Abstract
Obesity is characterized as an accumulation of adipose tissue mass represented by chronic, low-grade inflammation. Obesity-derived inflammation involves chemokines as important regulators contributing to the pathophysiology of obesity-related diseases such as cardiovascular disease, diabetes and some cancers. The obesity-driven chemokine network is poorly understood. Here, we identified the profiles of chemokine signature between human preadipocytes and adipocytes, using PCR arrays and qRT-PCR. Both preadipocytes and adipocytes showed absent or low levels in chemokine receptors in spite of some changes. On the other hand, the chemokine levels of CCL2, CCL7-8, CCL11, CXCL1-3, CXCL6 and CXCL10-11 were dominantly expressed in preadipocytes compared to adipocytes. Interestingly, CXCL14 was the most dominant chemokine expressed in adipocytes compared to preadipocytes. Moreover, there is significantly higher protein level of CXCL14 in conditioned media from adipocytes. In addition, we analyzed the data of the chemokine signatures in adipocytes obtained from healthy lean and obese postmenopausal women based on Gene Expression Omnibus (GEO) dataset. Adipocytes from obese individuals had significantly higher levels in chemokine signature as follows: CCL2, CCL13, CCL18-19, CCL23, CCL26, CXCL1, CXCL3 and CXCL14, as compared to those from lean ones. Also, among the chemokine networks, CXCL14 appeared to be the highest levels in adipocytes from both lean and obese women. Taken together, these results identify CXCL14 as an important chemokine induced during adipogenesis, requiring further research elucidating its potential therapeutic benefits in obesity.
Keywords: Chemokine, Adipogenesis, Obesity, Human adipocyte
INTRODUCTION
Adipose tissue is composed of adipocytes, stromal vascular fraction such as preadipocytes at various stage of differentiation, fibroblasts, vascular endothelial cells and heterogeneous population of immune cells (1). The obese state is now acknowledged as the main contributor to chronic low-grade inflammation (2). Recently, obesity has become a global epidemic and poses a large health burden as it is associated with excess adiposity that leads to an array of comorbidities such as type 2 diabetes, stroke, coronary heart disease and cancer (3,4). Obesity is an abnormal increase in adipose tissue mass leading to chronic inflammation and dysregulated adipocytokines such as proinflammatory mediators (CXCL8, IL-1, IL-6, TNF), angiogenic factors (VEGF) and metabolic regulators (leptin, adiponectin) (5). Interestingly, adipose tissue mass has an architectural organization in which metabolic cell such as adipocytes are in close proximity to macrophages, creating continuous interactions between immune and metabolic responses (5). It is now established that this interface contributes to the inflammatory response associated with obesity (6). Adipose tissue, a biologically-active endocrine tissue, serves as energy storage and secretes various proinflammatory cytokines and chemokines (7). Growing evidence suggests that cytokine and chemokine synthesis by human primary adipocytes may play a role in systemic inflammation, which describes the obese state (8,9).
Chemokines are chemotactic cytokines that instigate chemo-attraction, thus proinflammatory chemokines are produced in response to inflammation (10). There are four classified groups of chemokines according to the location of the conserved cysteine residues: XCL, CCL, CXCL and CX3CL, where X is any amino acid residue (11). Chemokines bind specifically to chemokine receptors which are seven-transmembrane spanning G proteincoupled cell-surface receptors followed by their own functional roles via the intracellular signaling pathway such as immune surveillance and inflammation (12). In this study, we used human preadipocytes and adipocytes and identified the profiles of chemokine signature, providing a better understanding into the complex interactions of the chemokine network between immune cells and adipocytes implicated in the pathogenesis of obesity-derived chronic low-grade inflammation.
MATERIALS AND METHODS
Reagents
The PCR array for customized human chemokines (CAPH10008) and SYBR® Green Master Mix (cat #: 330503) were purchased from SABiosciences/Qiagen (Frederick, MD, USA). Specific PCR primers for chemokines were obtained from Eurofins MWG Operon (Huntsville, AL, USA). All liquid culture media such as FBS (cat #: 26140), calf serum (cat #: 16010159), antibiotic-antimycotic solution (cat #: 15240096), and DMEM (cat #: 11965) were acquired from Invitrogen/Thermo Fisher Scientific Inc. (Waltham, MA, USA).
