Abstract
Design
Persistent latently infected CD4+ T cells represent a major obstacle to HIV eradication. Histone deacetylase inhibitors (HDACis) are a proposed activation therapy. However, off-target effects on expression in host immune cells are poorly understood. We hypothesized that HDACi-modulated genes would be best identified with dose-response analysis.
Methods
Resting primary CD4+ T cells were treated with 0.34, 1, 3, or 10 μM of the HDACi, SAHA, for 24 hours and subjected to microarray gene expression analysis. Genes with dose-correlated expression were filtered to identify a subset with consistent up or downregulation at each SAHA dose. Histone modifications were characterized in 6 SAHA dose-responsive genes by chromatin immunoprecipitation (ChIP-RT-qPCR).
Results
A large number of genes were shown to be up (N=657) or downregulated (N=725) by SAHA in a dose-responsive manner (FDR p-value < 0.05, fold change ≥ |2|). Several genes (CTNNAL1, DPEP2, H1F0, IRGM, PHF15, and SELL) are potential in vivo biomarkers of SAHA activity. SAHA dose-responsive genes included transcription factors, HIV restriction factors, histone methyltransferases, and host proteins that interact with HIV. Pathway analysis suggested net downregulation of T cell activation with increasing SAHA dose. Histone acetylation was not correlated with host gene expression, but plausible alternative mechanisms for SAHA-modulated gene expression were identified.
Conclusions
Numerous genes in CD4+ T cells are modulated by SAHA in a dose-responsive manner, including genes that may negatively influence HIV activation from latency. Our study suggests that SAHA influences gene expression through a confluence of several mechanisms, including histone modification, and altered expression and activity of transcription factors.
Keywords: Suberoylanilide hydroxamic acid, dose-response, transcriptome, histone deacetylase inhibitor, HIV, primary CD4+ T cells
Introduction
The unsolved challenge for HIV treatment is to eradicate persistent latent reservoirs in infected individuals maintained on combination antiretroviral therapy (ART). “Shock and Kill” approaches have been proposed as cure strategies for HIV[1]. Treatment with drugs that activate HIV out of latency (shock), combined with continued ART to prevent the spread of infection, may allow the host immune system to eliminate latently infected cells (kill)[2]. Numerous drugs have been proposed to provide the shock in a shock and kill strategy towards an HIV cure, including: histone deacetylase inhibitors (HDACis), Protein Kinase C agonists (e.g., Ingenol derivatives and Bryostatin), Bromodomain inhibitors (e.g., JQ1), among others[3]. HDACis have been the most scrutinized class of compounds and includes suberoylanilide hydroxamic acid (SAHA, also known as vorinostat), which is approved by the FDA for the treatment of cutaneous T cell lymphoma[4].
The ability of SAHA to activate HIV has been assessed in two separate clinical trials (ClinicalTrials.gov registry numbers NCT01319383 and NCT01365065) with several others currently recruiting (NCT02475915, NCT02336074 and NCT01249443). Analysis of data from the completed trials indicates that SAHA induces the production of cell associated HIV RNA in the vast majority HIV-infected subjects but activation of HIV varies from donor to donor, and a concurrent increase in plasma HIV RNA associated with virions is difficult to detect[5–7]. Encouragingly, two other HDACis, panobinostat and romidepsin, may have greater potency for activating HIV[8, 9], and are currently being assessed in clinical trials (NCT01933594 and NCT01680094, respectively).
However, the off-target effects of these compounds are even less understood than for SAHA. Therefore, the full extent of SAHA’s clinical utility in a shock and kill strategy remains to be evaluated. Current clinical trials assess shock compounds in isolation and future studies are required to assess compounds in combinations. Due to SAHA’s ability to activate HIV transcription, it is plausible that SAHA will be included in future combination therapies in conjunction with compounds that can relieve the inhibition that is restricting virion production[7].
HDACis cause hyperacetylation of histones by preventing deacetylation, leading to chromatin unwinding and recruitment of transcriptional machinery[10]. HIV transcription is thought to occur at least partly through this mechanism at sites where HIV is integrated in the host genome, but activation may also involve non-histone targets, and is not yet fully understood[11]. It has been hypothesized that HDACis alter the expression of host genes that influence HIV activation, but the identities of these host genes are largely unknown[11]. The non-specific nature of histone acetylation suggests that widespread effects on host gene expression are likely to occur, so it is also important to investigate the potential for off-target side effects of SAHA in immunocompromised patients. Furthermore, the optimal dosing regimen for SAHA has not yet been determined for HIV patients. The currently recommended oral dose of 400 mg, once daily, results in a peak serum concentration of 1.2 μM[4]. Intravenous dosing results in peak serum concentration of 9.1 μM, and was surprisingly better tolerated than oral dosing in clinical trials for cancer[12].
Our laboratory previously reported minimal off-target effects on host gene expression for primary CD4+ T cells treated for 24 hours with 0.34 μM SAHA[13]. Only 37 of 1880 significantly modulated genes had expression changes above 2-fold. However, SAHA-modulated gene expression has not previously been examined over the entire concentration range relevant to HIV cure strategies. Our goal was to identify SAHA dose-responsive genes that could be used as in vivo biomarkers of SAHA activity. Primary CD4+ T cells were treated with 0.34, 1, 3, or 10 μM of SAHA. Genes consistently up or downregulated in a dose-dependent manner were identified. Host cell functions affected by these genes may have implications for the efficacy and safety of therapies for both HIV and cancer.
