Abstract
Eight strains isolated from the stems of Lippia sidoides were identified as belonging to Lactococcus lactis, a bacterial species considered as “generally recognized as safe”. Their capacity to solubilize/mineralize phosphate was tested in vitro with different inorganic and organic phosphorus (P) sources. All strains were able to solubilize calcium phosphate as an inorganic P source, and the best result was observed with strain 003.41 which solubilized 31 % of this P source. Rock phosphate, a mined rock containing high amounts of phosphate bearing minerals, was solubilized by five strains. When calcium phytate was the organic P source used, the majority of the strains tested showed phosphate mineralization activity. Moreover, all strains were able to solubilize/mineralize phosphate from poultry litter, a complex P source containing inorganic and predominantly organic P. The presence of genes coding for phytase and alkaline phosphatase was searched within the strains studied. However, only gene sequences related to alkaline phosphatase (phoA and phoD) could be detected in the majority of the strains (excepting strain 006.29) with identities varying from 67 to 88 %. These results demonstrate for the first time the potential of L. lactis strains for phosphate solubilization/mineralization activity using a broad spectrum of P sources; therefore, they are of great importance for the future development of more safe bioinoculants with possible beneficial effects for agriculture.
Keywords: Phosphate solubilization, Phosphate mineralization, Generally recognized as safe (GRAS), Lactococcus lactis, Endophytes, Lippia sidoides Cham.
Background
The species Lactococcus lactis belongs to the Firmicutes phylum and its strains are characterized by the production of lactic acid as the main end product of carbohydrate metabolism. L. lactis is one of the most important microorganisms in the dairy industry (Ward et al. 2002), and has “generally recognized as safe” (GRAS) status (Wessels et al. 2004). Although L. lactis is found in a wide range of environments and even in a great variety of traditional food products (Salama et al. 1995; Velly et al. 2014), this species has been recurrently found in different parts of various plants. They have already been isolated from the interior of aquatic plants (Chen et al. 2012), from stems of Eucalyptus (Procópio et al. 2009), from baby corn and fresh green peas (Alemayehu et al. 2014) and from the inner tissues and leaves of sugarcane (Beneduzi et al. 2013; Cock and de Stouvenel 2006, respectively). Likewise, da Silva et al. (2013) isolated from the Brazilian essential oil producing plant Lippia sidoides Cham. (pepper-rosmarin) eight bacterial endophytes identified as L. lactis. Recently, Golomb and Marco (2015) showed that certain strains of L. lactis are well adapted for growth on plants.
Strains growing in association with plants may exert positive effects on plant growth directly or indirectly through one or more mechanisms. For example, these strains can fix nitrogen, synthesize phytohormones and vitamins, improve nutrient uptake, solubilize inorganic phosphate and mineralize organic phosphate, among other direct benefits to the plant. Indirectly, some bacterial strains can produce siderophores and/or synthesize antimicrobial compounds decreasing or even preventing the deleterious effects of pathogenic microorganisms (Dobbelaere et al. 2003). However, to the best of our knowledge, only very few studies are available considering strains of L. lactis as potential plant growth promoters. Somers et al. (2007) and Grönemeyer et al. (2012) described the isolation of L. lactis strains showing significant plant growth promoting activity in greenhouse trials with cabbage and from agricultural crops in the Kavango region of Namibia, respectively. The ability to solubilize tricalcium phosphate has been demonstrated in L. lactis by growing the strains MH5-2 and MH5-5, isolated from mahangu, on bromophenol blue-containing Pikovskaya agar (Grönemeyer et al. 2012). However, their ability to solubilize/mineralize phosphate converting different sources of insoluble phosphates into available forms for plant has not been explored so far.
Phosphorus (P) is considered the second most important nutrient for plant nutrition. It is implicated in different plant processes and as an integral component of several plant structures such as phospholipids, proteins and nucleic acids (Balemi and Negisho 2012). Although P is abundant in soil, this nutrient is usually unavailable for plant uptake because it readily forms insoluble complexes with aluminium, iron, calcium and/or magnesium in soil (Vance et al. 2003; Altier et al. 2013). It is estimated that crop productivity is limited by P deficiency on more than 40 % of the world arable lands (Vance 2001). To maintain the crop yield, huge amounts of P containing fertilizers are usually applied worldwide. Moreover, global P reserves are being depleted at a higher rate demanding high energy costs and resulting in deleterious environmental impacts, such as eutrophication and carbon emissions (Balemi and Negisho 2012; Sharma et al. 2013).
