Abstract
In vivo bioluminescent imaging (BLI) permits the visualization of engineered bioluminescence from living cells and tissues to provide a unique perspective toward the understanding of biological processes as they occur within the framework of an authentic in vivo environment. The toolbox of in vivo BLI includes an inventory of luciferase compounds capable of generating bioluminescent light signals along with sophisticated and powerful instrumentation designed to detect and quantify these light signals non-invasively as they emit from the living subject. The information acquired reveals the dynamics of a wide range of biological functions that play key roles in the physiological and pathological control of disease and its therapeutic management. This mini review provides an overview of the tools and applications central to the evolution of in vivo BLI as a core technology in the preclinical imaging disciplines.
Keywords: bioluminescence, optical imaging, in vivo imaging, luciferase, luciferin
Introduction
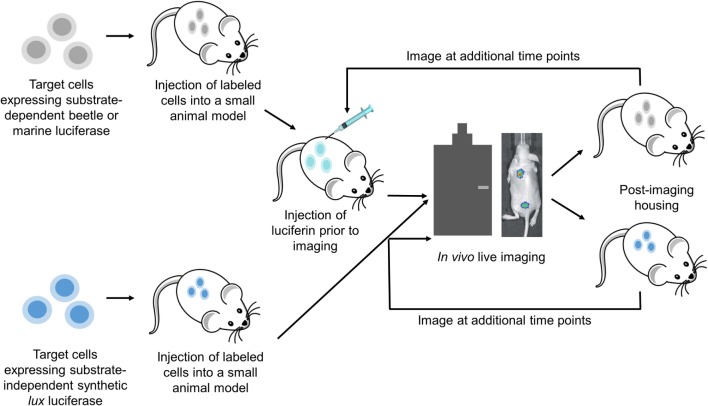
In vivo bioluminescent imaging (BLI) enables the visualization of biological processes as they occur within the living subject. The information obtained is unprecedented in its ability to elucidate biology beyond the boundaries of the conventional in vitro assay, where the complex interactions of a living system are all but ignored. In vivo BLI uses the luciferase family of proteins to create signature bioluminescent outputs that are then externally captured by advanced cameras (Figure 1). Luciferases operate in tandem with their luciferin substrates to generate light via an oxidation decarboxylation reaction that forms an excited state intermediate that releases energy in the form of photons as it returns to its ground state. In nature, bioluminescence is generated by various bacteria, fungi, protozoa, dinoflagellates, and higher-order terrestrial and marine organisms, with the firefly being the most recognized example. Molecular biology has enabled the genes involved in bioluminescent light reactions to be isolated, manipulated, and reapplied toward applications, such as in vivo BLI, where bioluminescence as an optical emission signature exhibits certain unique imaging advantages. Among the most critical is a superior signal-to-noise ratio due to cells and tissues emitting virtually no intrinsic bioluminescence, thus effectively eliminating background interference when probing for a bioluminescent signal within the intricate milieu of a living entity. However, detecting bioluminescence at depths beyond a few centimeters inside of a living animal remains challenging because light signals must be obtained and evaluated after passing through host tissue that absorbs, attenuates, and scatters their emissions (1). This has currently constrained in vivo BLI to small animal models, such as mice and rats, with service primarily limited to preclinical imaging applications. This mini review provides an overview of the luciferases currently being applied in in vivo imaging applications along with the toolbox of approaches that continue to expand the capabilities of in vivo BLI.
Figure 1.
In vivo BLI uses advanced camera imaging systems to visualize live animal subjects as they express bioluminescence from targeted cells and tissues, thereby allowing fundamental biological processes to be monitored non-invasively. Advances in the in vivo BLI field have created luciferase proteins with expanded wavelength emission profiles, stronger and more stable signal generation, substrate-independent real-time expression, and proximity-based expression characteristics that are providing innovative tools for preclinical diagnostics, drug discovery, and toxicology research.
Luciferases for BLI Applications
Luciferases applied in in vivo BLI include those derived from beetles, bacteria, and various marine species (Table 1), with the firefly luciferase (FLuc) being the most widely used. FLuc requires d-luciferin (a heterocyclic carboxylic acid), ATP, and molecular oxygen for light production. At a pH between 7.5 and 8.5, FLuc catalyzes the reaction between d-luciferin and ATP to form luciferyl-adenylate, which in the presence of oxygen then undergoes an oxidative decarboxylation reaction to form CO2, AMP, and oxyluciferin. Initially formed as an excited state intermediate, oxyluciferin quickly returns to its ground state and releases energy in the form of light (2). In addition to FLuc, other beetle luciferases, such as the green (CBG) and red (CBR) click beetle luciferases, emerald luciferase (ELuc), and stable red luciferase (SRL), also utilize d-luciferin as their substrate. However, despite this common substrate, these luciferases emit light of different wavelengths (Table 1).
Table 1.
The inventory of luciferases for in vivo BLI applications.