Human preadipocytes and adipocytes
The human preadipocytes and adipocytes were purchased from Cell Applications Inc. (San Diego, CA, USA). Preadipocytes and adipocytes were cultured in preadipocyte basal medium and adipocytes maintenance medium, respectively, according to manufacturer's instructions. Human preadipocyte and fully differentiated adipocytes were visualized under a digital inverted microscope (EVOS, Advanced Microscopy Group, Bothell, WA, USA) at 20 X magnification.
PCR array and qRT-PCR
After isolating total RNA and eliminating genomic DNA, the expression of chemokine ligands and receptors were analyzed using customized human chemokine PCR array (SABiosciences/Qiagen) on a Bio-Rad CFX96 real-time PCR instrument (Hercules, CA, USA). Briefly, reverse transcription reactions were performed at 42℃ for 15 min followed by 94℃ for 5 min. According to manufacturers' instructions, a real-time PCR was performed using a Bio-Rad CFX96 under the following two-step cycling program: 1 cycle at 95℃ for 10 min, 40 cycles at 95℃ for 15 sec and at 60℃ for 1 min. Data analysis was performed using PCR Array Data Analysis Software (http://www.sabiosciences.com/RTPCR.php) provided by SABiosciences/Qiagen. Changes in chemokine expression were confirmed using specific PCR primer sets described in Table I Values for PCR array are the means from duplicate experiments, while experiments for qRT-PCR were performed at least in triplicate.
Table I. Primers used in qRT-PCR analysis.
| Chemokines | Accession No. | Primers (sense/antisense) | CDS No. |
|---|---|---|---|
| CCL2 | NM_002982 | 5-TAGGAAGATCTCAGTGCAGAGG-3 | 120-217 |
| 5-CTTGGCCACAATGGTCTTGA-3 | |||
| CCL7 | NM_006273 | 5-ACTTCTGTGTCTGCTGCTC-3 | 17-144 |
| 5-CAGCCTCTGCTTAGGGATTT-3 | |||
| CCL8 | NM_005623 | 5-GGAGAGCTACACAAGAATCACC-3 | 144-260 |
| 5-TGGAATCCCTGACCCATCT-3 | |||
| CXCL6 | NM_002993 | 5-TTTACGCGTTACGCTGAGAG-3 | 153-279 |
| 5-TCCAGACAAACTTGCTTCCC-3 | |||
| CXCL10 | NM_001565 | 5-CTGTACCTGCATCAGCATTAGTA-3 | 87-224 |
| 5-GACATCTCTTCTCACCCTTCTTT-3 | |||
| CXCL14 | NM_004887 | 5-CTGCGAGGAGAAGATGGTTATC-3 | 186-306 |
| 5-CAGGCGTTGTACCACTTGA-3 | |||
| β-actin | X00351 | 5-CCTCATGAAGATCCTCACCG-3 | 605-712 |
| 5-CCATCTCTTGCTCGAAGTCC-3 |
CDS, Coding DNA Sequence.
Enzyme-linked Immunosorbent Assay (ELISA)
The level of secreted CXCL14 from conditioned media of preadipocytes and adipocytes media was assessed by enzyme-linked immunosorbent assay (ELISA) kit from Ray Biotech (Norcross, GA, USA) according to the manufacturer's instructions. The optical density of each well was determined using a microplate reader with 450 nm of wavelength. Assays were calibrated using serial dilutions of recombinant human CXCL14 (Ray Biotech).
