Abstract
The HIV-1 fusion peptide, comprising 15 to 20 hydrophobic residues at the N terminus of the Env-gp41 subunit, is a critical component of the virus-cell entry machinery. Here, we report the identification of a neutralizing antibody, N123-VRC34.01, which targets the fusion peptide and blocks viral entry by inhibiting conformational changes in gp120 and gp41 subunits of Env required for entry. Crystal structures of N123-VRC34.01 liganded to the fusion peptide, and to the full Env trimer, revealed an epitope consisting of the N-terminal eight residues of the gp41 fusion peptide and glycan N88 of gp120, and molecular dynamics showed that the N-terminal portion of the fusion peptide can be solvent-exposed. These results reveal the fusion peptide to be a neutralizing antibody epitope and thus a target for vaccine design.
Type 1 viral fusion machines, including HIV-1 envelope glycoprotein (Env), mediate virus entry through structural rearrangements that drive virus-cell membrane fusion (1–3). The hydrophobic N-terminal region of the gp41 transmembrane subunit (the fusion peptide), which is liberated by cleavage of the envelope precursor, is a critical element in this process because it directly interacts with the target-cell membrane in both intermediate and postfusion states (1–3). In the prefusion state, sequestration of the HIV-1 Env fusion peptide has been thought essential to avoid its being snared by the viral membrane or forming a hydrophobic aggregate with other fusion peptides. Published structures of trimeric HIV-1 Env in its prefusion closed state indicated a surface-exposed fusion peptide located in the membrane-proximal quartile of the viral spike (4–6); however, substantial disorder of the N-terminal portion of the fusion peptide in both antibody-bound and ligand-free crystal structures (6, 7) made exposure of the fusion peptide unclear.
A chronically HIV-1-infected individual, donor N123 (8, 9), displayed potent serum neutralization (fig. S1A); however, the epitope specificity of the serum-neutralizing antibodies could not be clearly categorized (fig. S1, B and C). We performed antigen-specific single memory B cell sorting with the BG505 SOSIP.664 trimer (6, 10, 11); among 92 antigen-specific B cells were 7 members of the clonal lineage N123-VRC34 (named for donor “N123” and antibody lineage “VRC34,” with specific clone “x” referred to as VRC34.“x”) (Fig. 1A and fig. S2). The most potent member of the clonal family (VRC34.01) neutralized 16 out of 22 HIV-1 Env-pseudoviruses, including BG505 (fig. S3), and further neutralized 49% of 208 HIV-1 strains (fig. S4 and database S1). VRC34.01 and clonal members bound to BG505 SOSIP.664, but not BG505 gp120 monomer in enzyme-linked immunosorbent assay (ELISA) (Fig. 1B and fig. S5A), and VRC34.01-04, but not VRC34.05-07, bound to cell-surface BG505 trimer (fig. S5, B and C). In competition ELISA, VRC34.01 was partially inhibited by antibody PGT151 (12) (fig. S6), and on a panel of 28 glycan mutants of strain BG505, VRC34.01 displayed a profile distinct from PGT151 (fig. S7). VRC34.01 neutralization was reduced by elimination of the N88 glycan and was enhanced by glycan mutations N611Q and N611D. There was minimal effect on VRC34.01 neutralization when viruses were grown in the presence of glycosylation inhibitors kifunensine, swainsonine, or in GnTI−/− cells (fig. S8), suggesting that VRC34.01 recognition does not require complex glycosylation. Similar to PGT151 (13), VRC34.01 stained 293T cells expressing wild-type JR-FL Env, but not its uncleaved mutant (Fig. 1C). Overall, these results indicated that VRC34.01 targets a unique trimer-specific, cleavage-dependent epitope that involved glycan N88.
Fig. 1. HIV-1-Env fusion peptide is targeted by antibody N123-VRC34.01.
