Abstract
Mammalian chromosome ends are protected by nucleoprotein structures called telomeres. Telomeres ensure genome stability by preventing chromosome termini from being recognized as DNA damage. Telomere length homeostasis is inevitable for telomere maintenance because critical shortening or over-lengthening of telomeres may lead to DNA damage response or delay in DNA replication, and hence genome instability. Due to their repetitive DNA sequence, unique architecture, bound shelterin proteins, and high propensity to form alternate/secondary DNA structures, telomeres are like common fragile sites and pose an inherent challenge to the progression of DNA replication, repair, and recombination apparatus. It is conceivable that longer the telomeres are, greater is the severity of such challenges. Recent studies have linked excessively long telomeres with increased tumorigenesis. Here we discuss telomere abnormalities in a rare recessive chromosomal instability disorder called Fanconi Anemia and the role of the Fanconi Anemia pathway in telomere biology. Reports suggest that Fanconi Anemia proteins play a role in maintaining long telomeres, including processing telomeric joint molecule intermediates. We speculate that ablation of the Fanconi Anemia pathway would lead to inadequate aberrant structural barrier resolution at excessively long telomeres, thereby causing replicative burden on the cell.
Keywords: telomere maintenance, Fanconi anemia, DNA repair, DNA joint molecule intermediates
1. Introduction
Telomeres are a paradox
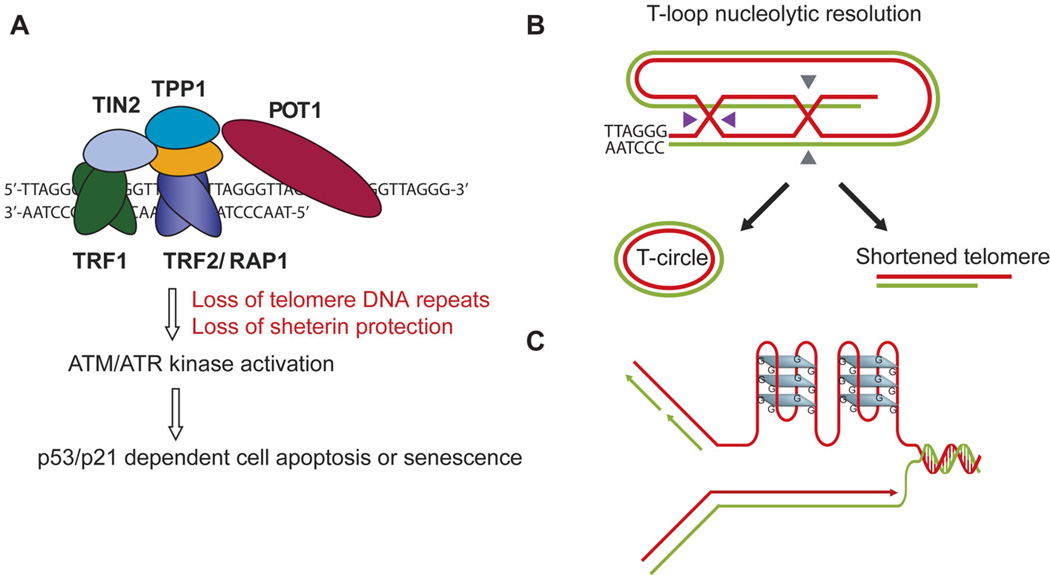
Telomeres are chromosome end nucleoprotein structures consisting of short tandem DNA repeats (5'-TTAGGG-3' in humans and mice) and inherently associated proteins, called the shelterin complex (TRF1, TRF2, POT1, TPP1, TIN2, and RAP1) (Figure 1A). Telomeres ensure genome stability by capping chromosome termini thereby preventing them from being recognized as broken DNA ends. Such protection of mammalian telomeres has been attributed to (i) the shelterin complex, where TRF2 and POT1 particularly have direct well-defined roles in protecting telomeres from activating ATM and ATR kinase pathways (Figure 1A); (ii) unusual structures such as the ‘T-loop’ configuration, where the 3'-telomeric overhang is tucked away in a loop (Figure 1B); and (iii) other non-shelterin accessory proteins with known functions in DNA repair (including helicases and nucleases that can process/remove unusual structural impediments in telomeric DNA) [1]. Paradoxically, some of the abovementioned features that enable telomeres to protect DNA ends may also act as impediments in its own maintenance thereby making DNA metabolism processes at telomeres a challenging task. Telomere maintenance entails several components, including length homeostasis (telomerase or ALT-sponsored [2, 3]) and replication-recombination-repair. In addition, their G-rich sequence also makes telomeres more susceptible to oxidative DNA damage and formation of replication blocking Gquadruplex structures (G4) [4] (Figure 1C).