| Luciferase | Luciferin substrate | Peak emission (nm) (25°C) | Reference | Examples of in vivo BLI applications |
|---|---|---|---|---|
| Beetle luciferases | ||||
| Photinus pyralis (firefly; FLuc, ffluc, or luc) and its enhanced variants (effLuc, luc2) | d-luciferin | 560 | (3–5) | • Detection of cancer cells and evaluation of tumor treatment (3, 5) • Imaging of neural precursor cell migration to glioma tumor (6) • Imaging of T cell migration to tumors (7) • Split luciferase assay to image apoptosis in response to chemotherapy and radiotherapy in a glioma model (8) |
| Red-shifted firefly luciferase PRE9 | d-luciferin | 620 | (9) | |
| Pyrearinus termitilluminans (click beetle emerald; ELuc) | d-luciferin | 538 | (10) | |
| Pyrophorus plagiophthalamus (click beetle red; CBR) | d-luciferin | 615 | (11) | |
| Pyrophorus plagiophthalamus (click beetle green; CBG) | d-luciferin | 540 | (12) | |
| Phrixothrix hirtus (railroad worm stable luciferase red; SLR) | d-luciferin | 630 | (13) | |
| Bacterial luciferases | ||||
| Aliivibrio fischeri, Vibrio harveyi, and Photorhabdus luminescens lux for bacterial expression | FMNH2 + long-chain aliphatic aldehyde | 490 | (14, 15) | • Simultaneous imaging of lux-labeled bacterial trafficking to FLuc-tagged tumor (14) • Substrate-free imaging of human cells in vivo (16) • Substrate-free real-time imaging of bacterial infection of human cells (17) |
| Synthetic lux for mammalian expression | FMNH2 + long-chain aliphatic aldehyde (self-supplied by luxCDEfrp) | 490 | (18, 19) | |
| Marine luciferases | ||||
| Renilla reniformis (RLuc) and its enhanced variants (RLuc8 and RLuc8.6–535) | Coelenterazine | 482–535 | (20–22) | • RLuc multiplexed with FLuc to monitor tumor regression in response to therapeutic genes delivery by neural precursor cells (6) • VLuc multiplexed with FLuc and RLuc to track delivery of therapeutic genes into brain tumor (23) • BRET assay to detect tumor metastasis (24) |
| Gaussia princeps (GLuc) and its mutants (I90L, 8990, 90115, Monsta, etc.) | Coelenterazine | 482–503 | (25, 26) | |
| Metridia longa (MLuc7, MLuc164) | Coelenterazine | 486–498 | (27, 28) | |
| Aequorea victoria (aequorin) | Coelenterazine | 470 | (29) | |
| Vargula hilgendorfii (VLuc) | Vargulin | 462 | (30) | |
| Cypridina noctiluca (CLuc) | Cypridina | 460 | (31) | |
| Oplophorus gracilirostris (NanoLuc) | Furimazine | 460 | (32) | |
| Benthosema pterotum (BP) | Coelenterazine | 475 | (7) | |
Within the bacterial genera, bioluminescence from Photobacterium and Aliivibrio/Vibrio are typically applied. These systems encode the lux gene cassette, which includes the luxAB genes encoding a heterodimeric bacterial luciferase and the luxCDE genes encoding a fatty acid synthetase/reductase complex that generates a long-chain fatty aldehyde substrate from endogenous intracellular metabolites. Marine bioluminescent bacteria also possess a luxG/frp gene that encodes a flavin reductase that recycles reduced flavin mononucleotide (FMNH2) for the luciferase reaction. Similar to beetle luciferases, bacterial luciferase generates light in an ATP-dependent manner in the presence of long-chain aldehyde, FMNH2, and molecular oxygen (33). Distinctive to bacterial bioluminescence is the ability of cells expressing the full luxCDABE gene cassette to produce light autonomously without the need for exogenous luciferin by self-supplying the aldehyde and FMNH2 substrates. While bacterial bioluminescence is traditionally employed to label bacterial pathogens for in vivo real-time infection tracking due to its prokaryotic origin, the lux cassette has been synthetically optimized for autonomous bioluminescent expression in eukaryotic organisms, allowing substrate-free in vivo imaging of mammalian cells (18).
The remaining luciferases include those isolated from marine invertebrates. Unlike other luciferases, marine luciferases utilize their luciferin substrate and molecular oxygen to generate light in an ATP-independent fashion. There is also no common luciferin substrate for all marine luciferases. While Gaussia (GLuc), Renilla (RLuc), and Metridia (MLuc) luciferases share the same substrate coelenterazine, Cypridina (CLuc) and Vargula (VLuc) luciferases catalyze their reactions using cypridina and vargulin, respectively. Some marine luciferases, including GLuc and MLuc, are naturally secreted outside of the cell, thus allowing bioluminescent detection without cell lysis (25, 27).
The Toolbox of BLI Approaches
Mutated and Synthetic Luciferase
The high utility and common limitations shared by luciferases has made them especially attractive targets for synthetic modification. One of the first major synthetic luciferase modifications was a polymutated variant of RLuc, which incorporated eight independent single amino acid changes to increase protein stability and improve light output (20). This mutated variant allowed for improved function during serum exposure in small animals, provided a facile means for conjugating luciferase protein to various ligands (34), and has recently been used for conjugation to immunoglobulin G proteins for visualizing antigen–antibody reactions (35).
Perhaps the most valuable synthetic luciferase modifications for in vivo BLI have been those that shift the luciferases’ emission signal further into the red spectrum, thereby improving signal penetration through living tissue. To overcome the naturally blue-shifted emission wavelength of RLuc, Loening et al. (21) generated a library of active site mutations and identified multiple variants with emission spectra peaks ranging from 475 to 547 nm. Branchini et al. (36) employed a similar approach with FLuc that shifted its native 557 nm emission peak to 617 nm.
Leveraging the proteomic sequences of known luciferases, Kim and Izumi (37) applied a consensus sequence-driven mutagenesis strategy to identify amino acids common to copepod luciferases and arranged these sequences under the constraints suggested by a statistical coupling analysis (38) to mimic the natural evolutionary constraints of the proteins. Using this strategy, they designed artificial luciferases (ALucs) that retained favorable emission wavelengths in the 515–548 nm range. This strategy enables the synthetic generation of alternative classes of luciferases for the continued expansion of BLI beyond those found in nature.