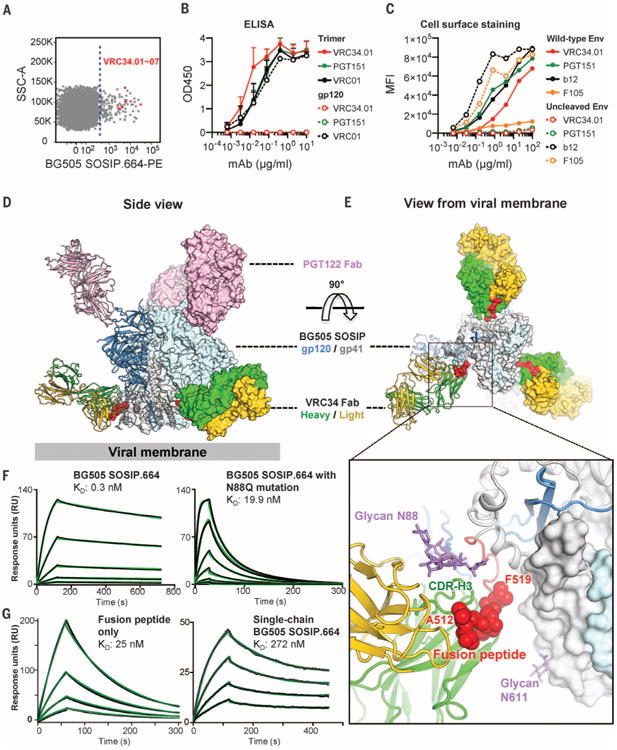
(A) Flow cytometric sorting of CD19+/IgG+/IgM− B cells from donor N123 that bind the BG505 SOSIP.664-PE probe. Cells are shown in dot plot format, with events to the right of the dotted line sorted. Red dots indicate VRC34 lineage B cells; SSC, side scatter. (B) Antibody binding to BG505 SOSIP.664 trimer and gp120 monomer. Mean and SD of three independent experiments shown. (C) Antibody staining of 293Tcells with surface expression of wild-type or uncleaved JR-FL Env trimer. MFI, median fluorescence intensity. Data are from one of three independent experiments. (D and E) Ternary complex crystal structure comprising Env trimer (BG505 SOSIP.664) and Fabs PGT122 and VRC34.01. One Env protomer and interacting Fabs is shown in ribbon representation; the rest of the complex is shown in surface representation, with the fusion peptide highlighted in red. (D) Side view. (E) View from the viral membrane. (Inset) An expanded view of the Env-VRC34.01 interface. The fusion peptide (red) and glycan N88 (purple) comprise the VRC34.01 epitope. N-terminal residues recognized by VRC34.01 depicted as red spheres and glycan N88 as purple sticks. (F) Fab VRC34.01 binding to BG505 SOSIP.664 trimer (left panel) and to a glycan N88 knock-out mutant (N88Q, right panel) measured by SPR. Three-fold serial dilutions from 200 nM Fab injected onto the captured trimers. (G) VRC34.01 binding to fusion peptide (left panel) and to single-chain BG505 SOSIP.664 trimer (right panel), as measured by SPR. Fab VRC34.01 Fab injected in two-fold serial dilutions starting from 128 nM (left panel) and 3200 nM (right panel). For (F) and (G), three independent experiments were performed, with data from one representative experiment shown. Black lines indicate experimental data and green lines global fit to a Langmuir 1:1 binding model.
To define further VRC34 recognition, we crystallized a ternary complex formed by antigen-binding fragment (Fab) VRC34.01, BG505 SOSIP.664, and Fab PGT122, a glycan-V3-directed antibody (4, 6, 14). Diffraction data extended to 4.3 Å resolution, and we determined (15) and refined the ternary complex structure (Fig. 1D and table S1). Each gp120-gp41 protomer bound a single VRC34.01 Fab and a single PGT122 Fab (Fig. 1, D and E). VRC34.01 recognized an epitope in the gp120-gp41 interface consisting primarily of the fusion peptide on gp41 (residues 512 to 519, 55% interactive surface area) and glycan N88 on gp120 (26% interactive surface area) (Fig. 1E and table S2) (16).