Figure 1. Consequences of telomere dysfunction and telomeric unique structural and architectural features.
(A) Telomeres, bound by the shelterin protein complex cap the chromosome ends against NHEJ, HR, DNA damage signaling, and nuclease degradation. Dysfunctional telomeres can arise due to loss of telomeric DNA repeats or loss of protection of shelterin, which activates ATM or ATR kinase pathways, leading to cell apoptosis and cellular senescence. (B–C) Telomeres present a challenging landscape for DNA metabolism, owing to their T-loop architecture (B) and G-rich sequence that makes it a hotspot for secondary G4 structure formation (C). Resolution of the double HJ in the T-loop would generate a shortened telomere and a circular telomeric DNA [60], a mechanism used for ‘trimming’ long telomeres in some human cells (B).
Chromosomal instability and predisposition to cancer are the common links between FA and telomere dysfunction
Dysregulation of telomere maintenance such as defective length homeostasis (leading to critical shortening or over-lengthening), loss in protective function of shelterin proteins, and other defects in telomere biology leads to ATM/ATR kinase-involved DNA damage response or delays in DNA replication, resulting in genome instability, cell proliferation defects, cellular senescence or cell apoptosis (Figure 1A) [5, 6]. In humans, telomere attrition is associated with replicative cell senescence in culture, ageing populations, and environmental and lifestyle factors that contribute to ageing and ageing-related diseases. Telomere attrition is also linked to human disorders, e.g. dyskeratosis congenita (DC), aplastic anemia, and idiopathic pulmonary fibrosis [7–9]. A rare recessive disorder that results in aplastic anemia is Fanconi Anemia (FA) that is characterized by bone marrow failure, congenital abnormalities, increased susceptibility to cancer, and sensitivity to DNA interstrand crosslinking agents [10–12]. Interestingly, FA individuals are also reported to have (i) telomere loss/break in peripheral leukocytes; (ii) increased end-to-end telomere fusions; and (iii) overall shorter telomeres [13–16]. Proposed molecular mechanisms for this shortening include direct breaks at telomere sequences; replicative shortening; and accumulation of breaks due to defective DNA repair at telomeres and impaired response to oxidative stress [17–22]. Although currently there is lack of experimental evidence, as discussed in this review, our current knowledge alludes to a potential direct role of the FA pathway in telomere maintenance.
2. FA proteins in telomere maintenance
The FA pathway
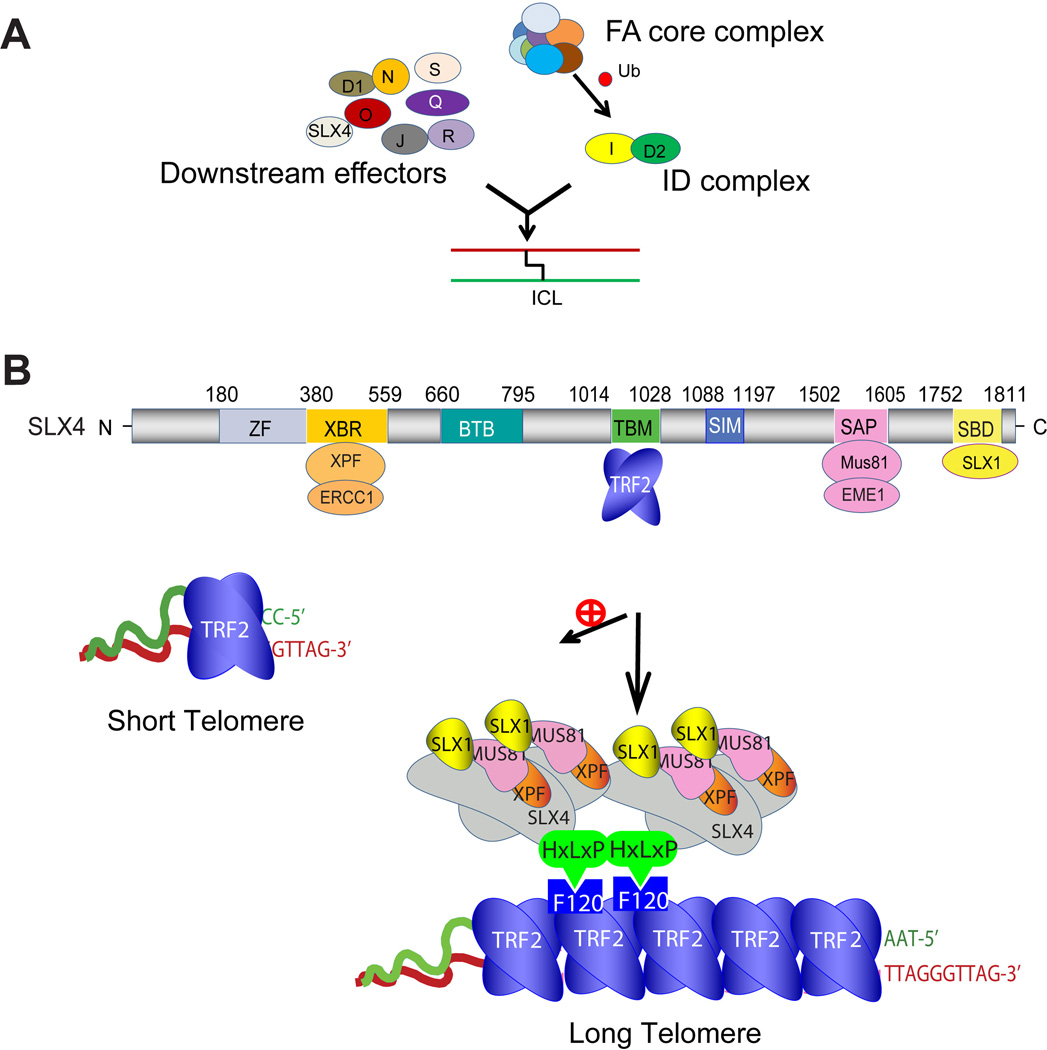