Methods
Isolation of primary CD4+ T cells
Human peripheral blood was drawn according to institutional review board approved protocols from 9 healthy HIV-seronegative donors by venipuncture and collected into sodium heparin containing tubes. Peripheral blood mononuclear cells (PBMCs) were collected from fresh whole blood by centrifugation. Primary CD4+ T cells were isolated from PBMCs with RosetteSep CD4+ Isolation Kits according to manufacturer protocol (Stem Cell Technologies). CD4+ T cells were incubated at 37°C, 5% CO2 overnight in RPMI-1640 with 5% human serum AB. CD4+ T cell purity and activation were assessed with flow cytometry. All samples included in microarray analysis had >95% purity with fewer than 10% activated cells (i.e., <10% expressing HLA-DR).
SAHA treatment of primary CD4+ T cells
CD4+ T cells were aliquoted into 6-well plates; 5 million cells per well in 2 mL of media (RPMI-1640 with 5% human serum AB). These cells were treated with 0.34 μM, 1 μM, 3 μM, or 10 μM SAHA (Merck) in 0.1% DMSO, or with only 0.1% DMSO, and incubated at 37°C, 5% CO2 for 24 hours.
Microarray analysis of gene expression
RNA was extracted from treated cells using Qiagen RNeasy kits according to manufacturer protocol. RNA integrity numbers (RINs) were determined with the Agilent 2100 Bioanalyzer (Agilent Technologies). RNA samples submitted for microarray analysis (N=6) had RINs between 7.9 and 9.0 with an average RIN of 8.5. cRNA was generated and hybridized to IlluminaHT-12 v3 BeadChips (48,803 probes). Expression data were extracted with GenomeStudio (Illumina); genes with undetectable expression were excluded from analysis. The lumi package[14] in bioconductor was used to transform (variance-stabilizing) and normalize (robust spline) raw expression data. Genes significantly modulated across SAHA doses were identified using a likelihood ratio test in the R package Isogene GX[15]. The familywise error rate was controlled using Bonferroni-adjusted p-values (p < 0.05). A subset of SAHA dose-responsive genes was identified and used in all subsequent analyses; included genes had consistent trends of up or downregulation across each increasing SAHA dose, and greater than 2-fold change in expression when comparing the highest dose (10 μM) to DMSO-treated controls. Unless specified, all analyses were corrected for multiple comparisons using the Benjamini-Hochberg method[16] with an FDR-corrected p-value ≤ 0.05 considered significant. Gene expression data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/): accession number GSE66994.
Gene ontology (GO), pathway, and protein interaction network analysis
GO terms significantly over-represented for SAHA dose-responsive genes were identified using the BiNGO plugin for Cytoscape[17]. KEGG and BIOCARTA biological pathways significantly over-represented for SAHA dose-responsive genes were identified and characterized with DAVID[18, 19]. A direct protein interaction network (PIN) using a database of literature-curated protein-protein and protein-DNA interactions was constructed for SAHA dose-responsive genes using Metacore (Thomson Reuters).
Real-Time Quantitative PCR (RT-qPCR)
Microarray expression data was confirmed by RT-qPCR for the top SAHA dose-responsive gene in samples (5 SAHA treatments in each of 6 donors), as described previously[13]. Briefly, RNA was reverse transcribed using qScript™ cDNA SuperMix (Quanta Biosciences). Expression was confirmed for the 3 most upregulated (CTNNAL1, H1F0, IRGM) and 3 most downregulated (DPEP2, PHF15, SELL) SAHA dose-responsive genes (Supplemental Table 1) with TaqMan® assays according to manufacturer protocols (TaqMan® Universal Master Mix II, ABI Prism 7900HT; Life Technologies). Expression data were normalized to RPL27; a gene previously determined to be non-responsive to SAHA[13]. The 2−ΔΔCt method[20] was used to determine fold change SAHA treated samples.
Chromatin Immunoprecipitation (ChIP)
Three additional donors were recruited for ChIP analysis of SAHA-induced histone modifications. CD4+ T cells were isolated and treated with SAHA as described above. Chromatin was isolated and processed using ChIP-IT High Sensitivity kits (Active Motif) according to manufacturer’s instructions, except as noted. DNA was cross-linked to protein by a 5-minute incubation in fresh 1% formaldehyde and sheared to an average size of 500–1500 base pairs by sonication in 0.1% SDS lysis buffer (Covaris M220 Sonicator; 6 minutes, 6% duty factor, 200 counts per burst, 75 watts). DNA complexes were immunoprecipitated with ChIP-grade antibodies directed against total histone H3 (H3), acetylated-H3 Lysine 9 (H3K9a), acetylated-total histone H4 (H4a), methylated-H3K4 (H3K4m), methylated-H3K27 (H3K27m), and RNA polymerase II (Pol II). Following cross-link reversal and DNA purification, RT-qPCR was conducted using primers and probes (Supplemental Table 1) designed to span regions where histone modifications are known to occur; these regions were identified using the UCSC Genome Browser histone track (http://genome.ucsc.edu/)[21]. Chromatin concentration was normalized to non-IP input control chromatin, and then histone acetylation or methylation for 6 SAHA dose-responsive test genes (CTNNAL1, DPEP2, H1F0, IRGM, PHF15, SELL) was normalized to total H3. The 2−ΔΔCt method was used to calculate enrichment of histone modifications; this value was log2 transformed for plotting. The significance of SAHA-modulated changes in histone modification was determined with a one-tailed, paired t-test (p ≤ 0.05). For each SAHA dose, RNA was isolated from treated donor samples for RT-qPCR validation of SAHA dose-responsiveness in test genes, as described above.
