- Movie 4, time-lapse AFM images of rp(H)M180 adsorption at 62.5 μg/mL (AVI)
- Movie 5, time-lapse AFM images of rp(H)M180 adsorption at 125 μg/mL (AVI)
- Movie 6, time-lapse AFM images of rp(H)M180 adsorption at 250 μg/mL (AVI)
Notes
The authors declare no competing financial interest.
References
- 1.Lowenstam HA, Weiner S. On Biomineralization. Oxford University Press; New York: 1989. [Google Scholar]
- 2.Brash JL. Protein Adsorption at the Solid-Solution Interface in Relation to Blood Material Interactions. Acs Sym Ser. 1987;343:490–506. [Google Scholar]
- 3.Horbett TA, Brash JL. Proteins at Interfaces - Current Issues and Future-Prospects. Acs Sym Ser. 1987;343:1–33. [Google Scholar]
- 4.Fleischer CC, Payne CK. Nanoparticle-Cell Interactions: Molecular Structure of the Protein Corona and Cellular Outcomes. Acc Chem Res. 2014;47(8):2651–2659. doi: 10.1021/ar500190q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowe S, O’Brien-Simpson NM, Connal LA. Antibiofouling polymer interfaces: poly(ethylene glycol) and other promising candidates. Polym Chem. 2015;6(2):198–212. [Google Scholar]
- 6.Tonga GY, Moyano DF, Kim CS, Rotello VM. Inorganic nanoparticles for therapeutic delivery: Trials, tribulations and promise. Curr Opin Colloid Interface Sci. 2014;19(2):49–55. doi: 10.1016/j.cocis.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tonga GY, Saha K, Rotello VM. 25th Anniversary Article: Interfacing Nanoparticles and Biology: New Strategies for Biomedicine. Adv Mater. 2014;26(3):359–370. doi: 10.1002/adma.201303001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norde W. Adsorption of Proteins from Solution at the Solid-Liquid Interface. Adv Colloid Interface Sci. 1986;25(4):267–340. doi: 10.1016/0001-8686(86)80012-4. [DOI] [PubMed] [Google Scholar]
- 9.Brash JL. Behavior of proteins at interfaces. Curr Opin Colloid Interface Sci. 1996;1(5):682–688. [Google Scholar]
- 10.Fincham AG, Moradian-Oldak J, Simmer JP. The Structural Biology of the Developing Dental Enamel Matrix. J Struct Biol. 1999;126:270. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 11.Moradian-Oldak J, Leung W, Fincham AG. Temperature and pH-Dependent Supramolecular Self-Assembly of Amelogenin Molecules: A Dynamic Light-Scattering Analysis. J Struct Biol. 1998;122:320. doi: 10.1006/jsbi.1998.4008. [DOI] [PubMed] [Google Scholar]
- 12.Moradian-Oldak J, Paine ML, Lei YP, Fincham AG, Snead ML. Self-assembly properties of recombinant engineered amelogenin proteins analyzed by dynamic light scattering and atomic force microscopy. J Struct Biol. 2000;131(1):27–37. doi: 10.1006/jsbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- 13.Moradian-Oldak J, Simmer JP, Lau EC, Sarte PE, Slavkin HC, Fincham AG. Detection of monodisperse aggregates of a recombinant amelogenin by dynamic light-scattering. Biopolymers. 1994;34(10):1339–1347. doi: 10.1002/bip.360341006. [DOI] [PubMed] [Google Scholar]
- 14.Fang PA, Conway JF, Margolis HC, Simmer JP, Beniash E. Hierarchical self-assembly of amelogenin and the regulation of biomineralization at the nanoscale. Proc Natl Acad Sci U S A. 2011;108(34):14097–102. doi: 10.1073/pnas.1106228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Supramolecular Assembly of Amelogenin Nanospheres into Birefringent Microribbons. Science. 2005;307:1450. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- 16.Bromley KM, Kiss AS, Lokappa SB, Lakshminarayanan R, Fan D, Ndao M, Evans JS, Moradian-Oldak J. Dissecting amelogenin protein nanospheres: characterization of metastable oligomers. J Biol Chem. 2011;40:34643–34653. doi: 10.1074/jbc.M111.250928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bidlack FB, Huynh C, Marshman J, Goetze B. Helium ion microscopy of enamel crystallites and extracellular tooth enamel matrix. Front Physiol. 2014;5:395. doi: 10.3389/fphys.2014.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fincham AG, Moradian-Oldak J, Diekwisch TG, Lyaruu DM, Wright JT, Bringas P, Jr, Slavkin HC. Evidence for Amelogenin “Nanospheres” as Functional Components of Secretory-Stage Enamel Matrix. J Struct Biol. 1995;115:50. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- 19.Paine ML, White SN, Luo W, Fong H, Sarikaya M, Snead ML. Regulated gene expression dictates enamel structure and tooth function. Matrix Biol. 2001;20:273. doi: 10.1016/s0945-053x(01)00153-6. [DOI] [PubMed] [Google Scholar]
- 20.Robinson C, Fuchs P, Weatherell JA. The appearance of developing rat incisor enamel using a freeze fracturing technique. J Cryst Growth. 1981;53(1):160–165. [Google Scholar]