Mechanistically, the FA syndrome is caused by mutations in genes involved in repair of DNA inter-strand crosslinks (ICLs), with about 19 gene products identified till date [10–12, 23](Figure 2A). Eight FA proteins (FANCA/B/C/E/F/G/L/M) form the FA core complex, and along with the ATR checkpoint kinase, regulate ubiquitination of the heterodimeric FANCD2-I (ID) complex that is recruited to the ICL lesion. The ID complex acts as a platform to coordinate repair activities at the ICL site with several downstream FA proteins (FANCP/J/D1/N/O/S/R/Q and FAN1). This includes bringing multiple nucleases at the lesion via their assembly on FANCP (or SLX4) (discussed below), thus creating nucleolytic incisions at the site, which are then repaired by homologous recombination (HR).
Figure 2. Schematic showing FA pathway and domain mapping of human SLX4 protein.
(A) FA pathway proteins. The FA core complex regulates monoubiquitination of the heterodimeric ID complex that is recruited to the ICL lesion. The ID complex coordinates repair activities at the ICL site with several downstream FA proteins. (B). Human SLX4 protein. ZF, ubiquitin-binding zinc finger domain; XBR, XPF-binding region; BTB, Bric-a-brac, Tramtrack and Broad complex domain; TBM, TRF2-binding motif; SIM: SUMO-Interacting Motif; SAP, SAP motif, MUS81-binding region; SBD, SLX1-binding domain. SLX4 preferentially localizes to long telomeres that face greater challenges of DNA replication and alternate structure resolution.
ID complex players involved in telomere maintenance
The shared genome instability phenomenon in telomere dysfunction and FA suggests possible connection(s) between FA proteins and telomere function. Mammalian telomeres are maintained either via extension by the nucleoprotein complex telomerase or via an alternate lengthening of telomeres (ALT) mechanism that relies on HR to synthesize new telomeric DNA. The key FA player FANCD2 has been shown to colocalize with the inherent telomeric protein TRF1 in ALT cells, in a FANCA, FANCL, and ATR-dependent manner, and depletion of FANCA and FANCD2 causes telomere loss and decrease in telomere sister chromatid exchange [24], suggesting a role for monoubiquitinated FANCD2 in ALT telomere maintenance through telomeric HR. TRF1 that directly binds to double-stranded telomeric DNA is believed to prevent replication defects in telomeres by recruiting the G4 structure resolving helicases BLM and RTEL1 [25, 26]. However, TRF1 ribosylation by the telomere-associated poly(ADP-ribose) polymerase Tankyrase 1 (binds to TRF1) displaces TRF1 from telomeric DNA [27, 28]. FANCD2 has been shown to interact with Tankyrase 1 and inhibit TRF1 ribosylation. In turn, FANCD2 deficiency increases ribosylation of TRF1 and its displacement from telomeres [29], that may then lead to increased replication challenges at telomeres. Thus, ribosylation-mediated control of TRF1 affinity for telomeric DNA may be one possible mechanism where FANCD2 ensures telomere stability. TRF1 also physically blocks the SLX4-nuclease complex from nucleolytically resolving the T-loop [30]. Thus, TRF1 removal from telomeres may promote nucleolytic resolution of telomeric joint molecule intermediates. Indeed, FANCD2 deficiency leads to formation of extrachromosomal telomeric structures [29], supporting this probability.