Predicted transcription factor binding site analysis
The DiRE web server (http://dire.dcode.org) was used to identify transcription factor binding sites enriched in SAHA-modulated genes[22]. Predicted key transcription factors were ranked by an “importance” score reflecting a transcription factor’s association with genes that are co-expressed following treatment, as well as its occurrence in candidate regulatory elements. Predicted transcription factors with an importance score ≥ 0.1 were incorporated into protein networks with SAHA-modulated genes, as described above.
Results
SAHA dose-response genes
To examine the effect of SAHA on host function in CD4+ T-cells, we utilized microarray analysis to identify 3,477 genes with a SAHA dose-responsive trend. This set was filtered to retain only genes with a fold change ≥ 2 or ≤ −2 and a consistent trend in expression across each increasing SAHA dose. This filter identified 1,382 genes (Supplemental Table 2) whose expression was consistently responsive to dose, of which 657 were upregulated and 725 downregulated. This higher confidence subset was used in all analyses of SAHA modulated gene expression, and is referred to hereafter as “dose-responsive genes”.
Our previous study in primary CD4+ T cells exposed to 0.34 μM SAHA for 24 hours identified 1,847 differentially expressed genes[13]. Only 29% of these genes were found to be dose-responsive across the concentration range of SAHA used in this study. This suggests that not all differentially expressed genes at this low dose of SAHA are truly dose-responsive, indicating the risks of relying on a single dose to identify gene expression biomarkers for SAHA response.
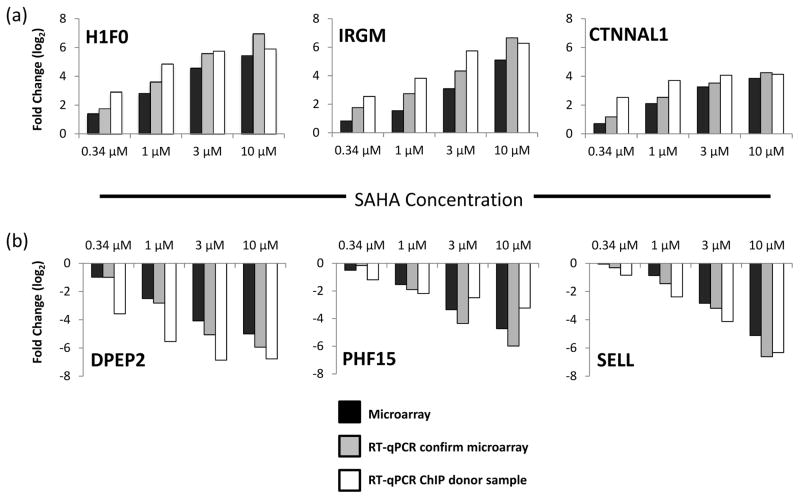
RT-qPCR analysis of microarray samples (Figure 1) confirmed dose-responsive changes in expression and consistent trends of up or downregulation in all six test genes across each tested SAHA dose. These genes represent gene expression biomarkers of SAHA, which are relevant over the concentration range examined here (0.34 to 10 μM).
Figure 1. RT-qPCR confirmation of SAHA dose-responsive gene expression.
Gene expression changes induced by SAHA were confirmed by RT-qPCR in the same RNA samples used for microarray analysis (Donors 1–6). RT-qPCR analysis was also used to confirm expression changes for the six most strongly SAHA dose-responsive genes in primary CD4+ T cells used in ChIP analysis (Donors 7–9). (a) The top 3 dose-responsive upregulated genes, and (b) the top 3 dose-responsive downregulated genes. Abbreviations are as follows: CTNNAL1, catenin (cadherin-associated protein), alpha-like 1; H1f0, H1 histone family, member 0; IRGM, immunity-related GTPase family, M; DPEP2, Dipeptidase 2; PHF15, PHD finger protein 15; SELL, Selectin L
SAHA dose-responsive genes associated with HIV replication
Literature-based searches identified host dose-responsive genes with prior evidence of involvement in HIV activation or replication. A number of SAHA dose-responsive genes (summarized in Table 1) have functions relating to histone methylation, transcription, HIV restriction, HIV protein interaction and cell surface receptors, which on the whole likely enhance HIV activation.
Table 1.
SAHA dose-responsive genes with potential roles in HIV activation or replication1.
|
Category
|
Gene
|
Maximum fold change (10 μM SAHA)
|
|---|---|---|
| Restriction Factors | EV12B | −17.6 |
| TRIM22 | −7.7 | |
| APOBEC3G | −5.5 | |
| APOBEC3F | −3.1 | |
| HIV protein interactors | IRGM | 34.3 |
| HSP70 (HSPA2) | 7.7 | |
| Histone methyltransferases | EZH2 | 13.3 |
| SETD3 | 5.2 | |
| SMYD2 | 5.2 | |
| SETD8 | 3.0 | |
| SETDB1 | 2.9 | |
| SETD1A | 2.5 | |
| SETD1B | −5.2 | |
| SMYD4 | −4.6 | |
| SETMAR | −3.9 | |
| MLL5 | −3.3 | |
| EHMT2 | −3.1 | |
| Transcription Factors | MYC | −10.9 |
| LEF1 | −5.7 | |
| ETS1 | −3.1 | |
| CEBPB | −2.4 | |
| Cell surface molecules | CCL5 (RANTES) | −15.6 |
| CD11a (ITGAL) | −13.1 |
Fold change was calculated by comparing expression between the 10 μM dose of SAHA and the untreated control.