- 21.Moradian-Oldak J, Goldberg M. Amelogenin supra-molecular assembly in vitro compared with the architecture of the forming enamel matrix. Cells Tissues Organs. 2005;181(3–4):202–218. doi: 10.1159/000091382. [DOI] [PubMed] [Google Scholar]
- 22.Simmer JP, Richardson AS, Hu YY, Smith CE, Hu JCC. A post-classical theory of enamel biomineralization··· and why we need one. Int J Oral Sci. 2012;4(3):129–134. doi: 10.1038/ijos.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarasevich BJ, Howard CJ, Larson JL, Snead ML, Simmer JP, Paine M, Shaw WJ. The nucleation and growth of calcium phosphate by amelogenin. J Cryst Growth. 2007;304(2):407–415. doi: 10.1016/j.jcrysgro.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang LJ, Guan XY, Du C, Moradian-Oldak J, Nancollas GH. Amelogenin promotes the formation of elongated apatite microstructures in a controlled crystallization system. J Phys Chem C. 2007;111(17):6398–6404. doi: 10.1021/jp0675429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beniash E, Simmer JP, Margolis HC. The effect of recombinant mouse amelogenins on the formation and organization of hydroxyapatite crystals in vitro. J Struct Biol. 2005;149(2):182–190. doi: 10.1016/j.jsb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Iijima M, Moriwaki Y, Takagi T, Moradian-Oldak J. Effects of bovine amelogenins on the crystal morphology of octacalcium phosphate in a model system of tooth enamel formation. J Cryst Growth. 2001;222(3):615–626. [Google Scholar]
- 27.Wen HB, Moradian-Oldak J, Fincham AG. Dose-dependent modulation of octacalcium phosphate crystal habit by amelogenins. J Dent Res. 2000;79(11):1902. doi: 10.1177/00220345000790111501. [DOI] [PubMed] [Google Scholar]
- 28.Fincham AG, Simmer JP. Amelogenin proteins of developing dental enamel. Ciba Foundation Symposium 205 - Dental Enamel. 1997;205:118–134. doi: 10.1002/9780470515303.ch9. [DOI] [PubMed] [Google Scholar]
- 29.Moradian-Oldak J, Tan J, Fincham AG. Interaction of amelogenin with hydroxyapatite crystals: an adherence effect through amelogenin molecular self-association. Biopolymers. 1998;46:225. doi: 10.1002/(SICI)1097-0282(19981005)46:4<225::AID-BIP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 30.Bartlett JD. Dental enamel development: proteinases and their enamel matrix substrates. ISRN dentistry. 2013;2013:1. doi: 10.1155/2013/684607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouropoulos N, Moradian-Oldak J. Analysis of hydroxyapatite surface coverage by amelogenin nanospheres following the langmuir model for protein adsorption. Calcif Tissue Int. 2003;72:599. doi: 10.1007/s00223-002-1099-1. [DOI] [PubMed] [Google Scholar]
- 32.Tarasevich BJ, Lea S, Bernt W, Engelhard M, Shaw WJ. Adsorption of amelogenin onto self-assembled and fluoroapatite surfaces. J Phys Chem B. 2009;113(7):1833–1842. doi: 10.1021/jp804548x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarasevich BJ, Lea S, Bernt W, Engelhard MH, Shaw WJ. Changes in the quaternary structure of amelogenin when adsorbed onto surfaces. Biopolymers. 2009;91(2):103–7. doi: 10.1002/bip.21095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen CL, Bromley KM, Moradian-Oldak J, De Yoreo JJ. In situ AFM study of amelogenin assembly and idsassembly dynamics on charged surfaces provides insights on matrix protein self-assembly. J Am Chem Soc. 2011;133:17406–17413. doi: 10.1021/ja206849c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hlady V, Buijs J, Jennissen HP. Methods for studying protein adsorption. Methods Enzymol. 1999;309:402. doi: 10.1016/s0076-6879(99)09028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snead ML, Lau EC, Zeichner-David M, Fincham AG, Woo SL, Slavkin HC. DNA Sequence for cloned cDNA for murine amelogenin reveal the amino acid sequence for enamel-specific protein. Biochem Biophys Res Commun. 1985;129:812. doi: 10.1016/0006-291x(85)91964-3. [DOI] [PubMed] [Google Scholar]
- 37.Bonde J, Bulow L. One-step purification of recombinant human emelogenin and use of amelogenin as a fusion partner. PLoS One. 2012;7:33269. doi: 10.1371/journal.pone.0033269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchko GW, Lin G, Tarasevich BJ, Shaw WJ. A solution NMR investigation into the impaired self-assembly properties of two murine amelogenins containing the point mutations T21I or P41T. Arch Biochem Biophys. 2013;537:217–224. doi: 10.1016/j.abb.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchko GW, Shaw WJ. Improved protocol to purify untagged amelogenin - Application to murine amelogenin containing the equivalent P70 T point mutation observed in human amelogenesis imperfecta. Protein Expression Purif. 