FA core complex members involved in telomere maintenance
Some effects of depletion of FA core complex genes have been reported. Fancg-deficient mice show no signs of telomere dysfunction in both hematopoietic and nonhematopoietic cell lineages, even in presence of extensive genomic stress such as ICL-inducing agent mitomycin C [31]. Although Fancc-deficiency also shows no telomere dysfunction in mouse cells with intrinsically long telomeres, it does accelerate telomere attrition in high turn-over hematopoietic cells and regulates short telomere-initiated telomere HR in the absence of telomerase [32]. Thus, FANCC may aid in telomere length maintenance under replicative pressure and during HR-mediated ALT.
Role of downstream FA proteins in telomere biology
FANCJ is a helicase that helps maintain genome stability by facilitating unhindered progression of replication, possibly via resolution of G4 DNA structures. Although novel proteomic methods have detected FANCJ at telomeres [33], its significance in telomere biology is unclear. G4 structures at telomeres can potentially affect not only telomere replication, but also access/function of telomerase and telomere 3'-overhang binding shelterin proteins POT1/TPP1. Because G4 removal is crucial for smooth progression of telomere metabolism, further exploration of FANCJ’s role in telomere maintenance in FA individuals will be interesting.
SLX4 (mutated in FA patients) is a Swiss army knife that assembles and coordinates a genome maintenance toolkit, functioning in diverse pathways that include ICL repair, DNA replication, processing of HR intermediates, and telomere maintenance [34]. The multi-domain architecture of SLX4 (Figure 2B) enables it to not only bind to a wide range of DNA repair proteins, but also orchestrate their delivery and activities at the target site, each function mediated by one or more specific domain(s) of SLX4. For example, direct interaction of SLX4 with structure-specific endonucleases (SSEs) SLX1, MUS81-EME1, and XPF-ERCC1 is mediated by the SLX4SBD (SLX1-binding domain), SLX4SAP (SAP motif, MUS81-binding region), and SLX4XBR (XPF-binding region) domains, respectively [35–38]. Although it lacks any catalytic activity, SLX4 coordinates dispatch and activity of its associated nucleases [39–41]. Other domains of SLX4 implicated in orchestration of its various functions in the right context and location include SLX4ZF (ubiquitin-binding zinc finger domain, implicated in ICL repair), SLX4SIMs (SUMO-interacting motifs, implicated in managing replication stress) [42–44], and SLX4BTB (Bric-a-brac, Tramtrack and Broad complex domain) [12, 45, 46].
A possible role for SLX4 in telomere maintenance was first suggested when the SLX4-complex isolated from human cells was shown to contain TRF2 [35]. Subsequently, crystallographic, cellular and biochemical studies revealed that SLX4 and its associated nucleases is preferentially recruited to long telomeres via direct interaction between a unique HxLxP (x, any amino acid) motif within the telomere binding motif (TBM) of SLX4 and a docking site at the TRF homology (TRFH) domain of TRF2 (Figure 2B) [38, 47]. The SLX4-nuclease complex is required for multiple aspects of telomere maintenance, including negative regulation of telomere length, regulation of telomere recombination, and prevention of telomere replication defects (manifested as fragile telomeres) [30, 38, 47]. The molecular mechanism behind the role of SLX4 in telomere maintenance likely involves the ability of the SLX4-associated nucleases to process and remove alternate DNA structures such as Holliday Junction (HJ) and T-loop. It has been shown that in vitro, the nucleases SLX1 and MUS81 nucleolytically resolve these structures, enabled by catalytic collaboration between them [30, 41]. In fact, the SLX4-nuclease complex employs the HR-dependent mechanism of telomere shortening, called ‘telomere trimming’, that entails resolution of the T-loop, leading to shorter telomeres and extrachromosomal telomeric circles (Figure 1B) [38, 48, 49].
It is tempting to speculate a link between SLX4 and the negative regulatory mechanisms of telomere length. TRF2 has been known to be a negative regulator of telomere length [50, 51], but the underlying mechanism is unclear. Because the localization of SLX4 to telomeres depends on protein levels of TRF2 [38], it is plausible that longer telomeres that are bound by more TRF2 recruit more SLX4. The double layered SLX4-TRF2 scaffold then assembles the nucleases, followed by shortening of long telomeres. However, it must be kept in mind that unregulated nucleolytic activity at telomeres would be dangerous for telomere and genome stability. It is believed that cell-cycle-based control of activity of the nucleases [41], alternate pathways of processing HR-intermediates, such as the helicase BLM, and inherent telomeric proteins such as TRF1/TRF2 may serve as a check on SLX4-complex-dependent nucleolytic activity at the telomeres [30, 41].
