Abstract
We report the principal characteristics of ‘Eisenbergiella massiliensis’ sp. nov. strain AT11 (CSURP = P2120, DSM = 101499) that was isolated from a stool sample collected after bariatric surgery of a 56-year-old obese French woman.
Keywords: Culturomics, Eisenbergiella massiliensis, human gastrointestinal microbiome, obesity, taxonomy
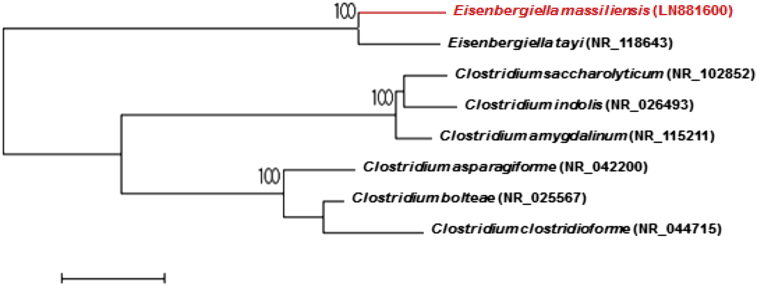
In April 2011, as part of a culturomics study [1], [2] to explore the gut microbiota from obese patients before and after bariatric surgery, we isolated a strain that did not correspond to any previously known species. The patient provided signed informed consent and the study was approved by the ethics committee of the Institut Federatif de Recherche IFR48 under number 09-022, 2010. The first growth was obtained after 21 days of culture in a blood culture bottle enriched with sheep blood and rumen medium under an anaerobic 37°C atmosphere as described elsewhere [3]. Agar-grown (Columbia agar + 5% sheep blood; bioMérieux, Marcy l’Etoile, France) grey colonies exhibited an irregular form with a mean diameter of 0.5 mm after 48–72 –h of culture. A bacterium whose spectrum could not be identified by our systematic matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF-MS) screening on a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) was identified by the 16S rRNA gene sequencing [4]. The 16S rRNA gene was sequenced using fD1-rP2 primers as previously described [5], using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France). It displays 97.7% sequence similarity with Eisenbergiella tayi strain B086562 (= LMG 27400 = DSM 26961 = ATCC BAA-2558), the phylogenetically nearest species with standing nomenclature (Fig. 1). This fact putatively classifies strain AT11 as a member of the genus Eisenbergiella within the Lachnospiraceae family in the phylum Firmicutes. However, this percentage remains lower than the 98.7% 16S rRNA gene sequence threshold recommended by Stackebrandt and Ebers to delineate a new species [6]. Strain AT11 exhibited a 16S rRNA sequence divergence of 2.24% with its phylogenetically closest species with standing in nomenclature [7]. The strain AT11 was catalase-positive and oxidase-negative. Cells are Gram-negative staining, non-motile, non-spore-forming with a mean diameter of 2 μm by electron microscopy. Based on phenotypic and phylogenetic characteristics we propose the creation of a new species named ‘Eisenbergiella massiliensis’ (ma.si.li.en′.sis. L. fem. adj. massiliensis, of Massilia, the Latin name of Marseille where ‘Eisenbergiella massiliensis’ was first isolated). Strain AT11 is the type strain of the new species ‘Eisenbergiella massiliensis’.
Fig. 1.
Phylogenetic tree showing the position of ‘Eisenbergiella massiliensis’ strain AT11 relative to other phylogenetically related species. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using the maximum-likelihood method within the MEGA software. Numbers at the nodes depict bootstrap percentage values obtained by repeating the analysis 500 times to yield a consensus tree. The bootstrap values of at least 95% were retained. The scale bar indicates a 1% nucleotide sequence divergence.
MALDITOF-MS Spectrum Accession Number
The MALDITOF-MS spectrum of this strain is available at http://www.mediterranee-infection.com/article.php?laref=256&titre=urms-database.
Nucleotide Sequence Accession Number
The 16S rRNA gene sequence was deposited in GenBank under Accession number LN881600.
Deposit in a Culture Collection
Strain AT11 was deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR) under number P2120.
Conflicts of Interest
No conflict of interest to declare.
Acknowledgements
The authors thank Karolina Griffiths for reviewing the English. This work was funded by the Mediterrannée-Infection Foundation.
References
- 1.Lagier J.-C., Armougom F., Million M., Hugon P., Pagnier I., Robert C. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 2.Lagier J.-C., Hugon P., Khelaifia S., Fournier P.-E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Togo A.H., Khelaifia S., Lagier J.-C., Caputo A., Robert C., Fournier P.-E. Noncontiguous finished genome sequence and description of Paenibacillus ihumii sp. nov. strain AT5. New Microbes New Infect. 2016;10:142–150. doi: 10.1016/j.nmni.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.-E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis Off Publ Infect Dis Soc Am. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 5.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stackebrandt A.N.D., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 7.Huson D.H., Richter D.C., Rausch C., Dezulian T., Franz M., Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]