GO and pathway analysis of SAHA dose-responsive genes
GO terms for biological processes related to T-cell activation, apoptosis, and chromatin organization were significantly over-represented (Supplemental Figure 1). Repeated analysis using only SAHA-upregulated dose-responsive genes primarily retained GO terms related to chromatin organization (Supplemental Figure 2). In contrast, analysis using only downregulated genes identified terms related to T-cell activation and apoptosis (Supplemental Figure 3). Pathway mapping of dose-responsive genes identified significant pathways only in downregulated genes (FDR q-value < 0.05)[23]; In downregulated genes, significant KEGG and BIOCARTA pathways were focused on T-cell receptor signaling, apoptosis, and cytotoxic T-lymphocyte mediated immune response (Table 2). Further analysis of expression patterns for genes associated with the T-cell receptor signaling pathway (KEGG) suggests a possible SAHA-induced net decrease in T-cell activation (Supplemental Figure 4). There was no consistent trend in apoptosis-related gene expression, so the net effect of SAHA on apoptosis could not be predicted (data not shown). This is consistent with previous results showing no cytopathic effects from SAHA-induced HIV reactivation[24].
Table 2.
Pathway categories significantly enriched (q-value ≤ 0.05) for SAHA dose-responsive genes.
| Category | Pathway1 | Genes | q-value2 |
|---|---|---|---|
| T cell receptor signaling/activation | Lck and Fyn tyrosine kinases in initiation of TCR activation (BIOCARTA) | CD3D, CD3E, LCK, CD247, ZAP70, CD4 | 2.38E-04 |
| Activation of Csk Inhibits Signaling through TCR (BIOCARTA) | CD3D, CD3E, LCK, CD247, ZAP70, CD4 | 1.12E-03 | |
| T-Cell Receptor Signaling Pathway (KEGG) | PTPN6, CARD11, CD3D, CD3E, CD40LG, MAPK13, LCK, CD247, ZAP70, CD4, CD28 | 3.38E-03 | |
| The Co-Stimulatory Signal During T-Cell Activation | CD3D, CD3E, LCK, CD247, ITGB2 | 1.44E-02 | |
| T-Cell Receptor and CD3 Complex | CD3D, CD3E, CD247 | 3.69E-02 | |
| Role of Tob in T-Cell Activation | CD3D, CD3E, CD247, CD28 | 4.55E-02 | |
| Apoptosis | HIV Induced T-Cell Apoptosis (BIOCARTA) | CD3D, CD3E, CD247, CD4, CD28 | 1.30E-03 |
| Cytotoxic T cell function | T-Cytotoxic Cell Surface Molecules (BIOCARTA) | ITGAL, CD3D, CD3E, CD247, ITGB2, CD28 | 2.66E-04 |
| CTL Mediated Immune Response Against Target Cells (BIOCARTA) | ITGAL, CD3D, CD3E, CD247, ITGB2 | 5.31E-03 | |
| Cell surface molecules | T-Helper Cell Surface Molecules (BIOCARTA) | ITGAL, CD3D, CD3E, CD247, ITGB2, CD4, CD28 | 2.36E-05 |
| Adhesion Molecules on Lymphocyte (BIOCARTA) | ITGAL, CD44, SELL, ITGB2, ITGA4 | 1.30E-03 | |
| Monocyte and its Surface Molecules (BIOCARTA) | ITGAL, CD44, SELL, ITGB2, ITGA4 | 2.18E-03 | |
| Neutrophil and its Surface Molecules (BIOCARTA) | ITGAL, CD44, SELL, ITGB2 | 1.10E-02 | |
| Cell Adhesion Molecules (BIOCARTA) | ITGAL, CD40LG, SELL, ICAM3, NLGN2, ITGB2, CD4, ITGA4, CD226, CD28 | 2.85E-02 | |
| Immune Function | Primary Immunodeficiency (KEGG) | CD3D, CD3E, CD40LG, LCK, ZAP70, CD4 | 3.69E-02 |
Pathways identified with DAVID v6.7; analysis utilized curated pathway databases KEGG and BIOCARTA.
q-value: FDR-adjusted p-value (Storey, 2003).
SAHA-induced histone modification
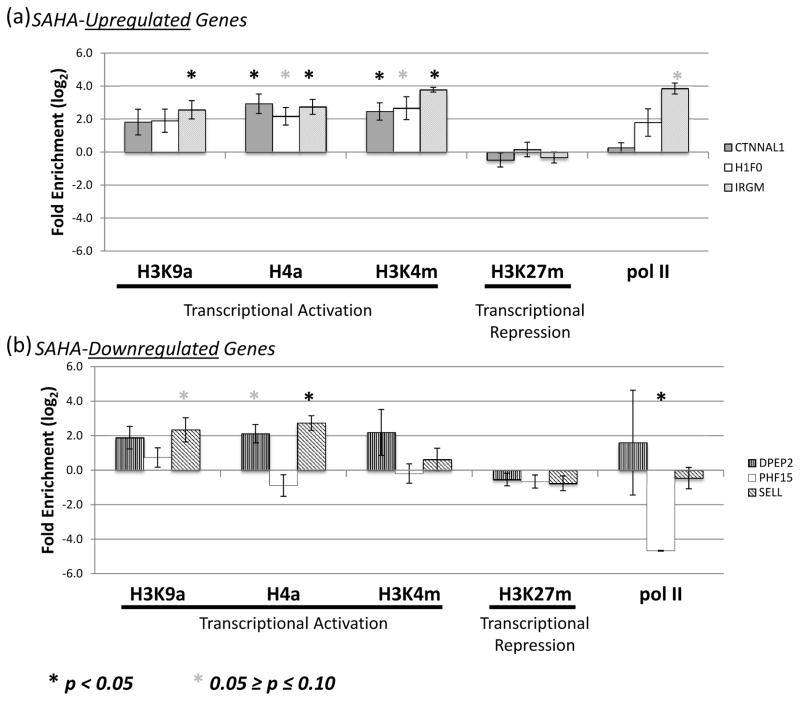
We conducted ChIP-RT-qPCR assays in primary CD4+ T cells to determine whether histones were hyperacetylated (H3K9a and H4a) as a consequence of SAHA treatment. Having identified numerous SAHA-modulated histone methyltransferases (HMTs, Table 1), ChIP-RT-qPCR also examined histone methylation associated with gene activation (H3K4m) and repression (H3K27m). H3K4-methylation was selected because 5 of 11 HMTs modulated by SAHA are known to methylate this site[25–29]. The most strongly upregulated HMT, EZH2, methylates H3K27, causing transcriptional repression[30]. Histone modifications were studied in the three most upregulated genes and three most downregulated genes. Expression of these genes was confirmed in ChIP donor samples, demonstrating expression patterns similar to previously tested microarray samples (Figure 1).