Synthetic Luciferin Analogs
For in vivo BLI to occur under the majority of luciferase/luciferin combinations, the luciferin substrate must first be injected into the animal and then diffuse to where the luciferase-expressing cells are located. This series of events can be challenging. The biodistribution of luciferin substrates in small animals is not homogenous, individual eukaryotic cells are limited in their ability to freely take up substrate, and the mere presence of the luciferin substrate represents a chemical contaminant that may unknowingly introduce experimental artifacts and/or toxicological side effects. Synthetic luciferins with properties better tuned to the in vivo environment are being developed to address some of these problems. Craig et al. (39) created some of the early chemically modified (esterified) d-luciferin analogs designed for improved cellular uptake kinetics and consequent near sixfold increases in bioluminescence output. However, for in vivo imaging, the focus has transitioned to red-shifting the emission wavelength for improved tissue penetration. This has resulted in aminoluciferin analogs, such as cyclic aminoluciferins and seleno-d-aminoluciferins, with wavelength emissions around 600 nm (40). Unfortunately, the majority of these analogs yield lower light intensities than conventional luciferin, although the CycLuc1 substrate reported by Evans et al. (41) does demonstrate superior photon flux under non-saturating substrate conditions. Infra-luciferin (λmax = 706 nm), a π-conjugated analog (λmax = 675 nm), and CycLuc10 (λmax = 648 nm) have successfully shifted their wavelengths even further into the far-red regions (42–44). However, maintaining elevated photon yields remains challenging, although the increased efficiency of signal penetration at these longer wavelengths does offer heightened resolution.
Multiplexed BLI
In multiplexed BLI, the subject is tagged with multiple luciferases that utilize different substrates, which are injected sequentially to trigger each bioluminescent signal to enable simultaneous monitoring of multiple biological processes. Common luciferase combinations include d-luciferin-activated beetle luciferase and coelenterazine-activated marine luciferase. The selectivity and specificity of luciferin substrates ensures minimal cross talk. This approach has been applied to monitor gene expression and promoter activities (45), mesenchymal stem cell differentiation (46), and cell migration and tumor apoptosis (6, 47). A triple BLI system consisting of the FLuc/d-luciferin, GLuc/coelenterazine, and VLuc/vargulin pairs has also been reported for simultaneous monitoring of three distinct biological events in an orthotopic brain tumor model (23). However, using multiple substrates inevitably introduces biases due to differential substrate biodistribution and uptake in animal tissues. Meanwhile, multiple substrate injections can be stressful for the animal and introduce potential operational errors. These drawbacks can be alleviated by utilizing a single substrate to simultaneously initiate multiple luciferases that emit light of separable colors. A common approach is to employ one luciferase with a green emission spectrum and a second luciferase emitting a more red-shifted wavelength. Upon a single-substrate application, both luciferases are activated, and the resulting green and red light signal can be spectrally resolved using appropriate detection systems. Beetle luciferases activated by d-luciferin, including FLuc, CBG, and CBR, are currently the most common reporters used for single-substrate multicolor BLI applications (13, 48–51). However, for in vivo applications, this arrangement is still constrained due to increased absorption and attenuation of the shorter wavelength (green) light compared to that of the red-shifted signal in animal tissues, which introduces potential detection biases.
Split Luciferases
Split luciferases, or luciferase fragments, are unique tools for probing protein–protein interactions. Instead of using the complete enzyme, the luciferase protein is split into a C-terminus fragment and an N-terminus fragment that are not capable of catalyzing the bioluminescent reaction on their own. In split luciferase complementation assays, each luciferase fragment is attached to each partner of the interacting peptides, domains, and/or full proteins. Upon interaction of the proteins of interest, the luciferase fragments are brought to a close proximity to form a complete and functional enzyme that produces bioluminescence when a luciferin substrate is available (52). Luciferase fragment complementation imaging can be designed to directly identify interacting protein pairs (53) and to indirectly report protein–protein interactions induced by various biological processes, such as binding of intracellular messengers (e.g., cyclic AMP and Ca2+) (54, 55), protein kinase activities (56, 57), caspase-3-mediated apoptosis (8), and activation and/or inhibition of disease-related cell signaling pathways (58, 59). Multiple luciferases can also be used for multiplexed examination of complex interactions involving multiple protein partners simultaneously in the same subject (12, 60). For improved in vivo applications, novel split sites and modifications of the luciferase enzyme are being continuously identified to enhance their characteristics (i.e., decreased basal activity, increased specificity, improved signal-to-noise ratio) (61).
Caged Luciferin
The caged luciferin reporter system uses a luciferin substrate that has been modified such that it cannot interact with its complementary luciferase to generate bioluminescence until an enzymatic cleavage event occurs (62). Lugal (d-luciferin-O-β-galactoside) is one example of a caged luciferin that only actively interacts with FLuc upon removal of its galactoside moiety by β-galactosidase. Thus, the bioluminescent reporter cell remains “dark” even after the addition of the Lugal substrate, with the co-addition of β-galactosidase being required to ultimately initiate light emission. Using this strategy, one cell (the reporter cell) can be designed to express FLuc, while another cell (the activator cell) expresses β-galactosidase. If the two cells are in close proximity, then the β-galactosidase released from the activator cell cleaves the Lugal to initiate light emission from the reporter cell. As the distance between these two cells increases, the intensity of the light response correspondingly decreases. For example, this has enabled in vivo bioluminescent visualization of tumor metastasis in mouse models, where β-galactosidase-expressing hematopoietic cells distributed throughout a mouse functionally activated luciferase-expressing breast cancer cells that had metastasized from a tumor implant (63). Due to Lugal being somewhat non-selective under biological conditions, other caged luciferin substrates have been developed that operate under a number of more selective enzymatic reaction schemes (β-lactamase, alkaline phosphatase, nitroreductase) (64, 65).