3. Concluding remarks
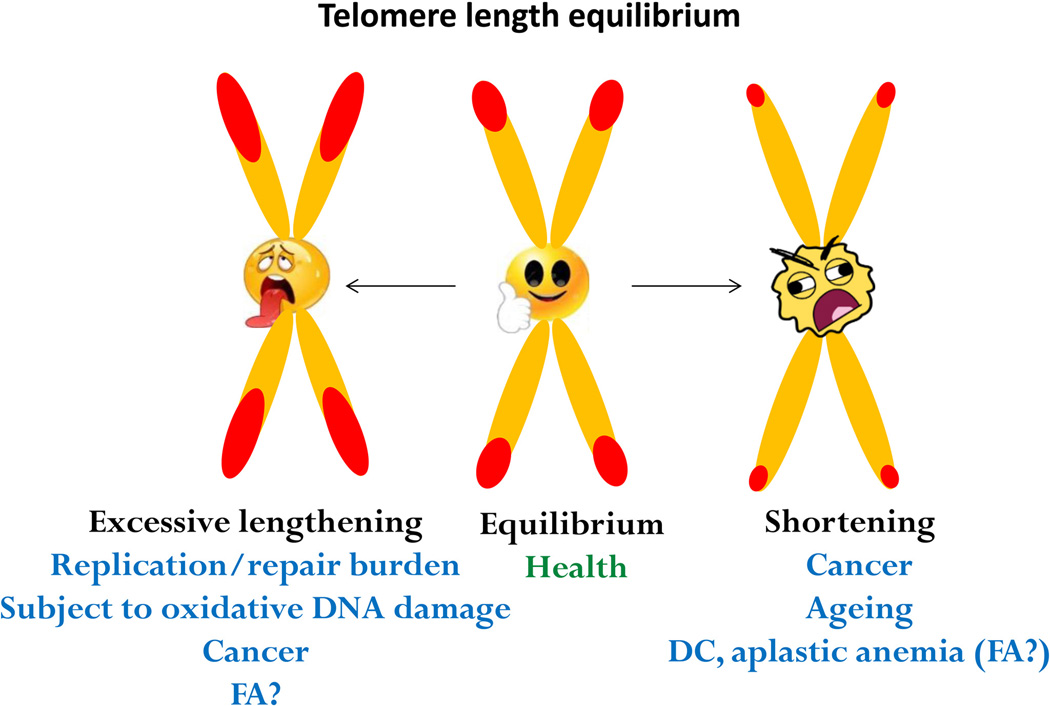
Maintaining telomere length at or near equilibrium (in a species-specific manner) is a critical aspect of telomere maintenance (Figure 3) [2, 52]. Dysregulation of telomere length homeostasis features in several inherited bone marrow failure syndromes including DC [7–9]. Short telomeres in DC are not limited to blood, but are also present in fibroblasts and buccal cells [53], and telomere erosion is believed to have played a role in pathogenesis of the disease [7–9]. Although short telomeres have been reported in FA leukocytes as well, the telomeres are not as abnormally short as in DC [53]. There exists lack of mutations in genes directly involved in telomere biology in FA patients [54]. Since FA-deficient cells are hypersensitive to oxygen [55], telomere defects in FA-deficient cells may be secondary effects of increased oxidative damage at telomeres. It remains to be determined if FA-deficient cells harbor steady-state level of oxidative DNA lesions at telomeres.
Figure 3. Telomere length homeostasis is inevitable for proper cellular function.
Loss of telomere length equilibrium associates with human ageing, cancer, and inherited genetic disorders.