The fusion peptide and glycan N88 formed a contiguous surface on HIV-1 Env, with the heavy and light chains of VRC34.01 oriented perpendicular to this surface, so that both heavy and light chains were involved in binding both fusion peptide and glycan N88 (Fig. 1E). The first eight residues of fusion peptide (17) were embedded in a hydrophobic groove formed by complementarity-determining regions (CDRs) H1, H2, H3, L1, and L3, consistent with VRC34.01 neutralization of strain BG505 being knocked out by site-directed mutations in this region (fig. S9) (18). Glycan N88 was recognized by a pocket formed by CDRs L1, L2, and H3 of VRC34.01 (fig. S10) (19). Surface plasma resonance (SPR) studies of the N88Q variant of BG505 SOSIP.664 indicated removal of glycan N88 to decrease VRC34.01 affinity from 0.3 nM to 19.9 nM (Fig. 1F). The ability of VRC34.01 to recognize the N88Q mutant with nM affinity indicated its interaction with fusion peptide to dominate the energetics of interaction; a more substantial decrease in affinity (to 272 nM) was observed with a single-chain version of BG505, indicating the charged N terminus to be critical in VRC34.01 recognition (Fig. 1G). Consistent with the ternary complex crystal structure, negative-stain electron microscopy (EM) of Fab VRC34.01 and BG505 SOSIP.664 trimer revealed a three-antibody to one-trimer stoichiometry (fig. S11).
We next cocrystallized Fab VRC34.01 with the N-terminal portion of the fusion peptide (residues 512 to 520). Diffraction data extended to 1.5 Å resolution, and we determined and refined the peptide-Fab structure (Fig. 2A and table S1). Well-defined electron density extended from 512 to 518, with the side chain of F519 partially ordered and residue L520 completely disordered (fig. S12). A shallow groove spanning all of the CDRs except L2 held the fusion peptide in extended conformation, with residues A512, V513, I515, and V518 fitting snugly into four hydrophobic pockets spanning the VRC34.01 paratope (Fig. 2A) (20). We also determined the ligand-free structure of VRC34.01 (table S1); no significant change in VRC34.01 paratope occurred upon fusion-peptide engagement (fig. S13).
Fig. 2. Fusion-peptide sequence variation is a dominant determinant of neutralization resistance to VRC34.01.
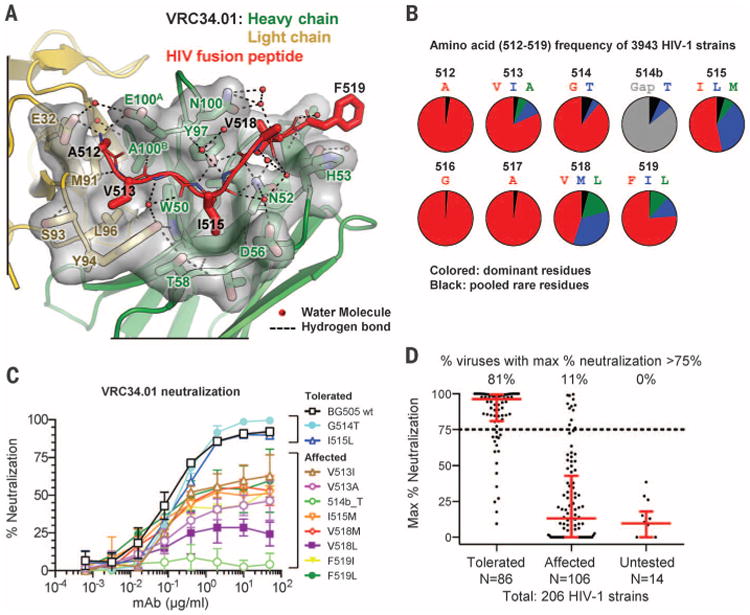
(A) Crystal structure of complex between Fab VRC34.01 and a synthetic fusion peptide (comprising residues 512 through 520, red) is shown in ribbon representation with interface residues as sticks and the molecular surface of VRC34.01-peptide interactive residues in gray. Hydrogen bonds are indicated by dotted lines. The side chains of N52CDR H2 and Y97CDR H3 were hydrogen-bonded with fusion peptide residue 515 to 517, and side chains of E32CDR L1, E100ACDR H3 formed electrostatic interactions with the free amine group of residue A512, anchoring the N terminus of fusion peptide. The last residue of the synthetic fusion peptide with ordered electron density, F519, stacked with the side chain of H53CDR H2. (B) Frequency of N-terminal amino acids of the fusion peptide. Red, most prevalent; blue, second-most prevalent; green, third-most prevalent; black, pooled rare residues; gray, sequence gap. (C) VRC34.01 neutralization of BG505 Env