Data analysis from GEO dataset
Data analysis was performed using microarray data sets deposited in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database under accession number GSE44000. Raw microarray data for chemokine network were RNA expression levels in subcutaneous adipose biopsies obtained from 7 lean (BMI<25) and 7 obese (BMI>30) postmenopausal women closely matched for age and NCEP III metabolic syndrome parameters exclusive of waist circumference (13). We employed Gitools 2.2.3 (http://www.gitools.org) based on Oracle Java 7, an open-source tool to perform Genomic Data Analysis and Visualization as interactive heat-maps (14).
Statistical analysis
Data values were expressed as the mean±SEM. The mean values for each group were analyzed and compared using paired Student's t test. Differences were considered statistically significant at p≤0.05.
RESULTS AND DISCUSSION
Obesity is known to alter metabolic and endocrine functions in adipose tissue followed by obesity-associated comorbidities. Inflammation in adipose tissue appears to be a key mechanism to explain the development of insulin resistance, as an important factors that leads to adipose tissue dysfunction (2). Cumulative evidence suggests that chemokines are one of important factors implicated in development of pathophysiological processes linking obesity and insulin resistance (10). In the present study, we characterized profiles of the chemokine network in human preadipocytes and adipocytes.
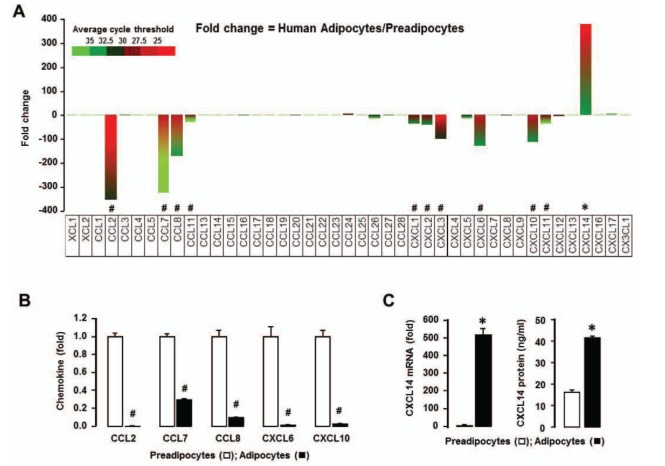
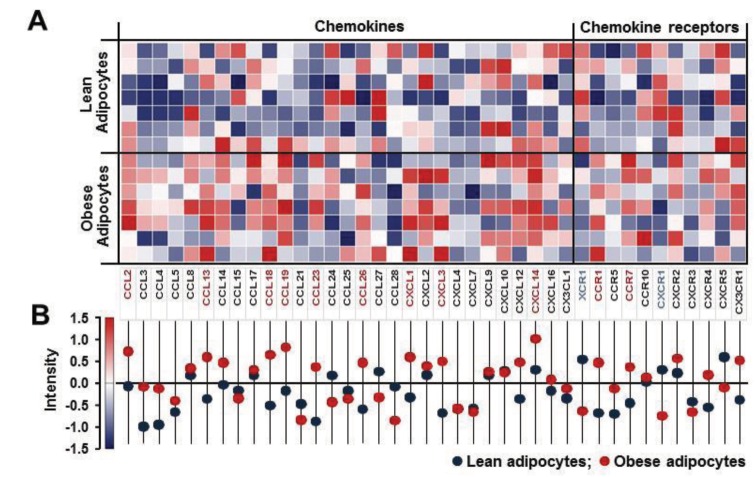
Morphologically, human preadipocytes revealed long, thin and flatted cells without detectable fat organelles while the matured human adipocytes had lipid accumulation in spherical form (Fig. 1A). Overall expression levels of chemokine receptors were absent or low (Fig. 1B), wherein adipocytes decreased CCR10 and increased XCR1, CXCR3, CXCR5 and CX3CR1 compared to preadipocytes (Fig. 1B). Our chemokine receptor signature indicates that chemokine receptors are unlikely to contribute to adipogenesis from preadipocytes to adipocytes. Meanwhile, there were significant differences in the chemokine signatures between preadipocyte and adipocyte. The dominant chemokines of preadipocytes were CCL2, CCL7-8, CCL11, CXCL1-3, CXCL6 and CXCL10-11 (Fig. 2A and 2B), which appears to be downregulated during adipogenesis. Interestingly, adipocytes expressed dominantly CXCL14 (Fig. 2A and 2C), indicating its important role in adipogenesis. In parallel, the ELISA data also revealed significantly higher protein level of CXCL14 in conditioned media of adipocyte compared to preadipocyte (Fig. 2C). Based on GEO dataset, chemokine signature in adipocytes obtained from healthy lean and obese postmenopausal women revealed that adipocytes from obese women had significantly higher levels in the following chemokines: CCL2, CCL13, CCL18-19, CCL23, CCL26, CXCL1, CXCL3 and CXCL14, compared to those from lean women (Fig. 3A and 3B). In particular, CXCL14 appeared to be expressed the highest levels in adipocytes from both lean and obese women (Fig. 3B). Based on the significant increase of CXCL14 in obese women compared to lean women (Fig. 3B), further study requires clarifying the positive relationship between CXCL14 levels and the degree of obesity to confirm CXCL14 as a biomarker for obesity.