Histone markers of transcriptional activation (H3K9a, H4a, and H3K4m) were consistently enriched, often attaining significance in the three SAHA-upregulated genes (Figure 2a). Unexpectedly, histone acetylation was also detected in 2 of 3 SAHA-downregulated genes, DPEP2 and SELL (Figure 2b). H3K4-methylation was not significantly different from vehicle in any of the downregulated genes. Trends in pol II enrichment were generally consistent with the known patterns of expression for these genes; however, significance was reached only for IRGM (increased pol II, SAHA-upregulated) and PHF15 (decreased pol II, SAHA-downregulated). H3K27-methylation was not correlated with SAHA-modulated gene expression. The enrichment patterns of histone modifications associated with transcriptional activation were consistent with gene upregulation, but did not explain SAHA-induced gene downregulation.
Figure 2. ChIP-RT-qPCR analysis of histone modifications for the top six genes SAHA dose-responsive genes.
Histone modifications and Pol II occupancy were examined in the promoters of the (a) 3 most upregulated (CTNNAL1, H1F0, IRGM) and (b) 3 most downregulated (DPEP2, PHF15, SELL) SAHA dose-responsive genes when comparing a SAHA treatment of 10μM to vehicle. Fold enrichment of immunoprecipitated chromatin upon SAHA treatment is plotted on the log2 scale with error bars representing the standard error of the mean. Significant enrichment was assessed in a paired t-test and reported at the 0.1 (*) and 0.05 (**) cut-offs. Abbreviations are as follows: H3K9a, acetylated H3 Lysine 9; H4a, acetylated total Histone H4; H3K4m, methylated H3K4; H3K27m, methylated H3K27; Pol II, RNA polymerase II.
SAHA dose-responsive transcription factor modulation
A direct protein interaction network (Supplemental Figure 5) containing SAHA dose-responsive genes identified key hubs through which SAHA may influence expression of additional genes; as summarized in Table 3. The most connected hub in the protein network was the transcription factor, c-Myc. Other large hubs were mostly downregulated and corresponded to transcription factors or coactivators (C/EBPbeta, p300, ETS1 and STAT1). Several of these hubs (e.g., c-Myc, EZH2, and CDK2) were also among the most connected hubs in our previous study summarizing differential expression in primary CD4+ T cells treated with 0.34 μM SAHA[13].
Table 3.
Major hubs (>20 edges) in the protein interaction network of SAHA dose-responsive genes combined with transcription factors predicted to bind in the promoters of SAHA modulated genes.
| Protein Hub | # Edges | Function | Response to SAHA |
|---|---|---|---|
| c-Myc | 294 | Transcription Factor | Downregulated |
| SP1* | 156 | Transcription Factor | N/A** |
| p300 | 94 | Histone Acetyl Transferase | Downregulated |
| C/EBPbeta | 63 | Transcription Factor | Downregulated |
| EZH2 | 60 | Histone Methyl Transferase | Upregulated |
| STAT1 | 53 | Transcription Factor | Upregulated |
| CBP | 49 | Generic Enzyme | Downregulated |
| p21 | 48 | Binding Protein | Upregulated |
| SP3 | 44 | Transcription Factor | Downregulated |
| ETS1* | 42 | Transcription Factor | Downregulated |
| AML1 (RUNX1) * | 41 | Transcription Factor | Downregulated |
| CDK2 | 40 | Protein Kinase | Downregulated |
| SMAD3 | 35 | Transcription Factor | Upregulated |
| TAL1* | 35 | Transcription Factor | N/A** |
| Lck | 33 | Protein Kinase | Downregulated |
| MYOD* | 32 | Transcription Factor | N/A** |
| Bcl-2 | 30 | Binding Protein | Downregulated |
| ZAP70 | 30 | Protein Kinase | Downregulated |
| Caspase-1 | 28 | Protease | Downregulated |
| STAT6* | 28 | Transcription Factor | Downregulated |
| HDAC3 | 27 | Histone Deacetylase | Upregulated |
| ATM | 26 | Protein Kinase | Downregulated |
| SHP-1 | 23 | Protein Phosphatase | Downregulated |
| Lef-1 | 22 | Transcription Factor | Downregulated |
| E2F3 | 21 | Transcription Factor | Downregulated |
Predicted transcription factors (DiRE, http://dire.dcode.org)
Expression not modulated by SAHA
We also explored the possibility that SAHA might modulate activity of transcription factors through other mechanisms (e.g., transcription factor acetylation). Candidate transcription factors were identified among SAHA-responsive genes using DiRE[22], to find enrichment in consensus transcription factor binding sites within regulatory elements of co-expressed genes. Transcription factor analysis identified 13 total candidate transcription factors among SAHA dose-responsive genes: 8 among upregulated and 5 among downregulated genes. The highest confidence predictions are shown in Supplemental Figure 6. SAHA treatment decreased expression of three predicted transcription factors (ETS1, STAT6, and the RUNX1 component of Core Binding Factor) with enrichment for consensus binding sites in downregulated genes; all three were hubs in the PIN of SAHA-modulated genes. Furthermore, when SAHA-non-responsive, DiRE-predicted transcription factors were analyzed for interactions with SAHA dose-responsive genes, the resulting PIN had SP1, TAL1, and MYOD emerging as major hubs; with curated evidence of acting on 156, 35, and 30 SAHA-modulated proteins, respectively (Table 3).