Recent studies have linked abnormally long telomeres to tumorigenesis [6, 8, 56–59]. Interestingly, both FANCD2 and SLX4 localize to telomeres and are required for maintaining long telomeres in ALT cells. Long telomeres face greater challenges and may seek extra attention from genomic DNA repair proteins. This potentially can impede timely progression of DNA replication and repair. The impact of this burden, particularly with respect to FA disease (or in other developmental disorders), may be manifested in a cell-specific manner. Thus, investigating telomere integrity in cell lineages other than leukocytes (such as germline, stem cells, and progenitor cells) in FA individuals may shed meaningful insight into the role of telomere biology in FA. We postulate that the FA pathway constitutes an important layer of telomere maintenance in these cell lineages, where long telomeres are subject to oxidative DNA damage and secondary structure formation. Disruption of FA pathway may thus alter telomere length homeostasis, contributing to hematopoiesis and oncology.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annual review of genetics. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 3.Pickett HA, Reddel RR. Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nature structural & molecular biology. 2015;22:875–880. doi: 10.1038/nsmb.3106. [DOI] [PubMed] [Google Scholar]
- 4.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 6.Zheng YL, Zhang F, Sun B, Du J, Sun C, Yuan J, Wang Y, Tao L, Kota K, Liu X, Schlegel R, Yang Q. Telomerase enzymatic component hTERT shortens long telomeres in human cells. Cell cycle. 2014;13:1765–1776. doi: 10.4161/cc.28705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calado RT, Young NS. Telomere diseases. The New England journal of medicine. 2009;361:2353–2365. doi: 10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanley SE, Armanios M. The short and long telomere syndromes: paired paradigms for molecular medicine. Current opinion in genetics & development. 2015;33:1–9. doi: 10.1016/j.gde.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savage SA, Alter BP. The role of telomere biology in bone marrow failure and other disorders. Mech Ageing Dev. 2008;129:35–47. doi: 10.1016/j.mad.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nature reviews. Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 11.Moldovan GL, D'Andrea AD. How the fanconi anemia pathway guards the genome. Annual review of genetics. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y, Spitz GS, Veturi U, Lach FP, Auerbach AD, Smogorzewska A. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood. 2013;121:54–63. doi: 10.1182/blood-2012-07-441212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ball SE, Gibson FM, Rizzo S, Tooze JA, Marsh JC, Gordon-Smith EC. Progressive telomere shortening in aplastic anemia. Blood. 1998;91:3582–3592. [PubMed] [Google Scholar]
- 14.Leteurtre F, Li X, Guardiola P, Le Roux G, Sergere JC, Richard P, Carosella ED, Gluckman E. Accelerated telomere shortening and telomerase activation in Fanconi's anaemia. Br J Haematol. 1999;105:883–893. doi: 10.1046/j.1365-2141.1999.01445.x. [DOI] [PubMed] [Google Scholar]
- 15.Hanson H, Mathew CG, Docherty Z, Mackie Ogilvie C. Telomere shortening in Fanconi anaemia demonstrated by a direct FISH approach. Cytogenet Cell Genet. 2001;93:203–206. doi: 10.1159/000056985. [DOI] [PubMed] [Google Scholar]
- 16.Callen E, Samper E, Ramirez MJ, Creus A, Marcos R, Ortega JJ, Olive T, Badell I, Blasco MA, Surralles J. Breaks at telomeres and TRF2-independent end fusions in Fanconi anemia. Human molecular genetics. 2002;11:439–444. doi: 10.1093/hmg/11.4.439. [DOI] [PubMed] [Google Scholar]
- 17.Dokal I. Fanconi's anaemia and related bone marrow failure syndromes. Br Med Bull. 2006;77–78:37–53. doi: 10.1093/bmb/ldl007. [DOI] [PubMed] [Google Scholar]
- 18.Adelfalk C, Lorenz M, Serra V, von Zglinicki T, Hirsch-Kauffmann M, Schweiger M. Accelerated telomere shortening in Fanconi anemia fibroblasts--a longitudinal study. FEBS letters. 2001;506:22–26. doi: 10.1016/s0014-5793(01)02869-1. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Leteurtre F, Rocha V, Guardiola P, Berger R, Daniel MT, Noguera MH, Maarek O, Roux GL, de la Salmoniere P, Richard P, Gluckman E. Abnormal telomere metabolism in Fanconi's anaemia correlates with genomic instability and the probability of developing severe aplastic anaemia. Br J Haematol. 2003;120:836–845. doi: 10.1046/j.1365-2141.2003.04225.x. [DOI] [PubMed] [Google Scholar]