Figure 1. Chemokine receptor signature in human preadipocytes and adipocytes. (A) Representative images of human adipocytes during adipogenesis. Human preadipocyte and differentiated adipocytes were visualized under a digital inverted microscope at 20 X magnification. Human preadipocytes show long, thin and flatted cells, while human adipocytes show differentiated fat cells in spherical form. (B) Comparison of chemokine ligands in preadipocytes vs. adipocytes. A PCR array for chemokine receptors was performed by using total RNA isolated from nondifferentiated (preadipocytes) and differentiated cells (adipocytes). Different colors indicate the average cycle threshold with expressions that ranged from >35 to <25. Expression levels of chemokine receptors were defined as absent (>35), low (30-35) and high (<30) on average threshold cycles. Chemokines with a >2-fold increase (*) or decrease (#) were recognized as the major differences between preadipocytes and adipocytes.
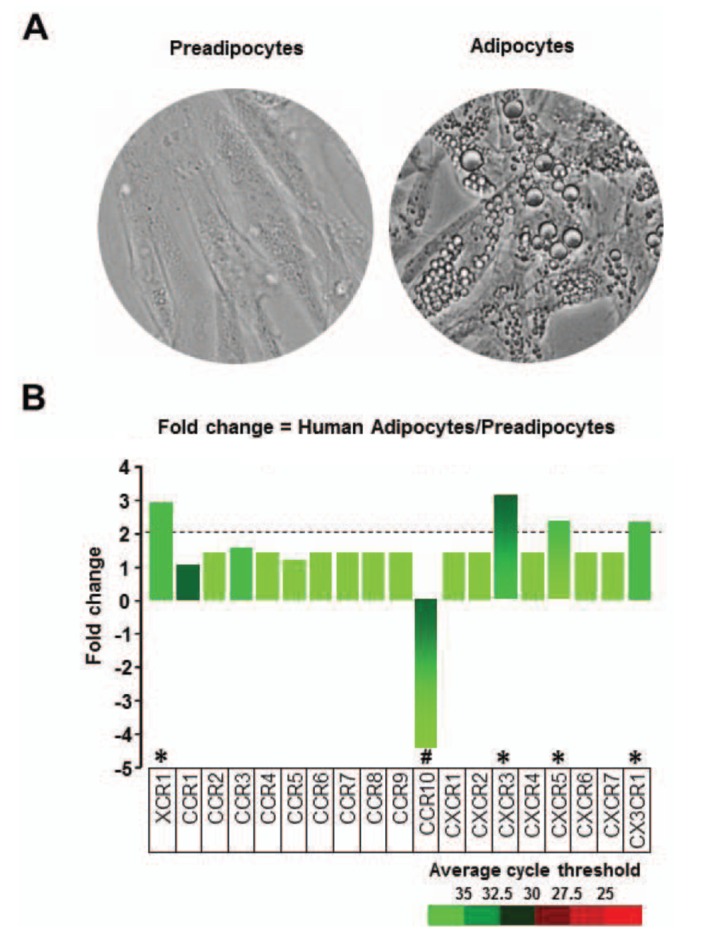
Figure 2. Chemokine ligand signature in human preadipocytes and adipocytes. (A) Comparison of chemokine ligands in preadipocytes and adipocytes using PCR array containing complementary sequences for human chemokine ligand genes. Different colors indicate the average cycle threshold with expressions that ranged from >35 to <25. Expression levels of chemokines were defined as absent (>35), low (30-35) and high (<30) on average threshold cycles. Chemokines with a >2-fold increase (*) or decrease (#) were recognized as the major differences between preadipocytes and adipocytes. (B) Changes in mRNA expression of CCL2, CCL7, CCL8, CXCL6, CXCL10 and (C) CXCL14 were confirmed using specific PCR primer sets by qRT-PCR. Fold changes were calculated as a relative value after setting the first sample of adipocytes as a control group (1.0). Also, secreted CXCL14 in conditioned media from preadipocytes and adipocytes was assessed by ELISA. Experiments values for PCR array are the means from duplicate experiments, while experiments for qRT-PCR were performed at least in triplicate. All data value are presented as mean±SEM. * and # indicates significant increase or decrease (p≤0.05), respectively when tested with Student's t test.