Phosphate solubilizing/mineralizing bacteria are an environmental friendly alternative to the use of chemical P fertilizers (Richardson and Simpson 2011). Moreover, bacterial groups with GRAS status are of great interest as potential bioinoculants with low chances of causing plant or human diseases (Rosenblueth and Martínez-Romero 2006; Mendes et al. 2013). Therefore, the main purpose of this study was to test the L. lactis strains previously isolated from L. sidoides for their capacity of phosphate solubilization/mineralization of several inorganic and organic phosphate sources, such as calcium phosphate, aluminium phosphate, rock phosphate, phytate and poultry litter. The results obtained here meet the increased interest in the harnessing of microorganisms to support P cycling in agroecosystems.
Methods
Bacterial strains from Lippia sidoides Cham.
The bacterial strains (003.41, 006.27, 006.29, 006.30, 006.31, 104.3, 104.6 and 104.7), used in this study, have been previously isolated from the stems of three genotypes of L. sidoides (LSID003, LSID006 and LSID104) as described by da Silva et al. (2013). The different isolates were maintained both in BD™ Tryptic Soy Broth (TSB) agar-containing slants at room temperature and in TSB with 20 % glycerol at −80 °C.
DNA extraction, PCR amplification, sequencing and phylogenetic analysis based on 16S rRNA
Genomic DNA was extracted from all bacterial strains using the protocol presented in da Silva et al. (2013). A partial sequence of the 16S rRNA gene (~1400 bp) was obtained by PCR amplification using the pair of universal primers pA (5′-AGAGTTTGATCCTGGCTCAG-3′) and pH (5′-AAGGAGGTGATCCAGCCGCA-3′) and the conditions described in Massol-Deya et al. (1995). Negative controls (without DNA) were run in all amplifications. The PCR products were sequenced using Macrogen (South Korea) facilities, and the partial 16S rRNA gene sequences were identified using the BLAST-N tool (www.ncbi.nlm.nih.gov/blast) on the National Center for Biotechnology Information (NCBI) website using the GenBank non-redundant database. The resulting sequences were aligned to sequences from Lactococcus type strains available in the GenBank database using the CLUSTALW tool (Thompson et al. 1994). MEGA 6 software (Tamura et al. 2013) was used to construct a phylogenetic tree based on the 16S rRNA gene sequences using the neighbor-joining method and Jukes–Cantor distances. Bootstrap analyses were performed with 1000 repetitions, and only values higher than 50 % are shown in the phylogenetic tree. The sequences generated in this study were deposited in NCBI GenBank under accession numbers KU639597- KU639604.
PCR amplification of alkaline phosphatase and phytase encoding genes
Fragments of alkaline phosphatase and phytase genes were amplified by PCR using the pair of primers ALPS-R1101 (5′-GAGGCCGATCGGCATGTCG-3′) and ALPS-F730 (5′-CAGTGGGACGACCACGAGGT-3′; Sakurai et al. 2008) and BPP-F (5′-GACGCAGCCGAYGAYCCNGCNITNTGG-3′) and BPP-R (5′-CAGGSCGCANRTCIACRTTRTT-3′; Huang et al. 2009), respectively.
The products obtained were visualized by 1.4 % agarose gel electrophoresis in TBE buffer (Sambrook et al. 1989) at 80 V for 3–4 h at room temperature, and stained with ethidium bromide. The PCR products were then sequenced using Macrogen facilities, and the sequences were further compared to those previously deposited at the GenBank database using the BLAST-N tool.
Phosphate solubilization/mineralization tests in liquid medium
The ability of the isolates to solubilize inorganic and mineralize organic phosphates in liquid medium was tested as described by Nautiyal (1999). Four different P sources (calcium phosphate, aluminium phosphate, rock phosphate and poultry litter) were used. The chemical composition of the poultry litter was (per g kg−1): Ca2+—0.39, Mg2+—0.43, K+—21.1, P—7.0, Al3+—0.65, Na+—3.5, and 0.86 % of organic carbon. Bacterial cultures were grown in the National Botanical Research Institute’s phosphate growth medium (NBRIP—containing in 50 ml: glucose, 0.5 g; MgCl2·6H2O, 0.25 g; MgSO4·7H2O, 0.0125 g; KCl, 0.01 g and (NH4)2SO4, 0.005 g) with the addition of 0.25 g of each P source. The cultures were incubated at 28 °C for 10 days with shaking (180 rpm). After incubation, the pH was measured using a pH Meter (PHI510, Beckman Coulter, California, USA) and the concentration of soluble P was determined as described by Murphy and Riley (1962).