Bioluminescence Resonance Energy Transfer
Bioluminescence resonance energy transfer (BRET) pairs together two chromophores such that the emission spectra of one (the bioluminescent donor) activates the excitation spectra of the other (the fluorescent acceptor) (66). In its earliest configuration, it exploited the 482-nm bioluminescent emission of Renilla luciferase to activate an enhanced yellow fluorescent protein (EYFP), thereby “switching” the blue-green color of RLuc to a 527-nm yellow emission (67). The switching only occurs if the donor and recipient chromophores are properly oriented and situated in close proximity (≤10 nm apart), which enables BRET’s primary application as an indicator of protein–protein interactions via the attachment of the donor chromophore to one protein and the recipient chromophore to the other protein (68). BRET has since evolved to include other bioluminescent/fluorescent pairings, which, for in vivo applications, have centered on shifting the emission spectra more toward the red to far-red regions to improve tissue penetration (24, 69–71). BRET has also advanced beyond fluorescent proteins to include organic dye (72) and quantum dot conjugates (73).
Fluorescence by Unbound Excitation from Luminescence
Fluorescence by unbound excitation from luminescence (FUEL) is similar to BRET in that it uses the emission spectra of a bioluminescent donor to activate the excitation spectra of a fluorescent acceptor. However, whereas BRET requires the donor and acceptor to reside within an approximate 10 nm distance of each other, FUEL can theoretically occur at donor/acceptor distances separated by micrometers to centimeters (74, 75). FUEL takes advantage of the unfocused radiative dissemination of photons by luciferase-bearing entities to activate neighboring fluorescent light sources. In one of its earliest demonstrations, Escherichia coli cells expressing bacterial luciferase were placed in one quartz cuvette, while red-emitting quantum dots (QD705, Invitrogen) with overlapping excitation wavelengths were placed in a neighboring cuvette. Photons emitted by E. coli were shown to activate red-shifted fluorescence from QD705, with signal intensity being dependent on the distance separating the two cuvettes. Injection of bioluminescent bacteria and QD705 into mice showed similar activation of red fluorescence emission under in vivo BLI. FUEL may serve as a unique mechanism to gage coproximity of donors and acceptors, much like BRET, but across larger spans of space, for example, to discern interactions between tissues and organs separated on a mesoscopic scale.
Bioluminescence Assisted Switching and Fluorescence Imaging
Bioluminescence assisted switching and fluorescence imaging (BASFI) is another spin-off of BRET, wherein a bioluminescent donor activates a reversible photoswitchable fluorescent acceptor protein. Proof of concept for BASFI has been demonstrated using the pairing of the bioluminescent Rluc8 donor with the photoswitchable fluorescent protein DG1 acceptor (76). DG1 normally exists in its excited state, emitting green fluorescence at 450–550 nm, but can be switched to an off-state when exposed to wavelengths around 488 nm. A DG1–Rluc8 fusion construct was transfected into a human embryonic kidney cell, thereby endowing it with a green fluorescent phenotype. Exposing the cell to a 488 nm laser switched DG1 to its off-state, and the cell became “dark.” Addition of a coelenterazine methoxy substrate then activated Rluc8, whose 400 nm emission switched DG1 back to its on-state. In traditional BRET, this on-state is transient and short-lived. In BASFI, this on-state persists for as long as the donor bioluminescence is being provided, thereby enabling the accumulation of signal over time. This allows supply of the activation signal to be decoupled from measurement of the emission signal to potentially reduce background and increase sensitivity. BASFI still requires close association between the donor and acceptor (≤10 nm), so its primary application remains with studying protein–protein interactions.
Bioluminescent Enzyme-Induced Electron Transfer
The bioluminescent enzyme-induced electron transfer (BioLeT) concept uses luciferin analogs that have been modified to contain moieties of differing electron donating capacities and then using the ensuing electron transfer process as an on/off switch to modulate bioluminescent signal output. In its proof-of-concept format, aminoluciferin substrates were modified to contain benzene moieties of differing highest occupied molecular orbital (HOMO) energy levels (77, 78). Substrates containing benzene moieties with high HOMO energy levels, such as a diaminophenyl moiety, were shown to quench bioluminescence when added to a FLuc reaction, presumably due to the electron transfer process occurring much more rapidly than the light-emitting reaction. Substrates containing benzene moieties with low HOMO energy levels did not quench bioluminescent signal output. A diamino-phenylpropyl-aminoluciferin (DAL) substrate was ultimately developed as a BioLeT probe for the targeting of biological nitric oxide. Upon reaction with nitric oxide, the diaminophenyl moiety is converted into a benzotriazole moiety with a lower HOMO energy level, thus transitioning from minimal to a highly bioluminescent output in the presence of luciferase. The scheme was validated in vivo in a transgenic FLuc rat model intraperitoneally injected with DAL substrate followed by injection of a NOC7 compound that spontaneously released nitric oxide under physiological conditions. Nitric oxide accumulation was detected via increased bioluminescence emission as the diaminophenyl to benzotriazole conversion occurred within the rat. It is anticipated that the BioLeT process can be designed to target other biomolecules, such as singlet oxygen and metal ions, to assist in the real-time, non-invasive surveillance of a subject’s physiological state.
Conclusion
The superior signal-to-noise ratio due to the absence of intrinsic bioluminescence background in cells and animal tissues has made BLI an attractive tool for investigating biological processes as they occur in real-time in living animals. The past two decades have witnessed not only a bloom in the discovery and engineering of luciferases with improved expression and performance in mammalian cells but also the emergence and expansion of innovative applications of such luciferase reporters for in vivo imaging. Despite in vivo BLI currently being constrained to small animal models, it has increasingly become a promising tool in preclinical biomedical research to investigate real-time biological events in complex biological systems, and it is reasonable to expect that in vivo BLI will continue to play a crucial role in basic research, drug development, disease diagnosis, therapy management, and many other biomedical research and applications.