Figure 3. Profiles of chemokine network in adipose tissues between lean and obese postmenopausal women. (A) Expression levels of chemokine ligands and receptors in subcutaneous adipose tissues between lean and obese women using microarray data via Gitools. (B) Intensity of chemokine network for statistical analysis. All values are presented as mean. Red and blue letters in chemokine signature indicate significant increase or decrease in obese women (p≤0.05), respectively, as tested with Student's t test.
Studies on adipose tissue in obese mice demonstrated increased level of CXCL14 (15,16). High fat diet-fed CXCL14-deficient mice showed impaired white adipose tissue macrophage immobilization (17), suggesting the involvement of CXCL14 in macrophage recruitment in obesity. CXCL14 might recruit macrophages into adipose tissue, thus producing a wide range of cytokines, chemokines and growth factors to induce chronic inflammation that promotes obesity-associated diseases such as diabetes and cancer progression. Identification and characterization of still unknown CXCL14 receptor will be helpful in understanding the functional role and regulation of this adipocyte-secreted CXCL14 in obesityassociated comorbidities. In spite of downregulation of CCL2, CXCL1 and CXCL3 during adipogenesis (Fig. 2A), obese adipose tissues upregulated these chemokines (Fig. 3B). Because these chemokines are mainly the NF-κB driven chemokines (18,19,20), obesity may contribute to a proinflammatory microenvironment by potentiating the NF-κB driven chemokines via recruitment of immune cells. This study identifies the chemokine signature during adipogenesis that could lay groundwork for the development of efficient therapeutic strategies based on the prevention of low-grade inflammation, which defines the obese state. Our finding suggests a new plausible marker to restore chronic, low-grade inflammation, by suggesting a distinct functional role of CXCL14 from other proinflammatory chemokines.
ACKNOWLEDGEMENT
This research was supported, in whole or in part, by National Institutes of Health as the following grants: NIGMS SC1 089630 and R01ES024756 (E.L.), and NIAID SC1AI089073 and NCI SC1CA200519 (D.S.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
Abbreviations
- VEGF
vascular endothelial growth factor
- GEO
gene expression omnibus
Footnotes
CONFLICTS OF INTEREST: The authors declare that they have no conflicts of interest with the contents of this article.