- 20.Uziel O, Reshef H, Ravid A, Fabian I, Halperin D, Ram R, Bakhanashvili M, Nordenberg J, Lahav M. Oxidative stress causes telomere damage in Fanconi anaemia cells - a possible predisposition for malignant transformation. Br J Haematol. 2008;142:82–93. doi: 10.1111/j.1365-2141.2008.07137.x. [DOI] [PubMed] [Google Scholar]
- 21.Joksic I, Vujic D, Guc-Scekic M, Leskovac A, Petrovic S, Ojani M, Trujillo JP, Surralles J, Zivkovic M, Stankovic A, Slijepcevic P, Joksic G. Dysfunctional telomeres in primary cells from Fanconi anemia FANCD2 patients. Genome Integr. 2012;3:6. doi: 10.1186/2041-9414-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alter BP, Giri N, Savage SA, Rosenberg PS. Telomere length in inherited bone marrow failure syndromes. Haematologica. 2015;100:49–54. doi: 10.3324/haematol.2014.114389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong H, Nebert DW, Bruford EA, Thompson DC, Joenje H, Vasiliou V. Update of the human and mouse Fanconi anemia genes. Hum Genomics. 2015;9:32. doi: 10.1186/s40246-015-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan Q, Zhang F, Barrett B, Ren K, Andreassen PR. A role for monoubiquitinated FANCD2 at telomeres in ALT cells. Nucleic acids research. 2009;37:1740–1754. doi: 10.1093/nar/gkn995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmermann M, Kibe T, Kabir S, de Lange T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes & development. 2014;28:2477–2491. doi: 10.1101/gad.251611.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 28.Hsiao SJ, Smith S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie. 2008;90:83–92. doi: 10.1016/j.biochi.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Lyakhovich A, Ramirez MJ, Castellanos A, Castella M, Simons AM, Parvin JD, Surralles J. Fanconi anemia protein FANCD2 inhibits TRF1 polyADP-ribosylation through tankyrase1-dependent manner. Genome Integr. 2011;2:4. doi: 10.1186/2041-9414-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarkar J, Wan B, Yin J, Vallabhaneni H, Horvath K, Kulikowicz T, Bohr VA, Zhang Y, Lei M, Liu Y. SLX4 contributes to telomere preservation and regulated processing of telomeric joint molecule intermediates. Nucleic acids research. 2015;43:5912–5923. doi: 10.1093/nar/gkv522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franco S, van de Vrugt HJ, Fernandez P, Aracil M, Arwert F, Blasco MA. Telomere dynamics in Fancg-deficient mouse and human cells. Blood. 2004;104:3927–3935. doi: 10.1182/blood-2003-10-3626. [DOI] [PubMed] [Google Scholar]
- 32.Rhee DB, Wang Y, Mizesko M, Zhou F, Haneline L, Liu Y. FANCC suppresses short telomere-initiated telomere sister chromatid exchange. Human molecular genetics. 2011;19:879–887. doi: 10.1093/hmg/ddp556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y. Nuclease delivery: versatile functions of SLX4/FANCP in genome maintenance. Molecules and cells. 2014;37:569–574. doi: 10.14348/molcells.2014.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svendsen JM, Smogorzewska A, Sowa ME, O'Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, Fuchs RP, McGowan CH, Gaillard PH. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, Kanaar R, Ponting CP, Lilley DM, Rouse J. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Molecular cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 38.Wan B, Yin J, Horvath K, Sarkar J, Chen Y, Wu J, Wan K, Lu J, Gu P, Yu EY, Lue NF, Chang S, Liu Y, Lei M. SLX4 assembles a telomere maintenance toolkit by bridging multiple endonucleases with telomeres. Cell reports. 2013;4:861–869. doi: 10.1016/j.celrep.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castor D, Nair N, Declais AC, Lachaud C, Toth R, Macartney TJ, Lilley DM, Arthur JS, Rouse J. Cooperative control of holliday junction resolution and DNA repair by the SLX1 and MUS81-EME1 nucleases. Molecular cell. 2013;52:221–233. doi: 10.1016/j.molcel.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garner E, Kim Y, Lach FP, Kottemann MC, Smogorzewska A. Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell reports. 2013;5:207–215. doi: 10.1016/j.celrep.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyatt HD, Sarbajna S, Matos J, West SC. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Molecular cell. 2013;52:234–247. doi: 10.1016/j.molcel.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 42.Ouyang J, Garner E, Hallet A, Nguyen HD, Rickman KA, Gill G, Smogorzewska A, Zou L. Noncovalent Interactions with SUMO and Ubiquitin Orchestrate Distinct Functions of the SLX4 Complex in Genome Maintenance. Molecular cell. 2015;57:108–122. doi: 10.1016/j.molcel.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guervilly JH, Takedachi A, Naim V, Scaglione S, Chawhan C, Lovera Y, Despras E, Kuraoka I, Kannouche P, Rosselli F, Gaillard PH. The SLX4 Complex Is a SUMO E3 Ligase that Impacts on Replication Stress Outcome and Genome Stability. Molecular cell. 2015;57:123–137. doi: 10.1016/j.molcel.