Phosphate mineralization tests in solid medium
The ability of the isolates to mineralize organic phosphate was tested as described by Rosado et al. (1998). Calcium phytate was used as the P source in the culture medium (glucose 1 %, (NH4)2SO4 0.05 %, KCl 0.02 %, MgSO4·7H2O 0.01 %, calcium phytate 0.2 %, yeast extract 0.05 %, agar 1.5 % (w/v), MnSO4 5 mg l−1, FeSO4 5 mg l−1). The presence of clear zones around the bacterial colonies after incubation for 10 days at 32 °C was indicative of positive organic phosphate mineralization. The ratio between the diameters of each halo and the corresponding colony was determined as described by Hankin and Anagnostakis (1975).
Statistical analysis
The phosphate mineralization/solubilization within the L. lactis strains was compared with one-way ANOVA followed by Tukey’s pairwise test. In case of unequal variances Welch F test was used, instead of ANOVA. All statistical analyses were computed using PAST software (Hammer et al. 2001).
Results
Eight different bacterial strains (as determined by BOX-PCR) previously isolated from L. sidoides stems and molecularly identified as L. lactis (rrs gene fragment of about 800 bp; da Silva et al. 2013) were selected among the other isolates based on their potential for being GRAS. In this study, a phylogenetic tree based on their rrs gene sequences (approximately 1400 bp) was constructed and their previous identification was confirmed (Fig. 1).
Fig. 1.

Phylogenetic tree based on the rrs gene sequence (~1400 bp) showing the relationship between the Lactococcus strains studied here and the other members of the genus. The tree was constructed based on Jukes–Cantor distance using the neighbor joining method. Bootstrap analyses were performed with 1000 repetitions and only values higher than 50 % are shown. The Genbank accession numbers of the Lactococcus species are shown in parentheses
To test the P solubilization capacity of the eight selected strains, two chemically defined sources—calcium phosphate and aluminium phosphate—were first used (Fig. 2). All strains were able to solubilize calcium phosphate, varying from less than 2 to 31 % of this P source (strains 006.29 and 003.41, respectively). The other six strains tested showed intermediate results (18–29 %). The difference observed among the solubilization percentages obtained for strains 104.3, 006.31, 006.27, 006.30 and 104.7 were not statistically significant based on Tukey’s pairwise test (p < 0.05). Oppositely, either low solubilization percentages or no solubilization capacity (strains 006.29 and 006.30) were observed when aluminium phosphate was used as an inorganic P source (Fig. 2). The best result was observed with strain 104.3 which solubilized approximately 5 % of the phosphate available (Fig. 2).
Fig. 2.
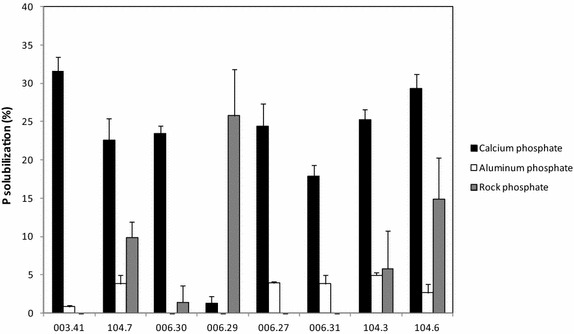
Percentages of solubilized phosphate using different P sources (calcium phosphate, aluminium phosphate and rock phosphate)
Rock phosphate, a mined rock that contains high amounts of phosphate bearing minerals, was also used to test the ability of the eight strains to solubilize P. In general, the strains tested were more efficient in solubilizing rock phosphate than aluminium phosphate, but less efficient when compared to calcium phosphate solubilization (Fig. 2). Surprisingly, the highest percentage (approximately 26 %) of rock P solubilized was achieved by the strain 006.29, which was unable to solubilize aluminium phosphate and presented a low solubilization percentage when calcium phosphate was used as a P source. Oppositely, the strains 003.41, 006.27 and 006.31 were unable to solubilize rock phosphate (Fig. 2).