References
- 1.Wagner M, Samdal Steinskog ES, Wiig H. Adipose tissue macrophages: the inflammatory link between obesity and cancer? Expert Opin Ther Targets. 2015;19:527–538. doi: 10.1517/14728222.2014.991311. [DOI] [PubMed] [Google Scholar]
- 2.Lafontan M. Adipose tissue and adipocyte dysregulation. Diabetes Metab. 2014;40:16–28. doi: 10.1016/j.diabet.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Crunkhorn S. Metabolic disorders: Breaking the links between inflammation and diabetes. Nat Rev Drug Discov. 2013;12:261. doi: 10.1038/nrd3988. [DOI] [PubMed] [Google Scholar]
- 4.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4:579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 5.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 6.Wellen KE, Hotamisligil GS. Obesity-induced inflammatory changes in adipose tissue. J Clin Invest. 2003;112:1785–1788. doi: 10.1172/JCI20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 8.Meijer K, de Vries M, Al-Lahham S, Bruinenberg M, Weening D, Dijkstra M, Kloosterhuis N, van der, van der, Kroesen BJ, Vonk R, Rezaee F. Human primary adipocytes exhibit immune cell function: adipocytes prime inflammation independent of macrophages. PLoS One. 2011;6:e17154. doi: 10.1371/journal.pone.0017154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y. Small lipid-binding proteins in regulating endothelial and vascular functions: focusing on adipocyte fatty acid binding protein and lipocalin-2. Br J Pharmacol. 2012;165:603–621. doi: 10.1111/j.1476-5381.2011.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chavey C, Lazennec G, Lagarrigue S, Clapé C, Iankova I, Teyssier J, Annicotte JS, Schmidt J, Mataki C, Yamamoto H, Sanches R, Guma A, Stich V, Vitkova M, Jardin-Watelet B, Renard E, Strieter R, Tuthill A, Hotamisligil GS, Vidal-Puig A, Zorzano A, Langin D, Fajas L. CXC ligand 5 is an adipose-tissue derived factor that links obesity to insulin resistance. Cell Metab. 2009;9:339–349. doi: 10.1016/j.cmet.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006;7:243. doi: 10.1186/gb-2006-7-12-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazennec G, Richmond A. Chemokines and chemokine receptors: new insights into cancer-related inflammation. Trends Mol Med. 2010;16:133–144. doi: 10.1016/j.molmed.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng T, Lyon CJ, Minze LJ, Lin J, Zou J, Liu JZ, Ren Y, Yin Z, Hamilton DJ, Reardon PR, Sherman V, Wang HY, Phillips KJ, Webb P, Wong ST, Wang RF, Hsueh WA. Class II major histocompatibility complex plays an essential role in obesity-induced adipose inflammation. Cell Metab. 2013;17:411–422. doi: 10.1016/j.cmet.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Llamas C, Lopez-Bigas N. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS One. 2011;6:e19541. doi: 10.1371/journal.pone.0019541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, Chen H. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nara N, Nakayama Y, Okamoto S, Tamura H, Kiyono M, Muraoka M, Tanaka K, Taya C, Shitara H, Ishii R, Yonekawa H, Minokoshi Y, Hara T. Disruption of CXC motif chemokine ligand-14 in mice ameliorates obesity-induced insulin resistance. J Biol Chem. 2007;282:30794–30803. doi: 10.1074/jbc.M700412200. [DOI] [PubMed] [Google Scholar]
- 18.Ping D, Boekhoudt GH, Rogers EM, Boss JM. Nuclear factor-kappa B p65 mediates the assembly and activation of the TNF-responsive element of the murine monocyte chemoattractant-1 gene. J Immunol. 1999;162:727–734. [PubMed] [Google Scholar]
- 19.Son DS, Parl AK, Rice VM, Khabele D. Keratinocyte chemoattractant (KC)/human growth-regulated oncogene (GRO) chemokines and pro-inflammatory chemokine networks in mouse and human ovarian epithelial cancer cells. Cancer Biol Ther. 2007;6:1302–1312. doi: 10.4161/cbt.6.8.4506. [DOI] [PubMed] [Google Scholar]
- 20.Kabir SM, Lee ES, Son DS. Chemokine network during adipogenesis in 3T3-L1 cells: Differential response between growth and proinflammatory factor in preadipocytes vs. adipocytes. Adipocyte. 2014;3:97–106. doi: 10.4161/adip.28110. [DOI] [PMC free article] [PubMed] [Google Scholar]