Author Contributions
TX and SR conceived, structured, and edited the mini review article. DC, WH, EM, and GS each wrote individual sections of the mini review article and critically revised it for intellectual content. All authors (TX, DC, WH, EM, GS, and SR) provided final approval of the version of the article submitted for publication, and they agreed to be accountable for all aspects of the work in regard to ensuring that questions pertaining to accuracy and/or integrity of any part of the work are appropriately investigated and resolved.
Conflict of Interest Statement
Drs. DC, GS, and SR have research-related financial interests in 490 BioTech, Inc. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewers (PW and ZC) and handling editor declared their shared affiliation, and the handling editor states that the process nevertheless met the standards of a fair and objective review.
Funding
The authors acknowledge research funding provided by the U.S. National Science Foundation under award numbers CBET-0853780, CBET-1530953, and CBET-1159344 and the U.S. National Institutes of Health under award numbers NIEHS-1R15ES023979-01, NIEHS-2R44ES022567-02, NIGMS-1R41GM116622-01, and NIGMS-1R43GM112241-01A1.
References
- 1.Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J Biomed Opt (2005) 10(4):41210. 10.1117/1.2032388 [DOI] [PubMed] [Google Scholar]
- 2.Fraga H, Fernandes D, Novotny J, Fontes R, da Silva JCG. Firefly luciferase produces hydrogen peroxide as a coproduct in dehydroluciferyl adenylate formation. Chembiochem (2006) 7(6):929–35. 10.1002/cbic.200500443 [DOI] [PubMed] [Google Scholar]
- 3.Rehemtulla A, Stegman LD, Cardozo SJ, Gupta S, Hall DE, Contag CH, et al. Rapid and quantitative assessment of cancer treatment response using in vivo bioluminescence imaging. Neoplasia (2000) 2(6):491–5. 10.1038/sj.neo.7900121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabinovich BA, Ye Y, Etto T, Chen JQ, Levitsky HI, Overwijk WW, et al. Visualizing fewer than 10 mouse T cells with an enhanced firefly luciferase in immunocompetent mouse models of cancer. Proc Natl Acad Sci U S A (2008) 105(38):14342–6. 10.1073/pnas.0804105105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JB, Urban K, Cochran E, Lee S, Ang A, Rice B, et al. Non-invasive detection of a small number of bioluminescent cancer cells in vivo. PLoS One (2010) 5(2):e9364. 10.1371/journal.pone.0009364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah K, Bureau E, Kim DE, Yang K, Tang Y, Weissleder R, et al. Glioma therapy and real-time imaging of neural precursor cell migration and tumor regression. Ann Neurol (2005) 57(1):34–41. 10.1002/ana.20306 [DOI] [PubMed] [Google Scholar]
- 7.Homaei AA, Mymandi AB, Sariri R, Kamrani E, Stevanato R, Etezad S-M, et al. Purification and characterization of a novel thermostable luciferase from Benthosema pterotum. J Photochem Photobiol B (2013) 125:131–6. 10.1016/j.jphotobiol.2013.05.015 [DOI] [PubMed] [Google Scholar]
- 8.Coppola JM, Ross BD, Rehemtulla A. Noninvasive imaging of apoptosis and its application in cancer therapeutics. Clin Cancer Res (2008) 14(8):2492–501. 10.1158/1078-0432.ccr-07-0782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang Y, Walczak P, Bulte JWM. Comparison of red-shifted firefly luciferase Ppy RE9 and conventional Luc2 as bioluminescence imaging reporter genes for in vivo imaging of stem cells. J Biomed Opt (2012) 17(1):016004. 10.1117/1.jbo.17.1.016004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakajima Y, Yamazaki T, Nishii S, Noguchi T, Hoshino H, Niwa K, et al. Enhanced beetle luciferase for high-resolution bioluminescence imaging. PLoS One (2010) 5(3):e10011. 10.1371/journal.pone.0010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang M, Anttonen KP, Cirillo SLG, Francis KP, Cirillo JD. Real-time bioluminescence imaging of mixed mycobacterial infections. PLoS One (2014) 9(9):e108341. 10.1371/journal.pone.0108341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villalobos V, Naik S, Bruinsma M, Dothager RS, Pan MH, Samrakandi M, et al. Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chem Biol (2010) 17(9):1018–29. 10.1016/j.chembiol.2010.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasunaga M, Nakajima Y, Ohmiya Y. Dual-color bioluminescence imaging assay using green- and red-emitting beetle luciferases at subcellular resolution. Anal Bioanal Chem (2014) 406(23):5735–42. 10.1007/s00216-014-7981-7 [DOI] [PubMed] [Google Scholar]