One of the most important mechanisms responsible for inorganic phosphate solubilization is the production of organic acids by bacterial P solubilizers. To detect the presence of acids during calcium phosphate solubilization, pH was measured after 10 days of culture incubation (just before the P solubilization measurements). The initial pH of the medium varied from 6.5 to 7.0. No straight correlation could be observed between acid production and calcium phosphate solubilization. For example, strains 003.41 and 006.29 showed the highest (315.6 mg l−1) and the lowest (13.3 mg l−1) phosphate solubilization values, respectively, but the pH values were almost the same as in the beginning of the culturing procedure in both strains (Fig. 3). A decrease in the pH values was observed for the remaining strains tested and the calcium phosphate solubilization values varied among them (Fig. 3).
Fig. 3.
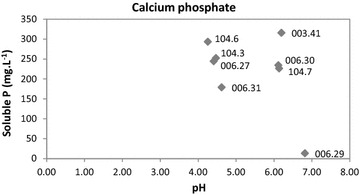
Correlation between the percentage of phosphate solubilized and the pH values of the media after 10 days of culture incubation. Calcium phosphate was used as the P source
Phosphate mineralization is another mechanism responsible for P availability in soil besides phosphate solubilization. Therefore, a semi-quantitative method using an organic chemical defined P source (calcium phytate) was used to test the phosphate mineralization capacity of the eight L. lactis strains. The majority of the strains tested (7 strains) showed phosphate mineralization activity, excepting strain 003.41. Some phosphate mineralizing strains showed differences in their phosphate mineralization capacity, which were statistically significant based on Tukey’s pairwise test (p < 0.05) (Fig. 4).
Fig. 4.
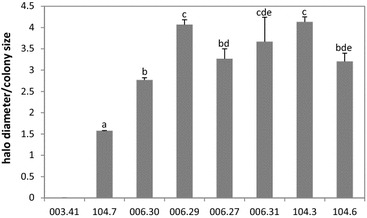
Phosphate mineralization activity of the different L. lactis strains determined by the semi-quantitative method described in the materials and methods. Calcium phytate was used as the P source. Different letters indicate significant differences of means in pairwise comparisons (Tukey test; p < 0.05)
Poultry litter, a complex P source containing inorganic and organic phosphates, was also included here in order to test the mineralization/solubilization activity of the eight selected strains. The highest activities were obtained with strains 006.30, 006.29, 006.27, 006.31 and 104.6 (Fig. 5). Based on Tukey’s pairwise test, no significant differences were found among these five strains and also among the remaining three strains—003.41, 104.7 and 104.3 (Fig. 5).
Fig. 5.
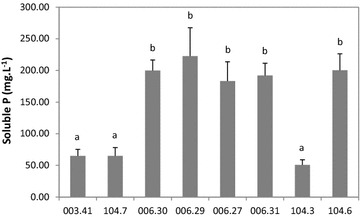
Phosphate mineralization/solubilization activity of the different L. lactis strains tested as described by Nautiyal (1999). Poultry litter was used as the P source. Different letters indicate significant differences of means in pairwise comparisons (Tukey test; p < 0.05)
The mineralization activity can be associated with the presence of enzymes such as phytase and/or alkaline phosphatase. The presence of genes coding for these proteins were searched within the genomes of the studied strains. When specific primers were used for phytase coding genes, no PCR products were observed. On the other hand, gene sequences related to alkaline phosphatase (phoA and phoD) could be detected in the majority of the strains (excepting strain 006.29) with identities varying from 67 to 88 %.
Discussion
Different endophytic bacteria are reported to increase the solubility of phosphorus and thus promote the growth of the host plants (Ryan et al. 2008). In this study, it was demonstrated for the first time that strains of Lactococcus lactis, a well-established group of GRAS bacteria, previously isolated from L. sidoides inner tissues were capable of mineralizing and/or solubilizing different P sources in vitro.
Strains belonging to the genera Pseudomonas and Bacillus are considered the most frequently found bacteria showing the capability of mineralizing and/or solubilizing phosphate in association with plants (Sharma et al. 2013). However, other genera such as Rhizobium (Sridevi et al. 2007), Azotobacter (Kumar et al. 2001), Enterobacter, Pantoea, Klebsiella (Chung et al. 2005), among other bacteria have also been reported as phosphate solubilizers/mineralizers. The few studies demonstrating the presence of Lactococcus able to mineralize and/or solubilize phosphate using a cultivation-dependent approach could be explained by either the low number of strains found inside the plants (or in the rhizosphere) or their slower growth compared to other bacterial genera. Inside the L. sidoides plants, Gammaproteobacteria appear to predominate which made it difficult to recover members of the bacterial community found in low numbers such as L. lactis (da Silva et al. 2013).