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez-Prieto R, Cuijpers SA, Luijsterburg MS, van Attikum H, Vertegaal AC. SUMOylation and PARylation cooperate to recruit and stabilize SLX4 at DNA damage sites. EMBO reports. 2015;16:512–519. doi: 10.15252/embr.201440017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. BioEssays : news and reviews in molecular, cellular and developmental biology. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 46.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome biology. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson JS, Tejera AM, Castor D, Toth R, Blasco MA, Rouse J. Localization-dependent and -independent roles of SLX4 in regulating telomeres. Cell reports. 2013;4:853–860. doi: 10.1016/j.celrep.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 49.Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. The EMBO journal. 2009;28:799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ancelin K, Brunori M, Bauwens S, Koering CE, Brun C, Ricoul M, Pommier JP, Sabatier L, Gilson E. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Molecular and cellular biology. 2002;22:3474–3487. doi: 10.1128/MCB.22.10.3474-3487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Molecular and cellular biology. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pickett HA, Reddel RR. The role of telomere trimming in normal telomere length dynamics. Cell cycle. 2012;11:1309–1315. doi: 10.4161/cc.19632. [DOI] [PubMed] [Google Scholar]
- 53.Gadalla SM, Cawthon R, Giri N, Alter BP, Savage SA. Telomere length in blood, buccal cells, and fibroblasts from patients with inherited bone marrow failure syndromes. Aging. 2010;2:867–874. doi: 10.18632/aging.100235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calado RT, Pintao MC, Rocha V, Falcao RP, Bitencourt MA, Silva WA, Jr, Gluckman E, Pasquini R, Zago MA. Lack of mutations in the human telomerase RNA component (hTERC) gene in Fanconi's anemia. Haematologica. 2004;89:1012–1013. [PubMed] [Google Scholar]
- 55.Pang Q, Andreassen PR. Fanconi anemia proteins and endogenous stresses. Mutat Res. 2009;668:42–53. doi: 10.1016/j.mrfmmm.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taboski MA, Sealey DC, Dorrens J, Tayade C, Betts DH, Harrington L. Long telomeres bypass the requirement for telomere maintenance in human tumorigenesis. Cell reports. 2012;1:91–98. doi: 10.1016/j.celrep.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robles-Espinoza CD, Harland M, Ramsay AJ, Aoude LG, Quesada V, Ding Z, Pooley KA, Pritchard AL, Tiffen JC, Petljak M, Palmer JM, Symmons J, Johansson P, Stark MS, Gartside MG, Snowden H, Montgomery GW, Martin NG, Liu JZ, Choi J, Makowski M, Brown KM, Dunning AM, Keane TM, Lopez-Otin C, Gruis NA, Hayward NK, Bishop DT, Newton-Bishop JA, Adams DJ. POT1 loss-of-function variants predispose to familial melanoma. Nature genetics. 2014;46:478–481. doi: 10.1038/ng.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi J, Yang XR, Ballew B, Rotunno M, Calista D, Fargnoli MC, Ghiorzo P, Bressac-de Paillerets B, Nagore E, Avril MF, Caporaso NE, McMaster ML, Cullen M, Wang Z, Zhang X, N.D.C.S.W. Group, N.D.C.G.R. Laboratory, G. French Familial Melanoma Study. Bruno W, Pastorino L, Queirolo P, Banuls-Roca J, Garcia-Casado Z, Vaysse A, Mohamdi H, Riazalhosseini Y, Foglio M, Jouenne F, Hua X, Hyland PL, Yin J, Vallabhaneni H, Chai W, Minghetti P, Pellegrini C, Ravichandran S, Eggermont A, Lathrop M, Peris K, Scarra GB, Landi G, Savage SA, Sampson JN, He J, Yeager M, Goldin LR, Demenais F, Chanock SJ, Tucker MA, Goldstein AM, Liu Y, Landi MT. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nature genetics. 2014;46:482–486. doi: 10.1038/ng.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calvete O, Martinez P, Garcia-Pavia P, Benitez-Buelga C, Paumard-Hernandez B, Fernandez V, Dominguez F, Salas C, Romero-Laorden N, Garcia-Donas J, Carrillo J, Perona R, Trivino JC, Andres R, Cano JM, Rivera B, Alonso-Pulpon L, Setien F, Esteller M, Rodriguez-Perales S, Bougeard G, Frebourg T, Urioste M, Blasco MA, Benitez J. A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li-Fraumeni-like families. Nature communications. 2015;6:8383. doi: 10.1038/ncomms9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doksani Y, de Lange T. The role of double-strand break repair pathways at functional and dysfunctional telomeres. Cold Spring Harb Perspect Biol. 2014;6:a016576. doi: 10.1101/cshperspect.a016576. [DOI] [PMC free article] [PubMed] [Google Scholar]