- 14.Cronin M, Akin AR, Collins SA, Meganck J, Kim JB, Baban CK, et al. High resolution in vivo bioluminescent imaging for the study of bacterial tumour targeting. PLoS One (2012) 7(1):e30940. 10.1371/journal.pone.0030940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y, Ramos RI, Cho JS, Donegan NP, Cheung AL, Miller LS. In vivo bioluminescence imaging to evaluate systemic and topical antibiotics against community-acquired methicillin-resistant Staphylococcus aureus-infected skin wounds in mice. Antimicrob Agents Chemother (2013) 57(2):855–63. 10.1128/aac.01003-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Close DM, Hahn RE, Patterson SS, Baek SJ, Ripp SA, Sayler GS. Comparison of human optimized bacterial luciferase, firefly luciferase, and green fluorescent protein for continuous imaging of cell culture and animal models. J Biomed Opt (2011) 16(4):047003. 10.1117/1.3564910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu T, Marr E, Lam H, Ripp S, Sayler G, Close D. Real-time toxicity and metabolic activity tracking of human cells exposed to Escherichia coli O157:H7 in a mixed consortia. Ecotoxicology (2015) 24(10):2133–40. 10.1007/s10646-015-1552-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu T, Ripp SA, Sayler GS, Close DM. Expression of a humanized viral 2A-mediated lux operon efficiently generates autonomous bioluminescence in human cells. PLoS One (2014) 9(5):e96347. 10.1371/journal.pone.0096347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Close DM, Patterson SS, Ripp SA, Baek SJ, Sanseverino J, Sayler GS. Autonomous bioluminescent expression of the bacterial luciferase gene cassette (lux) in a mammalian cell line. PLoS One (2010) 5(8):e12441. 10.1371/journal.pone.0012441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loening AM, Fenn TD, Wu AM, Gambhir SS. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Eng Des Sel (2006) 19(9):391–400. 10.1093/protein/gzl023 [DOI] [PubMed] [Google Scholar]
- 21.Loening AM, Wu AM, Gambhir SS. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat Methods (2007) 4(8):641–3. 10.1038/nmeth1070 [DOI] [PubMed] [Google Scholar]
- 22.Lorenz WW, Cormier MJ, Okane DJ, Hua D, Escher AA, Szalay AA. Expression of the Renilla reniformis luciferase gene in mammalian cells. J Biolumin Chemilumin (1996) 11(1):31–7. [DOI] [PubMed] [Google Scholar]
- 23.Maguire CA, Bovenberg MS, Crommentuijn MH, Niers JM, Kerami M, Teng J, et al. Triple bioluminescence imaging for in vivo monitoring of cellular processes. Mol Ther Nucleic Acids (2013) 2:e99. 10.1038/mtna.2013.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De A, Ray P, Loening AM, Gambhir SS. BRET3: a red-shifted bioluminescence resonance energy transfer (BRET)-based integrated platform for imaging protein-protein interactions from single live cells and living animals. FASEB J (2009) 23(8):2702–9. 10.1096/fj.08-118919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther (2005) 11(3):435–43. 10.1016/j.ymthe.2004.10.016 [DOI] [PubMed] [Google Scholar]
- 26.Kim SB, Suzuki H, Sato M, Tao H. Superluminescent variants of marine luciferases for bioassays. Anal Chem (2011) 83(22):8732–40. 10.1021/ac2021882 [DOI] [PubMed] [Google Scholar]
- 27.Markova SV, Golz S, Frank LA, Kalthof B, Vysotski ES. Cloning and expression of cDNA for a luciferase from the marine copepod Metridia longa – a novel secreted bioluminescent reporter enzyme. J Biol Chem (2004) 279(5):3212–7. 10.1074/jbc.M309639200 [DOI] [PubMed] [Google Scholar]
- 28.Markova SV, Larionova MD, Burakova LP, Vysotski ES. The smallest natural high-active luciferase: cloning and characterization of novel 16.5-kDa luciferase from copepod Metridia longa. Biochem Biophs Res Commun (2015) 457(1):77–82. 10.1016/j.bbrc.2014.12.082 [DOI] [PubMed] [Google Scholar]
- 29.Webb SE, Miller AL. Aequorin-based genetic approaches to visualize Ca2+ signaling in developing animal systems. Biochim Biophys Acta (2012) 1820(8):1160–8. 10.1016/j.bbagen.2011.12.008 [DOI] [PubMed] [Google Scholar]
- 30.Thompson EM, Adenot P, Tsuji FI, Renard JP. Real-time imaging of transcriptional activity in live mouse preimplantation embryos using a secreted luciferase. Proc Natl Acad Sci U S A (1995) 92(5):1317–21. 10.1073/pnas.92.5.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimomur O, Johnson FH, Masugi T. Cypridina bioluminescence – light-emitting oxyluciferin-luciferase complex. Science (1969) 164(3885):1299–300. 10.1126/science.164.3885.1299 [DOI] [PubMed] [Google Scholar]
- 32.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol (2012) 7(11):1848–57. 10.1021/cb3002478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waidmann MS, Bleichrodt FS, Laslo T, Riedel CU. Bacterial luciferase reporters: the Swiss army knife of molecular biology. Bioeng Bugs (2011) 2(1):8–16. 10.4161/bbug.2.1.13566 [DOI] [PubMed] [Google Scholar]
- 34.Wu JC, Sundaresan G, Iyer M, Gambhir SS. Noninvasive optical imaging of firefly luciferase reporter gene expression in skeletal muscles of living mice. Mol Ther (2001) 4(4):297–306. 10.1006/mthe.2001.0460 [DOI] [PubMed] [Google Scholar]
- 35.Farzannia A, Roghanian R, Zarkesh-Esfahani S, Nazari M, Emamzadeh R. FcUni-RLuc: an engineered Renilla luciferase with Fc binding ability and light emission activity. Analyst (2015) 140(5):1438–41. 10.1039/c4an01946f [DOI] [PubMed] [Google Scholar]
- 36.Branchini BR, Ablamsky DM, Davis AL, Southworth TL, Butler B, Fan F, et al. Red-emitting luciferases for bioluminescence reporter and imaging applications. Anal Biochem (2010) 396(2):290–7. 10.1016/j.ab.2009.09.009 [DOI] [PubMed] [Google Scholar]
- 37.Kim SB, Izumi H. Functional artificial luciferases as an optical readout for bioassays. Biochem Biophys Res Commun (2014) 448(4):418–23. 10.1016/j.bbrc.2014.04.128 [DOI] [PubMed] [Google Scholar]
- 38.Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R. Natural-like function in artificial WW domains. Nature (2005) 437(7058):579–83. 10.1038/nature03990 [DOI] [PubMed] [Google Scholar]