The eight L. lactis strains tested here were able to solubilize calcium phosphate and to solubilize/mineralize phosphate from poultry litter, a complex P source containing inorganic and predominantly organic P. Moreover, only one strain tested did not show any phosphate mineralization activity using calcium phytate as the organic P source. In fact, it has been previously demonstrated that other lactic acid bacteria isolated from sourdoughs exhibited a considerable phytate degrading capacity (De Angelis et al. 2003). The broad spectrum of phosphate utilization presented by these strains make them attractive as candidates for potential use in different types (alkaline, acidic or organic-rich) of soil, as suggested by Bashan et al. (2013). In addition, lactic acid produced by Lactococcus may have antimicrobial activity, exerting positive effects on plant growth indirectly. However, the isolates should be tested on a model plant as the ultimate test for potential phosphate solubilization (or mineralization) and/or for other plant growth promoting characteristics.
Inorganic phosphate solubilization by bacteria occurs mainly by the production of low molecular weight organic acids (Kpomblekou and Tabatabai 1994), and the lowering in pH of the medium suggests the release of these organic acids (Maliha et al. 2004). In this study, this was not the case for the strain 003.41 which showed the highest phosphate solubilization value (315.6 mg l−1) and a stable pH during the culturing procedure. Chelation of cations (Fe, Al and Ca) bound to phosphate, converting it into soluble forms may be other mechanism used for the release of phosphate (Sharma et al. 2013).
Organic phosphate mineralization by bacteria takes place due to the presence of particular enzymes. Plants are not able to acquire P directly from phytate (Richardson and Simpson 2011); therefore, phytase-producing microorganisms are the key drivers in regulating the mineralization of phytate in soil. In this study, none of the L. lactis strains tested seems to harbor phytase coding genes, as no PCR products were observed when specific primers were used. It is also possible that the primers used were not universal enough to target the phytase coding genes of the L. lactis strains studied here. Oppositely, gene sequences related to alkaline phosphatase (phoA and phoD) could be detected in seven of the strains tested. Alkaline phosphatases are more abundant in neutral and alkaline soils, and plants rarely produce large quantities of these enzymes (Sharma et al. 2013). Therefore, association of the L. lactis strains tested here with plants may be a potential niche for their use. Nevertheless, the presence of alkaline phosphatases coding genes only suggests that this can be one of the mechanisms of phosphate mineralization and further studies are necessary.
In conclusion, eight GRAS L. lactis strains are now available for future research to deeply understand their contribution to the cycling of P in soil–plant systems and for their eventual application under field conditions. The results presented here are of great importance as a first step for the development of more safe bioinoculants capable of mineralizing/solubilizing phosphate for the benefit of plants.
Acknowledgements
This study was supported by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ).
Competing interests
The authors declare that they have no competing interests.
Footnotes
Jackeline Rossetti Mateus de Lacerda and Thais Freitas da Silva have contributed equally to this study
Contributor Information
Jackeline Rossetti Mateus de Lacerda, Email: jacky.rossetti@gmail.com.
Thais Freitas da Silva, Email: thaisfs.bio@gmail.com.
Renata Estebanez Vollú, Email: revollu@yahoo.com.br.
Joana Montezano Marques, Email: jomonteza@yahoo.com.br.
Lucy Seldin, Phone: +55 21 39386741, Phone: +55 21 999897222, Email: lseldin@micro.ufrj.br.