pseudoviruses containing fusion-peptide mutations. Variants displaying more than 75% neutralization at highest antibody concentration were termed “tolerated”; variants showing less than 75% neutralization were termed “affected.” Mean and SD of three independent experiments are shown. (D) Maximum percentage neutralization by VRC34.01 against 206 HIV-1 Env pseudoviruses grouped according to their fusion-peptide sequences (25). Each dot represented one Env pseudovirus. Results were based on serial dilutions, starting at 50 μg/ml of VRC34.01. For each group, the percentage of viruses with a maximum neutralization greater than 75% is indicated on top, with the bar showing the median and interquartile range in red.
About 50% of HIV-1 isolates tested were resistant to VRC34.01 (fig. S4 and database S1). This resistance could arise from structural or sequence variation. Because glycan N88 is conserved in 98.6% of HIV-1 sequences (21), we analyzed the fusion-peptide sequence of 3943 HIV-1 sequences (22), which identified variation at several residues (Fig. 2B). We individually introduced less prevalent residues into the fusion peptide of the BG505-Env pseudovirus and assessed VRC34.01 neutralization. Several mutants displayed diminished neutralization, and a Thr insertion at position 514b led to complete resistance to VRC34.01 (Fig. 2C). Notably, most mutations led to incomplete neutralization, which could be potentially explained by the inability of VRC34.01 to remain tightly bound to the fusion peptide throughout the HIV-1 entry process (23, 24). We also analyzed the sequences and neutralization sensitivity of 208 Env-pseudoviruses. Two sequences lacked the glycosylation site at N88 and were resistant to VRC34.01 (database S2). The remaining 206 sequences, divided into three groups based on their fusion-peptide sequence (25), showed distinct VRC34.01 sensitivity (Fig. 2D and database S2). These results indicate that a dominant component of neutralization resistance to VRC34.01 stems from sequence variation of the fusion peptide, a corollary being that a majority of HIV-1 strains appear to be capable of assuming the solvent-exposed conformation of the fusion peptide observed in the VRC34.01-Env complex.
To delineate further the conformation of HIV-1 Env recognized by VRC34.01, we evaluated the interaction of VRC34.01 with cell-free virus and the contribution of this interaction to neutralization (Fig. 3A). These data indicated neutralization to be mediated primarily by binding to cell-free virus before cell surface attachment. Next we performed SPR assays to quantify VRC34.01 binding to soluble BG505 SOSIP.664 trimers (10), as well as a 201C-433C variant (DS-SOSIP.664), which had been disulfide-stabilized to prefer the prefusion closed Env conformation (7). VRC34.01 bound tightly to 2G12-captured SOSIP.664 and DS-SOSIP.664 trimers, as well as to CD4-Ig-captured DS-SOSIP.664 trimers (Fig. 3B and fig. S14). In contrast, VRC34.01 binding to CD4-captured SOSIP.664, which undergoes CD4-induced conformational opening, was substantially reduced. VRC34.01 thus appears to recognize a prefusion Env conformation before CD4-induced rearrangement.
Fig. 3. VRC34.01 engages prefusion closed HIV-1 Env and inhibits CD4-induced rearrangements.