Discussion
This study revealed large numbers of genes modulated by SAHA in a dose-dependent manner in primary CD4+ T cells. Six of the most dose-responsive genes were validated by RT-qPCR (CTNNALI, DPEP, H1F0, IRGM, PHF15, SELL), suggesting they could be useful in vivo biomarkers for monitoring SAHA activity in current (NCT02475915, NCT02336074 and NCT01249443) and future clinical trials, as well as in trials of other HDACis such as romidepsin and panobinostat (NCT01933594 and NCT01680094). Encouragingly, the SAHA gene expression biomarkers PH15 and H1F0 identified in this study appear to have broad applicability for other compartments and cell types (e.g., whole blood and PBMCs), in other species (e.g., mouse and rhesus macaques), and for other HDACis (e.g., panobinostat) (Drs. Richard Barnard and Bonnie Howell, Merck Inc., personal communication).
Acetylation of histones only partially explains the effects of SAHA on host cell gene expression [31–34]. ChIP-RT-qPCR based evaluation of histone modifications in the six most SAHA-dose responsive genes (Figure 2) demonstrated that increased histone acetylation was generally observed regardless of gene up or downregulation. The PIN of SAHA-modulated genes revealed several transcription factors that were major hubs (Table 3). Therefore, by altering expression or activity of transcription factors, it is plausible that SAHA influences secondary effects (including downregulation) on scores of other genes.
This study identified several SAHA dose-responsive host genes that may contribute to SAHA’s ability to facilitate HIV activation (Table 1). With respect to enhancing HIV activation, the protein products of IRGM and HSP70 have demonstrated direct interactions with HIV proteins to promote viral replication[35, 36] and were strongly upregulated by SAHA. Furthermore, several HIV restriction factors, APOBEC3G, APOBEC3F, EV12B and TRIM22[37] were downregulated in a dose-dependent manner by SAHA. Downregulation of MYC, the most connected hub in our PIN, is likely to be an important contributor to HIV activation by SAHA, as we have reported previously[13]. The transcription factor SP1 had predicted binding sites in a large proportion of upregulated genes and was a major hub when combined in a PIN with SAHA-modulated genes. Although no change in SP1 expression was detected, HDACi treatment can acetylate the SP1 protein, enhancing its ability to bind the HIV LTR and promote viral transcription[11]. Thus, SAHA may alter SP1 activity by direct acetylation without changing its expression, contributing to HIV activation. In contrast, with respect to repressing HIV activation, several transcription factors (ETS1, CEBPB, LEF1) shown to bind the HIV LTR and activate HIV transcription[38–40] were downregulated by SAHA. Furthermore, two cell surface molecules (CCL5/RANTES and ITGAL/CD11a) downregulated by SAHA are thought to enhance cell-to-cell transmission of the activated virus [41–43]. Finally, a number of histone methyltransferases were modulated by SAHA with half being upregulated and half downregulated (Table 1). This represents a heterogenous signal with respect to HIV activation where broad and specific methyltransferase inhibitors have been shown to activate HIV from latency [44, 45]. These observations support recent findings from the gene expression analysis of samples from clinical trials with SAHA suggesting that multiple mechanisms may contribute to HIV activation[7]. Beyond the ability to directly activate HIV through the relaxation of chromatin, SAHA may operate through indirect mechanisms by modulating host genes some of which appear to have positive and others negative roles with respect to HIV activation. For the majority of the genes listed in Table 1, SAHA induced changes in expression would likely favor HIV activation from latency. However, recent evidence suggests that HIV activation from latency may also require removal of an as yet unknown post-transcriptional block[46]; no obvious candidates for such an activity emerged from our results.
Analysis of functional categories and pathways associated with SAHA-modulated genes revealed categories related to T-cell activation and suggested possible downregulation of T-cell receptor activation. Several studies implicate T-cell activation as an essential component for efficient HIV infection and replication. Possible consequences of SAHA impeding T-cell activation may include decreased HIV transcription and translation, which may diminish eradication of infected cells during ART. This is consistent with a recent study in a primary CD4+ T cell latency model showing that SAHA treatment resulted in only a modest increase in HIV protein despite a 3-fold increase in HIV transcription[46]. Our data suggest that T-cell receptor activation may require particular scrutiny when evaluating dosing regimens for SAHA, or future analogues with increased bioavailability or potency.
The possible utilization of SAHA to activate latent HIV is still being evaluated in clinical trials, so it is too early to know whether it will have a key role in a shock and kill approach to an HIV cure. In the event that SAHA does not show the desired efficacy on its own, it may still be used as part of a combination therapy with other latency activating compounds. Our results indicated plausible alternative mechanisms of action for SAHA that may help to identify additional therapies or adjuvants that can be used to maximize the effectiveness of a cure strategy. This current study was focused on gene expression in primary resting CD4+ T cells because these cells are considered the primary reservoir of latent HIV infection in the peripheral blood. Future work will profile the effects of SAHA and other latency reversing agents in PBMCs, as well as other cell subsets and tissues to further elucidate the effects of SAHA.