References
- Alemayehu D, Hannon JA, McAuliffe O, Ross RP. Characterization of plant-derived lactococci on the basis of their volatile compounds profile when grown in milk. Int J Food Microbiol. 2014;172:57–61. doi: 10.1016/j.ijfoodmicro.2013.11.024. [DOI] [PubMed] [Google Scholar]
- Altier N, Beyhaut E, Pérez C. Bacteria in agrobiology: crop productivity. Berlin: Springer; 2013. pp. 167–184. [Google Scholar]
- Balemi T, Negisho K. Management of soil phosphorus and plant adaptation mechanisms to phosphorus stress for sustainable crop production: a review. J Soil Sci Plant Nutr. 2012;12:547–561. doi: 10.4067/S0718-95162012000200005. [DOI] [Google Scholar]
- Bashan Y, Kamnev AA, de Bashan LE. A proposal for isolating and testing phosphate-solubilizing bacteria that enhance plant growth. Biol Fertil Soils. 2013;49:1–2. doi: 10.1007/s00374-012-0756-4. [DOI] [Google Scholar]
- Beneduzi A, Moreira F, Costa PB, Vargas LK, Lisboa BB, Favreto R, Baldani JI, Passaglia LMP. Diversity and plant growth promoting evaluation abilities of bacteria isolated from sugarcane cultivated in the South of Brazil. Appl Soil Ecol. 2013;63:94–104. doi: 10.1016/j.apsoil.2012.08.010. [DOI] [Google Scholar]
- Chen W-M, Tang Y-Q, Mori K, Wu X-L. Distribution of culturable endophytic bacteria in aquatic plants and their potential for bioremediation in polluted waters. Aquat Biol. 2012;15:99–110. doi: 10.3354/ab00422. [DOI] [Google Scholar]
- Chung H, Park M, Madhaiyan M, Seshadri S, Song J, Cho H, Sa T. Isolation and characterization of phosphate solubilizing bacteria from the rhizosphere of crop plants of Korea. Soil Biol Biochem. 2005;37:1970–1974. doi: 10.1016/j.soilbio.2005.02.025. [DOI] [Google Scholar]
- Cock LS, de Stouvenel AR (2006) Lactic acid production by a strain of Lactococcus lactis subs lactis isolated from sugar cane plants. Eletron J Biotechnol 9:40–45. doi:10.2225/vol9-issue1-fulltext-10
- da Silva TF, Vollú RE, Jurelevicius D, Alviano DS, Alviano CS, Blank AF, Seldin L. Does the essential oil of Lippia sidoides Cham. (pepper-rosmarin) affect its endophytic microbial community? BMC Microbiol. 2013;13:29. doi: 10.1186/1471-2180-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis M, Gallo G, Corbo MR, McSweeney PLH, Faccia M, Giovine M, Gobbetti M. Phytase activity in sourdough lactic acid bacteria: purification and characterization of a phytase from Lactobacillus sanfranciscensis CB1. Food Microbiol. 2003;87:259–270. doi: 10.1016/S0168-1605(03)00072-2. [DOI] [PubMed] [Google Scholar]
- Dobbelaere S, Vanderleyden J, Okon Y. Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit Rev Plant Sci. 2003;22:107–149. doi: 10.1080/713610853. [DOI] [Google Scholar]
- Golomb BL, Marco ML. Lactococcus lactis metabolism and gene expression during growth on plant tissues. J Bacteriol. 2015;197:371–381. doi: 10.1128/JB.02193-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönemeyer JL, Burbano CF, Hurek T, Reinhold-Hurek B. Isolation and characterization of root-associated bacteria from agricultural crops in the Kavango region of Namibia. Plant Soil. 2012;356:67–82. doi: 10.1007/s11104-011-0798-7. [DOI] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4:9–18. [Google Scholar]
- Hankin L, Anagnostakis SL. The use of solid media for detection of enzymes production by fungi. Mycologia. 1975;67:597–607. doi: 10.2307/3758395. [DOI] [Google Scholar]
- Huang H, Shi P, Wang Y, Luo H, Shao N, Wang G, Yang P, Yao B. Diversity of beta-propeller phytase genes in the intestinal content of grass carp provides insight into the release of major phosphorus from phytate in nature. Appl Environ Microb. 2009;75:1508–1516. doi: 10.1128/AEM.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kpomblekou K, Tabatabai MA. Effect of organic acids on release of phosphorus from phosphate rocks. Soil Sci. 1994;158:442–453. doi: 10.1097/00010694-199415860-00006. [DOI] [Google Scholar]
- Kumar V, Behl RK, Narula N. Establishment of phosphate-solubilizing strains of Azotobacter chroococcumin the rhizosphere and their effect on wheat cultivars under greenhouse conditions. Microbiol Res. 2001;156:87–93. doi: 10.1078/0944-5013-00081. [DOI] [PubMed] [Google Scholar]