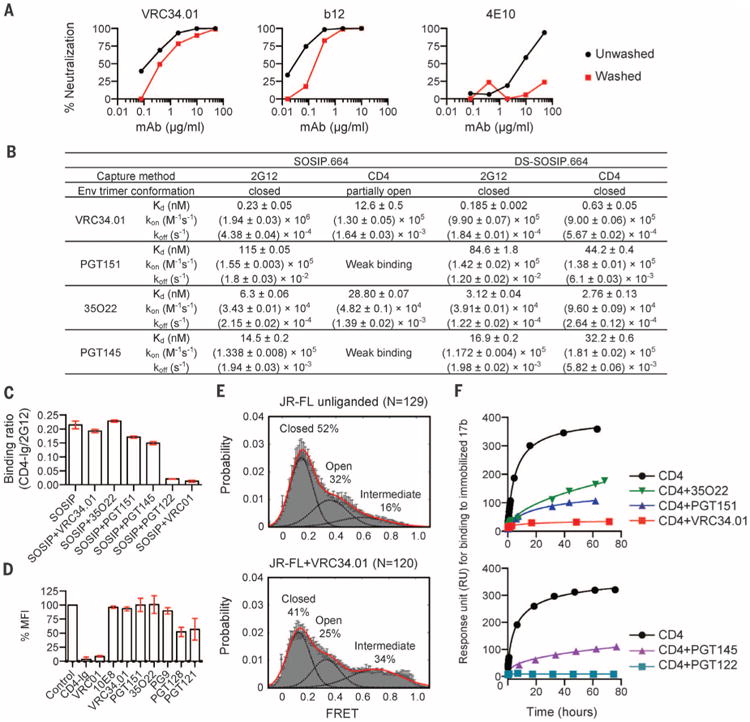
(A) Neutralization of JR-FL by indicated antibodies in which parallel virus-antibody mixtures were either pelleted and unbound antibody removed (washed, red lines) or not pelleted and antibody not removed (unwashed, black lines) before infection of TZM-bl cells. Representative data from one of three independent experiments are shown. The mean and SD inhibitory concentration (IC50 and IC80) values are shown in table S3. (B) Affinity of interaction of indicated antibodies with BG505 SOSIP.664 (10) or stabilized DS-SOSIP.664 (7). Standard errors from global fit of sensorgrams, obtained from independent injections of different concentrations of analyte, to a 1:1 Langmuir binding model are reported. Data sets are representative of at least two independent experiments. (C) Binding of BG505 SOSIP.664 to CD4, either alone or in the presence of antibodies (36). Mean and SD of three independent experiments are shown. (D) Influence of antibody on virus-cell attachment (37). Mean % MFI and SD of four independent experiments are shown. (E) smFRETanalysis of VRC34.01 incubation with JR-FL Env on native virions shows an increase in the high-FRET state, which is associated mechanistically with a required intermediate in the HIV-1 entry pathway (24, 38). N is the number of FRET traces analyzed. (F) Influence of antibody on the CD4-induced activation of BG505 SOSIP.664. Activation of SOSIP was monitored by assessing binding to the CD4-induced antibody, 17b, at different time points (39). Representative data of three independent experiments are shown.
We tested the ability of VRC34.01-bound BG505 SOSIP.664 to engage CD4 by preincubating the Env trimer with an excess of VRC34.01 and analyzed interactions with CD4-Ig or 2G12 by SPR. Preincubation with VRC34.01 showed little effect on CD4 engagement (Fig. 3C and fig. S15). Although VRC34.01 neutralized Env-pseudotyped JR-FL virions (Fig. 3A and fig. S16), incubation with these virions showed little effect on attachment to TZM-bl cells expressing surface CD4 (Fig. 3D). Single-molecule fluorescence resonance energy transfer (smFRET) of JR-FL Env molecules on native virions revealed VRC34.01 to increase the population of a partially open conformation (24), which is a required entry intermediate (Fig. 3E). This is in contrast to other broadly neutralizing antibodies that stabilize the prefusion closed conformation (6, 24). We also tested the effect of VRC34.01 on the CD4-induced formation of bridging sheet, which is required for co-receptor binding and virus entry (26, 27). We observed the CD4-induced formation of the bridging sheet formation–as detected by binding to antibody 17b–to be substantially inhibited by VRC34.01 (Fig. 3F and fig. S17). These results indicate that VRC34.01 recognizes cell-free virus and is able to engage the prefusion closed state while stabilizing Env in an intermediate state; VRC34.01 thus appears not to interfere with virus attachment to cell surface CD4 but to inhibit CD4-induced formation of the bridging sheet (26), which is required for co-receptor engagement and viral entry.