In summary, the primary outcome of this work was the identification of gene expression biomarkers of SAHA administration (CTNNALI, DPEP, H1F0, IRGM, PHF15, SELL). These results further indicated other possible mechanisms through which SAHA may affect activation of HIV from latency. Our data suggest that SAHA alters host gene expression through the combined action of multiple mechanisms, including histone acetylation, histone methylation, and altered expression and possible acetylation of transcription factors. These results may be used to identify targets for future knockdown or overexpression studies using in vitro HIV latency models to determine the contribution of host genes to the SAHA-induced activation of HIV.
Supplementary Material
Acknowledgments
Funding and support: This work was performed with the support of the Genomics and Sequencing Core and the Flow Cytometry Core at the UCSD Center for AIDS Research (AI36214), the Martin Delaney CARE Collaboratory (AI096113), the San Diego Veterans Medical Research Foundation, the James B. Pendleton Foundation, and NIH grants (AI08019 and AI104282), the Department of Veterans Affairs (VA), Veterans Health Administration, Office of Research and Development. Brian Reardon was supported by the Ruth L. Kirschstein National Research Service Award (NRSA) Institutional Research Training Grant (AI007384). SAHA was graciously provided by Merck and Co., Inc.
This work was performed with the support of the Genomics and Sequencing Core and the Flow Cytometry Core at the UCSD Center for AIDS Research (AI36214), the Martin Delaney CARE Collaboratory (AI096113), the San Diego Veterans Medical Research Foundation, the James B. Pendleton Foundation, and NIH grants (AI08019 and AI104282), the Department of Veterans Affairs (VA), Veterans Health Administration, Office of Research and Development. Brian Reardon was supported by the Ruth L. Kirschstein National Research Service Award (NRSA) Institutional Research Training Grant (AI007384). Unpublished results supporting wide applicability of SAHA biomarkers provided by Bonnie Howell and Richard Barnard at Merck and Co., Inc. SAHA was graciously provided by Merck and Co., Inc. The views expressed here are those of the authors and do not necessarily reflect the position or policy of the VA or the U.S. government. BR, NBB, DDR, CAS and CHW conceived and designed the study; BR, NBB, and AS performed the experiments; BR, CHW and AS analyzed the data; BR and CHW wrote the manuscript. All authors reviewed the manuscript and accepted it for publication. We would like to express our gratitude to the reviewers of this manuscript whose advice resulted in a higher quality publication. Finally, this manuscript is dedicated to the Community Advisory Board of the CARE Collaboratory (Lynda Dee, Jeff Taylor, Alasdair Burton, Tyler Fisher, Andy Kaytes, Damian Kelly, Jeffrey Mazo, David Palm, Tim Rogers, Jeff Sheehy, and Lorren Willenberg) whose partnership in the quest for finding a cure for HIV has been invaluable.
References
- 1.Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, et al. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet. 2005;366:549–555. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demonté D, Quivy V, Colette Y, Van Lint C. Administration of HDAC inhibitors to reactivate HIV-1 expression in latent cellular reservoirs: implications for the development of therapeutic strategies. Biochem Pharmacol. 2004;68:1231–1238. doi: 10.1016/j.bcp.2004.05.040. [DOI] [PubMed] [Google Scholar]
- 3.Archin NM, Margolis DM. Emerging strategies to deplete the HIV reservoir. Curr Opin Infect Dis. 2014;27:29–35. doi: 10.1097/QCO.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 5.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Archin NM, Bateson R, Tripathy MK, Crooks AM, Yang KH, Dahl NP, et al. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis. 2014;210:728–735. doi: 10.1093/infdis/jiu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, et al. Activation of HIV Transcription with Short-Course Vorinostat in HIV-Infected Patients on Suppressive Antiretroviral Therapy. PLoS Pathog. 2014;10:e1004473. doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasmussen TA, Schmeltz Søgaard O, Brinkmann C, Wightman F, Lewin SR, Melchjorsen J, et al. Comparison of HDAC inhibitors in clinical development: Effect on HIV production in latently infected cells and T-cell activation. Hum Vaccin Immunother. 2013;9 doi: 10.4161/hv.23800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei DG, Chiang V, Fyne E, Balakrishnan M, Barnes T, Graupe M, et al. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog. 2014;10:e1004071. doi: 10.1371/journal.ppat.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matalon S, Rasmussen TA, Dinarello CA. Histone deacetylase inhibitors for purging HIV-1 from the latent reservoir. Mol Med. 2011;17:466–472. doi: 10.2119/molmed.2011.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirakawa K, Chavez L, Hakre S, Calvanese V, Verdin E. Reactivation of latent HIV by histone deacetylase inhibitors. Trends Microbiol. 2013;21:277–285. doi: 10.1016/j.tim.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zain J, O’Connor OA. Targeting histone deacetyalses in the treatment of B- and T-cell malignancies. Invest New Drugs. 2010;28(Suppl 1):S58–78. doi: 10.1007/s10637-010-9591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beliakova-Bethell N, Zhang JX, Singhania A, Lee V, Terry VH, Richman DD, et al. Suberoylanilide hydroxamic acid induces limited changes in the transcriptome of primary CD4(+) T cells. AIDS. 2013;27:29–37. doi: 10.1097/QAD.0b013e32835b3e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 15.Pramana S, Lin D, Haldermans P, Shkedy Z, Verbeke T, Gohlmann H, et al. IsoGene: An R Package for Analyzing Dose-response Studies in Microarray Experiments. R Journal. 2010;2:5–12. [Google Scholar]