- 39.Craig FF, Simmonds AC, Watmore D, McCapra F, White MRH. Membrane-permeable luciferin esters for assay of firefly luciferase in live intact-cells. Biochem J (1991) 276:637–41. 10.1042/bj2760637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams ST, Jr, Miller SC. Beyond d-luciferin: expanding the scope of bioluminescence imaging in vivo. Curr Opin Chem Biol (2014) 21:112–20. 10.1016/j.cbpa.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans MS, Chaurette JP, Adams ST, Jr, Reddy GR, Paley MA, Aronin N, et al. A synthetic luciferin improves bioluminescence imaging in live mice. Nat Methods (2014) 11(4):393–5. 10.1038/nmeth.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iwano S, Obata R, Miura C, Kiyama M, Hama K, Nakamura M, et al. Development of simple firefly luciferin analogs emitting blue, green, red, and near-infrared biological window light. Tetrahedron (2013) 69(19):3847–56. 10.1016/j.tet.2013.03.050 [DOI] [Google Scholar]
- 43.Jathoul AP, Grounds H, Anderson JC, Pule MA. A dual-color far-red to near-infrared firefly luciferin analogue designed for multiparametric bioluminescence imaging. Angew Chem Int Ed Engl (2014) 53(48):13059–63. 10.1002/anie.201405955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mofford DM, Reddy GR, Miller SC. Aminoluciferins extend firefly luciferase bioluminescence into the near-infrared and can be preferred substrates over d-luciferin. J Am Chem Soc (2014) 136(38):13277–82. 10.1021/ja505795s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alcaraz-Perez F, Mulero V, Cayuela ML. Application of the dual-luciferase reporter assay to the analysis of promoter activity in Zebrafish embryos. BMC Biotechnol (2008) 8:81. 10.1186/1472-6750-8-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vilalta M, Jorgensen C, Degano IR, Chernajovsky Y, Gould D, Noel D, et al. Dual luciferase labelling for non-invasive bioluminescence imaging of mesenchymal stromal cell chondrogenic differentiation in demineralized bone matrix scaffolds. Biomaterials (2009) 30(28):4986–95. 10.1016/j.biomaterials.2009.05.056 [DOI] [PubMed] [Google Scholar]
- 47.Shah K, Tang Y, Breakefield X, Weissleder R. Real-time imaging of TRAIL-induced apoptosis of glioma tumors in vivo. Oncogene (2003) 22(44):6865–72. 10.1038/sj.onc.1206748 [DOI] [PubMed] [Google Scholar]
- 48.Hida N, Awais M, Takeuchi M, Ueno N, Tashiro M, Takagi C, et al. High-sensitivity real-time imaging of dual protein-protein interactions in living subjects using multicolor luciferases. PLoS One (2009) 4(6):e5868. 10.1371/journal.pone.0005868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitayama Y, Kondo T, Nakahira Y, Nishimura H, Ohmiya Y, Oyama T. An in vivo dual-reporter system of cyanobacteria using two railroad-worm luciferases with different color emissions. Plant Cell Physiol (2004) 45(1):109–13. 10.1093/pcp/pch001 [DOI] [PubMed] [Google Scholar]
- 50.Mezzanotte L, Que I, Kaijzel E, Branchini B, Roda A, Lowik C. Sensitive dual color in vivo bioluminescence imaging using a new red codon optimized firefly luciferase and a green click beetle luciferase. PLoS One (2011) 6(4):e19277. 10.1371/journal.pone.0019277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noguchi T, Ikeda M, Ohmiya Y, Nakajima Y. A dual-color luciferase assay system reveals circadian resetting of cultured fibroblasts by co-cultured adrenal glands. PLoS One (2012) 7(5):e37093. 10.1371/journal.pone.0037093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Azad T, Tashakor A, Hosseinkhani S. Split-luciferase complementary assay: applications, recent developments, and future perspectives. Anal Bioanal Chem (2014) 406(23):5541–60. 10.1007/s00216-014-7980-8 [DOI] [PubMed] [Google Scholar]
- 53.Neveu G, Cassonnet P, Vidalain PO, Rolloy C, Mendoza J, Jones L, et al. Comparative analysis of virus-host interactomes with a mammalian high-throughput protein complementation assay based on Gaussia princeps luciferase. Methods (2012) 58(4):349–59. 10.1016/j.ymeth.2012.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaihara A, Umezawa Y, Furukawa T. Bioluminescent indicators for Ca2+ based on split Renilla luciferase complementation in living cells. Anal Sci (2008) 24(11):1405–8. 10.2116/analsci.24.1405 [DOI] [PubMed] [Google Scholar]
- 55.Takeuchi M, Nagaoka Y, Yamada T, Takakura H, Ozawa T. Ratiometric bioluminescence indicators for monitoring cyclic adenosine 3’,5’-monophosphate in live cells based on luciferase-fragment complementation. Anal Chem (2010) 82(22):9306–13. 10.1021/ac102692u [DOI] [PubMed] [Google Scholar]
- 56.Fan-Minogue H, Cao ZW, Paulmurugan R, Chan CT, Massoud TF, Felsher DW, et al. Noninvasive molecular imaging of c-Myc activation in living mice. Proc Natl Acad Sci U S A (2010) 107(36):15892–7. 10.1073/pnas.1007443107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Macdonald-Obermann JL, Piwnica-Worms D, Pike LJ. Mechanics of EGF receptor/ErbB2 kinase activation revealed by luciferase fragment complementation imaging. Proc Natl Acad Sci U S A (2012) 109(1):137–42. 10.1073/pnas.1111316109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luker KE, Mihalko LA, Schmidt BT, Lewin SA, Ray P, Shcherbo D, et al. In vivo imaging of ligand receptor binding with Gaussia luciferase complementation. Nat Med (2012) 18(1):172–7. 10.1038/nm.2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takakura H, Hattori M, Takeuchi M, Ozawa T. Visualization and quantitative analysis of G protein-coupled receptor-beta-arrestin interaction in single cells and specific organs of living mice using split luciferase complementation. ACS Chem Biol (2012) 7(5):901–10. 10.1021/cb200360z [DOI] [PubMed] [Google Scholar]