We docked the VRC34.01-SOSIP.664 complex into EM-observed electron density (28), which positioned the N terminus of the fusion peptide ∼20 Å from the membrane (Fig. 4A). We carried out molecular dynamics simulations (29) over 500 ns on fully glycosylated BG505 SOSIP.664 with fusion peptide initially in the VRC34.01-bound conformation (Fig. 4B and fig. S18) (30); this analysis indicated residues 512 to 520 of the fusion peptide to be solvent-exposed, with the exposed residue range coinciding closely with the epitope recognized by VRC34.01 (Fig. 4B and fig. S18). Despite this flexibility, a 20 Å approach to the viral membrane (Fig. 4C), and intrinsic prefusion motion of HIV-1 Env (24), there did not seem to be an issue with the fusion peptide being trapped by the viral membrane or inducing trimer aggregation (31).
Fig. 4. Fusion peptide accessibility and position relative to viral membrane.
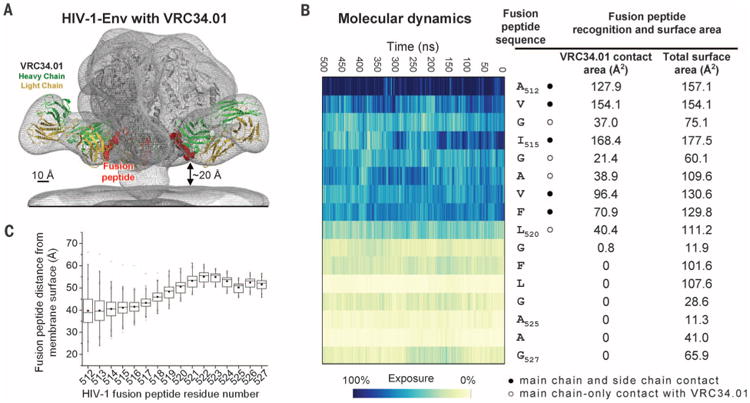
(A) Superposed EM map of the BG505 SOSIP.664-VRC34.01 complex and that of HIV-1 Env trimers reconstructed from their membrane-bound context (EMDB 5019 and 5022) (gray meshes). Fab VRC34.01 bound to BG505 SOSIP.664, as extracted from the ternary complex crystal structure, was docked and shown in ribbon representation with VRC34.01-contacting fusion peptide (512 to 519) in red spheres. (B) Fusion peptide in the context of a molecular dynamic simulation of fully glycosylated HIV-1 Env trimer. The degree of solvent exposure of each residue in the fusion peptide is depicted on a blue-to-yellow color scale, with antibody-contact surface and total surface area shown for context. (C) Range of distance to the viral membrane for each residue of the HIV-1 fusion peptide over the course of the molecular dynamics simulation. The median distance for each residue is represented as a horizontal line in each box. The mean value is represented as a small filled box. The height of each box is set by the 25th and 75th interquartile range, and the whiskers are determined by the 5th and 95th percentiles. The minimum and maximum distances for each residue over the length of the simulation is represented as “x.” See fig. S23 for an analysis of immune recognition of fusion peptide in other type 1 viral fusion machines.
VRC34.01 antibody recognition of the fusion peptide is shared to a limited degree by antibody PGT151, which binds in the vicinity of VRC34.01 (figs. S9 and S11). To provide further insight into the antibody-specific recognition of the fusion peptide during HIV-1 infection, we designed a VRC34-epitope scaffold (32, 33) (fig. S19 and database S3). This scaffold was used to assess 24 sera from a cohort of HIV-1-infected subjects. Ten sera displayed specific recognition of the fusion peptide (figs. S20 and S21), with four sera showing reduced neutralization when a fusion peptide-specific mutation G516A was introduced (fig. S22).