- 16.Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang dW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gotea V, Ovcharenko I. DiRE: identifying distant regulatory elements of co-expressed genes. Nucleic Acids Res. 2008;36:W133–139. doi: 10.1093/nar/gkn300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang HC, et al. Stimulation of HIV-1-Specific Cytolytic T Lymphocytes Facilitates Elimination of Latent Viral Reservoir after Virus Reactivation. Immunity. 2012;36:491–501. doi: 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sebastian S, Sreenivas P, Sambasivan R, Cheedipudi S, Kandalla P, Pavlath GK, et al. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc Natl Acad Sci U S A. 2009;106:4719–4724. doi: 10.1073/pnas.0807136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem. 2007;282:13419–13428. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 27.Tate CM, Lee JH, Skalnik DG. CXXC finger protein 1 restricts the Setd1A histone H3K4 methyltransferase complex to euchromatin. FEBS J. 2010;277:210–223. doi: 10.1111/j.1742-4658.2009.07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abu-Farha M, Lambert JP, Al-Madhoun AS, Elisma F, Skerjanc IS, Figeys D. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteomics. 2008;7:560–572. doi: 10.1074/mcp.M700271-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Gu B, Lee MG. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci. 2013;3:39. doi: 10.1186/2045-3701-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daly K, Shirazi-Beechey SP. Microarray analysis of butyrate regulated genes in colonic epithelial cells. DNA Cell Biol. 2006;25:49–62. doi: 10.1089/dna.2006.25.49. [DOI] [PubMed] [Google Scholar]
- 32.Rada-Iglesias A, Enroth S, Ameur A, Koch CM, Clelland GK, Respuela-Alonso P, et al. Butyrate mediates decrease of histone acetylation centered on transcription start sites and down-regulation of associated genes. Genome Res. 2007;17:708–719. doi: 10.1101/gr.5540007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peart MJ, Smyth GK, van Laar RK, Bowtell DD, Richon VM, Marks PA, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2005;102:3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Ruijter AJ, Meinsma RJ, Bosma P, Kemp S, Caron HN, van Kuilenburg AB. Gene expression profiling in response to the histone deacetylase inhibitor BL1521 in neuroblastoma. Exp Cell Res. 2005;309:451–467. doi: 10.1016/j.yexcr.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 35.Grégoire IP, Richetta C, Meyniel-Schicklin L, Borel S, Pradezynski F, Diaz O, et al. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog. 2011;7:e1002422. doi: 10.1371/journal.ppat.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agostini I, Popov S, Li J, Dubrovsky L, Hao T, Bukrinsky M. Heat-shock protein 70 can replace viral protein R of HIV-1 during nuclear import of the viral preintegration complex. Exp Cell Res. 2000;259:398–403. doi: 10.1006/excr.2000.4992. [DOI] [PubMed] [Google Scholar]
- 37.Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008;4:e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Posada R, Pettoello-Mantovani M, Sieweke M, Graf T, Goldstein H. Suppression of HIV type 1 replication by a dominant-negative Ets-1 mutant. AIDS Res Hum Retroviruses. 2000;16:1981–1989. doi: 10.1089/088922200750054710. [DOI] [PubMed] [Google Scholar]
- 39.Mukerjee R, Sawaya BE, Khalili K, Amini S. Association of p65 and C/EBPbeta with HIV-1 LTR modulates transcription of the viral promoter. J Cell Biochem. 2007;100:1210–1216. doi: 10.1002/jcb.21109. [DOI] [PubMed] [Google Scholar]
- 40.Sheridan PL, Sheline CT, Cannon K, Voz ML, Pazin MJ, Kadonaga JT, et al. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 41.Shattock RJ, Rizzardi GP, Hayes P, Griffin GE. Engagement of adhesion molecules (CD18, CD11a, CD45, CD44, and CD58) enhances human immunodeficiency virus type 1 replication in monocytic cells through a tumor necrosis factor-modulated pathway. J Infect Dis. 1996;174:54–62. doi: 10.1093/infdis/174.1.54. [DOI] [PubMed] [Google Scholar]
- 42.Roscic-Mrkic B, Fischer M, Leemann C, Manrique A, Gordon CJ, Moore JP, et al. RANTES (CCL5) uses the proteoglycan CD44 as an auxiliary receptor to mediate cellular activation signals and HIV-1 enhancement. Blood. 2003;102:1169–1177. doi: 10.1182/blood-2003-02-0488. [DOI] [PubMed] [Google Scholar]
- 43.He W, Neil S, Kulkarni H, Wright E, Agan BK, Marconi VC, et al. Duffy antigen receptor for chemokines mediates trans-infection of HIV-1 from red blood cells to target cells and affects HIV-AIDS susceptibility. Cell Host Microbe. 2008;4:52–62. doi: 10.1016/j.chom.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding D, Qu X, Li L, Zhou X, Liu S, Lin S, et al. Involvement of histone methyltransferase GLP in HIV-1 latency through catalysis of H3K9 dimethylation. Virology. 2013;440:182–189. doi: 10.1016/j.virol.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 45.Bouchat S, Gatot JS, Kabeya K, Cardona C, Colin L, Herbein G, et al. Histone methyltransferase inhibitors induce HIV-1 recovery in resting CD4(+) T cells from HIV-1-infected HAART-treated patients. AIDS. 2012;26:1473–1482. doi: 10.1097/QAD.0b013e32835535f5. [DOI] [PubMed] [Google Scholar]
- 46.Mohammadi P, di Iulio J, Muñoz M, Martinez R, Bartha I, Cavassini M, et al. Dynamics of HIV latency and reactivation in a primary CD4+ T cell model. PLoS Pathog. 2014;10:e1004156. doi: 10.1371/journal.ppat.1004156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.