- 60.Paulmurugan R, Tamrazi A, Massoud TF, Katzenellenbogen JA, Gambhir SS. In vitro and in vivo molecular imaging of estrogen receptor alpha and beta homo- and heterodimerization: exploration of new modes of receptor regulation. Mol Endocrinol (2011) 25(12):2029–40. 10.1210/me.2011-1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paulmurugan R, Gambhir SS. Combinatorial library screening for developing an improved split-firefly luciferase fragment-assisted complementation system for studying protein-protein interactions. Anal Chem (2007) 79(6):2346–53. 10.1021/ac062053q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Porterfield WB, Jones KA, McCutcheon DC, Prescher JAA. “Caged” luciferin for imaging cell-cell contacts. J Am Chem Soc (2015) 137(27):8656–9. 10.1021/jacs.5b02774 [DOI] [PubMed] [Google Scholar]
- 63.Sellmyer MA, Bronsart L, Imoto H, Contag CH, Wandless TJ, Prescher JA. Visualizing cellular interactions with a generalized proximity reporter. Proc Natl Acad Sci U S A (2013) 110(21):8567–72. 10.1073/pnas.1218336110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li J, Chen LZ, Du LP, Li MY. Cage the firefly luciferin! – a strategy for developing bioluminescent probes. Chem Soc Rev (2013) 42(2):662–76. 10.1039/c2cs35249d [DOI] [PubMed] [Google Scholar]
- 65.Vorobyeva AG, Stanton M, Godinat A, Lund KB, Karateev GG, Francis KP, et al. Development of a bioluminescent nitroreductase probe for preclinical imaging. PLoS One (2015) 10(6):e0131037. 10.1371/journal.pone.0131037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De A, Jasani A, Arora R, Gambhir SS. Evolution of BRET biosensors from live cell to tissue-scale in vivo imaging. Front Endocrinol (2013) 4:131. 10.3389/fendo.2013.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu Y, Piston DW, Johnson CH. A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc Natl Acad Sci U S A (1999) 96(1):151–6. 10.1073/pnas.96.1.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu X, Soutto M, Xie Q, Servick S, Subramanian C, von Arnim AG, et al. Imaging protein interactions with bioluminescence resonance energy transfer (BRET) in plant and mammalian cells and tissues. Proc Natl Acad Sci U S A (2007) 104(24):10264–9. 10.1073/pnas.0701987104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dragulescu-Andrasi A, Chan CT, De A, Massoud TF, Gambhir SS. Bioluminescence resonance energy transfer (BRET) imaging of protein-protein interactions within deep tissues of living subjects. Proc Natl Acad Sci U S A (2011) 108(29):12060–5. 10.1073/pnas.1100923108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bakayan A, Vaquero CF, Picazo F, Llopis J. Red fluorescent protein-aequorin fusions as improved bioluminescent Ca2+ reporters in single cells and mice. PLoS One (2011) 6(5):e19520. 10.1371/journal.pone.0019520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ke DC, Tu SC. Activities, kinetics and emission spectra of bacterial luciferase-fluorescent protein fusion enzymes. Photochem Photobiol (2011) 87(6):1346–53. 10.1111/j.1751-1097.2011.01001.x [DOI] [PubMed] [Google Scholar]
- 72.Wu C, Mino K, Akimoto H, Kawabata M, Nakamura K, Ozaki M, et al. In vivo far-red luminescence imaging of a biomarker based on BRET from Cypridina bioluminescence to an organic dye. Proc Natl Acad Sci U S A (2009) 106(37):15599–603. 10.1073/pnas.0908594106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee JO, Lim B, Kim Y-P. Bioluminescence resonance energy transfer nanoprobes for imaging. IEEE J Sel Top Quantum Electron (2014) 20(3):6801410. 10.1109/jstqe.2013.2283155 [DOI] [Google Scholar]
- 74.Dragavon J, Sinow C, Holland AD, Rekiki A, Theodorou I, Samson C, et al. A step beyond BRET: fluorescence by unbound excitation from luminescence (FUEL). J Vis Exp (2014) 87:e51549. 10.3791/51549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holland AD, Rückerl F, Dragavon JM, Rekiki A, Tinevez J-Y, Tournebize R, et al. In vitro characterization of fluorescence by unbound excitation from luminescence: broadening the scope of energy transfer. Methods (2014) 66(2):353–61. 10.1016/j.ymeth.2013.09.005 [DOI] [PubMed] [Google Scholar]
- 76.Zhang L, Xu F, Chen Z, Zhu X, Min W. Bioluminescence assisted switching and fluorescence imaging (BASFI). J Phys Chem Lett (2013) 4(22):3897–902. 10.1021/jz402128j [DOI] [Google Scholar]
- 77.Zhang TC, Du LP, Li MY. BioLeT: a new design strategy for functional bioluminogenic probes. Chin Chem Lett (2015) 26(8):919–21. 10.1016/j.cclet.2015.06.020 [DOI] [Google Scholar]
- 78.Takakura H, Kojirna R, Kamiya M, Kobayashi E, Komatsu T, Ueno T, et al. New class of bioluminogenic probe based on bioluminescent enzyme-induced electron transfer: BioLeT. J Am Chem Soc (2015) 137(12):4010–3. 10.1021/ja511014w [DOI] [PubMed] [Google Scholar]