In summary, the N terminus of the fusion peptide on HIV-1 is antibody-accessible, binding and neutralizing antibodies against it are elicited during natural infection, and broadly neutralizing antibodies recognize it either partially (anti-body PGT151) (34, 35) or more completely (antibody VRC34.01). Taken together, our results indicate the fusion peptide of the Env trimer to be a region of interest for vaccine design against HIV-1.
Supplementary Material
Acknowledgments
We thank the members of the Flow Cytometry Core, Vaccine Research Center, and especially R. Nguyen for his assistance in cell sorting. We thank members of the Structural Biology Section, Structural Bioinformatics Core Section, and Human Immunology Section, Vaccine Research Center, for discussions and comments on the manuscript. We thank J. Baalwa, D. Ellenberger, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1 Envelope plasmids used in our neutralization panel. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. Nucleotide sequences of VRC34.01-VRC34.07 variable regions are available under GenBank accession numbers KU711816 to KU711829. Atomic coordinates and structure factors of the reported crystal structures have been deposited with the Protein Data Bank (PDB) under accession codes 5I8C, 5I8E, and 5I8H. EM data have been deposited with the Electron Microscopy Data Bank under accession code EMD-8125. Support for this work was provided by the Intramural Research Programs of the Vaccine Research Center and Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, and from the International AIDS Vaccine Initiative's (IAVI's) Neutralizing Antibody Consortium. IAVI's work is made possible by support from many donors, including the Bill & Melinda Gates Foundation, the Ministry of Foreign Affairs of Denmark, Irish Aid, the Ministry of Finance of Japan, the Ministry of Foreign Affairs of the Netherlands, the Norwegian Agency for Development Cooperation (NORAD), the UK Department for International Development (DFID), and the U.S. Agency for International Development (USAID). The full list of IAVI donors is available at www.iavi.org. S.C.B. was supported by NIH grants R01GM079238 and P01GM56550. W.M. was supported by NIH grants R01GM116654 and P01GM56550. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38. The following patent applications are pending: U.S. Patent Application 13/202,351, Methods and Compositions for Altering Photopysical Properties of Fluorophores via Proximal Quenching (S.C.B.); U.S. Patent Application 14/373,402 Dye Compositions, Methods of Preparation, Conjugates Thereof, and Methods of Use (S.C.B.); and International and U.S. Patent Application PCT/US13/42249 Reagents and Methods for Identifying Anti-HIV Compounds (S.C.B. and W.M.). S.C.B. holds an equity interest in Lumidyne Technologies Corporation.
Footnotes
References and Notes
- 1.Colman PM, Lawrence MC. Nat Rev Mol Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 2.Eckert DM, Kim PS. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 3.Harrison SC. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Julien JP, et al. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyumkis D, et al. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pancera M, et al. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon YD, et al. Nat Struct Mol Biol. 2015;22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doria-Rose NA, et al. J Virol. 2010;84:1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doria-Rose NA, et al. J Virol. 2009;83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders RW, et al. PLOS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sok D, et al. Proc Natl Acad Sci U S A. 2014;111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blattner C, et al. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falkowska E, et al. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker LM, et al. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCoy AJ, et al. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Residue number for HIV-1 Env follows the standard HXB2 convention (www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/HXB2.html).
- 17.VRC34.01-bound portions of fusion peptide (residues 512 to 517) were disordered in previous HIV-1 Env structures, whereas residues 518 to 530 were ordered in the PGT122-35022-bound HIV-1 structure (PDB ID 4TVP) and assumed the same conformation as in the VRC34.01-bound structure (0.19 Å root mean square deviation).
- 18.Residues 512 to 519 (which correspond to the N terminus of the fusion peptide and are recognized by VRC34.01) do not interact with other portions of the HIV-1 Env. Residues 520 to 530 of the fusion peptide are embedded in a groove defined by gp41 residues 533 to 543 and 627 and by gp120 residues 45, 84 to 89, 492, and glycan N88.