- Maliha R, Samina K, Najma A, Sadia A, Farooq L. Organic acids production and phosphate solubilization by phosphate solubilizing microorganisms under in vitro conditions. Pak J Biol Sci. 2004;7:187–196. doi: 10.3923/pjbs.2004.187.196. [DOI] [Google Scholar]
- Massol-Deya AA, Odelson DA, Hickey RF, Tiedje JM. Bacterial community fingerprinting of amplified 16S and 16-23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA) In: Akkermans ADL, van Elsas JD, Bruijn FJ, editors. Molecular microbiology ecology manual 3.3.2. Dordrecht: Kluwer; 1995. [Google Scholar]
- Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- Murphy J, Riley JP. A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta. 1962;27:31–36. doi: 10.1016/S0003-2670(00)88444-5. [DOI] [Google Scholar]
- Nautiyal CS. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett. 1999;170:265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x. [DOI] [PubMed] [Google Scholar]
- Procópio REL, Araújo WL, Maccheroni W, Azevedo JL. Characterization of an endophytic bacterial community associated with Eucalyptus spp. Genet Mol Res. 2009;8:1408–1422. doi: 10.4238/vol8-4gmr691. [DOI] [PubMed] [Google Scholar]
- Richardson AE, Simpson RJ. Soil microorganisms mediating phosphorus availability. Plant Physiol. 2011;156:989–996. doi: 10.1104/pp.111.175448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado AS, Azevedo FS, Cruz DWG, Seldin L. Phenotypic and genetic diversity of Paenibacillus azotofixans strains isolated from rhizoplane or rhizosphere of different grasses. J Appl Microbiol. 1998;84:216–226. doi: 10.1046/j.1365-2672.1998.00332.x. [DOI] [Google Scholar]
- Rosenblueth M, Martínez-Romero E. Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact. 2006;19:827–837. doi: 10.1094/MPMI-19-0827. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Germaine K, Franks A, Ryan DJ, Dowling DN. Bacterial endophytes: recent developments and applications. FEMS Microbiol Lett. 2008;278:1–9. doi: 10.1111/j.1574-6968.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- Sakurai M, Wasaki J, Tomizawa Y, Shinano T, Osaki M. Analysis of bacterial communities on alkaline phosphatase genes in soil supplied with organic matter. Soil Sci Plant Nutr. 2008;54:62–71. doi: 10.1111/j.1747-0765.2007.00210.x. [DOI] [Google Scholar]
- Salama MS, Musafija-Jeknic T, Sandine WE, Giovannoni SJ. An ecological study of lactic acid bacteria: isolation of new strains of Lactococcus including Lactococcus lactis subsp. cremoris. J Dairy Sci. 1995;78:1004–1017. doi: 10.3168/jds.S0022-0302(95)76716-9. [DOI] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sharma SB, Sayyed RZ, Trivedi MH, Gobi TA. Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus. 2013;2:587. doi: 10.1186/2193-1801-2-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers E, Amke A, Croonenborghs A, van Overbeek LS, Vanderleyden J (2007) Lactic acid bacteria in organic agricultural soils. Poster presentation, Montpellier, Plant Research International
- Sridevi M, Mallaiah KV, Yadav NCS. Phosphate solubilization by Rhizobium isolates from Crotalaria species. J Plant Sci. 2007;2:635–639. doi: 10.3923/jps.2007.635.639. [DOI] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP. Symbiotic nitrogen fixation and phosphorus acquisition: plant nutrition in a world of declining renewable resources. Plant Physiol. 2001;127:390–397. doi: 10.1104/pp.010331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Ehde-Stone C, Allan D. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003;157:423–447. doi: 10.1046/j.1469-8137.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- Velly H, Renault P, Abraham AL, Loux V, Delacroix-Buchet A, Fonseca F, Bouix M. Genome sequence of the lactic acid bacterium Lactococcus lactis subsp. lactis TOMSC161, isolated from a nonscalded curd pressed cheese. Genome Announc. 2014;2(6):e01121-14. doi: 10.1128/genomeA.01121-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LJH, Davey GP, Heap HA, Kelly WJ. Lactococcus lactis. In: Roginski H, Fuquay JW, Fox PF, editors. Encyclopedia of dairy science. London: Elsevier; 2002. pp. 1511–1516. [Google Scholar]
- Wessels S, Axelsson L, Bech Hansen E, De Vuyst L, Laulund S, Lähteenmäki L, Lindgren S, Mollet B, Salminen S, von Wright A. The lactic acid bacteria, the food chain, and their regulation. Trends Food Sci Technol. 2004;15:498–505. doi: 10.1016/j.tifs.2004.03.003. [DOI] [Google Scholar]