2015;105:14. doi: 10.1016/j.pep.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uskokovic V, Castiglione Z, Cubas P, Zhu L, Li W, Habelitz S. Zeta-potential and Particle Size Analysis of Human Amelogenins. J Dent Res. 2010;89(2):149–153. doi: 10.1177/0022034509354455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao J, Battle KC, Pan H, Salter EA, Chien YC, Wierzbicki A, De Yoreo JJ. Energetic basis for the molecular-scale organization of bone. Proc Natl Acad Sci U S A. 2015;112(2):326–31. doi: 10.1073/pnas.1404481112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tas AC. Molten Salt Synthesis of Calcium Hydroxyapatite Whiskers. J Am Ceram Soc. 2001;84:295. [Google Scholar]
- 43.Aichmayer B, Margolis HC, Sigel R, Yamakoshi Y, Simmer JP, Fratzl P. The onset of amelogenin nanosphere aggregation studied by small-angle X-ray scattering and dynamic light scattering. J Struct Biol. 2005;151:239. doi: 10.1016/j.jsb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Hansen AL, Kay LE. Measurement of histidine pKa values and tautomer populations in invisible protein states. Proc Natl Acad Sci U S A. 2014;111:E1705–E1712. doi: 10.1073/pnas.1400577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heyda J, Mason PE, Jungwirth P. Attractive interactions between side chains of histidine-histidine and histidine-arginine-based cationic dipeptides in water. J Phys Chem B. 2010;114:8744–8749. doi: 10.1021/jp101031v. [DOI] [PubMed] [Google Scholar]
- 46.Sasaki S, Takagi T, Suzuki M. Cyclical Changes in Ph in Bovine Developing Enamel as Sequential Bands. Arch Oral Biol. 1991;36(3):227–231. doi: 10.1016/0003-9969(91)90090-h. [DOI] [PubMed] [Google Scholar]
- 47.Lacruz RS, Nanci A, Kurtz I, Wright JT, Paine ML. Regulation of pH During Amelogenesis. Calcif Tissue Int. 2010;86(2):91–103. doi: 10.1007/s00223-009-9326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lyman GE, Waddell WJ. Ph Gradients in Developing Teeth of Young Mice from Autoradiography of [Dmo-C-14] Am J Physiol. 1977;232(4):F364–F367. doi: 10.1152/ajprenal.1977.232.4.F364. [DOI] [PubMed] [Google Scholar]
- 49.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen EH, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276(34):31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 50.Beniash E, Metzler RA, Lam RSK, Gilbert PUPA. Transient amorphous calcium phosphate in forming enamel. J Struct Biol. 2009;166(2):133–143. doi: 10.1016/j.jsb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diekwisch TGH, Berman BJ, Gentner S, Slavkin HC. Initial enamel crystals are not spatially associated with mineralized dentin. Cell Tissue Res. 1995;279(1):149–167. doi: 10.1007/BF00300701. [DOI] [PubMed] [Google Scholar]
- 52.Nanci A, Bendayan M, Slavkin HC. Enamel protein-biosynthesis and secretion in mouse incisor secretory ameloblasts as revealed by high-resolution immunocytochemistry. J Histochem Cytochem. 1985;33(11):1153–1160. doi: 10.1177/33.11.4056379. [DOI] [PubMed] [Google Scholar]
- 53.Travis DF, Glimcher MJ. Structure + organization of + relationship between organic matrix + inorganic crystals of embryonic bovine enamel. J Cell Biol. 1964;23(3):447. doi: 10.1083/jcb.23.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Margolis HC, Beniash E, Fowler CE. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J Dent Res. 2006;85(9):775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- 55.Paine ML, Zhu DH, Luo W, Bringas P, Jr, Goldberg M, White SN, Lei YP, Sarikaya M, Fong HK, Snead ML. Enamel biomineralization defects result from alterations to amelogenin self-assembly. J Struct Biol. 2000;132(3):191–200. doi: 10.1006/jsbi.2000.4324. [DOI] [PubMed] [Google Scholar]
- 56.Martinez-Avila OM, Wu SP, Cheng YF, Lee R, Khan F, Habelitz S. Self-assembly of amelogenin proteins at the water-oil interface. Eur J Oral Sci. 2011;119:75–82. doi: 10.1111/j.1600-0722.2011.00907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martinez-Avila O, Wu SP, Kim SJ, Cheng YF, Khan F, Samudrala R, Sali A, Horst JA, Habelitz S. Self-assembly of filamentous amelogenin requires calcium and phosphate: from dimers via nanoribbons to fibrils. Biomacromolecules. 2012;13(11):3494–3502. doi: 10.1021/bm300942c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu JC-C, Chan H-C, Simmer SG, Seymen F, Richardson AS, Hu Y, Milkovich RN, Estrella NMRP, Yildirim M, Bayram M, Chen C-F, Simmer JP. Amelogenesis imperfecta in two families with defined amelx deletions in ARHGAP6. PLoS One. 2012;7(12):e52052. doi: 10.1371/journal.pone.0052052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.