- 19.Antibody residue numbers follow Kabat convention, and, for clarity, heavy-chain residues are followed by an “HC” subscript and light-chain residues by an “LC” subscript. In some cases, the CDR loops are specified to provide further clarity.
- 20.The pocket for A512 is defined by E32CDR L1, M91CDR L3, E100ACDR H3, and A100BCDR H3; the pocket for V513 is defined by S93CDR L3, Y94CDR L3, W50CDR H2, and A100BCDR H3; the pocket for I515 is defined by A33CDR H1, W50CDR H2, I51CDR H2, N52CDR H2, and Y97CDR H3; and the pocket for V518 is defined by G31CDR H1, N52CDR H2, and Y97CDR H3.
- 21.Huang J, et al. Nature. 2014;515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sequences of 3943 HIV-1 strains were downloaded from the HIV database (www.hiv.lanl.gov/content/index).
- 23.Kwong PD, et al. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 24.Munro JB, et al. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The following nomenclature is used: “Tolerated” Env sequences contained one or more mutations within the eight residue fusion-peptide sequence, which individually showed little effect on VRC34.01 neutralization. “Affected” sequences contained one or more residues (513I, 513A, 514b_T, 515M, 518M, 518L, 519I, or 519L) that diminished VRC34.01 neutralization. “Untested” sequences contained one or more untested residues plus tolerated residues.
- 26.Kwong PD, et al. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trkola A, et al. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harvey MJ, Giupponi G, Fabritiis GD. J Chem Theory Comput. 2009;5:1632–1639. doi: 10.1021/ct9000685. [DOI] [PubMed] [Google Scholar]
- 30.While 500 ns is not enough time to sample large-scale conformational transitions associated with the prefusion HIV-1 Env trimer, it should be sufficient to assess local motions of the fusion peptide.
- 31.When trimers are positioned into nearest-neighbor contact, only the N-terminal three residues of the fusion peptide can extend to contact a fusion peptide on a neighboring trimer. The N-terminal three residues of the fusion peptide are Ala-Val-Gly. Although the Ala-Val are somewhat hydrophobic, the positive charges of two N termini likely repel, preventing aggregation. In terms of close-by glycans, these may help to prevent aggregation, especially glycans N88, N611, and N618. However, it appears to be primarily the recessed nature of the fusion peptide, coupled to the fact that only eight residues of the fusion peptide are flexible (Fig. 4), that prevents aggregation.
- 32.Materials and methods are available as supplementary materials on Science Online.
- 33.Pancera M, et al. Nat Struct Mol Biol. 2013;20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JH, Ozorowski G, Ward AB. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The structure of PGT151 in complex with HIV-1 Env was recently reported (34). This structure shows PGT151 to recognize the fusion peptide in an exposed, extended conformation.
- 36.Trimer (or trimer-antibody complex) was injected for 60 s onto CD4-IgG (immunoglobulin G) captured on a Biacore CM5 chip coated with a monoclonal mouse anti-human IgG (Fc) antibody (GE Healthcare), and binding responses were measured 60 s after the injection ended. Binding to CD4-Ig normalized with binding to antibody 2G12, which was measured similarly on a parallel-flow cell.
- 37.TZM-bl cells were incubated with JR-FL Env pseudovirus in the presence of antibodies at 50 μg/ml or medium control and then stained with biotinylated 2G12 followed by phycoerythrin (PE)-conjugated streptavidin. MFI was obtained from at least 10,000 cell events and normalized to the medium control.
- 38.The HIV Env trimer is conformationally dynamic. Binding and smFRET experiments show that VRC34 do not bind to the open co-receptor binding-competent conformation. Thus, any molecule spontaneously in that conformation would not be neutralized. The conformation specificity of VRC34.01 for the closed and intermediate conformation may explain the observed incomplete neutralization.
- 39.Specifically, soluble SOSIP.664 trimers were incubated with soluble, two-domain CD4, either alone or following preincubation with Fab, and the incubated trimer samples were injected over a 17b-coupled surface for 30 s, with response units reported 10 s after the injection ended.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


